Abstract
The cleavage furrow is created by an actomyosin contractile ring that is regulated by small GTPase proteins such as Rac1 and RhoA. Guanine nucleotide exchange factors (GEFs) are positive regulators of the small GTPase proteins and have been implicated as important factors in regulating cytokinesis. However, it is still unclear how GEFs regulate the contractile ring during cytokinesis in mammalian cells. Here we report that a novel GEF, which is termed MyoGEF (myosin-interacting GEF), interacts with non-muscle myosin II and exhibits activity toward RhoA. MyoGEF and non-muscle myosin II colocalize to the cleavage furrow in early anaphase cells. Disruption of MyoGEF expression in U2OS cells by RNA interference (RNAi) results in the formation of multinucleated cells. These results suggest that MyoGEF, RhoA, and non-muscle myosin II act as a functional unit at the cleavage furrow to advance furrow ingression during cytokinesis.
Keywords: MyoGEF, non-muscle myosin II, RhoA, Rac1, cytokinesis, GTPase signaling
INTRODUCTION
The small GTPase proteins, such as RhoA, Rac1, and Cdc42, are key regulators of the actin-myosin cytoskeleton.1 RhoA stimulates the assembly of contractile actomyosin filaments and associated focal adhesion complexes.2 Rac1 induces the formation of lamel-lipodia and membrane ruffles, whereas Cdc42 induces filopodia.3 The activity of these small GTPase proteins is largely regulated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs). GEFs activate the small GTPase proteins by catalyzing the exchange of bound GDP for GTP, while GAPs inactivate the small GTPase proteins by increasing their low intrinsic GTPase activity.4 RhoA can activate Rho-kinase that in turn inhibits myosin phosphatase and it can also directly phosphorylate myosin regulatory light chains, resulting in an increase in myosin contractile activity.5,6
The balance between Rac1 and RhoA activity at the cleavage furrow is critical for the progression of cytokinesis, i.e., successful cytokinesis requires low Rac1 activity and/or high RhoA activity at the cleavage furrow.7 Activation of RhoA and an increase in myosin contractile activity are required for the initiation and progression of furrow ingression.8–11 Recent studies suggest that Ect2, a GEF that can activate RhoA, is required to initiate furrow ingression and the completion of cytokinesis.9,12,13 In contrast, activation of Rac1 inhibits cytokinesis.10,14 Therefore, repression of Rac1 activity is expected at the cleavage furrow during cytokinesis. Indeed, fluorescence resonance energy transfer (FRET) analysis has indicated that Rac activity decreases at the cleavage furrow during cytokinesis and this inactivation is abolished by the expression of a dominant-negative form of MgcRacGAP.10,15 MgcRacGAP (a GAP) is a component of centralspindlin and plays an important role in the regulation of cytokinesis.12,13 Collectively, coordinate regulation of Rac1 and RhoA activity at the cleavage furrow is critically important for the completion of cytokinesis.
In this article, we report the identification of a novel GEF that localizes to the cleavage furrow during cytokinesis. This GEF is termed MyoGEF (myosin-interacting GEF), because it can bind to non-muscle myosin II. MyoGEF can activate RhoA. Disruption of MyoGEF by RNA interference (RNAi) results in the formation of binucleated/multinucleated cells. Our results suggest that MyoGEF can modulate the activity of the small GTPase proteins and plays an important role in the regulation of cytokinesis.
MATERIALS AND METHODS
Plasmids and cell culture
The open reading frame for MyoGEF was amplified from human brain RNA (purchased from BD Biosciences) by RT-PCR. The primers used are as follows: 5′aagcctatgaaggcctttggtcctcca3′ (forward primer) and 5′gtcgactcatacctctgatgcggacag3′ (reverse primer). The PCR fragment was cloned into HindIII/SalI sites of a pEGFP-C3 vector, generating pEGFP-C3-MyoGEF (GFP-tagged MyoGEF). MyoGEF full-length cDNA fragment was also cloned into pCS3+MT and pcDNA4/HisMax (Invitrogen) to generate pCS3+MT-MyoGEF (Myc-tagged MyoGEF) and pcDNA4/HisMax-MyoGEF (His-tagged MyoGEF), respectively. The SmartPool siRNA against MyoGEF (NM_018173) was purchased from Dharmacon. Both siRNA and plasmids were transfected into HeLa Tet-On cells (BD biosciences) or U2OS cells by using Lipofectamine 2000 (Invitrogen) according to manufacturer’s instructions. HeLa Tet-On and U2OS cells were maintained in DMEM supplemented with 5% fetal bovine serum at 37°C under 5% CO2.
