Summary
The SH3 domain is a protein-protein interaction module commonly found in intracellular signaling and adaptor proteins. The SH3 domains of multiple endocytic proteins have been recently implicated in binding ubiquitin, which serves as a signal for diverse cellular processes including gene regulation, endosomal sorting, and protein destruction. Here we describe the solution NMR structure of ubiquitin in complex with an SH3 domain belonging to the yeast endocytic protein Sla1. The ubiquitin binding surface of the Sla1 SH3 domain overlaps substantially with the canonical binding surface for proline-rich ligands. Like many other ubiquitin-binding motifs, the SH3 domain engages the Ile44 hydrophobic patch of ubiquitin. A phenylalanine residue located at the heart of the ubiquitin-binding surface of the SH3 domain serves as a key specificity determinant. The structure of the SH3-ubiquitin complex explains how a subset of SH3 domains has acquired this non-traditional function.
Keywords: endocytosis, monoubiquitin signaling, ubiquitin-binding motif, protein-protein interactions, SH3
Introduction
The SH3 domain is a protein-protein interaction module found in many proteins in diverse eukaryotes. The best-characterized function of these domains, which were initially described in signaling proteins, is to bind proline-enriched sequences of the type P-x-x-P (x: any residue).1 The repertoire of proteins in which these domains are found as well as that of sequence and structural motifs recognized by these domains appear to be substantially broader than initially expected.2 The recent discovery of ubiquitin as a target of SH3 domains thus not only adds to the diversity but also brings two key players in cellular signaling into the same picture.3 Monoubiquitination of target proteins serves to signal internalization and sorting through the endocytic pathway and gene activation at the transcriptional level.4,5 More than a dozen distinct ubiquitin-binding motifs have been described that function in the direct recognition of the ubiquitin signal,6 but none of these motifs shares any overt similarity to SH3 domains. Similarly, ubiquitin does not contain any of the known SH3 binding helical motifs.2 To gain insights into the mode of ubiquitin binding by SH3 domains, we determined the solution structure of the third SH3 domain of the yeast Sla1 protein (henceforth designated Sla1 SH3−3) in complex with monoubiquitin. The Sla1 protein is localized at the cell cortex and is thought to couple actin dynamics with endocytosis by functioning as an adaptor interacting with multiple components of the actin cytoskeleton and the endocytic machinery besides engaging specific signals within transmembrane proteins.7 Sla1 orthologs exist in a broad range of species and the mammalian ortholog CIN85, which retains the ubiquitin binding function, also functions as an adaptor and is specifically implicated in coordinating the internalization and destruction of activated receptor tyrosine kinases.8
Results and Discussion
We had previously shown using solution NMR spectroscopy that like other ubiquitin-binding motifs Sla1 SH3−3 could bind ubiquitin specifically but with modest affinity (Kd ∼40 μM).3 To alleviate exchange-broadening effects associated with a subset of resonances at the protein-protein interface, the NMR structural studies were conducted at a slightly elevated temperature (45 °C; Kd ∼400 μM). Essentially all the resonances belonging to each protein in the complex were assigned using standard through-bond approaches and structures were determined using a combination of chemical shift-based torsion angle and nuclear Overhauser effect (NOE)-based distance restraints (Table 1). Structures of reasonable quality and precision and in agreement with input experimental data were obtained (Table 1 & Figure 1(a)).
Table 1.
