Summary
The progression of progenitors to oligodendrocytes requires proliferative arrest and the activation of a transcriptional program of differentiation. While regulation of cell cycle exit has been extensively characterized, the molecular mechanisms responsible for the initiation of differentiation remain ill-defined. Here, we identify the transcription factor Yin Yang1 (YY1) as a critical regulator of oligodendrocyte progenitor differentiation. Conditional ablation of yy1 in the oligodendrocyte lineage in vivo, induces a phenotype characterized by defective myelination, ataxia and tremor. At the cellular level, lack of YY1 arrests differentiation of oligodendrocyte progenitors after they exit from the cell cycle. At the molecular level, YY1 acts as a lineage-specific repressor of transcriptional inhibitors of myelin gene expression (Tcf4 and Id4), by recruiting histone deacetylase-1 to their promoters during oligodendrocyte differentiation.
Thus, we identify YY1 as an essential component of the transcriptional network regulating the transition of oligodendrocyte progenitors from cell cycle exit to differentiation.
INTRODUCTION
During development, oligodendrocytes derive from precursor cells residing throughout the neural axis (Pringle and Richardson, 1993; Noll and Miller, 1993). Their maturation into myelin-forming cells is a complex event that requires exit from the cell cycle and the activation of a transcriptional program leading to expression of myelin genes. It was originally proposed that oligodendrocyte progenitor differentiation is intrinsically regulated by a “timing” mechanism that links the number of cell divisions to growth arrest and the initiation of differentiation (Temple and Raff, 1986). This mechanism was regulated by mitogens (Calver et al., 1998) and molecularly characterized by the progressive accumulation of cell cycle inhibitors (Durand et al., 1997). It was shown that genetic ablation of cell cycle regulators impaired differentiation of progenitors into oligodendrocytes and promoted the persistence of a proliferative state (Casaccia-Bonnefil et al., 1997; Durand et al., 1998). However, over-expression studies with viral vectors expressing cell cycle inhibitors did not induce differentiation of progenitors, even though cells arrested at the G1/S transition (Tikoo et al., 1998; Tang et al., 1999; Ghiani and Gallo, 2001). Together these studies suggested that cell cycle exit was necessary but not sufficient to induce differentiation of progenitors into oligodendrocytes. Because oligodendrocyte differentiation is dependent on the bioavailability of transcriptional activators and the corresponding decrease of transcriptional inhibitors (Wegner 2000; Stolt et al., 2002; Gokhan et al., 2005; Ligon et al., 2006; Marin-Husstege et al., 2006; Liu et al., 2006), in this study we investigate the mechanisms at the interface between cell cycle exit and the initiation of a transcriptional program of differentiation.
Previous studies from our group demonstrated that global deacetylation of histone H3 is one of the first events detected during this transition. Pharmacological inhibitors of histone deacetylases prevented the expression of myelin genes in vitro (Marin-Husstege et al., 2002) and developmental myelination in vivo (Shen et al., 2005) and suggested that histone deacetylation was necessary for the initiation of the transcriptional program leading to myelin gene expression.
To identify possible molecular links between histone deacetylation and transcriptional regulation of myelin genes, we screened the promoters of genes regulated by TSA, an inhibitor of HDAC activity during oligodendrocyte differentiation for the presence of common binding motifs using Gene2Promoter software (Genomatix Software GmbH, Munich). This analysis revealed the presence of the consensus sequence NNCCATNN (Shrivastava and Calame, 1994; Yant et al., 1995) for the transcription factor Yin Yang1 (YY1) in the promoter region of over 30% of the genes down-regulated by histone deacetylation during oligodendrocyte differentiation. YY1 is a zinc-finger protein, with the ability to bind DNA (Seto et al., 1991) and function as chromatin modifier (Thomas and Seto, 1999; Liu and Shi, 2005). Genetic ablation of YY1 in mice resulted in peri-implatation lethality and suggested a role for YY1 in cell proliferation and differentiation (Donohoe et al., 1999).
Here we present the first in vivo analysis of yy1 conditional knockout (cko) mice in the oligodendrocyte lineage and in combination with an integrated in vitro approach, we define YY1 as a molecular regulator of the transition from progenitors into myelinating oligodendrocytes.
RESULTS
Conditional ablation of YY1 in the oligodendrocyte lineage impairs myelination
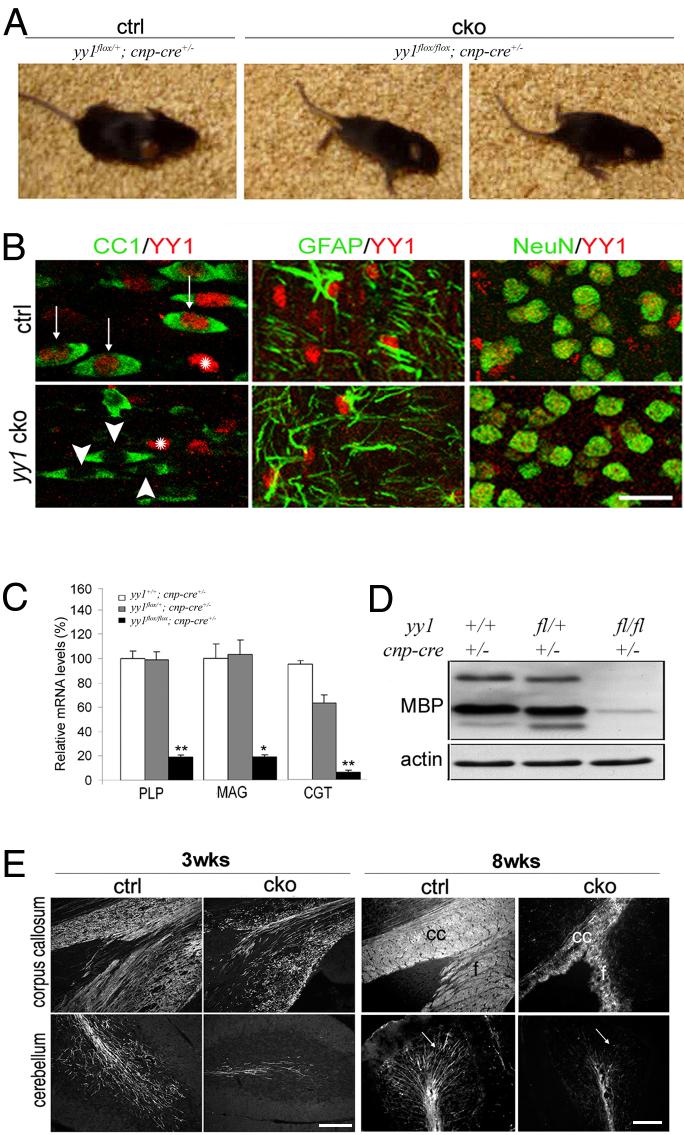
To assess the functional role of YY1 in oligodendrocyte differentiation in vivo, we generated yy1 cko mice by employing a Cre/lox strategy by crossing yy1flox/flox mice (Affar et al., 2006) with Cnp-cre line expressing the recombinase Cre from the oligodendrocyte lineage specific Cnp1 promoter (Lappe-Siefke et al., 2003). The resulting homozygous yy1flox/flox;cnp-cre+/− mice (yy1 cko; Fig. 1A) appeared normal at birth but developed shaking, ataxia, tremor and head wobbling by 14 days of age (Suppl.Video) that persisted throughout adulthood. The cell-specific ablation of yy1 gene in cells of the oligodendrocyte lineage was confirmed by double immunofluorescence using antibodies against YY1 and against markers for different cell types: CC1 for oligodendrocytes, GFAP for astrocytes and NeuN for neurons (Fig. 1B). The lack of YY1 immunoreactivity in CC1+ oligodendrocytes, but not in other cell types, indicated that the gene was efficiently excised in this lineage, although it was still expressed in astrocytes and neurons. Heterozygotes yy1flox/+;cnp-cre+/− mice did not show behaviour abnormalities and were undistinguishable from wild type mice. Because tremor and ataxia in yy1 cko mice were highly reminiscent of the shaking phenotype described in myelin-deficient mouse mutants (Nave et al., 1994; Griffiths, 1996), we asked whether lack of YY1 inhibited myelin formation. Quantitative RT-PCR revealed 80% lower levels of transcripts for proteolipid protein (PLP), myelin associated glycoprotein (MAG) and ceramide-galactosyl transferase (CGT) in the brains of p18 yy1 cko mice compared to yy1+/+;cnp-cre+/− controls (Fig. 1C). Decreased myelin proteins were also detected by western blot analysis and immunohistochemistry in the CNS of homozygous yy1 cko mice, but not in heterozygous mice (Fig. 1D-E). To determine whether this defective myelin gene expression persisted through adulthood, we analyzed the brain (Fig. 1E) and spinal cord (data not shown) of adult cko mice and control siblings. Defective myelination was still detected in 8 week-old yy1 mutant mice, thereby indicating the lack of compensatory mechanisms.
