Synopsis
Flow cytometry based immunophenotyping assays have become increasingly multi-parametric, concomitantly analyzing multiple cellular parameters. In order to maximize the quality of the information obtained, antibody conjugate panels need to be developed with care, including requisite controls at every step. Such an optimization procedure for multi-colour immunophenotyping assays is very time-consuming, but the value of having a reliable antibody conjugate panel that provides for sensitive detection of all molecules of interest justifies this time investment. The present chapter outlines important considerations and procedures to undertake for the successful design and development of multi-colour flow cytometry panels.
Keywords: Spillover, compensation
The importance of multi-colour flow cytometry for immunophenotyping and immunomonitoring is ever increasing in multiple applications. Today, both research and clinical laboratories addressing various immunological aspects heavily rely on flow cytometry for phenotypical and functional analyses of immune components in different disease settings, as well as in the healthy immune system. Flow cytometry is currently the only technological platform that can suitably analyze the complex components of the immune system – i.e., to separately characterize the dozens (or hundreds) of phenotypically and functionally distinct subsets of leukocytes, any of which might be clinically relevant. Multi-colour flow cytometry assays, therefore, need to be developed with great care in order to ensure reliability of data generated therewith. In this chapter, we will illustrate the process of generating and optimizing a complex immunophenotyping panel to address vaccine-induced T cell responses.
With the advent of instrumentation capable of measuring up to 20 parameters, new possibilities for performing more complete and more complex analyses have opened up. This has spurred the search for correlates of protection and correlates of disease (1), which are necessary for the rapid evaluation of immunological interventions. In addition, there is the possibility of identifying phenotypically unique cellular subsets to which distinct functional responses can be attributed; such assignments would possibly preclude, or at least reduce, the necessity of performing time-consuming functional assays for the evaluation of antigen-specific immune responses. This is based on the presumption that cells with a given phenotype will be an indication of a productive immune response in a given disease setting, while other phenotypes are indicative of inefficient responses or even of cellular activity leading to pathologies. It is noteworthy that the relationship between phenotype and desirability of the response may vary considerably in different disease contexts and tissue environments.
The complexity of the T-cell compartment, both phenotypically and functionally, is enormous. Unfortunately, virtually none of the T-cell subsets can be uniquely identified by the expression of one or even a few molecules, and generally the simultaneous discrimination of the expression patterns of four or more molecules is required (2). Thus, the advent of polychromatic flow cytometry (measuring 5 or more colours) has dramatically impacted on our ability to reveal T-cell biology.
However, the complexity in the design, implementation, and analysis of such experiments increases geometrically with the number of colours used; there are a wide range of hurdles and problems that must be addressed. In this chapter, we describe some of the more common ones, and lay out a general plan for succeeding with multi-colour experiments.
Considerations in choosing a flow cytometer
Most machines employed in research and diagnostics today typically measure 6 to 12 parameters. Although the present chapter is going to focus on the development of a 15-colour (17-parameter) panel, the process described applies to all polychromatic assays, whether 4 or 18 colours are being measured. Meticulous development of the antibody panel will always prove rewarding in the end.
Today’s most sophisticated flow cytometers can measure 20 parameters simultaneously – two physical parameters (cell size and granularity), as well as 18 fluorescences (Table 1). If polychromatic flow cytometry is to be used e.g. for immunophenotyping during vaccine studies, we warmly recommend acquiring such a high-end machine. Even though the possibility exists, the aim is not to develop 18-colour panels for all scientific questions. More importantly, the more fluorescence detectors an instrument incorporates, the more flexibility it will provide to researchers in respect to assay development, since detector numbers do not become a constraint. This increases the likelihood of being able to accommodate antibodies to all antigens desired to be evaluated in a particular assay, by giving the option of choosing from more antibody conjugates.
Table 1.
Detectors used in the VRC’s LSR II instruments and corresponding dyes
| Detector | Conjugates | Non-conjugates |
|---|---|---|
| FSC | -- | |
| SSC | -- | |
| V450 | Cascade Blue, Pacific Blue | DAPId, ViViDd, CFPpt |
| V545 | QD545O, Ax430, Pacific Orange | AquaBlued |
| V565 | QD565O | |
| V585 | QD585O | |
| V605 | QD605O | |
| V655 | QD655O | |
| V705 | QD705O | |
| V800 | QD800O | |
| B515 | FITC+++, Ax488 | CFSEc, GrViDd, GFPpt |
| B710 | PerCP, PerCP-Cy5.5++ | |
| G560 | PE+++, Ax532 | CMTMRc |
| G610 | PE-TR/ECD+, TR, Ax594, PE-Ax594 | EMAd, OrViDd, RFPpt |
| G660 | PE-Cy5++, PE-Ax647 | PId |
| G710 | PE-Cy5.5+, PE-Ax700 | |
| G780 | PE-Cy7++, PE-Ax750 | |
| R660 | APC++, Ax633, Ax647 | |
| R710 | APC-Cy5.5+, Ax660, Ax680, Ax700 | |
| R780 | APC-Cy7+, APC-Ax750, Ax750 |
Detector nomenclature: laser light exciting the fluorochrome is indicated by the letter (V: violet, 407 nm; B: blue, 488 nm; G: green, 532 nm; R: red, 633 nm), the subsequent number specifying the peak photon emission wavelength measured by the band-pass filter used in front of the respective photomultiplier tubes (PMT).
many conjugates are commercially available,
some conjugates are commercially available,
few conjugates are commercially available,
conjugates are made on a custom basis,
cellular dye,
dead cell marker dye,
protein tag.