NMR Structure Determination Statistics for the Sla1 SH3-3-Ubiquitin Complex
| Restraint Statistics | |
| NOE-based distance restraints | |
| Unambiguous NOE-based restraints | 2817 |
| Intraresidue | 1277 |
| Sequential (|i — j| = 1) | 445 |
| Medium-range (1 < |i — j| ≤ 4) | 211 |
| Intramolecular long-range (|i — j| > 4) | 756 |
| Intermolecular | 128 |
| Ambiguous NOE-based restraints | 589 |
| Intramolecular NOE-based restraints | 1503 SH3, 1775 ubiquitin |
| Hydrogen bonding distance restraints | 70 |
| Torsion angle restraints | 166 (83 ℱ, 83 ψ) |
| Structure Quality of NMR Ensemble | |
| Restraint satisfaction | |
| Root-mean-square differences for distance restraints | 0.012 ± 0.001 Å |
| Root-mean-square differences for torsion angle restraints | 0.124° ± 0.038° |
| Deviations from ideal covalent geometry | |
| Bond lengths | 0.003 ± 0.000 Å |
| Bond angles | 0.453° ± 0.011° |
| Impropers | 1.340° ± 0.087° |
| Ramachandran plot statistics | |
| Residues in most favored regions | 85.0% |
| Residues in allowed regions | 14.4% |
| Residues in disallowed regions | 0.6% |
| Average Atomic Root-Mean-Square Deviations from the Average Structure | |
| All atoms | 2.46 Å |
| All atoms except disordered regionsa | 1.39 Å |
| All atoms in secondary structural elementsb | 1.16 Å |
| Backbone atoms (N, Cα, C′) | |
| All residues | 1.96 Å |
| All residues excluding disordered regionsa | 0.77 Å |
| All residues in secondary structural elementsb | 0.59 Å |
: disordered regions include residues 350-354 and 414-420 of Sla1 and 74-76 of ubiquitin
: secondary structural elements include residues 356-360, 379-384, 391-396, 402-406, and 410-412 of Sla1 and 2-7, 12-16, 23-34, 41-45, 48-50, and 66-71 of ubiquitin
Figure 1.
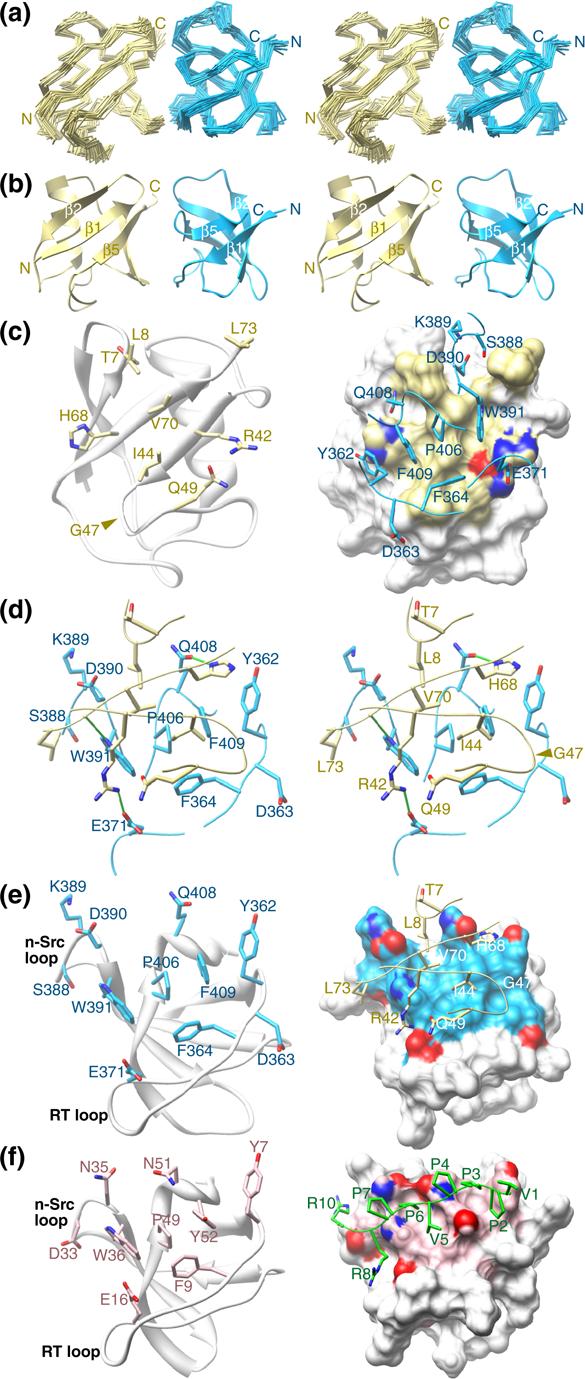
The Sla1 SH3−3 domain binds ubiquitin through a similar surface used for interactions with helical motifs. (a) Stereo views of the Cα trace following a best-fit superposition of backbone atoms in well-ordered regions corresponding to residues 355−413 of Sla1 and 1−73 of ubiquitin in the ensemble. The Sla1 SH3−3 domain is colored blue and ubiquitin is shown in yellow. The disordered regions corresponding to residues 350−354 and 414−420 of Sla1 and 74−76 of ubiquitin have been omitted for clarity. (b) Stereo views of the representative structure from the NMR ensemble of the Sla1 SH3−3–ubiquitin complex with the same coloring scheme as in panel (a). (c) Structure of ubiquitin in the complex shown in a cartoon representation along with the side chains deemed to be interacting with Sla1 SH3−3 (left panel). A view of the molecular surface of ubiquitin shown along with the ubiquitin-interacting side chains and the corresponding backbone segments of Sla1 SH3−3 (right panel). Oxygen and nitrogen atoms are shown in red and dark blue, respectively. (d) Stereo views of the Sla1 SH3−3–ubiquitin interface depicting the side chains of the interacting residues and the corresponding backbone segments. Intermolecular hydrogen bonding interactions are depicted as solid green lines. (e) Structure of Sla1 SH3−3 in the complex shown in a cartoon representation along with the ubiquitin interacting side chains (left panel). A view of the molecular surface of the SH3 domain shown along with the SH3-interacting side chains and the corresponding backbone segments of ubiquitin (right panel). (f) Structure of the Grb2 SH3-SOS peptide complex (PDB code: 1GBR10) with the SH3 domain presented in the same orientation as Sla1 SH3−3 in panel (e). The side chains of Grb2 deemed to be interacting with the proline-rich peptide are shown in light pink; oxygen and nitrogen atoms are shown in red and dark blue, respectively. The peptide backbone and side chains are colored in green (right).