Protein expression
GST-myosin-ROD was expressed in a baculovirus expression system and was purified using glutathione-conjugated agarose beads as described previously.22 ThioHis-tagged MyoGEF full-length protein was expressed in a bacterial expression system and was purified according to manufacturer’s instruction (Invitrogen).
Immufluorescent staining
The cells grown on coverslips were transfected with plasmids or siRNA as indicated. Twenty-four or 48 h after transfection, the cells were fixed in 3.7% paraformaldehyde for 15 min, permeabilized in 0.5% Triton X-100 for 10 min, blocked in 1% bovine serum albumin (BSA) for 1 h at 23°C, incubated with primary antibodies for 3 h at 23°C or overnight at 4°C, followed by incubation with secondary antibodies for 1 h at 23°C. Affinity-purified polyclonal rabbit antibodies against a carboxyl-terminal sequence of non-muscle myosin heavy chain II-A (NMHC II-A; 1:1000) were described previously.30 Monoclonal antibodies to beta-tubulin was purchased from Sigma. The secondary antibodies Alexa 594 goat anti-mouse IgG (1:1000), Alexa 594 goat anti-rabbit IgG (1:1000), and Alexa 350 goat anti-mouse IgG (1:500) were from Molecular Probes (Eugene, OR). Actin filaments were visualized by incubation with rhodamine-phalloidin (1:500; Molecular Probes) for 1 h at 23°C. The coverslips were mounted using a Prolong antifade kit (Molecular Probes). The images were collected using a Zeiss LSM 510 confocal microscope (Carl Zeiss, Thornwood, NY).
Immunoblot
Total cell protein from transfected cells was separated by SDS-6% PAGE, transferred to an Immobilon-P transfer membrane (Millipore, Bedford, MA), blocked in 5% non-fat milk, and incubated with primary antibodies for 3 h at 23°C or overnight at 4°C. The following primary antibodies were used: affinity-purified polyclonal rabbit antibodies against MyoGEF was generated by using the following peptide: MRGPHIIQLDTPLSASEV (1:500). Monoclonal antibodies to Myc (9E10; 1:1000) was purchased from Santa Cruz Biotechnology (Santa Cruz, CA); polyclonal rabbit antibodies specific for non-muscle myosin II-A (NMHC II-A; 1:5000). The blot was washed and then incubated with horseradish peroxidase-conjugated secondary antibodies (1:5,000; Santa Cruz) at room temperature for 2 h. The blots were visualized by SuperSignal West Pico Luminol/Enhancer solution (Pierce).
RhoA and Rac1 activation assays
The transfected cells were washed with phosphate-buffered saline, and then lysed in RIPA buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 2.5% Na-deoxycholate, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mM Na3VO4, 1 mM NaF). The same amount of total protein from clarified lysate was incubated with GST-PBD (p21-binding domain of human PAK-1) or GST-RBD (Rho binding domain of rhotekin) to precipitate GTP-bound Rac1 and GTP-bound RhoA, respectively, according to the manufacturer’s instructions (Upstate Biotechnology). Precipitated GTP-bound Rac1 or RhoA were resolved on a SDS-12% PAGE and immunoblotted using mono-clonal antibodies specific for Rac1 (1:1000; Upstate Biotechnology) and RhoA (1:200; Santa Cruz Biotechnology). Six percent of the cell lysate was also resolved in a SDS-12% PAGE and immunoblotted to measure the total amount of Rac1 or RhoA.
RESULTS
MyoGEF interacts with non-muscle myosin II
To identify novel GEFs that localize to the cleavage furrow and that are important for cytokinesis, we amplified cDNAs for novel GEFs by RT-PCR using total RNA from human brain tissue as template (Invitrogen). We designed the primers based on the cDNA sequences available in the database. The cDNA coding regions were fused to the 3′ end of a green fluorescent protein (GFP) gene and were transfected into HeLa cells. The transfected cells were then screened to determine whether the GFP-GEFs localized to the cleavage furrow in anaphase cells.
One of the GEFs (NM_018173) was found to localize to the cleavage furrow (see supplemental video 1) and was named myosin-interacting GEF (MyoGEF), because this novel GEF could directly bind to myosin II and colocalize with myosin II at the cleavage furrow during cytokinesis. MyoGEF contains 790 amino acids, and has a Dbl-homology (DH) domain and a pleckstrin homology (PH) domain (Fig. 1A). As shown in Figure 1B, confocal microscopy showed that GFP-MyoGEF localized to the central spindle (arrowhead in panel a) and cleavage furrow (arrows in panel a) in anaphase HeLa cells. Importantly, GFP-MyoGEF colocalizes with myosin II at the cleavage furrow (Fig. 1B, arrows in panels a, b, and c), suggesting that MyoGEF interacts with non-muscle myosin II. We have further confirmed that MyoGEF directly interacts with non-muscle myosin II. As shown in Figure 1C, purified GST-tagged myosin II-A rod region (GST-myosin-ROD) can bring down ThioHis-tagged MyoGEF full-length protein in a GST-pulldown assay (compare lane 2 with lane 3).
