Summary
MLL-containing complexes methylate histone H3 at lysine 4 (H3K4) and have been implicated in the regulation of transcription. However, it is unclear how MLL complexes are targeted to specific gene loci. Here, we show that the MLL2 complex associates with the hematopoietic activator NF-E2 in erythroid cells and is important for H3K4 trimethylation and maximal levels of transcription at the β-globin locus. Furthermore, recruitment of the MLL2 complex to the β-globin locus is dependent upon NF-E2 and coincides spatio-temporally with NF-E2 binding during erythroid differentiation. Thus a DNA-bound activator is important initially for guiding MLL2 to a particular genomic location. Interestingly, while the MLL2-associated subunit Ash2L is restricted to the β-globin locus control region 38 kb upstream of the βmaj-globin gene, the MLL2 protein spreads across the β-globin locus, suggesting a previously undefined mechanism by which an activator influences transcription and H3K4 trimethylation at a distance.
Introduction
Developmentally regulated patterns of gene expression are established through the opposing function of trithorax (TrxG) and polycomb (PcG) group proteins (Ringrose and Paro, 2004). PcG proteins have been shown to maintain a repressive chromatin structure, while TrxG proteins act to establish a chromatin structure permissive for transcription. While the exact mechanism by which these complex patterns of gene expression are established is unclear, epigenetic modification of histones appears to play an important role (Ringrose and Paro, 2004). Of particular interest, specific methylation events at histone H3 mark chromatin for either transcriptional activation or repression –methylation of histone H3 at lysine 4 (H3K4) marks genes located in an open chromatin structure, while methylation at lysine 9 and/or 27 target transcriptionally repressed genes (Margueron et al., 2005; Martin and Zhang, 2005; Ruthenburg et al., 2007; Shilatifard, 2006). In mammals, H3K4 methylation is mediated by the SET1 family of TrxG proteins, which includes 4 related MLL (mixed lineage leukemia)-like methyltransferases. All MLL-like proteins associate with an overlapping set of regulatory subunits to form the MLL complexes (Ruthenburg et al., 2007). While all core subunits are important for regulating the H3K4 methyltransferase activity, the TrxG protein Ash2L seems to be required for H3K4 trimethylation (H3K4me3) (Dou et al., 2006; Steward et al., 2006), a modification which in contrast to mono- and di-methylation, is strictly correlated with active genes in vivo (Bernstein et al., 2005). Despite extensive study, the mechanism that permits targeting of MLL complexes to gene loci has remained elusive (Slany, 2005). On one hand, MLL-like proteins interact with sequence-specific DNA-bound transcription factors (i.e. p53, Max, E2F6 (Dou et al., 2005), estrogen receptor (Mo et al., 2006), retinoic acid receptor (Goo et al., 2003) and LEF-1/TCF (Sierra et al., 2006)), suggesting a gene targeting mechanism through interactions with cell-specific DNA-bound transcription factors. On the other hand, several studies have reported an interaction between MLL and the Ser5-phosphorylated form of RNA polymerase II (Ser5P-Pol II) (Hughes et al., 2004; Milne et al., 2005a), suggesting that recruitment of MLL-like proteins could occur via interaction with the elongating Pol II. Consistent with this possibility, the genomic distribution of the H3K4me3 mark overlaps that of the early elongating Pol II (Milne et al., 2005a; Ng et al., 2003).
Here, we report the first study investigating the recruitment mechanism of a MLL2-containing methyltransferase complex to the β-globin locus during terminal erythroid differentiation. We show that the MLL2 complex associates with the hematopoietic activator NF-E2 in erythroid cells and is recruited to the β-globin locus in a NF-E2-dependent manner. This suggests that a DNA-bound activator is important initially for guiding MLL2 to a specific region of the genome. Interestingly, while recruitment of the MLL2 complex is limited to the β-globin Locus Control Region (LCR) located 38 kb upstream of the active βmaj-globin gene (Hardison et al., 1997), the MLL2 protein spreads across the β-globin locus. Importantly, this spreading increases during erythroid differentiation suggesting a mechanism by which a distal activator influences transcription and chromatin structure far downstream of its binding site. Finally, MLL2 is fully activated only when it reaches the coding region of the active β-maj globin gene, as trimethylation of H3-K4 is restricted to this part of the locus.