Figure 1. Conditional ablation of YY1 in the oligodendrocyte lineage in mice results in a trembling phenotype.
(A) Conditional yy1 knockout mice (cko), obtained by crossing yy1flox/flox with Cnp-cre mice displayed progressive tremor, ataxia and head wobbling during the second-third postnatal week. Examples of abnormal posturing and gait in the mutant mice (cko) at postnatal day 18 (p18) are shown in comparison with control siblings (ctrl). (B) Double immunostaining of cells in white matter tracts of p18 mouse sections stained for YY1 (red) and cell-specific markers (green): CC1 for oligodendrocytes, GFAP for astrocytes and NeuN for neurons. Note the specific deletion of YY1 in oligodendrocytes. Scale bar = 20 μm. (C) Quantitative real-time PCR of brain RNA from wild type (yy1+/+; cnp-cre+/-), heterozygotes (yy1flox/+; cnp-cre+/-) and conditional knockout (yy1flox/flox; cnpcre+/-) mice at p14. The levels of the indicated transcripts are normalized to GAPDH and the mRNA control levels are arbitrarily set as 100. The bar graph indicates decreased myelin gene transcripts in yy1 cko mice (black), but not in heterozygous siblings (gray) compared to controls (white) *p<0.05, **p<0.01. (D) Western blot analysis of protein lysates from the cortex of p14 mice revealed decreased myelin basic protein (MBP) expression in the mutants. Beta-actin serves as loading control. (E) Brain sagittal sections stained for PLP (3wks) and for MBP (8wks) reveal fewer myelinated fibers in the corpus callosum (cc), fornix (f) and cerebellum of yy1 cko mice compared to controls (ctrl). Scale bar = 100 μm.
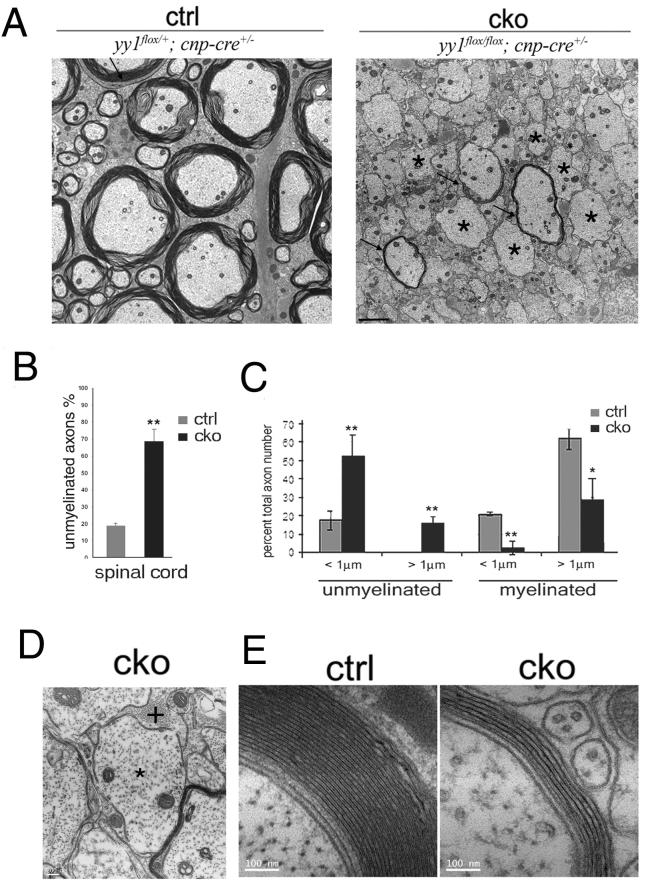
Defective myelination was confirmed by electron microscopy of the spinal cord, that revealed a 70% reduction of myelinated axons in yy1 cko mice compared to controls (Fig. 2A-C). The few myelinated axons in yy1 cko mice were characterized by thinner myelin sheaths (Fig. 2D) and higher g ratios (0.892±0.045) compared to control siblings (0.774±0.047, p<0.0001). Therefore, we concluded that the ablation of yy1 in oligodendrocyte progenitors significantly impaired developmental myelination.
Figure 2. Defective myelination in the spinal cord of yy1 conditional knockout mice.
(A) Ultrastructural analysis of spinal cord sections shows lack of myelin (asterisks) and thinner myelin sheaths (arrows) in p18 yy1 cko mice compared to controls. Scale bar= 2 μm. (B) Bar graph shows increased % of unmyelinated axons relative to total number of axons in the spinal cord of yy1 cko mice (black) compared to controls (gray). (C) Bar graph shows the relative distribution of unmyelinated and myelinated axons in relation to axonal diameter in the spinal cord of yy1 cko mice (black) and controls (gray) at p18 *p<0.05; **p<0.01. (D) Representative EM picture of a large yy1 cko spinal cord axon (diameter >1.5μm) that lacks myelin (*) and is in contact with an astrocytic process (+). Scale bar = 200 nm. (E) Representative EM pictures of myelinated axons of equivalent diameter in control and yy1 cko siblings. Scale bar = 100nm.
The phenotype detected in yy1 cko mice could be explained by hypothesizing a role for YY1 as transcriptional activator of myelin gene expression. To test this hypothesis, YY1 was over-expressed in the murine oligodendrocyte progenitor cell line Oli-neu (Jung et al., 1995), in the absence or presence of myelin gene promoters driving luciferase reporters. The increased levels of YY1 neither affected the levels of endogenous myelin transcripts, nor activated luciferase reporter activity from the transfected myelin promoters (Suppl. Fig S1). Similar results were obtained in rat derived CG4 cells, when the cells were cultured in the presence of mitogens (data not shown). However, when the cells were cultured in differentiation conditions, YY1 over-expression increased the activity of myelin gene promoters. Interestingly, YY1 increased not only the activity of promoters containing YY1 binding sites (i.e. PLP, Berndt et al., 2001), but also of myelin gene promoters (i.e. CGT and MBP) lacking consensus sequence for YY1 (Suppl. Fig. S1). Together these data suggested a more global role for YY1 in regulating myelin gene expression.
Impaired myelination in yy1 cko mice is due to arrested maturation of oligodendrocyte progenitors
A potential role for YY1 in the oligodendrocyte lineage, suggested by the hypomyelinating phenotype of yy1 cko mice was a deficit in the maturation of oligodendrocyte progenitors into myelin-forming cells. To test this hypothesis, we conducted a temporal immunophenotypic analysis of oligodendrocyte lineage cells in brain and spinal cord sections of control and mutant mice at postnatal day 2, 11 and 18 (Fig. 3 and Suppl. Fig. S2 and S3). These time points were chosen because they define the period of oligodendrocyte differentiation and developmental myelination (Bjelke and Seiger, 1989; Hamano et al., 1998). We used double immunofluorescence with antibodies specific for NG2 to identify progenitors and CC1 to identify mature oligodendrocytes. Confocal microscopy was used for image acquisition and the relative proportion of NG2+ and CC1+ cells was calculated as a percentage of the total oligodendrocyte lineage population. At p2, a similar proportion of NG2+ progenitors and CC1+ cells was detected in the spinal cord of conditional mutants and control siblings (Fig. 3, Suppl. Table 1). By p18, however, the oligodendrocyte lineage population of control mice was composed of 60.6±2.9% CC1+ cells while in yy1 cko mice only 21.6±1% of the cells were CC1+. Similar differences were observed in the developing corpus callosum (Suppl. Fig. S2). These results were confirmed by immunohistochemistry using antibodies specific for the progenitor marker PDGFRα and for the lipid sulfatide recognized by O4 (Suppl. Fig. S2). Therefore we concluded that YY1 plays a critical role in modulating the differentiation of progenitors into CC1+ oligodendrocytes.
Figure 3. Impaired oligodendrocyte progenitor differentiation in the spinal cord of yy1 conditional mutants.
(A) Confocal image of lumbar spinal cord sections from controls (ctrl) and yy1 cko mice at postnatal day 2, 11 and 18, stained with antibodies specific for NG2 (green) to identify progenitors, CC1 (red) to identify oligodendrocytes and DAPI (blue) to counterstain nuclei. Optical sections (Z = 1.0 μm; X = 12 μm) of confocal epifluorescence images were sequentially acquired and LSM software was used to merge images. Examples of NG2+/DAPI+ and CC1+/DAPI+cells selected for counting are shown in the boxed areas (arrowheads) while their relative position is shown at low magnification (arrows). Scale bar=50μm, 10μm in inserts. (B) Quantification of the data shown in panel A. The values indicate mean ± SD of cell counts obtained in 3-4 mice of each genotype per time point. (C) Co-localization of progenitor markers PDGFRα (red) and NG2 (green) in the spinal cord of yy1 cko mice at p18. Scale bar=20μm
Since YY1 had been previously reported to negatively regulate p53-induced apoptosis in lymphocytes (Sui et al., 2004, Gronroos et al., 2004), we asked whether the dramatic decrease of CC1+ cells detected in the yy1 cko mice could have been caused by increased apoptosis. TUNEL assay (Gavrieli et al., 1992) revealed a consistent, but statistically insignificant increase of total apoptotic cells in the corpus callosum of yy1 cko mice compared to control siblings (Suppl. Fig. S2). Similar results were obtained using immunohistochemistry and antibodies specific for the activated form of the apoptosis effector caspase-3 (Gown and Willingham, 2002). At all the developmental time points analyzed (p2, p11 and p18) we detected a statistically insignificant increase of total apoptotic cells in yy1 cko compared to sibling controls (Suppl. Fig. S4).