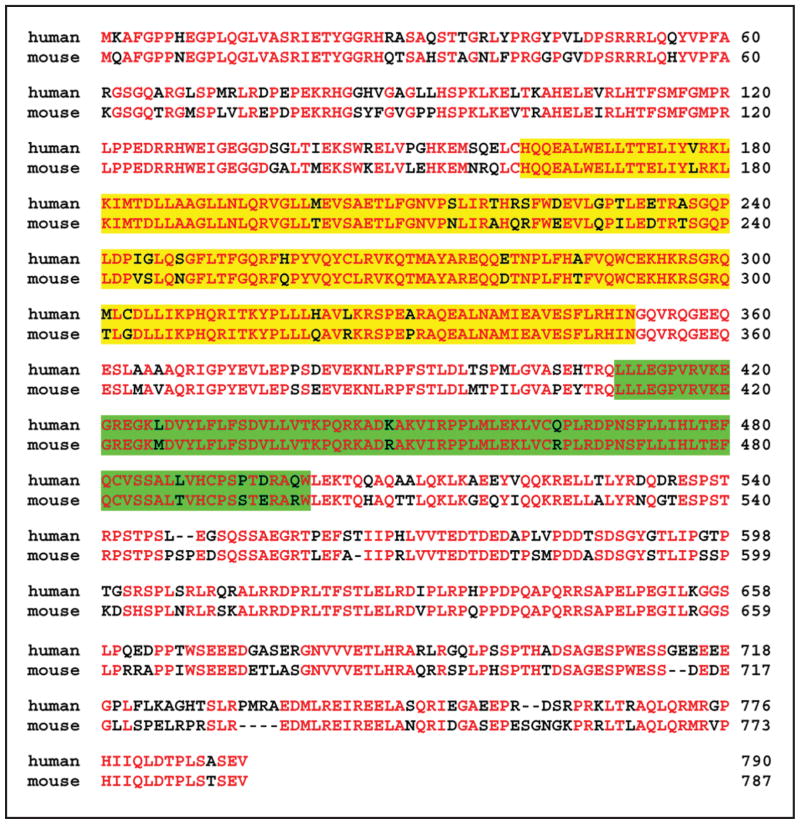
Figure 1.
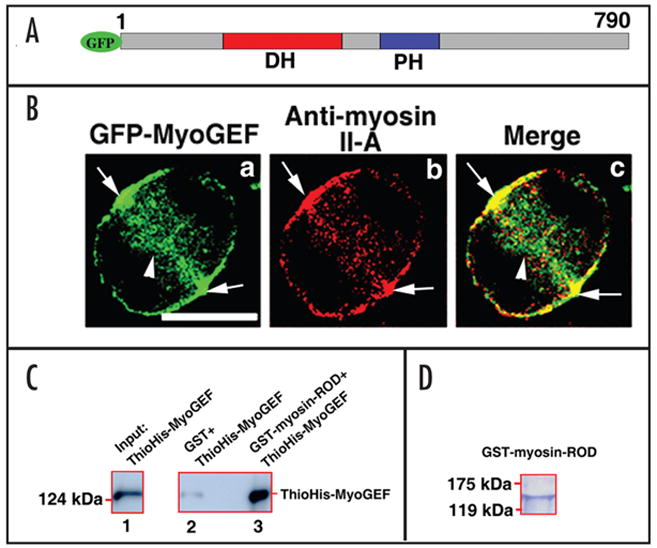
MyoGEF interacts with non-muscle myosin II-A. (A) Schematic diagram of GFP-tagged MyoGEF. The numbers represent the amino acids in MyoGEF. The Dbl-homology (DH) and pleckstrin homology (PH) domains are shown. (B) GFP-MyoGEF colocalizes with non-muscle myosin II-A at the cleavage furrow. HeLa cells were transfected with a plasmid encoding GFP-tagged MyoGEF (green). Twenty-four hours after transfection, the cells were fixed and stained with an antibody specific for non-muscle myosin II-A (red). Note that both GFP-MyoGEF and non-muscle myosin II-A localize to the cleavage furrow (arrows in a–c). Bar, 10 μm. (C) MyoGEF can bind to non-muscle myosin II. GST-myosin-ROD region (but not GST alone) can bring down ThioHis-MyoGEF (compare lane 2 with lane 3). ThioHis-MyoGEF protein is detected by immonublot analysis using a specific antibody for MyoGEF (lanes 1 and 3). (D) Coomassie blue staining gel of GST-myosin-ROD protein used in (C).