Table 1 illustrates the configuration of LSR II flow cytometers employed in our laboratories. Since all fluorochromes excited by the green laser (532 nm excitation) – with the exception of Ax594 – can also be measured off the blue laser (488 nm excitation), it is not absolutely necessary to install four lasers in this instrument. However, the green laser increases immunofluorescence sensitivity in comparison to the blue laser (3), and provides for the detection of an additional unique colour (i.e. Ax594, to obtain a total of 18).
Initial considerations
As increasing numbers of parameters are evaluated in single samples, the importance of adhering to a meticulous optimization strategy cannot be over-emphasized (4) Design and analysis of multi-colour assays becomes more complex and time-consuming as more parameters are included. In order to illustrate the process of designing and optimizing such an assay, we are going to discuss the steps undertaken for the development of a panel to be used to evaluate CD4+ and CD8+ T-cell responses in human peripheral blood mononuclear cell (PBMC) samples isolated from humans before or after receiving different anti-viral vaccines or treatments. Our goal was to evaluate putative T-cell activation in response to the respective vaccination or treatment. To this end, the analysis panel had to include antibodies specific for lymphocyte lineage markers (CD3, CD4, CD8), differentiation markers (CCR7, CD27, CD28, CD45RA), as well as for cytokines (IFN-γ, IL-2, TNF-α) and other activation markers (CD107a, possibly CD154) in order to identify the functionality of respective T-cell subpopulations.
We also wanted to include a viability marker in order to increase sensitivity for cytokine detection, as false-positive events due to antibody-conjugates that non-specifically bind to dead cells could produce misleading results. A number of dead cell markers are available, including DNA-intercalating dyes (EMA, PI, 7-AAD, DAPI), phosphatidylserine-binding reagents (annexin V) and amine-reactive dyes (UViD, ViViD, GrViD, OrViD). The latter are reportedly the most stable, allowing their use in combination with intracellular flow cytometry, which requires fixation and permeabilization of cells (5). Additionally, they outrank PI and annexin V in identifying the highest proportion of dead cells (5).
Flow cytometry enjoys widespread use in research and diagnostics, thanks in part to the growing number of fluorescent reagents that are available. While some of the more classical dyes used to label antibodies, such as phycoerythrin (PE), have broad emission spectra, some of the newer reagents have narrower emission spectra. This is important, for it minimizes spillover into other fluorescence detectors – a process that reduces sensitivity for measuring the desired fluorescence in those detectors. The recently introduced semiconductor nanocrystals known as quantum dots (Q-dots) offer a number of distinct fluorescent particles excited by the violet laser (6). However, Q-dot reagents do not work well for all antibodies and have been difficult to optimize for the detection of intracellular molecules. With all fluorochromes, but especially with Q-dots, one has to be aware that unbound particles can potentially interact with each other, forming aggregates. Such aggregates need to be minimized by subjecting the antibody dilutions to a quick high-speed spin, as they non-specifically label some cells.
Sensitivity is a serious issue when developing multi-colour panels. The sensitivity of a reagent is determined by three factors: the cell’s autofluorescence in that region of the spectrum, the performance of the antibody conjugate (brightness of the fluorochrome, expression level of the molecule), and the presence of other antibody conjugates attached to the same cell (that result in spillover fluorescence into the same detector). It is important to realize that not all fluorochromes provide the same sensitivity, due to substantial differences in brightness. Furthermore, including more reagents typically decreases the sensitivity of a given antibody conjugate because of the accumulated interference from spillover fluorescences.
In order to maintain the highest possible sensitivity for all antigens to be analyzed, the best reagent panel has to be identified by an iterative process of testing antibody conjugates and putative panels. At each stage, the performance of the panel is compared to the previous step, so as to know which interactions are causing any loss in sensitivity and thereby providing an indication as to whether or not the panel requires adjustment.
The grid
Initially, a list of antigens of interest is drawn up – the “wish list” of antigens to be included in the analysis. It is useful to include a ranking of importance in case not all of them will be able to be included in the final panel. The antigens are subsequently assigned to one of the following three categories:
Primary antigens are those that are well characterized and identify broad subsets of cells. Their expression is usually either “on” or “off” and used to gate on cellular subsets of interest (e.g. CD3, CD4, CD8, CD14, CD19, CD20).
Secondary antigens are also well characterized with relatively high molecular density per cell. However, their expression pattern is often a continuum (e.g. CD27, CD28, CD45RA/RO, Interferon-γ (IFN-γ), Perforin).
Tertiary antigens are expressed at low levels only (e.g. CD25, chemokine receptors). Uncharacterized antigens also fall into this category.
In order to be able to select the best antibody conjugate for every antigen of interest, one needs to test multiple reagents for each antigen. Our approach is to purchase many reagents for primary, less for secondary and only one or two (with the brightest fluorochromes, e.g. PE, allophycocyanin (APC)) for tertiary antigens. This is expensive, but the investment is well worth it, as it will allow the development of the ideal reagent panel for the research question at hand. Panels are often developed for the subsequent analysis of a large number of samples, so the total expenditure for antibodies will always be large. The gain from having a highly optimized panel will offset the information cost from testing only a limited number of initial conjugates; do not skimp on the testing and optimization phase!