To further define the effect of YY1 deletion on oligodendrocyte differentiation, we performed in vitro ablation experiments. Primary cultures of neonatal cortical oligodendrocyte progenitors, from yy1 flox/flox mice, were transduced with adenoviral vectors (Gil-Perotin et al., 2006) expressing the recombinase Cre from a CMV promoter (adeno-CMV-Cre). Untransduced cultures of yy1 flox/flox progenitors were used as controls. The efficient excision of yy1 was confirmed by immunocytochemistry (data not shown). Differentiation was induced by mitogen withdrawal and the progression along the lineage was followed using stage specific markers, including immunoreactivity for the late progenitor marker O4 and for the mature oligodendrocyte markers GalC, MBP and PLP (Fig. 4A). The proportion of immunoreactive cells in the YY1+ and in the YY1− population was quantified (Fig. 4B). After 3 days in differentiation conditions, more than 45% of control cultures became O4+, and after two additional days a large proportion of the cells displayed the characteristically branched morphology of mature oligodendrocytes and immunoreactivity for myelin proteins. In contrast, in Cre transduced cells, at day 3 only 26±3.1% of cells were characterized by simple morphology and O4+ immunoreactivity and by day 5 all cells lacked the expression of myelin gene products.
Figure 4. In vitro ablation of yy1 in oligodendrocyte progenitors prevents their differentiation into oligodendrocytes.
(A) Primary oligodendrocyte progenitors isolated from the cortex of neonatal yy1flox/flox mice were transduced with adenoviral vectors expressing the recombinase Cre (CMV-Cre). Forty-eight hours later the cells were differentiated by mitogen withdrawal. Immunoreactivity for O4 (green) was assessed 3 days later, while immunoreactivity for MBP, GalC or PLP (green) was assessed 5 days after transduction. Note that the Cre+ yy1flox/flox cells (red) progressed to the O4+ stage, but were unable to differentiate into myelin-expressing cells. Scale bar = 50μm. (B) Quantification of three distinct experiments performed in duplicate. * p<0.05, ***p<0.001.
Therefore, ablation of yy1 in oligodendrocyte progenitors was incompatible with their differentiation into myelin-forming cells.
Defective differentiation in yy1 cko mice is independent on impaired cell cycle exit
Since the majority of the cells in yy1 cko mice retained the progenitor markers NG2 and PDGFRα, it was conceivable that the arrested maturation of these cells was due to their abnormal persistence in the cell cycle. To test this hypothesis we assessed proliferation and cell cycle exit at several time points of post-natal development in yy1 cko mice and control siblings (Fig. 5). The proliferation rate was inferred by the relative proportion of cells in S-phase, calculated after a 1 hour in vivo pulse-labelling with the thymidine analogue bromodeoxyuridine (BrdU). Immunohistochemical analysis of sections of the neonatal spinal cord of yy1 conditional knockout and control mice revealed a similar progressive reduction of the proportion of BrdU labeled cells as the animals developed from p2 to p18 (Fig. 5A-B). Similar results were obtained in primary cultures of yy1flox/flox oligodendrocyte progenitors that were either not transduced or transduced with adeno-CMV-Cre, in order to ablate YY1 in vitro (Suppl. Fig. S5A). Consistently, over-expression of YY1 in immortalized Oli-neu progenitors did not change the proportion of BrdU labelled cells (Suppl. Fig. S5B) and did not affect the levels of cell cycle regulators (Suppl. Fig. 5C). Together these data suggest that YY1 does not affect cell division and that the NG2+ cells in the yy1 cko mice were not proliferating.
Figure 5. Lack of YY1 does not affect the ability of oligodendrocyte progenitors to exit from the cell cycle.
(A) Confocal image of spinal cord sections of p2 yy1 cko and control siblings (ctrl) in vivo labeled with BrdU for 1 hour. Immunohistochemistry with YY1 (red), BrdU (green) and DAPI (blue) revealed a similar distribution BrdU+ cells in mice of the two genotypes. Scale bar = 20μm. (B) Quantification of BrdU+ cells in white matter tracts of the spinal cord at the indicated time points; *p<0.05. (C) Representative images of spinal cord sections of control and cko mice stained with antibodies against Ki-67 (green) to identify cells in any phase of the cell cycle except for G0 and BrdU (red) to identify cells in S phase at the time of labeling. (D) Cell cycle exit index was calculated by dividing the number of BrdU+/Ki-67− cells, by the total number of BrdU+ cells. Scale bar = 20μm.
To further rule out the possibility that oligodendrocyte progenitors were unable to exit from the cell cycle in yy1 mutant mice, we performed double labeling with BrdU and Ki-67 to calculate the cell cycle exit index (Chenn and Walsh, 2002; Siegenthaler and Miller, 2005). Conditional yy1 mice and control siblings received a single BrdU injection and were sacrificed one hour later. The brains were sectioned and immunostained with antibodies specific for BrdU, to label a cohort of cells in S-phase, and for Ki-67, to label proliferating cells throughout the phases of the cell cycle, but not in G0. Double immunoreactive BrdU+/Ki-67+ cells measured the proportion of cycling cells, while the remaining BrdU+ /Ki-67− cells indicated cells that had exited from the cell cycle. The cell cycle exit index was calculated by dividing the number of BrdU+/Ki-67− cells by the total population of BrdU+ cells. Our results indicate that at p2, p11 and p18 the cell cycle exit index was similar in cko and control mice (Fig. 5C-D). From these data we conclude that lack of YY1 does not affect the ability of oligodendrocyte progenitors to exit from the cell cycle. In agreement with the similar apoptotic rate and cell cycle exit index, the total number of cells measured in yy1 cko mice was similar to the number measured in control siblings, at each of the developmental points analyzed (Suppl. Table I).
Thus, the defective differentiation of oligodendrocyte progenitor observed in the knockout mice could not be explained in terms of increased apoptosis or persistent proliferation.
YY1 repress the expression of myelin gene transcriptional inhibitors
To understand how YY1 regulates oligodendrocyte progenitor differentiation, we performed a gene expression profile study of postnatal day 2 cko and control brains, using Affymetrix microarrays. Among the genes whose expression was affected by YY1 deletion, we focused our analysis on those encoding for transcription factors and validated their expression by RT-PCR (Fig. 6A) and quantitative RT-PCR (Fig. 6B). We detected a nine fold increase of the transcription factor Tcf4, a four fold increase of the differentiation inhibitor Id4, and a two fold increase of the HMG-protein Sox11(Fig. 6B). No changes were detected in the expression levels of nestin or REST, a gene that was previously shown to be regulated by YY1 in mouse embryonic fibroblasts (Affar et al., 2006). A repressive role for YY1 on the transcriptional regulation of the transcription factors Tcf4 and Id4 was further supported by over-expression studies in oligodendrocyte progenitors (Fig. 6C).
Figure 6. YY1 is a repressor of oligodendrocyte differentiation inhibitors Id4 and Tcf4.
(A) Validation of gene expression profiling studies in the cortex of yy1 cko mice and control littermates at p2, by semi-quantitative (A) and quantitative (B) RT-PCR. The RNA levels were normalized to the levels of GAPDH and the values in the control mice were arbitrarily set as 1. *p<0.05. (C) The expression level of Tcf4 and Id4 was down-regulated in oligodendrocyte progenitors transfected with pCX-yy1-EGFP (YY1) and not in cells transfected with pCX-EGFP vector as control. (D) Luciferase activity of immortalized oligodendrocyte progenitors (Oli-neu cells) co-transfected with Tcf4, and either myelin (MBP-luc, CGT-luc) or astrocytes-specific (GFAP-luc) promoters driving luciferase reporters. The luciferase activity in pCDNA3 transfected controls was arbitrarily set as 100; ** p<0.01.