We sequenced the full-length MyoGEF cDNA and the nucleotide sequence matches the genomic DNA sequence. Comparison of human and mouse MyoGEF (NM_198604) shows that 74% of the amino acids are identical in both proteins (Fig. 2).
Figure 2.
Amino acid sequence alignment of human and mouse MyoGEF. Yellow and green highlight represent the DH and PH domains, respectively. Amino acid residues in red represent identical residues between human and mouse sequence. GenBank accession numbers for human and mouse MyoGEF are NM_018173 and NM_198604, respectively.
MyoGEF activates RhoA and induces myosin filament formation
To test whether MyoGEF can activate the small GTPase proteins RhoA and Rac1, HeLa cells were transfected with control siRNA or MyoGEF siRNA. As shown in Figure 3A, MyoGEF siRNA significantly reduces the endogenous MyoGEF protein level in transfected HeLa cells (upper panels, compare lane 1 with lane 2). RhoA activity in these transfected cells was then determined by a specific Rho activation assay.16,17 It has been well established that the Rho-GTP binding domain (RBD) of rhotekin can bind to the GTP-bound, active form (but not the GDP-bound, inactive form) of RhoA. Therefore, GST-RBD bound to glutathione agarose beads will only bring down the active (but not the inactive) form of RhoA in the Rho activation assay.18 As shown in Figure 3B, GST-RBD bound to glutathione agarose beads brings down significantly less active RhoA in HeLa cells transfected with MyoGEF siRNA (upper panels, compare lane1 with lane 2). It should be noted that the total RhoA protein level is not changed (Fig. 3B, compare lane 1 with lane 2 in the lower panel). These results suggest that MyoGEF has the capacity to activate RhoA. HeLa cells transfected with control siRNA or MyoGEF siRNA were also subjected to a specific Rac activation assay.16,17 It is also known that the p21 binding domain (PBD) of PAK-1 (p21-activating kinase-1) can bind to the GTP-bound, active form (but not the GDP-bound, inactive form) of Rac1. Therefore, GST-PBD bound to glutathione agarose beads will only bring down the active (but not the inactive) form of Rac1 in the Rac activation assay.19 As shown in Figure 3C, GST-PBD bound to glutathione agarose beads brings down a similar amount of active Rac1 in HeLa cells transfected with control or MyoGEF siRNA, suggesting that disruption of MyoGEF in HeLa cells does not affect the activation of Rac1 (upper panels, compare lane 1 and lane 2). Disruption of MyoGEF by RNAi does not affect the total Rac1 protein level (lower panels, compare lane 1 with lane 2).
Figure 3.
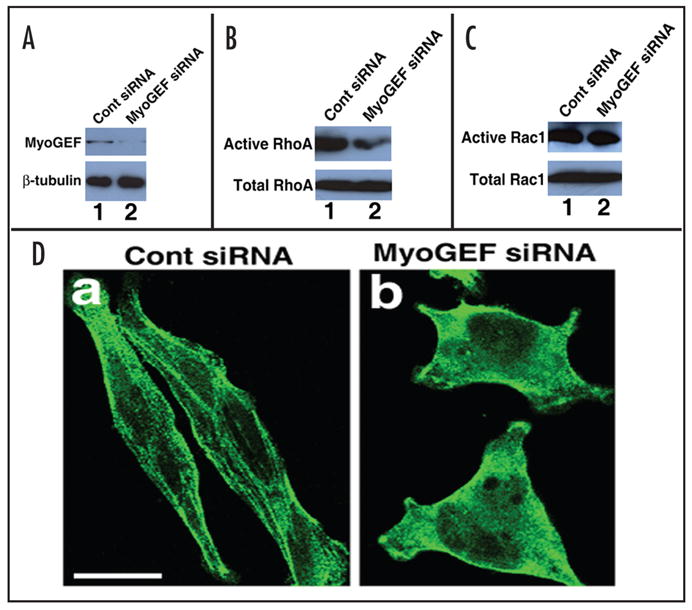
MyoGEF activates RhoA that in turn induces myosin filament formation. (A) Downregulation of MyoGEF protein in HeLa cells by RNA interference (RNAi). HeLa cells were transfected with control siRNA (lane1) or MyoGEF siRNA (lane 2). Foutry-eight hours after transfection, the transfected HeLa cells were subjected to an immunoblot analysis using antibodies specific for MyoGEF (upper panel) and beta-tubulin (lower panel). Note that MyoGEF siRNA significantly reduces MyoGEF (but not beta-tubulin) protein level (compare lane 1 with lane 2). (B) MyoGEF activates RhoA. The transfected HeLa cells in (A) were subjected to a specific RhoA activation assay. Note that inactivation of MyoGEF in Hela cells significantly reduces the active RhoA level (compare lane 1 with lane 2). (C) MyoGEF does not activate Rac1. The transfected HeLa cells in (A) were subjected to a specific Rac activation assay. Note that inactivation of MyoGEF in Hela cells does not affect the active Rac1 level (compare lane 1 with lane 2). (D) MyoGEF is important for myosin filament formation. The transfected HeLa cells in (A) were fixed and stained with an antibody specific for non-muscle myosin II-A. Note that inactivation of MyoGEF in HeLa cells significantly reduces myosin filaments. Bar, 10 μm.