Results
NF-E2 interacts with the histone H3K4 methyltransferase complex MLL2
The murine β-globin locus is comprised of four clustered β-like globin genes (Figure 3A), which are transcribed at specific developmental stages and only in erythroid cells (Hardison et al., 1997). Of particular interest is the hematopoietic transcriptional activator NF-E2/p45 (Andrews, 1998), which is required for erythroid differentiation and β-globin activation in Mouse ErythroLeukemia (MEL) cells and has been implicated in mediating H3K4 dimethylation at the β-globin locus (Kiekhaefer et al., 2004; Kiekhaefer et al., 2002). To decipher the mechanism by which NF-E2/p45 activates β-globin transcription at a distance, we used immunoprecipitation (IP) to purify cofactors associating with the endogenous NF-E2/p45 protein from erythroid cells. Numerous proteins (currently under investigation) co-immunoprecipitate with NF-E2/p45 in a specific manner. Among them, several subunits of the MLL complexes were identified by quantitative mass spectrometry, including WDR5 (identification score 100%) and HCF1 (identification score 100%). Furthermore, HCF1, WDR5 and Ash2L were detected in our previous mass spectrometry study of proteins interacting with the NF-E2/p45-dimerization partner MafK ((Brand et al., 2004) and data not shown). These 3 proteins Ash2L, WDR5 and HCF1 are conserved subunits of the MLL methyltransferase complexes (Ruthenburg et al., 2007). Western blots were performed to confirm these interactions and to determine whether NF-E2 also associates with other subunits of the MLL complexes. We detected 5 conserved subunits of MLL complexes in the elution of the NF-E2/p45 IP (Figure 1A), including Ash2L, WDR5 and RbBP5, which constitute the structural core of the complex (Dou et al., 2006) and the tumor suppressor menin, previously found in related methyltransferase complexes containing Ash2L (Hughes et al., 2004; Yokoyama et al., 2004). Interestingly, using several independently raised Abs, we could not detect MLL1 in the NF-E2/p45 IP, even though this gene is transcribed in MEL cells (not shown). However, we were able to show an interaction of NF-E2/p45 with its close homolog MLL2/TRX2 (also called MLL4) (Figure 1A). Importantly, MLL1 and MLL2 display a high degree of similarity resulting from gene duplication (FitzGerald and Diaz, 1999). To further confirm the association of NF-E2/p45 with the MLL2 complex, we performed reciprocal IPs using Abs against the endogenous MLL2 and Ash2L proteins in erythroid nuclear extracts. Both subunits of the NF-E2 complex (MafK and p45) were detected in the MLL2 and Ash2L IPs (Figure 1A, B) confirming the association of the activator NF-E2 with the MLL2 complex in differentiated erythroid cells. Notably, the acetyltransferase p300/CBP was also identified in the MLL2 and NF-E2/p45 IPs, consistent with previously published data ((Ernst et al., 2001) and (Brand et al., 2004; Hung et al., 2001), respectively). Interaction with this acetyltransferase has been shown to be important for the function of both MLL1 (Popovic and Zeleznik-Le, 2005) and NF-E2/p45 (Hung et al., 2001).
Figure 3. Recruitment of NF-E2/p45 and the MLL2 complex to the β-globin locus during erythroid differentiation analyzed by chromatin immunoprecipitation.