Both ubiquitin and the Sla1 SH3 domain adopt the well-characterized eponymous folds in the complex (Figure 1(b)). The five β-strands of Sla1 SH3−3 span residues 356−360, 379−384, 391−396, 402−406 and 410−412; a 310-helix separates strands β4 and β5. The polypeptide backbones of the two proteins in the complex are remarkably similar to those of the free forms with atomic root-mean-square deviations of 0.87 Å for ubiquitin9 and 0.88 Å for the SH3 domain (Figure 2(a); PDB codes 1UBQ and 1Z9Z, respectively). Similarly, only small conformational rearrangements upon complex formation are noted for the SH3 side chains (Figure 2(b)). Interestingly, two molecules are found in the asymmetric unit in the crystal structure of apo Sla1 SH3−3 with the molecules participating in homotypic interactions via the ubiquitin binding surface (PDB code: 1Z9Z). However, we find no evidence for homodimerization by NMR as the Sla1 SH3−3 domain spectra are characterized by narrow resonances that broaden somewhat upon complex formation with ubiquitin due to the increased molecular size of the complex (data not shown).
Figure 2.
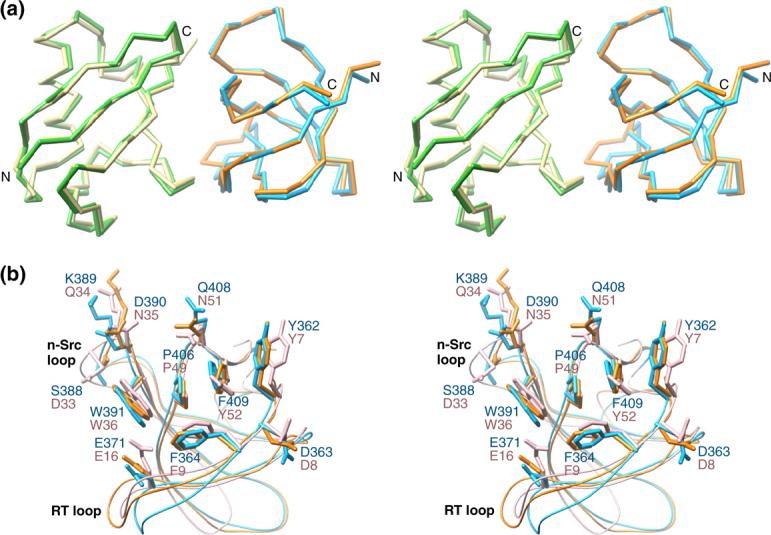
Economy in conformational rearrangements upon complex formation between Sla1 SH3−3 and ubiquitin and basis for specificity of SH3-ubiquitin interactions. (a) Stereo views of the Cα traces of Sla1 SH3−3 and ubiquitin following a best-fit superposition of backbone atoms corresponding to residues 357−415 of Sla1 and 1−73 of ubiquitin in the NMR structure of the complex with the corresponding segments in the crystal structures of the apo forms. The Sla1 SH3−3 domain is colored blue (complex) and orange (apo; PDB code: 1Z9Z) whereas ubiquitin is shown in yellow (complex) and green (apo; PDB code: 1UBQ9). (b) Stereo views of the ubiquitin-binding residues of Sla1 SH3−3 in the apo (orange) and ubiquitin-bound (blue) states and comparison with the equivalent residues in the Grb2 SH3 domain (light pink).10 The side chains are shown in stick representation whereas the backbones are shown as worms. Residues and important structural landmarks are annotated. Notice also the relatively modest changes in side chain conformation for Sla1 SH3−3 upon complex formation with ubiquitin.