RhoA can activate Rho-kinase that, in turn, inhibits myosin phosphatase and directly phosphorylates myosin regulatory light chains, resulting in an increase in myosin contractile activity and the formation of actin-myosin filaments.5,6 Therefore, we asked whether MyoGEF can regulate myosin filament formation. HeLa cells were transfected with control siRNA or MyoGEF siRNA. 48 h after transfection, the transfected cells were fixed and stained with antibody specific for non-muscle myosin II-A. As shown in Figure 3D, disruption of MyoGEF by RNAi significantly reduces myosin filament formation in HeLa cells (compare panel a with panel b), suggesting that MyoGEF can activate RhoA that, in turn, induces myosin filament formation.
Endogenous MyoGEF localizes to the spindle pole, central spindle and cleavage furrow
To elucidate the localization of endogenous MyoGEF protein during cytokinesis, we have developed a polyclonal antibody specific for MyoGEF using the following peptide as antigen: MRGPHI-IQLDTPLSASEV. Our results indicate that MyoGEF is expressed in HeLa and U2OS cells. As shown in Figure 4A, the specific polyclonal antibody recognizes endogenous MyoGEF in HeLa (lane 1 and 2) and U2OS (lane 4) cells as well as GFP-MyoGEF (lane 2) and His-MyoGEF (lane 3) expressed in HeLa cells. In lane 3, endogenous MyoGEF and His-MyoGEF overlapped and thereby appeared as a single band. Immunofluorescence microscopy using the polyclonal antibody specific for MyoGEF shows that MyoGEF localizes to the spindle pole, central spindle, and cleavage furrow in U2OS cells. As shown in (Fig. 4B), arrowheads indicate the localization of MyoGEF at the spindle pole in metaphase (panels a–c), anaphase (panels d–f), and telophase (panels g–i), while arrows indicate the localization MyoGEF at the central spindle (d–e) as well as cleavage furrow (g–h).
Figure 4.
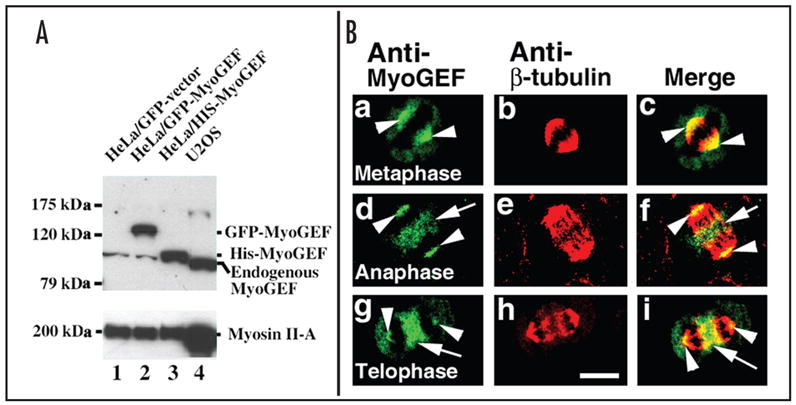
Endogenous MyoGEF localizes to the spindle pole, central spindle, and cleavage furrow. (A) Characterization of a polyclonal antibody specific for MyoGEF. An immunoblot analysis shows that a polyclonal antibody against MyoGEF recognizes the endogenous, GFP-tagged, and His-tagged MyoGEF in HeLa cells (upper panel, lanes 1, 2, and 3) as well as endogenous MyoGEF in U2OS cells (upper panel, lane 4). (B) Localization of endogenous MyoGEF during cytokinesis. U2OS cells growing on coverslips were fixed in 10% trichloroacetic acid and stained with antibodies specific for MyoGEF (green) and beta-tubulin (red). Note that MyoGEF localizes to the spindle pole in metaphase (a–c), to the spindle pole and central spindle in anaphase (d–f), and to the spindle pole and cleavage furrow in telophase (g–i). Arrowheads in a–i indicate the localization of MyoGEF to the spindle pole. Arrows in d–f indicate the localization of MyoGEF to the central spindle. Arrows in g–i indicate the localization of MyoGEF to the cleavage furrow. Bar, 10 μm.