Id4 is a well characterized inhibitor of oligodendrocyte progenitor differentiation and myelin gene expression (Kondo and Raff, 2000; Samanta and Kessler 2004; Marin-Husstege et al., 2006). Tcf4 is a critical downstream effector of Wnt signalling (Cho and Dressler, 1998; Tetsu and McCormick, 1999), but its role in the oligodendrocyte lineage has not been characterized. To define the functional role of Tcf4 in the oligodendrocyte lineage, we co-expressed Tcf4 with myelin gene promoters (i.e. MBP, CGT) driving luciferase reporter genes. Co-transfection of Tcf4 with luciferase reporter driven by the astrocyte-specific promoters GFAP was used as control. Interestingly, Tcf4 significantly decreased the activity of myelin gene promoters compared to vector transfected controls, but did not affect the activity of the GFAP promoter (Fig. 6D).
Together, these data support a role for YY1 in oligodendrocyte progenitor differentiation that is dependent on the down-regulation of transcriptional inhibitors of myelin genes.
Lineage-specific effect of YY1 in oligodendrocyte progenitor differentiation
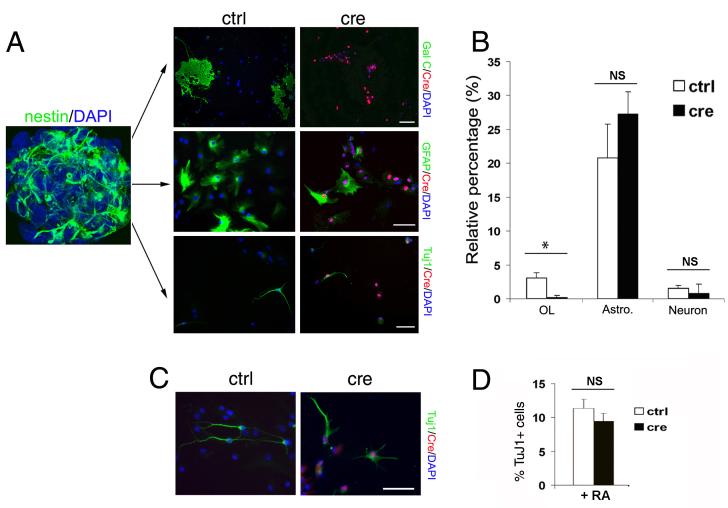
The effect of YY1 ablation on oligodendrocyte progenitor maturation raised the question of the specificity of this transcription factor for the oligodendrocyte lineage. In other words, it was important to define whether the function of YY1 in oligodendrocyte progenitor differentiation was specific or whether YY1 was a transcription factor that is universally required for the differentiation into any neural lineage. We therefore, performed in vitro ablation of YY1 in multipotential nestin+ precursors and then allowed them to differentiate into distinct lineages (Fig. 7). Nestin+ precursors were generated from the subventricular zone of neonatal yy1 flox/flox mice and grown in suspension as neurospheres (Fig. 7A). After dissociation, the cells were transduced with adeno-CMV-Cre and yy1 excision was confirmed by immunocytochemistry (data not shown). Untransduced cultures were used as controls. Differentiation was induced by culturing the cells in a medium permissive for differentiation along distinct lineages (Belachew et al., 2003) and the cells were analyzed by immunocytochemistry, using markers specific for astrocytes (i.e. GFAP), oligodendrocytes (i.e. GalC) and neurons (i.e. TuJ1) (Fig. 7A). The number of immunoreactive cells in each lineage that was either YY1+ or YY1− was counted and expressed as percentage of the total population of cells (Fig. 7B). Our results showed a decrease of newly generated oligodendrocytes in cells lacking YY1 but no statistically significant effect on the generation of neurons or astrocytes (Fig. 7B). To further evaluate the effect of yy1-deletion on neuronal differentiation, we repeated the experiment and cultured the cells in conditions promoting neuronal differentiation (i.e. in the presence of retinoic acid, Wang et al., 2005). Also in this case, yy1-deletion did not significantly affect neurogenesis from neonatal precursor cells (Fig. 7C-D).
Figure 7. In vitro ablation of YY1 in nestin+ precursor cells impairs oligodendrocyte differentiation in a cell type specific manner.
(A) Subventricular zone (SVZ) cells were isolated from neonatal yy1flox/flox mice and cultured as nestin+ (green) neurospheres. Dissociated neurospheres without (ctrl) or with adenovirus-CMV-Cre transduction (cre) were allowed to differentiate into oligodendrocytes, astrocytes and neurons. Seven days later the cultures were stained for Cre (red) and for the lineage specific markers (green): Gal C (oligodendrocytes), GFAP (astrocytes) and Tuj1 (neurons). Scale bar=50μm. (B) Bar graphs indicate the percentage of immunoreactive cells relative to total cell number. * p<0.05. (C) The experiment was repeated in medium supplemented with retinoic acid (RA) to promote neuronal differentiation. (D) Bar graphs show that even in this culture condition no significant difference was detected between control and yy1- cells.
We therefore hypothesized that the lineage-specific role of YY1 in the oligodendrocyte lineage included the transcriptional regulation of the inhibitory transcription factors Tcf4 and Id4. To test this hypothesis, YY1 was over-expressed in progenitors and the cells were later allowed to differentiate either into oligodendrocytes or into astrocytes. The RNA was isolated and the relative levels of the transcripts were measured by quantitative real time PCR (Fig. 8A). Consistent with a specific role of YY1 in the oligodendrocyte lineage, the levels of Tcf4 and Id4 were decreased only when cells differentiated into oligodendrocytes, but not when they differentiated into astrocytes (Fig. 8A). To understand the molecular basis of this YY1 dependent lineage-specific repression of Tcf4 and Id4, we performed co-immunoprecipitation with YY1 and histone deacetylases (i.e. HDACs), the chromatin modifying enzymes involved in transcriptional repression and previously shown to be critical for oligodendrocyte differentiation (Marin-Husstege et al., 2002; Shen et al., 2005). YY1 was immunoprecipitated from protein extracts generated from undifferentiated progenitors and from cells induced to differentiate along the oligodendrocyte lineage. As control for lineage specificity, we also included extracts from cells induced to differentiate into astrocytes by BMP4 treatment. The association with histone deacetylases (i.e. HDAC) was assayed by western blot, using antibodies specific for HDAC isoforms 1 and 2 (Fig. 8B). The association of YY1 with HDAC was weakly detected in undifferentiated progenitors and astrocytes, and was enhanced in differentiating oligodendrocytes (Fig. 8B). Next we examined the functional status of YY1 in relation to the differentiative state of progenitors, using YY1 TransLucent Reporter Vector (Panomics, CA). This vector contains a luciferase reporter under the control of an enhancer element containing multiple YY1 binding sites, upstream of the minimal Herpes Simplex thymidine kinase promoter (Dai et al., 2005). After transfection of this reporter vector in undifferentiated progenitors, cells were exposed to mitogenic, astrogliogenic or oligodendrogliogenic stimuli and luciferase activity was measured. In differentiating oligodendrocytes the Luciferase activity was 3 fold higher than in astrocytes (Fig. 8C). Therefore, the functional ability of YY1 to bind DNA and form protein complexes with HDAC were greater when progenitors differentiated into oligodendrocytes rather than into astrocytes.
Figure 8. The YY1-dependent repression of Tcf4 and Id4 is lineage-specific and mediated by recruitment of HDAC1 to their promoters.
(A) Quantitative RT-PCR of Tcf4, Id4 and REST in oligodendrocyte progenitors transfected with pCX-EGFP (vector) or pCX-yy1-EGFP (YY1) and induced to differentiate into astrocytes (astro) or oligodendrocytes (oligo). The transcript levels of each gene in vector transfected cells was arbitrarily set as 100; *p<0.05. (B) Co-immunoprecipitation of YY1 and HDAC-1 and -2. Western blot analysis of whole cell lysates (lysates) and YY1 immunoprecipitated protein extracts (IP:YY1) derived from undifferentiated progenitors (prog) and cells differentiated into oligodendrocytes (diff prog) or astrocytes (astro). (C) YY1 activity, measured by TransLucent vector reporter system. Note the increased activity of YY1 in cultures of progenitors differentiating into oligodendrocytes (diff. prog.) compared to undifferentiated cells (prog.) or cells differentiating into astrocytes (astro). **p<0.01. (D) Chromatin immunoprecipitation (ChIP) of samples isolated from progenitors (prog), differentiating oligodendrocytes (diff) or astrocytes (astro) and immunoprecipitated with antibodies against YY1 and HDAC1. The diagram shows the Tcf4 promoter with the relative position of the YY1 consensus sequences (black boxes) and the regions of DNA (roman numerals) amplified by specific primer pairs (arrows). Input DNA was used as positive control, while ChIP in the absence of antibodies or amplification of immunoprecipitated chromatin with primers for region IV were used as negative controls. (E) ChIP of samples isolated in the same conditions described above. The diagram shows the Id4 promoter, with the position of the YY1 consensus sequence (black box) and the regions of the promoter (roman numerals) amplified by specific primer pairs (arrows). YY1 was bound to the Id4 promoter in progenitors, but it recruited HDAC1 only when cells differentiated into oligodendrocytes. No binding was observed in progenitors differentiating into astrocytes. (F) Model of oligodendrocyte progenitor differentiation as two step event. First, proliferating progenitors exit from the cell cycle and remain in an undifferentiated state characterized by high levels of transcriptional inhibitors (Id4, Tcf4) and lack of myelin gene expression. As the progenitors begin differentiating, repressive complexes containing YY1 and HDAC1 are recruited to the promoter of these inhibitors. The decreased levels of these inhibitory molecules allow myelin gene expression to begin.