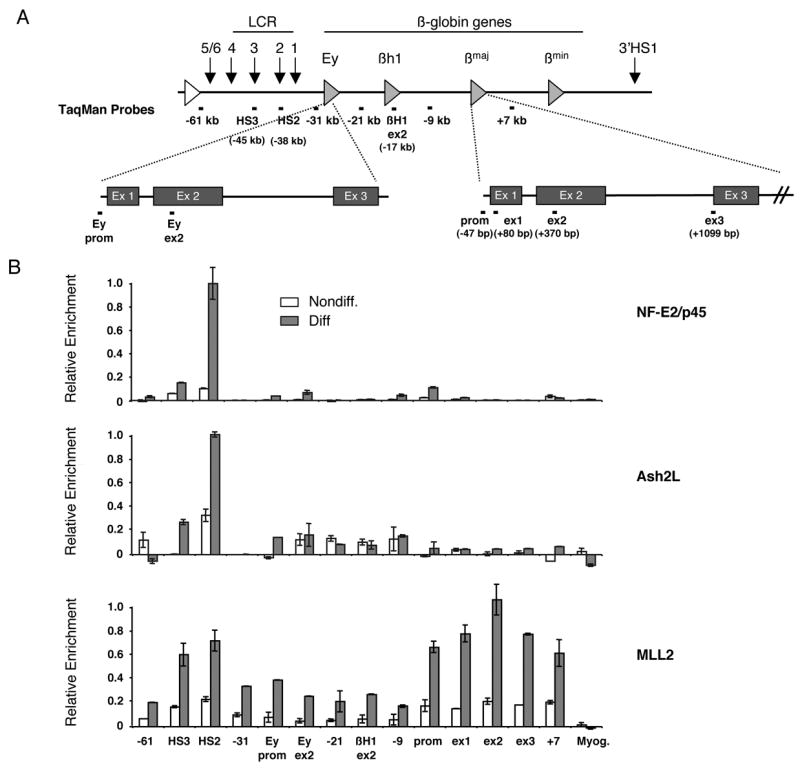
(A) Schematic representation of the murine β-globin locus. Shaded triangles represents the β-like globin genes. The white triangle represents the inactive olfactory receptor gene. Vertical arrows represent DNAse I hypersensitive sites (HSs). The βmaj-globin and Ey genes are shown in details below the locus with their 3 exons (ex). Positions of the TaqMan probes used to reveal the chromatin immunoprecipitations (ChIPs) by real-time qPCR are indicated relative to the βmaj-globin transcription start site (set to +1). Myog. indicates the muscle-specific gene myogenin.
(B) ChIP analysis of NF-E2/p45, Ash2L and MLL2 binding to the β-globin locus during erythroid differentiation.
ChIPs were performed in MEL cells before (Nondiff.) and after (Diff.) induction of erythroid differentiation. Average values of qPCR duplicates are expressed as a function of the highest enrichment on the locus with error bars corresponding to standard deviations. Each ChIP experiment was performed at least twice independently.
Figure 1. NF-E2 associates with the H3K4 methyltransferase complex MLL2 in differentiated erythroid nuclear extracts.
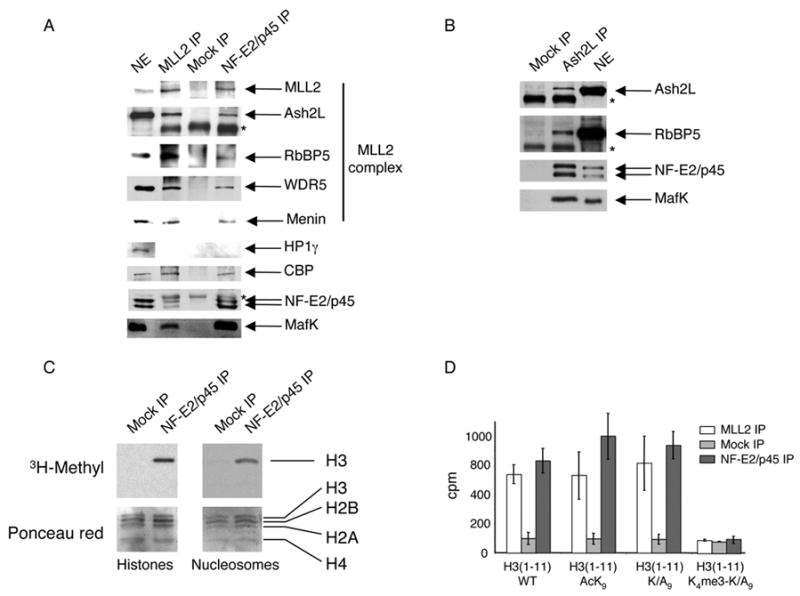
(A) Western blot analysis of proteins immunoprecipitated from an erythroid nuclear extract using Abs against MLL2 and NF-E2/p45. A mock IP using normal rabbit IgG was used as a negative control. Abs used for western blot are indicated on the right. *, Ab heavy chain
(B) Western blot analysis of proteins immunoprecipitated from an erythroid nuclear extract using Abs against Ash2L. *, Ab heavy chain
(C) NF-E2/p45 and Mock immunoprecipitated proteins were used in an in vitro methylation assay in the presence of [3H] SAM and equal amounts of purified core histones (left) or nucleosomes (right) as a substrate. The top panel is an autoradiograph of the bottom panel (stained with Ponceau red).