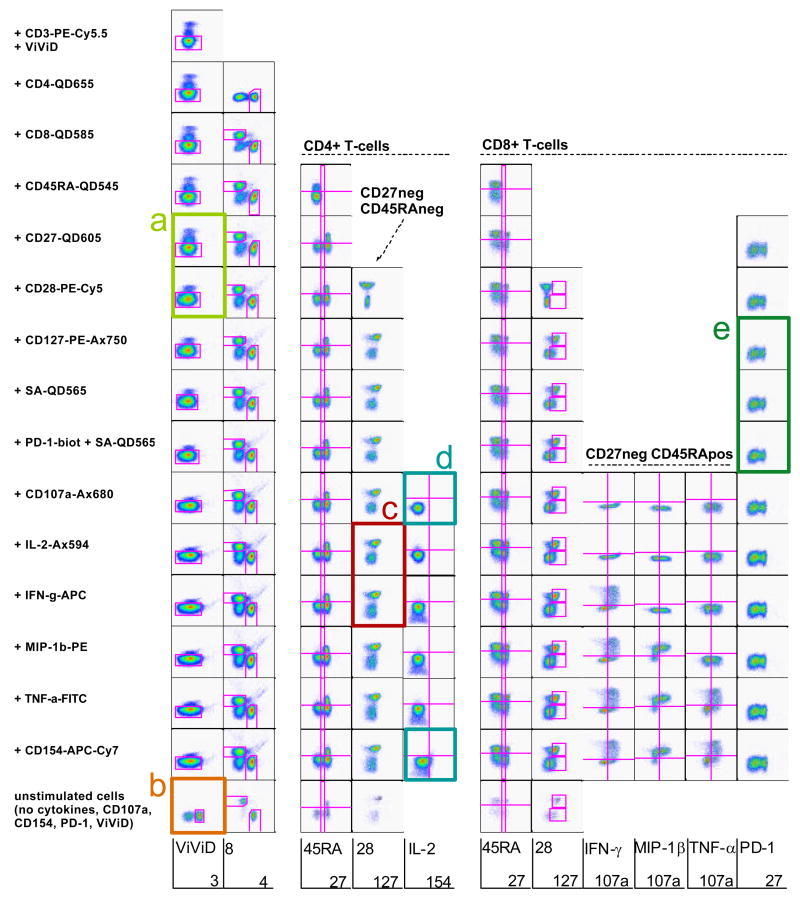
Before proceeding, all antibodies need to be carefully titrated and the optimal titre recorded. All antibody conjugates – including disparate antibody clones and different dyes read in single detectors – available for the antigens of interest are then screened systematically in single-colour experiments. For those antigens that are known to be expressed on small subsets of cells only, samples are specifically stained for those subsets in order to facilitate subsequent analysis of the antibody screening results. After carefully gating on single lymphocytes, dot plots are created to compare the performance of the various reagents. Results are grouped by detectors and dyes and the best antibody conjugate chosen for each detector and antigen is then inserted into a grid (Figure 1). Such a grid, where dot plots are sorted by antigen vs. detector, is extremely useful for subsequently selecting possible antibody combinations to be tested in potential multi-colour panels.
Figure 1. The antigen/detector grid.
The indicated antigens are read on the y-axis. For CCR7 and CD27, CD45RA expression was co-analyzed (x-axis), while for CD45RA, CCR7 was co-analyzed. For CD107a, CD4 expression is shown on the x-axis, while for CD154, IFN-γ, IL-2, MIP-1β and TNF-α, CD8 is illustrated on the x-axis. Orange frames highlight those antibodies included in the antibody panel to be tested, while green frames mark the detectors that are not used in the panel and could potentially be exploited to include further markers in the assay. Blue frames indicate those antigens not included in the selected antibody panel. Numbers on the right hand side of the table indicate primary (1°), secondary (2°) and tertiary (3°) antigens.
In the process of allocating candidate detectors for the different antigens, those antigens where only few antibody conjugates are available and/or those where good labelling is not easily obtained are considered first. Generally, bright fluorochromes, such as PE and APC, are preferentially chosen for weak antigens where only low numbers of molecules are expressed per cell or those that are expressed by only a very small fraction of cells (tertiary antigens). Starting with the most difficult antigen (few antibody conjugate options, expressed on a small subpopulation of T-cells only), here CD107a, the options are compared and the optimal staining identified and marked. This implies blocking the detector chosen for CD107a for other antigens, which can easily be visualized using the grid. This process is repeated for all tertiary antigens, then all secondary antigens until finishing with primary antigens such as CD3, CD4 and CD8 that are expressed by relatively large proportions of cells and at a high molecular density and where typically a large selection of antibody conjugates are available. Ideally, a number of different panels (perhaps half a dozen or more) evaluating different combinations of these reagents will be tested in the first rounds.
Evaluating candidate antibody panels
In order to evaluate which, if any, of the selected putative antibody panels will provide reliable staining of all antigens of interest, cells are incubated not only with the full sets of antibodies making up a panel, but also with subsets thereof. This permits the identification of those antibody conjugates that create problems of interference (i.e. reduced sensitivity). Ideally, a basic roster of antibodies is first identified that marks major subsets, such as CD4+ and CD8+ T-cells. In subsequent samples, the remaining antibodies are added one at a time, in order to evaluate the effect of each added reagent to the overall staining pattern. During this process, it makes sense to add antibodies in the sequential order that they will be required for gating purposes during the analysis. Alternate antibody conjugates can rapidly be tested in this manner, leading to the gradual build-up of the final antibody panel.