We then asked whether HDAC1 was recruited to YY1 binding sites [NNCCATNN (Shrivastava and Calame, 1994; Yant et al., 1995)] in the promoter region of Tcf4 and Id4 (Fig. 8D and 8E). Chromatin immunoprecipitation (ChIP) was performed using antibodies against YY1 or HDAC1 and the DNA was amplified using primers flanking the promoter regions containing YY1 binding sites. Immunoprecipitation without antibodies and amplification with primers in regions lacking YY1 consensus sequences were used as negative controls. We detected increased YY1 and HDAC1 recruitment to the same regions of the Tcf4 promoter containing YY1 binding sites in differentiating cells compared to undifferentiated cells (Fig. 8D). This was not observed when progenitors differentiated into astrocytes (Fig. 8D). A similar mechanism was detected for the YY1-dependent regulation of the Id4 promoter (Fig. 8E). Also in this case the enhanced recruitment of HDAC1 to the YY1 binding sites was observed when progenitors differentiated into oligodendrocytes, but not when they differentiated into astrocytes (Fig. 8E).
From these combined in vitro and in vivo evidence, YY1 emerges as a lineage-specific modulator of the transcriptional network leading to oligodendrocyte differentiation (Fig. 8F).
Discussion
Oligodendrocyte progenitor differentiation is regulated by intrinsic and extrinsic mechanisms
Myelination of the CNS is dependent on the correct execution of a genetic program of oligodendrocyte differentiation that culminates with the timely and coordinated expression of several myelin genes. It is well established that differentiation of oligodendrocytes from progenitors requires exit from the cell cycle and the initiation of a transcriptional program leading to myelin gene expression (Bogler et al., 1990; Casaccia-Bonnefil and Liu, 2003). However, the events occurring during this transition remain unidentified. This study identifies a novel function for the transcription factor YY1 as a molecule regulating the early stages of oligodendrocyte progenitor differentiation after cell cycle exit (Fig. 8F).
Original studies in clonal cultures of oligodendrocyte progenitors purified from the optic nerve had suggested the existence of a mechanism linking the activation of a differentiation program to the exit from the cell cycle (Temple and Raff, 1986; Hart et al., 1989). This concept was supported by the evidence that mitogens prevent oligodendrocyte progenitor differentiation (Gard and Pfeiffer, 1993; Barres et al., 1994; Calver et al., 1998; Nakatsuji and Miller 2001), and that the ability of oligodendrocyte progenitors to timely differentiate is delayed in animals with genetic deletion of cell cycle regulators (Casaccia-Bonnefil et al., 1997 &1999; Durand et al., 1997 & 1998). Together these studies led to a model of lineage progression as binary state, such that a progenitor could either proliferate while remaining undifferentiated or progress to post-mitotic differentiated oligodendrocyte. However, the induction of cell cycle exit was not sufficient, per se, to induce oligodendrocyte progenitor differentiation (Tikoo et al., 1998; Tang et al. 1999). Therefore, “differentiation” was redefined as the linear succession of sequential states, from proliferative and undifferentiated cells to quiescent progenitors to mature oligodendrocytes (Pfeiffer et al., 1993), each defined by a complex balance of positive (i.e. Sox10, Mash1) and negative (i.e. Id4, Hes5) regulators (Gokhan et al., 2005; Liu et al., 2006).
Previous studies from our laboratory defined HDAC activity as necessary during the transition between cell cycle exit and the initiation of a developmental program of differentiation (Marin-Husstege et al., 2002; Shen et al., 2005). In this study we identify YY1 as the molecule that recruits HDAC to the promoter of transcriptional inhibitors of myelin genes (i.e. Id4, Tcf4). This YY1-mediated repressive event occurs only when progenitors differentiate into oligodendrocytes, but not when they become astrocytes.
YY1 deletion impairs oligodendrocyte progenitor differentiation without affecting cell cycle exit
The most prominent feature of in vivo ablation of YY1 in the oligodendrocyte lineage is the detection of a shaking phenotype characterized by ultrastructural evidence of defective developmental myelination, due to impaired differentiation of oligodendrocyte progenitors. In the spinal cord, small caliber axons show complete lack of myelin and a small percentage of large caliber axons are wrapped by thin myelin sheaths. At the biochemical level, defective myelination in the yy1 cko mice is associated with lower levels of myelin proteins and transcripts compared to sibling controls and inefficient differentiation of progenitors into myelinating oligodendrocytes.
In the CNS of control mice, progenitors differentiate into myelinating oligodendrocytes during the first three weeks of postnatal development. Differentiation can be assessed by detecting a progressive decline in the number of cells expressing progenitor markers and the corresponding increase in the proportion of differentiated cells. In yy1 mutants, we detect a greater proportion of cells expressing progenitor markers that is paralleled by a smaller proportion of mature oligodendrocytes.
A potential explanation for defective oligodendrocyte progenitor differentiation was the possibility that progenitors were retained in a proliferative state due to lack of YY1. However, the ability of progenitors to exit from the cell cycle and the number of cells in S-phase are not affected by YY1 levels. In addition, the increased absolute number of progenitors during the first postnatal days, a distinctive feature of defective differentiation due to persistent proliferation (Casaccia-Bonnefil et al., 1997), is not detected in yy1 cko mice. Rather, the differences between yy1 cko mice and control siblings are best detected at later developmental time-points, coincident with the acquisition of a differentiated phenotype.
Based on this cumulative evidence, we conclude that YY1 modulates oligodendrocyte progenitor differentiation independent of cell division.
YY1-dependent pathways modulating myelination
We have previously discussed the importance of positive and negative regulators of myelin gene expression in defining the transcriptional network that defines the myelinating phenotype. In this study, the experimental results identify the role of YY1 in oligodendrocyte differentiation as lineage-specific repressor of genes that inhibit myelin gene expression, including the repressive HLH protein Id4 and the HMG protein Tcf4. Higher levels of Id4 and Tcf4 were detected in the brains of yy1 cko mice compared to control siblings, and lower levels of Id4 and Tcf4 were detected in YY1 over-expressing cells compared to the vector transfected controls. The role of Id4 as an inhibitor of oligodendrocyte progenitor differentiation and myelin gene expression has been previously described (Kondo and Raff, 2000; Samanta and Kessler 2004; Marin-Husstege et al., 2006). The role of Tcf4 as inhibitor of oligodendrocyte differentiation is less well defined. Tcf4 is a member of the Tcf/Lef family of HMG box transcription factors, and it is highly expressed in the developing central nervous system (Cho and Dressler, 1998; Korinek et al., 1998). Tcf4 is functionally up-regulated by the Wnt signaling pathway (Korinek et al., 1998), which has been shown to inhibit oligodendrocyte differentiation (Shimizu et al., 2005) and down-regulated by the Shh pathway which favours oligodendrogliogenesis (Ishibashi and McMahon, 2002). Other studies indicated that the levels of Tcf4 are up-regulated by the inhibition of HDAC activity (Saegusa et al., 2005; Yamaguchi et al., 2005). Taken together with our observation that Tcf4 represses the promoter activities of myelin gene mbp and cgt, this identifies Tcf4 as a novel inhibitor of oligodendrocyte differentiation. Therefore YY1 acts as repressor of transcriptional inhibitors of the bHLH (i.e. Id4) and HMG (i.e. Tcf4) family and enhances progenitor differentiation and myelin gene expression. This explains also why YY1 over-expression increased the luciferase activity of reporters driven not only by the PLP-promoter (which contains YY1 consensus sequences), but also that of reporters driven by MBP or CGT promoters (which lack YY1 consensus sequences) when the cultures were exposed to differentiation conditions.
An alternative possibility was recently suggested by the report that YY1 can also regulate the promoter of BACE1 (beta-site amyloid precursor protein-cleaving enzyme 1) in primary neurons and astrocytes (Nowak et al. 2006). BACE1 has been shown to influence myelination through cleavage of neuregulins (Willem et al., 2006; Hu et al., 2006). Even though the levels of BACE were not down-regulated in the yy1 cko (data not shown), we cannot exclude the possibility that neuronal or astrocytic regulation of BACE1 levels by YY1 could influence myelination in normal development.
From these data, we conclude that the effect of YY1 on myelin gene expression is indirect and is mediated by the recruitment of the chromatin modifier enzyme HDAC to YY1 binding sites in the promoter of transcriptional inhibitors of myelin gene expression.