Disruption of MyoGEF leads to the formation of multinucleated cells
Our results revealed that MyoGEF localized to the cleavage furrow (Figs. 1B and 4B) and could activate RhoA (Fig. 3B). The small GTPase proteins have been implicated as important regulators of cytokinesis.7 RhoA is a key regulator of actomyosin contractility and plays an important role in the progression of cytokinesis.20 Inactivation of RhoA leads to defects in cytokinesis in a cell type specific fashion.10 MyoGEF not only localized to the cleavage furrow (Fig. 1B and Fig. 4B), but also showed activity towards RhoA (Fig. 3B), suggesting that MyoGEF plays a role in regulating cytokinesis. Our results indicated that MyoGEF is important for the completion of cytokinesis. As shown in Figure 5, disruption of MyoGEF by siRNA in U2OS cells significantly reduces MyoGEF protein level (Fig. 5A), leading to an increase in the percentage of binucleated/multinucleated cells (Fig. 5B and C). This result further confirms that MyoGEF can activate RhoA (but not Rac1), because RhoA (but not Rac1) activity is critically important for the initiation and progression of cytokinesis.10,13
Figure 5.
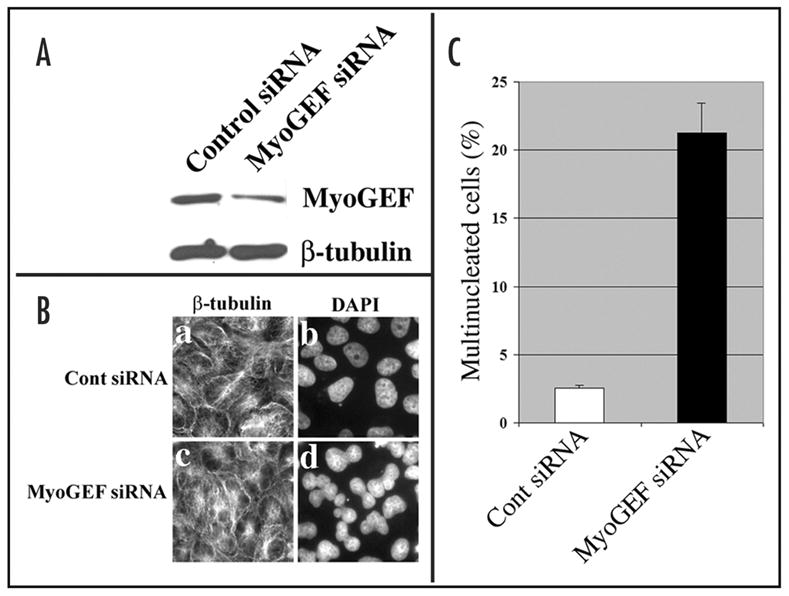
Disruption of MyoGEF in U2OS cells leads to the formation of binucleated/multinucleated cells. (A) Immunoblot analysis shows that siRNA specific for MyoGEF could specifically reduce MyoGEF protein levels in U2OS cells. (B) U2OS cells were transfected with control siRNA (panels a and b) and MyoGEF siRNA (panels c and d). The transfected cells were stained with anti-β-tubulin antibody (panels a and c) and DAPI (panels b and d). (C) Nuclei were identified by DAPI staining and were counted in U2OS transfected with control or MyoGEF siRNAs.
DISCUSSION
Furrow ingression is believed to be powered by the mechanical force generated by the interaction between actin and myosin filaments.21 Non-muscle myosin II localizes to the cleavage furrow during cell division and is established to be one of the critical components of the contractile ring.11,21–25 Disruption of myosin II in mammalian cells and in Dictyostelium lead to the formation of multinucleated cells.26–29 The actomyosin cytoskeleton is regulated by the small GTPase proteins that, in turn, are controlled by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs).1,2 RhoA is a key regulator of actomyosin contractility10,15,20 and has been shown to be critical for the initiation and completion of cytokinesis.8–11 Our results have demonstrated MyoGEF can activate RhoA and plays an important role in regulating cytokinesis. MyoGEF also interacts with non-muscle myosin II and colocalizes with non-muscle myosin II at the cleavage furrow. Our results suggest that interaction between MyoGEF and myosin II would allow myosin II to be locally activated at the cleavage furrow, leading to the progression of cytokinesis.