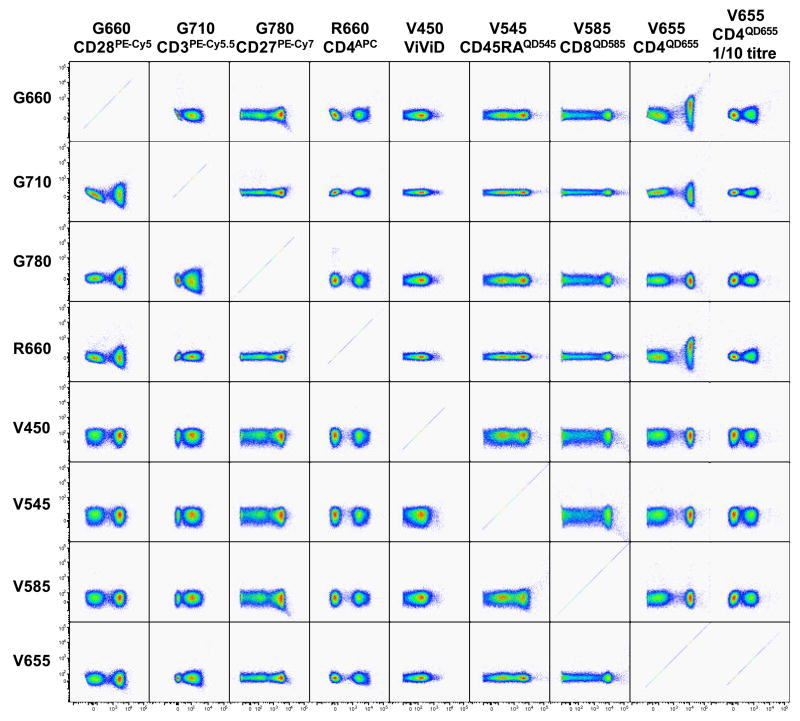
In the example illustrated in Figure 2, antibody conjugates are added one at a time, and dot plots created accordingly to depict the labelling achieved with the respective reagent, as well as any putative effect on other reagents included in the sample. Figure 3A illustrates a decrease in sensitivity for the dead cell marker ViViD upon addition of CD28PE-Cy5. As in the present example ViViD was added at the same time as the antibody conjugates, there are two possible explanations for this phenomenon: 1) it might indicate that there is something in the CD28PE-Cy5 antibody preparation (e.g. amines, free protein) that interferes with the ViViD reaction, or 2) the fact that this antibody conjugate was used at a relatively high concentration (25 μl in100 μl) was sufficient to dilute the ViViD enough to reduce its staining intensity. A solution to this is to perform separate incubations for the ViViD and antibody conjugates specific for surface markers.
Figure 2. Progressive construction of a multi-colour analysis panel.
In order to determine whether antibody conjugates selected for a multi-colour panel produce signal interference, a series of samples were labelled with subsets of the antibodies making up the panel. The first sample contained one antibody conjugate only (plus ViViD to exclude dead cells), while each subsequent sample contained one more antibody conjugate. Each row depicts plots from a single sample, and the antibody conjugate added at each point is indicated at the left of each respective row. For each sample, dot plots were created that follow the gating strategy to be used during analysis as far as it can be predicted. This allows the identification of antibody conjugates that create signal interferences with other antibody conjugates or simply do not result in sufficient detection sensitivity in the present antibody combination.
Figure 3. Problem solving.
The coloured boxes from the panel evaluation illustrated in Fig. 2 are enlarged here in order to better visualize the problems identified.
These pseudo-colour plots also indicate that the CD3-labeling was not successful, or at least not bright enough to allow distinction of CD3pos and CD3neg cells. This was due to the Cytostim (anti-CD3 and anti-CD28 antibody preparation from BD Biosciences) stimulation providing strong activatory stimuli to all T-cells resulting in severe down-regulation of the CD3-molecule. Parallel labelling of an unstimulated PBMC sample with the same antibody mixture shows that the antibody conjugate does indeed perform well (Figure 3B). The patient samples we are interested in will ultimately be stimulated with peptide before flow cytometric analysis, and it is expected that peptide stimulation does not induce such an extreme CD3 down-regulation, although this will have to be verified before commencing the study. Note that this illustrates an important aspect of panel development: at all phases, putative panels must be tested in exactly the conditions of the final assay; there are aspects of experimental procedures which may impact on sensitivity that are not revealed by simply labelling untreated fresh cells.
An apparent loss of sensitivity in the CD28 detection is illustrated in Figure 3C, which depicts CD127 (x-axis) against CD28 (y-axis) expression on CD4pos CD27neg CD45RAneg T-cells both before and after addition of an IFN-γAPC conjugate. In the upper panel (no IFN-γAPC), CD28pos and CD28neg cells are clearly separated, whereas in the lower panel (IFN-γAPC added) the negative population appears to be scattered towards the positive population, hampering unambiguous distinction of these two cellular fractions. A pseudo-colour plot illustrating CD28 vs. IFN-γ expression revealed that, in this case, APC was not sufficiently compensated, resulting in significant spillover into other detectors, most notably into the G660, which is dedicated to measuring CD28PE-Cy5 in the present panel. The problem was that the APC fluorescence achieved by labelling CompBeads with IFN-γAPC was less bright than that obtained in the PBMC samples. Therefore, the software was not able to properly compensate the signals, as it did not have any information on the behaviour of signals brighter than those in the compensation control sample. This spillover could be corrected by adjusting the compensation matrix. Once corrected, the CD28 separation was significantly improved, although there was still loss of sensitivity brought about by the inclusion of IFN-γAPC in the panel. This loss is due to the spillover-induced spreading of the negative population in the G660 detector (specific for PE-Cy5) (for a full explanation of this phenomenon, see (7). The long-term solution for this specific issue is to include a cell-based compensation control for this detector, ensuring that the compensation control is at least as bright as the experimental sample. For other detectors, the bead-based compensation controls were sufficiently bright and resulted in appropriate compensation calculations.
Figure 3D highlights that there was very weak IL-2 detection, as well as only limited detection of CD154 expression even though the CD154 specific antibody conjugate was added to the stimulation culture, which is the best labelling strategy for detection of this molecule on activated CD4+ T-cells (8). It would be expected that a larger fraction of CD4+ cells would be producing IL-2 and at a higher level than indicated by the present labelling. Other CD154 and IL-2 specific antibody conjugates will have to be evaluated in the context of this antibody panel in order to identify better combinations.