The effect of YY1 on oligodendrocyte progenitor differentiation is lineage-specific
This study supports a novel and cell-type specific role of YY1 in the oligodendrocyte lineage. In differentiating myoblasts (Lee et al., 1994, Latinkic et al., 2004) and keratinocytes (Xu et al., 2004), YY1 prevents the expression of differentiation genes and is down-regulated during differentiation (Walowitz et al., 1998). In contrast, in oligodendrocyte lineage cells YY1 facilitates differentiation of progenitors and its expression remains constant throughout development. Importantly, in vitro ablation of YY1 in nestin+ precursor cells prevents generation of oligodendrocytes but not other lineages. Finally, the transcriptional targets of YY1 are not equivalent in distinct cell types. In mouse embryonic fibroblasts, for instance, YY1 has been reported to modulate the expression levels of REST, a repressor of neuronal fate (Affar et al., 2006; Ballas et al., 2005), but this was not detected in our study. The experimental evidence provided in this study supports the role of YY1 as lineage-specific activator of HDAC enzymatic activity during oligodendrocyte progenitor differentiation. This interpretation is based on the association of YY1 with the chromatin modifier enzyme HDAC1 only when progenitors differentiate into oligodendrocytes, but not when they differentiate into astrocytes. The lineage-specific formation of YY1/HDAC1 protein complexes was also associated with a 3-fold greater DNA binding activity of YY1 in progenitors differentiating into oligodendrocytes compared to astrocytes. Finally, these data are consistent with the detection of YY1/HDAC1 repressive complexes in the promoter regions of Tcf4 and Id4 only during differentiation of progenitors into oligodendrocytes. Therefore, although previous studies had already reported the constitutive association of YY1 with HDAC1 in Hela (Ya et al., 2001) and in erythroleukemia cell lines, our study is the first one that demonstrates an inducible and lineage-specific association between YY1 and HDAC1 in oligodendrocyte progenitors.
These results improve our understanding of oligodendrocyte development by providing a mechanistic insight into the critical transition between cell cycle exit and initiation of a transcriptional program of differentiation.
Experimental Procedures
Animals
All the mice used in this study were handled according to protocols approved by the Institutional IACUC committee. Conditional yy1 mutants were generated by crossing Yy1flox/flox mice (Affar et al, 2006) with Cnp1-cre mice (Lappe-Siefke et al., 2003), as detailed in Supplemental Experimental Procedures.
Antibodies and plasmids
Immunohistochemistry, immunocytochemistry and image processing
Cells and sections were stained overnight with primary antibodies as previously described (Marin-Husstege et al., 2006). After incubation with fluorophore conjugated secondaries, immunoreactivity was analyzed, using the Zeiss LSM 510 Meta confocal laser-scanning microscope. Image acquisition and quantification can be found in Supplemental Experimental Procedures.
BrdU incorporation, Caspase3 immunohistochemistry and TUNEL assays
Mice received 10 mg/kg BrdU injection 1 hr before sacrifice. After perfusion and cryopreservation, brains were sectioned and stained as previously described (Shen et al., 2005). To identify the apoptotic cells, terminal deoxynucleotidyl transferase-mediated dUTP end labeling (TUNEL) assay and immunohistochemistry using antibody against activated cleaved 17/19kD caspase-3 were performed (Supplemental Experimental Procedures).
Electron microscopy
Yy1 conditional mutant mice and control siblings were deeply anesthetized and transcardially perfused with 0.9% NaCl followed by a 0.1M Millonig's solution containing 4% formaldehyde and 5% glutaraldehyde (pH 7.3). The mice were post-fixed for 2 weeks in the same fixative solution at 4° C. Following the post-fixation, tissue samples from cervical spinal cord and corpus callosum at the level of the fornix were harvested and processed for standard transmission electron microscopic analysis as previously described (Dupree et al., 1998; Marcus et al., 2006). For quantification details see Supplemental Experimental Procedures.
Neurosphere culture of SVZ-derived multipotential progenitors
The lateral SVZ was dissected from yy1flox/flox neonatal mice and dissociated with Papain. Approximately 1-2×104 SVZ cells (one brain) per well of 24-well plate were cultured to form neurospheres and grown in the presence of EGF and bFGF. After 7 days, the neurospheres were mechanically dissociated and differentiated for 7 additional days into several lineages (Supplemental Experimental Procedures).
Oligodendrocyte progenitor culture
Oligodendrocyte progenitors were isolated from postnatal day 1 cortex of rat or yy1flox/flox mice and immunoselected using A2B5 antibodies (for rat cells) and NG2 antibodies (for mouse cells) and secondary antibodies conjugated with magnetic beads (Supplemental Experimental Procedures). Progenitors were cultured in the presence of bFGF and PDGF while oligodendrocyte differentiation was induced by culturing the cells in the absence of mitogens (ODM) and astrocytic differentiation by culturing the cells in BMP4 (100ng/ml). Viral transduction, plasmid transfection protocols and Luciferase assays can be found in Supplemental Experimental Procedures.
Quantitative and semi-quantitative reverse transcriptase PCR
Mouse brains or cell pellets were homogenized in Trizol® Reagent and RNA was isolated following manufacturer's instruction. Quantitative real-time PCR was performed using Stratagene SYBR Green PCR master mix in Stratagene MX4000 multiplex quantitative PCR system (Supplemental Experimental Procedures).
Western blot and immunoprecipitation
Proteins from the brain were extracted as described in Liu et al., 2006 and immunoprecipitation was conducted using standard biochemical procedures (Supplemental Experimental Procedures).
Chromatin immunoprecipitation
Chromatin was isolated from 1×107 oligodendrocyte progenitors (Jung et al., 1995) that were cultured either in growth medium or in oligodendrocyte differentiating medium, or induced to astrocytic lineage by BMP4 treatment. After immunoprecipitation with 5μg anti-YY1 antibody (mouse monoclonal sc-7341X, Santa Cruz) or 5μg of anti-HDAC1 antibody (rabbit polyclonal, Affinity BioReagents, Inc) chromatin was reverse cross-linked using EZ ChIP kit (Upstate Biotechnology). Proteins were digested with proteinase K and the recovered DNA was purified using QIAGEN QIAquick PCR purification kit and subjected to PCR amplification as previously described (Liu et al., 2006). Details in Supplemental Experimental Procedures
Statistical Methods
Results are expressed as mean ± standard deviation (SD) and statistically analyzed using two tailed Student's t tests. P of <0.05 was considered to be statistically significant. *p< 0.05, **p<0.01, ***p<0.001.