The Sla1 SH3−3–ubiquitin complex interface extends over an area averaging 470 Å2 (± 36 Å2) in the NMR ensemble. Like many other ubiquitin-binding motifs,6 the SH3 domain targets a primarily hydrophobic ubiquitin surface comprising Leu8, Ile44, Val70, and Leu73 surrounded by an array of polar residues (Figure 1(c) & 1(d)). The complementary surface on the SH3 domain is dominated by hydrophobic residues including Tyr362, Phe364, Trp391, Pro406, and Phe409 that form a relatively smooth, shallow groove (Figure 1(e)). The complex appears to be stabilized by intermolecular hydrogen bonding interactions between the side chains of His68 and Gln408, Arg42 and Glu371, and between the backbone carbonyl of Leu71 and the side chain of Trp391 and by electrostatic interactions involving Arg42 of ubiquitin and Glu367 and Glu371 of Sla1 SH3−3 (Figure 1(d) & Figure 3 (a)). We note that these non-bonding interactions were consistently detected in over 60% of the conformers in the NMR ensemble and were inferred from the structures resulting from the structure determination protocol rather than being explicitly entered as experimentally-derived restraints. These interactions are consistent with previous mutagenesis studies that implicated a subset of these residues in stabilizing the complex including Ile44 and Val70 of ubiquitin and Tyr362, Phe364, Trp391, Gln408, and Phe409 of Sla1 SH3−3 as mutations of any of these residues to alanine were found to severely diminish the SH3-ubiquitin interaction (Figure 3).3
Figure 3.
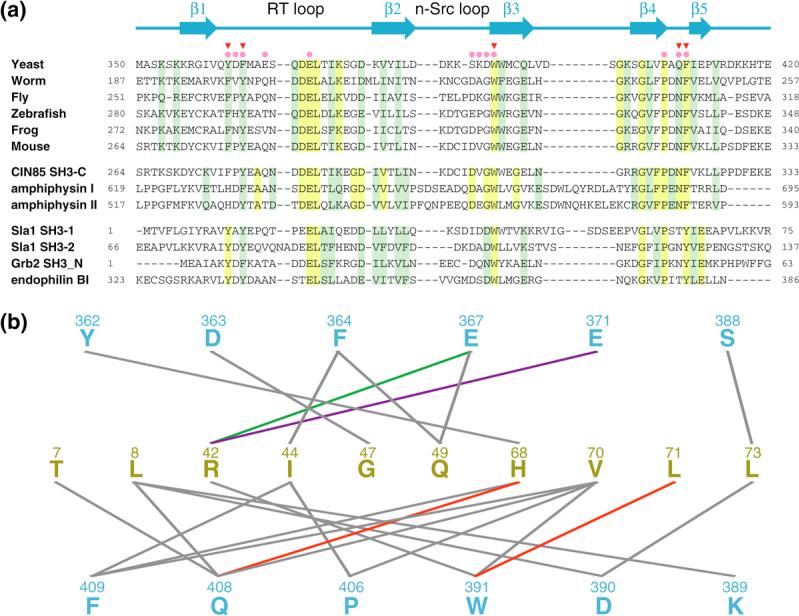
Sequence conservation and non-covalent interactions mediated by residues of SH3 domains with ubiquitin-binding activity. (a) CLUSTAL W-guided multiple sequence alignment of a selection of SH3 domains. The proteins were grouped into three functional classes including Sla1 orthologs (top), proteins with demonstrable ubiquitin-binding activity (middle) and those lacking this activity (bottom).3 Conserved and invariant residues in each group are shaded green and yellow, respectively. The solid pink circles and red inverted triangles denote Sla1 SH3−3 residues deemed to contact ubiquitin in the NMR structure and those residues that when mutated to alanine diminish monoubiquitin binding,3 respectively. A cartoon depicting the location of secondary structural elements in Sla1 SH3−3 is shown on top. (b) A catalog of non-covalent intermolecular interactions in the Sla1 SH3−3-ubiquitin complex detected in ≥60% of conformers in the NMR ensemble.27 Sla1 SH3−3 and ubiquitin residues are colored in blue and yellow, respectively. The lines connect interacting residues. Line colors indicate the type of interaction (green: electrostatic; red: electrostatic; purple: salt bridge; gray: hydrophobic).