Lastly, the present experiment did not allow any conclusion as to whether the reagents chosen for PD-1 detection provide adequate detection sensitivity in conjunction with the other reagents included in the panel (Figure 3E). The reason for this is that PD-1 is typically expressed at very low levels on T-cells from healthy individuals, its expression being upregulated on memory CD8+ T-cells specific for poorly controlled chronically persistent viruses, such as human immunodeficiency virus (HIV) (9). The panel would, therefore, need to be re-evaluated on PBMC from an HIV-infected individual or on cells that were stimulated for several days.
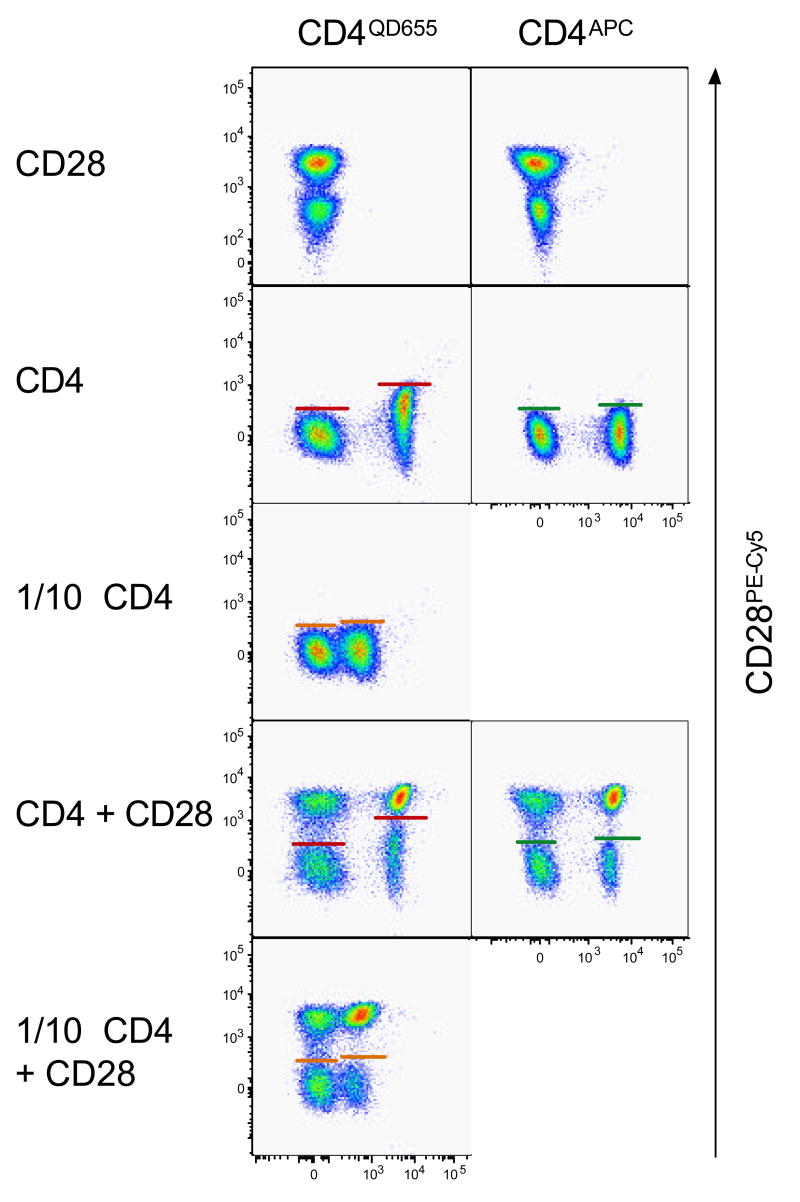
It is important to note that “spreading errors” (a consequence of imprecise measurement and compensation (7)) can highly differ between fluorochrome-antigen pairs. Labelling antigens such as CD3, CD4, CD8 and CD45 that are highly expressed on lymphocytes will often result in bright signal intensities. A QD655-labeled anti-CD4 antibody (measured by the V655 detector, see Table 1) will demonstrate significant spreading error in the G660 detector, and to a lesser extent in the R660 detector, even after the mean fluorescence intensities (MFI) of CD4neg and CD4pos cells have been correctly aligned (Figure 4A). This is due to the fact that all three detectors (V655, G660 and R660) measure very similar wavelengths (640 nm to 680 nm). However, the G660 detector is affected more severely by very bright signals that should be measured in the V655 detector than the R660 detector. As a consequence, the minimal fluorescence intensity of PE-Cy5-signals (on the G660 detector) detectable on CD4neg and CD4pos cells are very different (Figure 4B) (3, 10). This also holds true for other antibody and fluorochrome combinations, respectively. It is therefore advisable to closely monitor spreading errors in other detectors and where possible to prevent this effect by changing the antibody combination (Figure 4B). In the present example, reducing the anti-CD4QD655 antibody titre by 1/10th eliminated the dramatic spreading error in the G660 detector, while still providing a good separation between CD4neg and CD4pos cells.
Figure 4. Spreading error of fluorescence signals.
A. A spreading error analysis is performed on samples labelled with single antibody conjugates. For every sample, the detector relevant for the antibody conjugate (x-axis) added is plotted against all other detectors to be used in the experiment (y-axis). This allows to rapidly detect spreading errors that would reduce sensitivity in other detectors, such as obtained with CD4QD655. Diluting the reagent by 1/10 obliterates this problem. B. Spreading error of QD655pos cells reduces the sensitivity of the G660 detector for PE-Cy5pos signals on QD655pos cells (red lines). The severe spreading error observed with previously titrated amounts of anti-CD4QD655 (V655) in the G660 detector can be corrected by either using a lower titre of the anti-CD4QD655 antibody or by replacing it with another anti-CD4 conjugate, such as an anti-CD4APC antibody (R660).