(D) Eleven amino-acids peptides corresponding to the N-term part of histone H3 that are either wild-type (WT), acetylated at lysine 9 (AcK9), mutated at position 9 (Mut. K/A9) or mutated at lysine 9 and trimethylated at lysine 4 (K4me3-K/A9) were used as a substrate for methylation by NF-E2/p45-, mock-, or MLL2- associated proteins and the radioactivity was counted (in cpm) after a filter binding assay. Error bars correspond to the standard deviation of 3 counts of 1 min. each. The experiment was performed at least twice independently.
To determine whether the MLL2 complex associating with NF-E2 is active, NF-E2 interacting proteins isolated on beads were used to perform an in vitro methylation assay with either purified or nucleosomal histones as a substrate. We found that NF-E2-associated proteins specifically methylate histone H3 in both free and nucleosomal contexts (Figure 1C). Furthermore, 11 amino acid peptides corresponding to the N-term part of histone H3, that are either wild type, acetylated or mutated at position 9 are efficiently methylated by NF-E2-associated proteins. Meanwhile, a peptide trimethylated at H3K4 is a poor substrate for methylation by these proteins (Figure 1D). In combination, these results demonstrate that H3K4 is the methylation site of NF-E2-associated proteins.
The MLL2 complex is recruited to the β-globin locus in a NF-E2-dependent manner
To determine whether NF-E2 helps recruit the MLL2 complex to the β-globin locus, we used Abs recognizing Ash2L and MLL2 to perform chromatin immunoprecipitations (ChIPs) in the NF-E2/p45-null murine erythroleukemia cell line CB3 (Kotkow and Orkin, 1995; Lu et al., 1994) and in a rescued CB3 cell line expressing exogenous NF-E2/p45 in which β-globin transcription is partially restored (Figure 2A,B). We observed that Ash2L and MLL2 are not bound to the β-globin LCR in the NF-E2-null CB3 cell line while they are recruited to the β-globin LCR upon re-expression of NF-E2/p45 (Figure 2D). Thus the MLL2 complex is recruited to the β-globin locus in a NF-E2-dependent manner. In addition, it has been shown that Ser5P-Pol II is bound to the LCR in the absence of NF-E2/p45 in CB3 cells (Johnson et al., 2001), which eliminates the possibility that the MLL2 complex could be recruited indirectly through an interaction between NF-E2 and Pol II.
Figure 2. Recruitment of the MLL2 complex and H3K4 trimethylation at the β-globin locus are dependent upon NF-E2/p45.
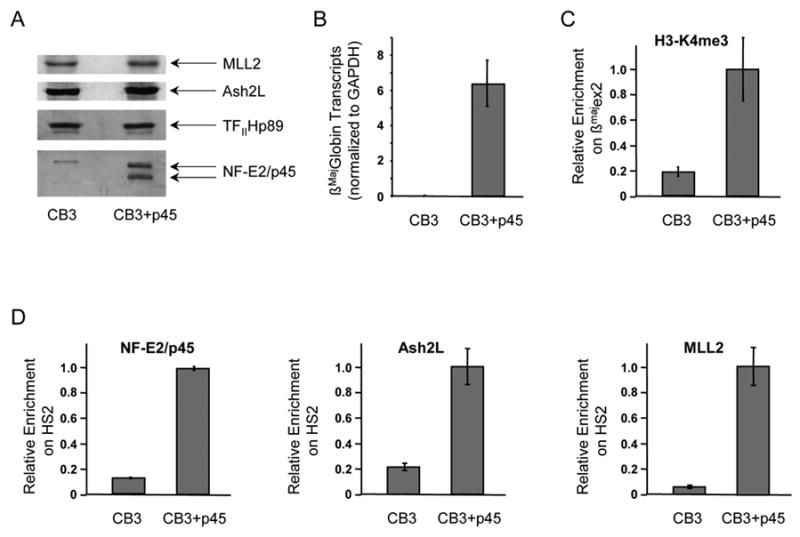
(A) Western blot analysis of nuclear extracts prepared from the NF-E2/p45-null mouse erythroid cell line CB3 and from a rescued clone derived from this cell line, which expresses exogenous NF-E2/p45 (CB3+p45). CB3 cells and the rescued clone were induced to differentiate along the erythroid lineage by exposure to DMSO.