It should be noted that Ect2, another GEF, has been implicated in the regulation of cytokinesis. Recent studies showed that inactivation of Ect2 by RNAi in HeLa cells interfered with myosin contractile ring assembly.12,13 Why does inactivation of MyoGEF also lead to the defect in cytokinesis in the presence of Ect2? The localization of Ect2 and MyoGEF during cytokinesis appears to be different. Ect2 localizes to the central spindle. In contrast, MyoGEF localizes to the spindle pole, central spindle, and the cleavage furrow. Therefore, MyoGEF and Ect2 might utilize different mechanisms to regulate myosin contractile ring assembly at different stages of cytokinesis, and the coordinate regulation of MyoGEF and Ect2 might be essential for completion of cytokinesis.
In conclusion, we have demonstrated that a newly identified guanine nucleotide exchange factor, termed MyoGEF, can interact directly with non-muscle myosin II. Our results also provide strong evidence to support our conclusion that MyoGEF can activate the small GTPase protein RhoA that, in turn, induces myosin filament formation. Further, MyoGEF colocalizes with non-muscle myosin II at the cleavage furrow and plays an important role in the regulation of cytokinesis.
Acknowledgments
Accession numbers: NM_018173 and NM_198604
This publication was made possible by NIH Grant Number P20 RR-17708 from the National Center for Research Resources. This work was also supported by National Institutes of Health Grants k22 HL071542-01 (to Q.W.). This is contribution 06-116-J from the Kansas Agricultural Experiment Station, Manhattan, Kansas.
Footnotes
NOTE
Supplemental Video S1 can be found at: http://www.landesbioscience.com/journals/cc/wuCC5-11-vidS1.mov. Localization of GFP-tagged MyoGEF in HeLa cells during cytokinesis. HeLa cells were transfected with a plasmid encoding GFP-tagged MyoGEF by using Lipofectamine 2000 (Invitrogen). Twenty-four hours after transfection, the live HeLa cells expressing GFP-tagged MyoGEF were imaged by using a fluorescence Olympus X170 microscope equipped with a cooled CCD camera.
References
- 1.Burridge K, Wennerberg K. Rho and Rac take center stage. Cell. 2004;116:167–79. doi: 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 2.Ridley AJ, Hall A. The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell. 1992;70:389–99. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 3.Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin microspikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–52. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackay DJ, Hall A. Rho GTPases. J Biol Chem. 1998;273:20685–8. doi: 10.1074/jbc.273.33.20685. [DOI] [PubMed] [Google Scholar]
- 5.Matsui T, Amano M, Yamamoto T, Chihara K, Nakafuku M, Ito M, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Rho-associated kinase, a novel serine/threonine kinase, as a putative target for small GTP binding protein Rho. Embo J. 1996;15:2208–16. [PMC free article] [PubMed] [Google Scholar]
- 6.Kimura K, Ito M, Amano M, Chihara K, Fukata Y, Nakafuku M, Yamamori B, Feng J, Nakano T, Okawa K, Iwamatsu A, Kaibuchi K. Regulation of myosin phosphatase by Rho and Rho-associated kinase (Rho-kinase) Science. 1996;273:245–8. doi: 10.1126/science.273.5272.245. [DOI] [PubMed] [Google Scholar]
- 7.D’Avino PP, Savoian MS, Glover DM. Cleavage furrow formation and ingression during animal cytokinesis: A microtubule legacy. J Cell Sci. 2005;118:1549–58. doi: 10.1242/jcs.02335. [DOI] [PubMed] [Google Scholar]
- 8.Bement WM, Benink HA, von Dassow G. A microtubule-dependent zone of active RhoA during cleavage plane specification. J Cell Biol. 2005;170:91–101. doi: 10.1083/jcb.200501131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamijo K, Ohara N, Abe M, Uchimura T, Hosoya H, Lee JS, Miki T. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol Biol Cell. 2006;17:43–55. doi: 10.1091/mbc.E05-06-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshizaki H, Ohba Y, Parrini MC, Dulyaninova NG, Bresnick AR, Mochizuki N, Matsuda M. Cell type-specific regulation of RhoA activity during cytokinesis. J Biol Chem. 2004;279:44756–62. doi: 10.1074/jbc.M402292200. [DOI] [PubMed] [Google Scholar]