Sample preparation
The number of cells per sample very much depends on the scientific question addressed, e.g. whether one is interested in analyzing total T-cells or only a subset of T-cells. The total number of cells acquired will need to be estimated to include sufficient events for the cell population of interest in order to provide clearly definable subpopulations and results that can be statistically evaluated.
Most antibodies bind well at room temperature or 4°C, the two most commonly used incubation temperatures for flow cytometry assays. In our laboratory, cells are routinely labelled at room temperature. However, in some cases incubation at higher (37°C) temperatures has been found to increase an antibody’s binding to its antigen. Chemokines and chemokine receptors have been found to be exceptionally sensitive to variations in cell isolation and staining techniques (11). The optimal protocol for most, but not all, of these molecules was found to be lymphocyte enrichment over a Ficoll-gradient followed by labelling at 37°C. A good example for one such chemokine receptor is CCR5. Conversely, other antigens, such as CXCR6, do not tolerate incubations at 37°C very well, resulting in severely reduced or even abrogated detection (11).
In panels incorporating antibodies to both extracellular and intracellular proteins, a sequential labelling protocol needs to be applied. If a dead cell marker dye such as an amine-reactive dye is to be included, we suggest to first incubate the samples with the dead cell marker dye before proceeding with cell surface marker labelling, fixation/permeabilization, and finally labelling of intracellular markers. It is of note that not all dead cell marker dyes are compatible with the fixation/permeabilization process required for intracellular labelling (12). Similarly, some tandem dyes are not completely spectroscopically stable to the fixation/permeabilization procedure required for the labelling of intracellular markers, leading to apparent compensation errors. Whether this applies to any reagents included in putative antibody panels can be determined by comparing two samples labelled with the same master mix of antibody conjugates specific for all cell surface markers to be included in each panel and submitting one of these samples to fixation/permeabilization prior to acquisition on the flow cytometer. Comparison of reagent performance in the two samples will rapidly indicate potential incompatibilities of individual reagents with such a procedure, and, therefore, with concomitant detection of intracellular proteins. To circumvent the problem of treatment induced down-modulation of selective surface molecules preventing their detection by the conventional cell surface labelling (e.g. CD3 down-regulation upon T-cell stimulation, see Figure 3A), they can be labelled intracellularly with the same two-step protocol used for detection of cytokines. However, the antibody conjugate selected for extracellular labelling might not perform well when used for intracellular labelling, as it appears that not all fluorochromes are applicable in antibody conjugates used for the detection of intracellular proteins (data not shown).
Correct compensation
Any flow cytometry experiment making use of more than one colour needs to include samples labelled with single antibody conjugates incorporating the relevant fluorochromes for compensation. Attention has to be paid that the positive signals in the compensation tubes are at least as bright as the brightest signal in test samples for the corresponding fluorochromes. Also, one should always use the same fluorochrome for compensation as used in the samples, not merely one of all possible fluorochromes for a given detector (e.g. PE-Cy5, or PE-Ax647 for G660), as emission spectra and, therefore, spillover into other detectors that will have to be compensated for, can differ significantly between the various fluorochromes and dyes.
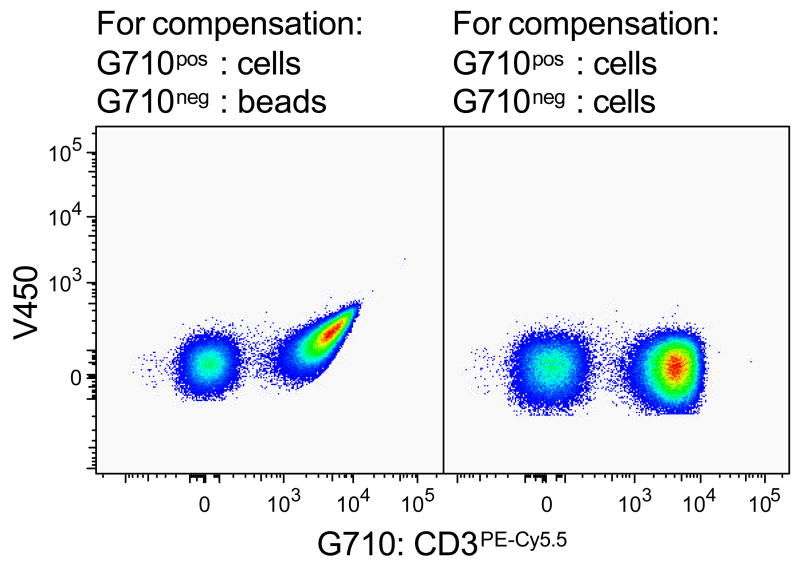
Compensation controls can be prepared by labelling either cells or antibody-capture beads (CompBeads) with antibody conjugates. Ideally, anti-mIgGκ coated CompBeads should be used for anti-human antibody conjugates, as all events will be positive and signal peaks will typically be narrow. However, this is not possible when the antibody conjugate of choice is not a mIgGκ clone, in which case cells will be labelled with the reagent in question. It is mandatory to use the same antibody conjugate carrier for negative and positive controls of each respective detector, as cells have very different autofluorescence properties than CompBeads – i.e. unlabeled cells have to be used as the negative control for compensation samples where cells are labelled as a positive control and not CompBeads and vice versa. If this is not respected, samples will not be properly compensated (Figure 5).
Figure 5. Compare like with like for compensation.
Both pseudo-colour plots depict the same PBMC sample labelled with anti-CD3PE-Cy5.5 antibody. PE-Cy5.5 fluorescence was compensated using this very sample. However, for the left dot plot unlabeled CompBeads were used as the PE-Cy5.5neg control, while unlabeled PBMC were used for the right dot plot.