Supplementary Material
Acknowledgments
This work is dedicated to Mr. R. Rosa for inspiration and constant support. Grant support from NIH NINDS NS42925 (PCB), NIHGM53874 (YS), NMSS RG 3957 (PCB) and MSRF (PCB). We thank Dr. Nowakowski for Genomatix Software, Ms Jiang and Kuo for help. Dr. Nishiyama and Wight for helpful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Affar el B, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. Essential dosage-dependent functions of the transcription factor yin yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26:3565–3581. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Salas LM, Benitez-Hess ML, Dipaolo JA. YY-1 and c-Jun transcription factors participate in the repression of the human involucrin promoter. Int J Oncol. 2005;26:259–266. doi: 10.3892/ijo.26.1.259. [DOI] [PubMed] [Google Scholar]
- Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Barres BA, Lazar MA, Raff MC. A novel role for thyroid hormone, glucocorticoids and retinoic acid in timing oligodendrocyte development. Development. 1994;120:1097–1108. doi: 10.1242/dev.120.5.1097. [DOI] [PubMed] [Google Scholar]
- Belachew S, Chittajallu R, Aguirre AA, Yuan X, Kirby M, Anderson S, Gallo V. Postnatal NG2 proteoglycan-expressing progenitor cells are intrinsically multipotent and generate functional neurons. J Cell Biol. 2003;161:169–186. doi: 10.1083/jcb.200210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt JA, Kim JG, Tosic M, Kim C, Hudson LD. The transcriptional regulator Yin Yang 1 activates the myelin PLP gene. J Neurochem. 2001;77:935–942. doi: 10.1046/j.1471-4159.2001.00307.x. [DOI] [PubMed] [Google Scholar]
- Bjartmar C, Hildebrand C, Loinder K. Morphological heterogeneity of rat oligodendrocytes: electron microscopic studies on serial sections. Glia. 1994;11:235–244. doi: 10.1002/glia.440110304. [DOI] [PubMed] [Google Scholar]
- Bjelke B, Seiger A. Morphological distribution of MBP-like immunoreactivity in the brain during development. Int J Dev Neurosci. 1989;7:145–164. doi: 10.1016/0736-5748(89)90065-8. [DOI] [PubMed] [Google Scholar]
- Bogler O, Wren D, Barnett SC, Land H, Noble M. Cooperation between two growth factors promotes extended self-renewal and inhibits differentiation of oligodendrocyte-type-2 astrocyte (O-2A) progenitor cells. Proc Natl Acad Sci U S A. 1990;87:6368–6372. doi: 10.1073/pnas.87.16.6368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calver AR, Hall AC, Yu WP, Walsh FS, Heath JK, Betsholtz C, Richardson WD. Oligodendrocyte population dynamics and the role of PDGF in vivo. Neuron. 1998;20:869–882. doi: 10.1016/s0896-6273(00)80469-9. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Tikoo R, Kiyokawa H, Friedrich V, Jr., Chao MV, Koff A. Oligodendrocyte precursor differentiation is perturbed in the absence of the cyclin-dependent kinase inhibitor p27Kip1. Genes Dev. 1997;11:2335–2346. doi: 10.1101/gad.11.18.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Hardy RJ, Teng KK, Levine JM, Koff A, Chao MV. Loss of p27Kip1 function results in increased proliferative capacity of oligodendrocyte progenitors but unaltered timing of differentiation. Development. 1999;126:4027–4037. doi: 10.1242/dev.126.18.4027. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Liu A. Relationship between cell cycle molecules and onset of oligodendrocyte differentiation. J Neurosci Res. 2003;72:1–11. doi: 10.1002/jnr.10565. [DOI] [PubMed] [Google Scholar]
- Chenn A, Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- Cho EA, Dressler GR. TCF-4 binds beta-catenin and is expressed in distinct regions of the embryonic brain and limbs. Mech Dev. 1998;77:9–18. doi: 10.1016/s0925-4773(98)00131-2. [DOI] [PubMed] [Google Scholar]
- Dai Y, Rahmani M, Dent P, Grant S. Blockade of histone deacetylase inhibitor-induced RelA/p65 acetylation and NF-kappaB activation potentiates apoptosis in leukemia cells through a process mediated by oxidative damage, XIAP downregulation, and c-Jun N-terminal kinase 1 activation. Mol Cell Biol. 2005;25:5429–5444. doi: 10.1128/MCB.25.13.5429-5444.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19:7237–7244. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupree JL, Coetzee T, Suzuki K, Popko B. Myelin abnormalities in mice deficient in galactocerebroside and sulfatide. J Neurocytol. 1998;27:649–659. doi: 10.1023/a:1006908013972. [DOI] [PubMed] [Google Scholar]
- Durand B, Gao FB, Raff M. Accumulation of the cyclin-dependent kinase inhibitor p27/Kip1 and the timing of oligodendrocyte differentiation. Embo J. 1997;16:306–317. doi: 10.1093/emboj/16.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Fero ML, Roberts JM, Raff MC. p27Kip1 alters the response of cells to mitogen and is part of a cell-intrinsic timer that arrests the cell cycle and initiates differentiation. Curr Biol. 1998;8:431–440. doi: 10.1016/s0960-9822(98)70177-0. [DOI] [PubMed] [Google Scholar]
- Galvagni F, Cartocci E, Oliviero S. The dystrophin promoter is negatively regulated by YY1 in undifferentiated muscle cells. J Biol Chem. 1998;273:33708–33713. doi: 10.1074/jbc.273.50.33708. [DOI] [PubMed] [Google Scholar]
- Gard AL, Pfeiffer SE. Glial cell mitogens bFGF and PDGF differentially regulate development of O4+GalC- oligodendrocyte progenitors. Dev Biol. 1993;159:618–630. doi: 10.1006/dbio.1993.1269. [DOI] [PubMed] [Google Scholar]
- Ghiani CA, Eisen AM, Yuan X, DePinho RA, McBain CJ, Gallo V. Neurotransmitter receptor activation triggers p27(Kip1) and p21(CIP1) accumulation and G1 cell cycle arrest in oligodendrocyte progenitors. Development. 1999;126:1077–1090. doi: 10.1242/dev.126.5.1077. [DOI] [PubMed] [Google Scholar]
- Ghiani C, Gallo V. Inhibition of cyclin E-cyclin-dependent kinase 2 complex formation and activity is associated with cell cycle arrest and withdrawal in oligodendrocyte progenitor cells. J Neurosci. 2001;21:1274–1282. doi: 10.1523/JNEUROSCI.21-04-01274.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil-Perotin S, Marin-Husstege M, Li J, Soriano-Navarro M, Zindy F, Roussel MF, Garcia-Verdugo JM, Casaccia-Bonnefil P. Loss of p53 induces changes in the behavior of subventricular zone cells: implication for the genesis of glial tumors. J Neurosci. 2006;26:1107–1116. doi: 10.1523/JNEUROSCI.3970-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhan S, Marin-Husstege M, Yung SY, Fontanez D, Casaccia-Bonnefil P, Mehler MF. Combinatorial profiles of oligodendrocyte-selective classes of transcriptional regulators differentially modulate myelin basic protein gene expression. J Neurosci. 2005;25:8311–8321. doi: 10.1523/JNEUROSCI.1850-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths IR. Myelin mutants: model systems for the study of normal and abnormal myelination. Bioessays. 1996;18:789–797. doi: 10.1002/bies.950181005. [DOI] [PubMed] [Google Scholar]
- Hall A, Giese NA, Richardson WD. Spinal cord oligodendrocytes develop from ventrally derived progenitor cells that express PDGF alpha-receptors. Development. 1996;122:4085–4094. doi: 10.1242/dev.122.12.4085. [DOI] [PubMed] [Google Scholar]
- Hamano K, Takeya T, Iwasaki N, Nakayama J, Ohto T, Okada Y. A quantitative study of the progress of myelination in the rat central nervous system, using the immunohistochemical method for proteolipid protein. Brain Res Dev Brain Res. 1998;108:287–293. doi: 10.1016/s0165-3806(98)00063-7. [DOI] [PubMed] [Google Scholar]
- Hart IK, Richardson WD, Bolsover SR, Raff MC. PDGF and intracellular signaling in the timing of oligodendrocyte differentiation. J Cell Biol. 1989;109:3411–3417. doi: 10.1083/jcb.109.6.3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassig CA, Schreiber SL. Nuclear histone acetylases and deacetylases and transcriptional regulation: HATs off to HDACs. Curr Opin Chem Biol. 1997;1:300–308. doi: 10.1016/s1367-5931(97)80066-x. [DOI] [PubMed] [Google Scholar]
- Hu X, Hicks CW, He W, Wong P, Macklin WB, Trapp BD, Yan R. Bace1 modulates myelination in the central and peripheral nervous system. Nat Neurosci. 2006;9:1520–1525. doi: 10.1038/nn1797. [DOI] [PubMed] [Google Scholar]
- Ishibashi M, McMahon AP. A sonic hedgehog-dependent signaling relay regulates growth of diencephalic and mesencephalic primordia in the early mouse embryo. Development. 2002;129:4807–4819. doi: 10.1242/dev.129.20.4807. [DOI] [PubMed] [Google Scholar]
- Jung M, Kramer E, Grzenkowski M, Tang K, Blakemore W, Aguzzi A, Khazaie K, Chlichlia K, von Blankenfeld G, Kettenmann H, et al. Lines of murine oligodendroglial precursor cells immortalized by an activated neu tyrosine kinase show distinct degrees of interaction with axons in vitro and in vivo. Eur J Neurosci. 1995;7:1245–1265. doi: 10.1111/j.1460-9568.1995.tb01115.x. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. The Id4 HLH protein and the timing of oligodendrocyte differentiation. Embo J. 2000;19:1998–2007. doi: 10.1093/emboj/19.9.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korinek V, Barker N, Willert K, Molenaar M, Roose J, Wagenaar G, Markman M, Lamers W, Destree O, Clevers H. Two members of the Tcf family implicated in Wnt/beta-catenin signaling during embryogenesis in the mouse. Mol Cell Biol. 1998;18:1248–1256. doi: 10.1128/mcb.18.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe-Siefke C, Goebbels S, Gravel M, Nicksch E, Lee J, Braun PE, Griffiths IR, Nave KA. Disruption of Cnp1 uncouples oligodendroglial functions in axonal support and myelination. Nat Genet. 2003;33:366–374. doi: 10.1038/ng1095. [DOI] [PubMed] [Google Scholar]