The amino-terminal Grb2 SH3 domain represents the closest homologue of Sla1 SH3−3 (37% sequence identity) for which a high-resolution structure has been described.10 A comparison of the structures of the Sla1 SH3−3–ubiquitin complex with that of the Grb2 SH3 domain in complex with a proline-rich sequence reveals substantial overlap between the ligand-binding surfaces of the two SH3 domains (Figure 1(f)). The vast majority of the ligand-binding residues within the two domains are either identical or conserved and this feature is shared with other SH3 domains whose structures in complex with proline-containing peptides have been described.2 Unlike Sla1 SH3−3, the Grb2 SH3 domain does not exhibit discernible ubiquitin-binding activity,3 so what attribute(s) confers a particular SH3 domain with the ability to bind ubiquitin? Intriguingly, the side chain conformations of the ligand-binding residues are similar for the Grb2 and Sla1 SH3 domains (Figure 2(b)), implying that the functional differences are unlikely to originate from distinct conformational preferences. A key affinity and specificity determinant for ubiquitin-binding was previously postulated to reside with a phenylalanine residue (Phe409 of Sla1).3 This residue is located at the heart of the hydrophobic interface and is shielded from the solvent in the SH3-ubiquitin complex (Figure 1(d)). The Grb2 SH3 domain harbors a tyrosine residue at the equivalent position (Figures 1(e), 1(f) and 2(b)) and mutation of Sla1 Phe409 to tyrosine was found to abrogate ubiquitin binding.3 The most likely explanation based on the structure of the Sla1 SH3−3-ubiquitin complex is that the introduction of a hydroxyl group results in unfavorable steric clashes between this moiety and Ile44 of ubiquitin. This is probably exacerbated by the absence of suitable hydrogen bonding donor and acceptor groups near the hydroxyl group. Predictably, a phenylalanine rather than a tyrosine is consistently detected at this position in the third SH3 domain of Sla1 orthologs including mammalian CIN85 and in other SH3 domains possessing ubiquitin-binding activity (Figure 3(a)). Conversely, a tyrosine residue is consistently detected in SH3 domains lacking ubiquitin-binding activity. Interestingly, the phenylalanine does not preclude Sla1 SH3−3 from binding proline-rich ligands,3 consistent with previous structural analyses of other SH3 domains. The Sla1 SH3 domain thus provides a striking example of how disparate functions can be accommodated within a common interaction surface through subtle changes in sequence.
Although the phenylalanine residue is an important affinity and specificity determinant for ubiquitin-binding, it is likely that the full or near-full complement of non-covalent interactions described for the Sla1 SH3−3-ubiquitin complex is essential for efficient binding. Indeed, mutating Tyr64 to a phenylalanine in the Sla1 SH3−1 domain failed to produce a protein competent to bind ubiquitin.3 One explanation for this observation could be the lack of a Sla1 SH3−1 residue equivalent to Gln408 capable of hydrogen bonding interactions (Figure 3(b)). Similar considerations could apply to other phenylalanine-harboring SH3 domains that might fail to bind ubiquitin.
The pervasiveness of ubiquitin-binding motifs in endocytic proteins underscores the importance of this signal in endocytosis. It is intriguing that multiple endocytic proteins harbor SH3 domains with ubiquitin-binding activity while simultaneously retaining their ability to bind proline-rich ligands, suggesting a dynamic interplay between these two processes that warrants further investigation. Finally, the availability of detailed molecular pictures of all the key domains for the Sla1/CIN85 protein in complex with their physiologically relevant targets sets the stage for a comprehensive functional analyses of this evolutionarily conserved protein. Intriguingly, all four domains including the three SH3 domains and the SHD1 domain unique to Sla1 share structural similarity but exhibit considerable diversity in the way they engage their targets, that are themselves structurally varied.11,12 This could be a general theme for adaptor proteins involved in intracellular signaling.