Controls
It is of utmost importance to include all necessary controls from the beginning of a project, as not having the proper controls can markedly reduce the impact of an otherwise complete study. There are three categories of controls: 1) instrument set-up and validation, 2) gating controls, and 3) biological controls.
Instrument validation should be performed on a daily basis and stability of the system verified before each individual experiment. This consists in adjusting instrument parameters (e.g. voltages for individual detectors, optical filters) to ensure that its performance is stable over time and that measurements obtained on different days can be compared for analytical purposes. This is a prerequisite in order to validate data comparison during long-term studies. To evaluate the machine before acquiring experimental samples, we perform a quality control by measuring a sample of fluorescent beads that emit at known fluorescence intensities in all wavelengths measured by the photomultiplier tubes (PMTs) installed. Additionally, for every fluorochrome employed in the panel a singly labelled sample (cells or beads) needs to be acquired so that spillover fluorescence into other PMTs can subsequently be calculated by the analysis software for proper spectral compensation (see above).
Even after the flow cytometer has been set-up and validated and the ideal antibody panel finalized, gating controls and biological controls are crucial to be included in each experiment. Gating controls are also referred to as “fluorescence-minus-one” (FMO) controls, as all antibodies are used for the labelling of FMO controls except one – typically one that is directed against a weak antigen. Such FMO controls should be included for antigens of unknown expression, and tertiary antigens (see above).
Relevant biological controls will differ between research projects. For example, if cells are subjected to stimulation or other treatment prior to labelling and flow cytometric analysis, untreated, “co-stimulation only” controls, or samples having been subjected to a control treatment are included. In a time-course experiment, a “time zero” sample will be required in order to determine the baseline measurement, and when studying patient samples it is usually instructive to study corresponding samples from healthy donors.
Analysis and display of multi-colour flow cytometric data
First of all, the scientific question needs to be extremely well defined. With polychromatic flow cytometry one can easily get overwhelmed with the multi-faceted results. It is impossible, and not instructive, to look at every single sub-population defined by every possible combination of the antibodies used. Instead, one needs to determine ahead of time what it is that is going to be examined and use a consecutive gating strategy in order to obtain clear and interpretable data. Nevertheless, data can always be reanalyzed under a different perspective at a later time point if another scientific question comes up.
Primary analysis should be performed using plots with a “logicle” (biexponential) scale, which permit correct visualization of fluorescence signals of high and low intensities as well as those of negative values (10, 13).
Traditional dot plots or variations thereof (pseudo-colour plots, heat plots, zebra plots etc.), as well as logicle plots, are not necessarily the best way of representing multi-colour flow cytometry results. Nevertheless, it is useful to show a single example of the gating strategy applied, before presenting the digested results in a more unifying representation. There are a number of possible ways to collectively illustrate the findings obtained from large data sets. There is no gold standard, since different types of representations might be better suited for one type of analysis than another.
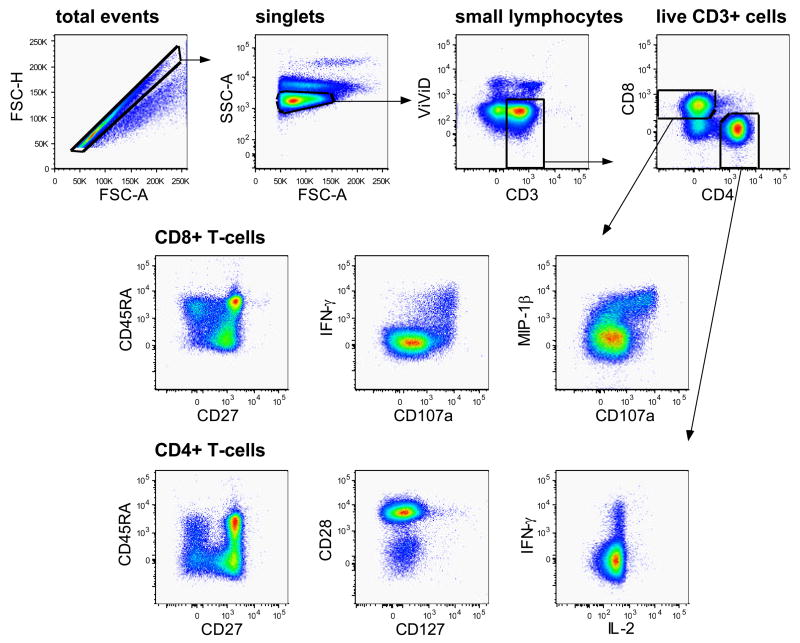
As mentioned above, an example of the gating strategy applied for analysis of the data should always be included in a publication, even as supplementary data available online. This is an integral part of the data presented, as it aids understanding of the final analyses as well as comparison with other data sets from independent publications. In the hierarchical gating strategy applied in our example (Figure 6) we first selected singlets as defined by having a similar height (FSC-H) and area (FSC-A) measurement in the forward scatter. Next, a gate was set around the small lymphocytes and if a dump channel were included, cells positive for unwanted antigens would be gated out at this point, before gating on live T-cells (ViViDneg CD3pos). These were then divided into CD4+ and CD8+ subsets, and each subset was separately analyzed for activation markers and immunological response proteins. It is important to note that gates for activation and response markers need to be created separately for individual cellular lineages or subsets. As an example, gates for CD45RA expression will be very different on CD4+ and CD8+ T-cells. The publication of such a gating tree allows for others to more fully evaluate the results presented, as well as paving the way for the reproduction of the experiments in other laboratories.
Figure 6. Gating strategy.
The sequential gating strategy applied for the analysis of CD4+ and CD8+ T-cell responses is illustrated by the gates and arrows. Healthy donor PBMC were incubated with anti-CD3 and anti-CD28 antibodies in the presence of CD107aAx680 and monensin.