- Latinkic BV, Cooper B, Smith S, Kotecha S, Towers N, Sparrow D, Mohun TJ. Transcriptional regulation of the cardiac-specific MLC2 gene during Xenopus embryonic development. Development. 2004;131:669–679. doi: 10.1242/dev.00953. [DOI] [PubMed] [Google Scholar]
- Lee YM, Lee SC. Transcriptional activation of the alpha-1 acid glycoprotein gene by YY1 is mediated by its functional interaction with a negative transcription factor. DNA Cell Biol. 1994;13:1029–1036. doi: 10.1089/dna.1994.13.1029. [DOI] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. Embo J. 2006;25:4833–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HF, Shi Y. In: Yin Yang 1. Zinc Finger Proteins: from Atomic Contact to Cellular Function. Iuchi S, Kuldell N, editors. Landers Biosciences and Kluwer Academic Press; Austin, TX: 2005. pp. 182–193. [Google Scholar]
- Marcus J, Honigbaum S, Shroff S, Honke K, Rosenbluth J, Dupree JL. Sulfatide is essential for the maintenance of CNS myelin and axon structure. Glia. 2006;53:372–381. doi: 10.1002/glia.20292. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, He Y, Li J, Kondo T, Sablitzky F, Casaccia-Bonnefil P. Multiple roles of Id4 in developmental myelination: predicted outcomes and unexpected findings. Glia. 2006;54:285–296. doi: 10.1002/glia.20385. [DOI] [PubMed] [Google Scholar]
- Mason JL, Langaman C, Morell P, Suzuki K, Matsushima GK. Episodic demyelination and subsequent remyelination within the murine central nervous system: changes in axonal calibre. Neuropathol Appl Neurobiol. 2001;27:50–58. doi: 10.1046/j.0305-1846.2001.00301.x. [DOI] [PubMed] [Google Scholar]
- Miller RH. Dorsally derived oligodendrocytes come of age. Neuron. 2005;45:1–3. doi: 10.1016/j.neuron.2004.12.032. [DOI] [PubMed] [Google Scholar]
- Nakatsuji Y, Miller RH. Control of oligodendrocyte precursor proliferation mediated by density-dependent cell cycle protein expression. Dev Neurosci. 2001;23:356–363. doi: 10.1159/000048719. [DOI] [PubMed] [Google Scholar]
- Nave KA. Neurological mouse mutants and the genes of myelin. J Neurosci Res. 1994;38:607–612. doi: 10.1002/jnr.490380602. [DOI] [PubMed] [Google Scholar]
- Noll E, Miller RH. Oligodendrocyte precursors originate at the ventral ventricular zone dorsal to the ventral midline region in the embryonic rat spinal cord. Development. 1993;118:563–573. doi: 10.1242/dev.118.2.563. [DOI] [PubMed] [Google Scholar]
- Nowak K, Lange-Dohna C, Zeitschel U, Gunther A, Luscher B, Robitzki A, Perez-Polo R, Rossner S. The transcription factor Yin Yang 1 is an activator of BACE1 expression. J Neurochem. 2006;96:1696–1707. doi: 10.1111/j.1471-4159.2006.03692.x. [DOI] [PubMed] [Google Scholar]
- Pfeiffer SE, Warrington AE, Bansal R. The oligodendrocyte and its many cellular processes. Trends Cell Biol. 1993;3:191–197. doi: 10.1016/0962-8924(93)90213-k. [DOI] [PubMed] [Google Scholar]
- Pringle NP, Richardson WD. A singularity of PDGF alpha-receptor expression in the dorsoventral axis of the neural tube may define the origin of the oligodendrocyte lineage. Development. 1993;117:525–533. doi: 10.1242/dev.117.2.525. [DOI] [PubMed] [Google Scholar]
- Raff MC, Lillien LE. Differentiation of a bipotential glial progenitor cell: what controls the timing and the choice of developmental pathway? J Cell Sci Suppl. 1988;10:77–83. doi: 10.1242/jcs.1988.supplement_10.6. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross SE, Greenberg ME, Stiles CD. Basic helix-loop-helix factors in cortical development. Neuron. 2003;39:13–25. doi: 10.1016/s0896-6273(03)00365-9. [DOI] [PubMed] [Google Scholar]
- Saegusa M, Hashimura M, Kuwata T, Hamano M, Okayasu I. Upregulation of TCF4 expression as a transcriptional target of beta-catenin/p300 complexes during trans-differentiation of endometrial carcinoma cells. Lab Invest. 2005;85:768–779. doi: 10.1038/labinvest.3700273. [DOI] [PubMed] [Google Scholar]
- Seto E, Shi Y, Shenk T. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature. 1991;354:241–245. doi: 10.1038/354241a0. [DOI] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Kagawa T, Wada T, Muroyama Y, Takada S, Ikenaka K. Wnt signaling controls the timing of oligodendrocyte development in the spinal cord. Dev Biol. 2005;282:397–410. doi: 10.1016/j.ydbio.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Shrivastava A, Calame K. An analysis of genes regulated by the multi-functional transcriptional regulator Yin Yang-1. Nucleic Acids Res. 1994;22:5151–5155. doi: 10.1093/nar/22.24.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spassky N, Heydon K, Mangatal A, Jankovski A, Olivier C, Queraud-Lesaux F, Goujet-Zalc C, Thomas JL, Zalc B. Sonic hedgehog-dependent emergence of oligodendrocytes in the telencephalon: evidence for a source of oligodendrocytes in the olfactory bulb that is independent of PDGFRalpha signaling. Development. 2001;128:4993–5004. doi: 10.1242/dev.128.24.4993. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Schlierf A, Lommes P, Hillgartner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, Wegner M. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell. 2006;11:697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, Shi Y. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117:859–872. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Tang XM, Beesley JS, Grinspan JB, Seth P, Kamholz J, Cambi F. Cell cycle arrest induced by ectopic expression of p27 is not sufficient to promote oligodendrocyte differentiation. J Cell Biochem. 1999;76:270–279. doi: 10.1002/(sici)1097-4644(20000201)76:2<270::aid-jcb10>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Temple S, Raff MC. Clonal analysis of oligodendrocyte development in culture: evidence for a developmental clock that counts cell divisions. Cell. 1986;44:773–779. doi: 10.1016/0092-8674(86)90843-3. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Thomas MJ, Seto E. Unlocking the mechanisms of transcription factor YY1: are chromatin modifying enzymes the key? Gene. 1999;236:197–208. doi: 10.1016/s0378-1119(99)00261-9. [DOI] [PubMed] [Google Scholar]
- Tikoo R, Casaccia-Bonnefil P, Chao MV, Koff A. Changes in cyclin-dependent kinase 2 and p27kip1 accompany glial cell differentiation of central glia-4 cells. J Biol Chem. 1997;272:442–447. doi: 10.1074/jbc.272.1.442. [DOI] [PubMed] [Google Scholar]
- Tikoo R, Osterhout DJ, Casaccia-Bonnefil P, Seth P, Koff A, Chao MV. Ectopic expression of p27Kip1 in oligodendrocyte progenitor cells results in cell-cycle growth arrest. J Neurobiol. 1998;36:431–440. [PubMed] [Google Scholar]
- Tokumoto YM, Apperly JA, Gao FB, Raff MC. Posttranscriptional regulation of p18 and p27 Cdk inhibitor proteins and the timing of oligodendrocyte differentiation. Dev Biol. 2002;245:224–234. doi: 10.1006/dbio.2002.0626. [DOI] [PubMed] [Google Scholar]
- Walowitz JL, Bradley ME, Chen S, Lee T. Proteolytic regulation of the zinc finger transcription factor YY1, a repressor of muscle-restricted gene expression. J Biol Chem. 1998;273:6656–6661. doi: 10.1074/jbc.273.12.6656. [DOI] [PubMed] [Google Scholar]
- Wang S, Sdrulla A, Johnson JE, Yokota Y, Barres BA. A role for the helix-loop-helix protein Id2 in the control of oligodendrocyte development. Neuron. 2001;29:603–614. doi: 10.1016/s0896-6273(01)00237-9. [DOI] [PubMed] [Google Scholar]
- Wang TW, Zhang H, Parent JM. Retinoic acid regulates postnatal neurogenesis in the murine subventricular zone-olfactory bulb pathway. Development. 2005;132:2721–2732. doi: 10.1242/dev.01867. [DOI] [PubMed] [Google Scholar]
- Willem M, Garratt AN, Novak B, Citron M, Kaufmann S, Rittger A, DeStrooper B, Saftig P, Birchmeier C, Haass C. Control of peripheral nerve myelination by the beta-secretase BACE1. Science. 2006;314(5799):664–6. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- Xu X, Kawachi Y, Nakamura Y, Sakurai H, Hirota A, Banno T, Takahashi T, Roop DR, Otsuka F. Yin-yang 1 negatively regulates the differentiation-specific transcription of mouse loricrin gene in undifferentiated keratinocytes. J Invest Dermatol. 2004;123:1120–1126. doi: 10.1111/j.0022-202X.2004.23492.x. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M, Tonou-Fujimori N, Komori A, Maeda R, Nojima Y, Li H, Okamoto H, Masai I. Histone deacetylase 1 regulates retinal neurogenesis in zebrafish by suppressing Wnt and Notch signaling pathways. Development. 2005;132:3027–3043. doi: 10.1242/dev.01881. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr Opin Genet Dev. 2003;13:143–153. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- Yant SR, Zhu W, Millinoff D, Slightom JL, Goodman M, Gumucio DL. High affinity YY1 binding motifs: identification of two core types (ACAT and CCAT) and distribution of potential binding sites within the human beta globin cluster. Nucleic Acids Res. 1995;23:4353–4362. doi: 10.1093/nar/23.21.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21:5979–5991. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.