Methods
Production of the Sla1 SH3−3 domain and ubiquitin
The Saccharomyces cerevisiae Sla1 SH3−3 domain corresponding to residues 350−420 was expressed as a recombinant protein and purified as described previously.3 The identity of the protein was confirmed by electrospray ionization mass spectrometry. Sla1 SH3−3 protein samples uniformly labeled with 15N and/or 13C isotopes were produced using the same procedure except that cells were grown in M9 minimal medium containing 15N-ammonium sulfate and/or 13C-D-glucose (Spectra Stable Isotopes, Columbia, MD) respectively. Unlabeled and uniformly 15N- and 15N,13C-labeled samples of Saccharomyces cerevisiae ubiquitin were produced as described previously.13,14
NMR samples
All NMR samples were prepared in 20 mM sodium phosphate buffer (pH 6), 2 mM DTT-d10, and 0.2% (w/v) NaN3. An equimolar complex of 15N,13C-labeled Sla1 SH3−3 and unlabeled ubiquitin was generated by titrating Sla1 SH3−3 with ubiquitin until a 1:1 ratio was attained. The progress of the titration was monitored by recording one-dimensional (1D) 1H and two-dimensional (2D) 1H-15N correlated spectra. An equimolar 15N,13C-labeled ubiquitin-unlabeled Sla1 SH3−3 complex was generated in a similar manner. Protein concentrations were measured spectrophotometrically15 and the concentrations of the NMR samples were ∼0.9 mM.
NMR spectroscopy and structure determination
NMR data were acquired on a Varian Inova 600 MHz spectrometer at 45 °C. NMR data processing and analysis were performed using an in-house modified version of Felix 98.0 (Accelrys) and NMRView.16,17 Backbone and side chain 1H, 15N, and 13C resonances for the Sla1 SH3−3 domain and ubiquitin were assigned by analyzing three-dimensional (3D) HNCACB, C(CO)NH-TOCSY, HNCO, H(CCO)NH-TOCSY, and HCCH-COSY spectra.18,19 Aromatic proton resonances were assigned from 2D 15N,13C-double-half-filtered TOCSY spectra and 3D HCCH-COSY and HCCH-TOCSY spectra.20,21
For structure determination, backbone ℱ and ψ torsion angle restraints were derived from an analysis of Hα, Cα, Cβ, C′ and backbone 15N chemical shifts using TALOS.22 Restraints were imposed only for those residues that exhibited TALOS reliability scores of 9. NOE-based distance restraints for each component of the binary complex were derived from 3D 15N-edited NOESY (mixing time, τm = 80 ms) and 3D 13C-filtered, 13C-edited NOESY (τm = 120 ms) spectra recorded in H2O, 3D aliphatic 13C-edited NOESY (τm = 65 ms) and 2D 15N,13C-double-half-filtered NOESY (τm = 60 ms) spectra recorded in D2O. Intermolecular NOEs were assigned manually and given upper bounds of 6 Å. All other NOEs were calibrated and assigned iteratively and automatically by ARIA (version 1.2)23,24 and were checked manually between successive rounds of refinement. Structures were calculated using ARIA in conjunction with CNS.25 Structures were calculated from extended conformations as starting models. A total of 80 structures were computed in the final iteration, 40 of which were refined in the presence of a shell of explicit water solvent, and the 20 structures with the lowest restraint energies, restraint violations, and RMS deviations from ideal covalent geometry were selected for structural analysis. The final structures were analyzed using PROCHECK, MONSTER, and CNS.25-27 Molecular images were generated using CHIMERA.28
Accession code
The RCSB PDB accession code for the coordinates and the restraint tables used for the calculation of the Sla1 SH3−3–ubiquitin complex is 2JT4.