Note that CD3 molecules were visualized by intracellular labelling. After gating on single-cells, analysis was narrowed down on small lymphocytes. Live CD3+ T-cells were identified and further subdivided into CD4+ and CD8+ T-cells. These were subsequently analyzed for activation markers and cytokines of interest. Some pseudo-colour plots for possible antigen combinations are illustrated.
Summary
Multicolor experiments can be frustratingly difficult to implement and optimize; however, it is extremely important to spend as much time as possible trying out variations and understanding in detail the impact of every reagent (and every labelling step) on the distribution of fluorescences in the final experiment. The resulting data sets will be far more interpretable and provide a rich source of information regarding the experimental conditions. While panel development is still largely empirical, we hope that heuristic approaches and experience will soon translate into algorithms that can be used to assist in the development of such panels.
Acknowledgments
This work is supported by the Intramural Research Program of the NIH, Vaccine Research Center, NIAID. The authors declare no conflict of interest.
Footnotes
Abbreviations: 7-AAD, 7-aminoactinomycin D; APC, allophycocyanin; Ax…, Alexa; CFP, cyan fluorescent protein; CFSE, carboxyfluorescein diacetate, succinimidyl ester; CMTMR, 5-(and-6)-(((4-chloromethyl) benzoyl) amino) tetramethylrhodamine; Cy… - cyanin; DAPI, 4′,6-diamidino-2-phenylindole dihydrochloride; ECD, energy-coupling dye; EMA, ethidium monoazide; FITC, fluorescein isothiocyanate; FMO, fluorescence-minus-one; FSC-A, forward scatter area; FSC-H, forward scatter height; GFP, green fluorescent protein; GrViD, green fluorescent amine reactive viability dye; HIV, human immunodeficiency virus; IFN, interferon; MFI, mean fluorescence intensity; MIP, macrophage inflammatory protein; OrViD, orange fluorescent amine reactive viability dye; PBMC, peripheral blood mononuclear cells; PD-1, programmed death-1; PE, phycoerythrin; PerCP, peridinin chlorophyll protein; PI, propidium iodide; PMT, photomultiplier tube; Q-dots, quantum dots; RFP, red fluorescent protein; TR, texas red (also known as ECD); UviD, ultraviolet excitable amine reactive viability dye; ViViD, violet fluorescent amine reactive viability dye.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Yolanda D. Mahnke, Visiting Fellow, ImmunoTechnology Section, Vaccine Research Center, NIAID, NIH, Bethesda, MD.
Mario Roederer, Senior Investigator, ImmunoTechnology Section, Vaccine Research Center, NIAID, NIH, Bethesda, MD.
References
- 1.Heeney JL, Plotkin SA. Immunological correlates of protection from HIV infection and disease. Nat Immunol. 2006 Dec;7(12):1281–1284. doi: 10.1038/ni1206-1281. [DOI] [PubMed] [Google Scholar]
- 2.Roederer M, Brenchley JM, Betts MR, De Rosa SC. Flow cytometric analysis of vaccine responses: how many colors are enough? Clin Immunol. 2004 Mar;110(3):199–205. doi: 10.1016/j.clim.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 3.Perfetto SP, Roederer M. Increased immunofluorescence sensitivity using 532 nm laser excitation. Cytometry A. 2007 Jan 2; doi: 10.1002/cyto.a.20358. [DOI] [PubMed] [Google Scholar]
- 4.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol. 2004 Aug;4(8):648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 5.Perfetto SP, Chattopadhyay PK, Lamoreaux L, et al. Amine reactive dyes: an effective tool to discriminate live and dead cells in polychromatic flow cytometry. J Immunol Methods. 2006 Jun 30;313(1–2):199–208. doi: 10.1016/j.jim.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 6.Chattopadhyay PK, Price DA, Harper TF, et al. Quantum dot semiconductor nanocrystals for immunophenotyping by polychromatic flow cytometry. Nat Med. 2006 Aug;12(8):972–977. doi: 10.1038/nm1371. [DOI] [PubMed] [Google Scholar]
- 7.Roederer M. Spectral compensation for flow cytometry: visualization artifacts, limitations, and caveats. Cytometry. 2001 Nov 1;45(3):194–205. doi: 10.1002/1097-0320(20011101)45:3<194::aid-cyto1163>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 8.Chattopadhyay PK, Yu J, Roederer M. A live-cell assay to detect antigen-specific CD4+ T cells with diverse cytokine profiles. Nat Med. 2005 Oct;11(10):1113–1117. doi: 10.1038/nm1293. [DOI] [PubMed] [Google Scholar]
- 9.Petrovas C, Casazza JP, Brenchley JM, et al. PD-1 is a regulator of virus-specific CD8+ T cell survival in HIV infection. J Exp Med. 2006 Oct 2;203(10):2281–2292. doi: 10.1084/jem.20061496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol. 2006 Jul;7(7):681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 11.Berhanu D, Mortari F, De Rosa SC, Roederer M. Optimized lymphocyte isolation methods for analysis of chemokine receptor expression. J Immunol Methods. 2003 Aug;279(1–2):199–207. doi: 10.1016/s0022-1759(03)00186-8. [DOI] [PubMed] [Google Scholar]
- 12.O’Brien MC, Bolton WE. Comparison of cell viability probes compatible with fixation and permeabilization for combined surface and intracellular staining in flow cytometry. Cytometry. 1995 Mar 1;19(3):243–255. doi: 10.1002/cyto.990190308. [DOI] [PubMed] [Google Scholar]
- 13.Parks DR, Roederer M, Moore WA. A new “Logicle” display method avoids deceptive effects of logarithmic scaling for low signals and compensated data. Cytometry A. 2006 Jun;69(6):541–551. doi: 10.1002/cyto.a.20258. [DOI] [PubMed] [Google Scholar]