- 11.Dean SO, Rogers SL, Stuurman N, Vale RD, Spudich JA. Distinct pathways control recruitment and maintenance of myosin II at the cleavage furrow during cytokinesis. Proc Natl Acad Sci USA. 2005;102:13473–8. doi: 10.1073/pnas.0506810102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao WM, Fang G. MgcRacGAP controls the assembly of the contractile ring and the initiation of cytokinesis. Proc Natl Acad Sci USA. 2005;102:13158–63. doi: 10.1073/pnas.0504145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuce O, Piekny A, Glotzer M. An ECT2-centralspindlin complex regulates the localization and function of RhoA. J Cell Biol. 2005;170:571–82. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.D’Avino PP, Savoian MS, Glover DM. Mutations in sticky lead to defective organization of the contractile ring during cytokinesis and are enhanced by Rho and suppressed by Rac. J Cell Biol. 2004;166:61–71. doi: 10.1083/jcb.200402157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshizaki H, Ohba Y, Kurokawa K, Itoh RE, Nakamura T, Mochizuki N, Nagashima K, Matsuda M. Activity of Rho-family GTPases during cell division as visualized with FRET-based probes. J Cell Biol. 2003;162:223–32. doi: 10.1083/jcb.200212049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei Q, Adelstein RS. Pitx2a expression alters actin-myosin cytoskeleton and migration of HeLa cells through Rho GTPase signaling. Mol Biol Cell. 2002;13:683–97. doi: 10.1091/mbc.01-07-0358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu BP, Burridge K. Vav2 activates Rac1, Cdc42, and RhoA downstream from growth factor receptors but not beta1 integrins. Mol Cell Biol. 2000;20:7160–9. doi: 10.1128/mcb.20.19.7160-7169.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. Embo J. 1999;18:578–85. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benard V, Bohl BP, Bokoch GM. Characterization of rac and cdc42 activation in chemoat-tractant-stimulated human neutrophils using a novel assay for active GTPases. J Biol Chem. 1999;274:13198–204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 20.Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol. 2005;15:371–7. doi: 10.1016/j.tcb.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Satterwhite LL, Pollard TD. Cytokinesis. Curr Opin Cell Biol. 1992;4:43–52. doi: 10.1016/0955-0674(92)90057-j. [DOI] [PubMed] [Google Scholar]
- 22.Wei Q, Adelstein RS. Conditional expression of a truncated fragment of nonmuscle myosin II-A alters cell shape but not cytokinesis in HeLa cells. Mol Biol Cell. 2000;11:3617–27. doi: 10.1091/mbc.11.10.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maupin P, Pollard TD. Arrangement of actin filaments and myosin-like filaments in the contractile ring and of actin-like filaments in the mitotic spindle of dividing HeLa cells. J Ultrastruct Mol Struct Res. 1986;94:92–103. doi: 10.1016/0889-1605(86)90055-8. [DOI] [PubMed] [Google Scholar]
- 24.Egelhoff TT, Brown SS, Spudich JA. Spatial and temporal control of nonmuscle myosin localization: Identification of a domain that is necessary for myosin filament disassembly in vivo. J Cell Biol. 1991;112:677–88. doi: 10.1083/jcb.112.4.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–4. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 26.De Lozanne A, Spudich JA. Disruption of the Dictyostelium myosin heavy chain gene by homologous recombination. Science. 1987;236:1086–91. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 27.Bao J, Jana SS, Adelstein RS. Vertebrate nonmuscle myosin II isoforms rescue small interfering RNA-induced defects in COS-7 cell cytokinesis. J Biol Chem. 2005;280:19594–9. doi: 10.1074/jbc.M501573200. [DOI] [PubMed] [Google Scholar]
- 28.Burns CG, Larochelle DA, Erickson H, Reedy M, De Lozanne A. Single-headed myosin II acts as a dominant negative mutation in Dictyostelium. Proc Natl Acad Sci USA. 1995;92:8244–8. doi: 10.1073/pnas.92.18.8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burns CG, Reedy M, Heuser J, De Lozanne A. Expression of light meromyosin in Dictyostelium blocks normal myosin II function. J Cell Biol. 1995;130:605–12. doi: 10.1083/jcb.130.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips CL, Yamakawa K, Adelstein RS. Cloning of the cDNA encoding human nonmuscle myosin heavy chain-B and analysis of human tissues with isoform-specific antibodies. J Muscle Res Cell Motil. 1995;16:379–89. doi: 10.1007/BF00114503. [DOI] [PubMed] [Google Scholar]