Acknowledgements
We thank Hisae Matsuura and Richard Kang for the initial development of the project, Yongbo Zhang for assistance with NMR data collection, the NIH for grant support to L.H. and I.R. and the Lurie Cancer Center for supporting structural biology research at Northwestern. I.R. is a Scholar of the Leukemia and Lymphoma Society.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mayer BJ. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–63. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 2.Li SS. Specificity and versatility of SH3 and other proline-recognition domains: structural basis and implications for cellular signal transduction. Biochem J. 2005;390:641–53. doi: 10.1042/BJ20050411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stamenova SD, French ME, He Y, Francis SA, Kramer ZB, Hicke L. Ubiquitin binds to and regulates a subset of SH3 domains. Mol Cell. 2007;25:273–84. doi: 10.1016/j.molcel.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hicke L. Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 5.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–72. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 6.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–72. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ayscough KR. Coupling actin dynamics to the endocytic process in Saccharomyces cerevisiae. Protoplasma. 2005;226:81–8. doi: 10.1007/s00709-005-0107-5. [DOI] [PubMed] [Google Scholar]
- 8.Dikic I. CIN85/CMS family of adaptor molecules. FEBS Lett. 2002;529:110–5. doi: 10.1016/s0014-5793(02)03188-5. [DOI] [PubMed] [Google Scholar]
- 9.Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 Å resolution. J Mol Biol. 1987;194:531–44. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]
- 10.Wittekind M, Mapelli C, Lee V, Goldfarb V, Friedrichs MS, Meyers CA, Mueller L. Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts. J Mol Biol. 1997;267:933–52. doi: 10.1006/jmbi.1996.0886. [DOI] [PubMed] [Google Scholar]
- 11.Jozic D, Cardenes N, Deribe YL, Moncalian G, Hoeller D, Groemping Y, Dikic I, Rittinger K, Bravo J. Cbl promotes clustering of endocytic adaptor proteins. Nat Struct Mol Biol. 2005;12:972–9. doi: 10.1038/nsmb1000. [DOI] [PubMed] [Google Scholar]
- 12.Mahadev RK, Di Pietro SM, Olson JM, Piao HL, Payne GS, Overduin M. Structure of Sla1p homology domain 1 and interaction with the NPFxD endocytic internalization motif. EMBO J. 2007;26:1963–71. doi: 10.1038/sj.emboj.7601646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci U S A. 1996;93:861–6. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang RS, Daniels CM, Francis SA, Shih SC, Salerno WJ, Hicke L, Radhakrishnan I. Solution structure of a CUE-ubiquitin complex reveals a conserved mode of ubiquitin binding. Cell. 2003;113:621–630. doi: 10.1016/s0092-8674(03)00362-3. [DOI] [PubMed] [Google Scholar]
- 15.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–26. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 16.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Structural analyses of CREB-CBP transcriptional activator-coactivator complexes by NMR spectroscopy: implications for mapping the boundaries of structural domains. J Mol Biol. 1999;287:859–65. doi: 10.1006/jmbi.1999.2658. [DOI] [PubMed] [Google Scholar]
- 17.Johnson BA. Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol. 2004;278:313–52. doi: 10.1385/1-59259-809-9:313. [DOI] [PubMed] [Google Scholar]
- 18.Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13C/15N-enriched proteins. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- 19.Ferentz AE, Wagner G. NMR spectroscopy: a multifaceted approach to macromolecular structure. Q Rev Biophys. 2000;33:29–65. doi: 10.1017/s0033583500003589. [DOI] [PubMed] [Google Scholar]
- 20.Otting G, Wuthrich K. Heteronuclear filters in two-dimensional [1H,1H]-NMR spectroscopy: combined use with isotope labelling for studies of macromolecular conformation and intermolecular interactions. Q Rev Biophys. 1990;23:39–96. doi: 10.1017/s0033583500005412. [DOI] [PubMed] [Google Scholar]
- 21.Lohr F, Ruterjans H. Novel pulse sequences for the resonance assignment of aromatic side chains in 13C-labeled proteins. J Magn Reson B. 1996;112:259–68. doi: 10.1006/jmrb.1996.0140. [DOI] [PubMed] [Google Scholar]
- 22.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 23.Linge JP, Habeck M, Rieping W, Nilges M. ARIA: automated NOE assignment and NMR structure calculation. Bioinformatics. 2003;19:315–6. doi: 10.1093/bioinformatics/19.2.315. [DOI] [PubMed] [Google Scholar]
- 24.Linge JP, Habeck M, Rieping W, Nilges M. Correction of spin diffusion during iterative automated NOE assignment. J Magn Reson. 2004;167:334–42. doi: 10.1016/j.jmr.2004.01.010. [DOI] [PubMed] [Google Scholar]
- 25.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Pt 5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 26.Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–86. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 27.Salerno WJ, Seaver SM, Armstrong BR, Radhakrishnan I. MONSTER: inferring non-covalent interactions in macromolecular structures from atomic coordinate data. Nucleic Acids Res. 2004;32:W566–8. doi: 10.1093/nar/gkh434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]