(B) NF-E2/p45 expression in the CB3 cell line partially restores β-globin transcription. Production of βmaj-globin mRNA transcripts after erythroid differentiation was measured by RT-qPCR in the NF-E2/p45 null CB3 cell line and in a rescued NF-E2/p45-expressing CB3 clone. βmaj-globin transcripts were normalized for the amount of GAPDH transcripts. Average values of RT-qPCR triplicates are indicated with error bars corresponding to standard deviations.
(C) H3K4me3 enrichment at the βmaj-globin gene was measured by chromatin immunoprecipitation (ChIP) in the absence (CB3) vs presence (CB3+p45) of NF-E2/p45
(D) NF-E2/p45, Ash2L and MLL2 binding to the β-globin locus were analyzed by ChIP in the absence (CB3) vs presence (CB3+p45) of NF-E2/p45.
TaqMan probes used to reveal the ChIPs by real-time qPCR are the same as in Figure 3. Average values of qPCR duplicates are expressed as a function of the highest enrichment on the locus with error bars corresponding to standard deviations. Each experiment was performed at least twice independently.
To further analyze the recruitment of the MLL2 complex to the β-globin locus during erythroid differentiation, we used MEL cells, which can be induced to undergo most subsequent steps of normal erythroid differentiation, including a greater than 100 fold activation of β-globin transcription (Sawado et al., 2001). Ash2L and MLL2 binding were analyzed by ChIP before and after induction of erythroid differentiation in MEL cells (Figure 3). TaqMan probes recognizing 14 sequences located within representative regions of the β-globin locus (e.g. upstream of the LCR, at the LCR, within the inactive Ey and βH1 genes, within the active βmaj-globin gene and at locations between these genes- Figure 3A) were used to detect the binding of NF-E2/p45, Ash2L and MLL2 by quantitative PCR (qPCR). Consistent with previous results (Brand et al., 2004; Forsberg et al., 2000; Sawado et al., 2001), we observed that during differentiation NF-E2/p45 is recruited primarily to the HS2 site of the LCR, where NF-E2-DNA binding sites are located (Figure 3B). In addition, we found that Ash2L, MLL2 and the DPY-30 subunit of the MLL2 complex are also recruited to the β-globin locus during erythroid differentiation (Figure 3B and data not shown). Interestingly, while recruitment of Ash2L and DPY-30 parallels that of NF-E2/p45 (mostly limited to the LCR), binding of MLL2 after differentiation extends well beyond the LCR, spanning the entire β-globin locus (Figure 3B and data not shown). This enrichment profile is reminiscent of MLL1 binding to active genes (Guenther et al., 2005; Scacheri et al., 2006) in that it is more prominent within the 5′ coding region of the βmaj-globin gene (exon 2). Furthermore, our detailed analysis of MLL2 binding across the β-globin locus reveals that MLL2 is also recruited to additional regions outside the gene coding area and the LCR, including the region upstream of the LCR and the intervening region between the LCR and the active βmaj-globin gene, which spans the 2 inactive embryonic/fetal β-like globin genes Ey/βH1. In contrast, MLL2 does not bind to the inactive muscle-specific gene myogenin. While we note that the MLL2 binding signal is lower in areas outside the LCR and the βmaj-globin gene, it is still significant and specific. Indeed, the signals are ~20 fold higher than on the myogenin control. In addition, MLL2 binding increases significantly during erythroid differentiation (Figure 3B) and is further reduced in response to MLL2 knockdown (Supplementary Figure 1). Therefore, our results suggest a mechanism in which the methyltransferase MLL2 spreads across the β-globin gene during terminal erythroid differentiation. The apparent increased binding of MLL2 at the LCR and the active βmaj-globin gene may result from MLL2 “stalling” in specific areas of the β-globin locus and more efficient spreading in others, perhaps dependent on different chromatin architectures.
H3K4me3 and H3K9ac are restricted to the 5′ region of the active βmaj-globin gene
Having shown that the MLL2 H3K4 methyltransferase is recruited to the β-globin locus, we next used ChIP to examine whether the H3K4me3 mark is enriched within this locus during erythroid differentiation (Figure 4). A strong increase of H3K4me3 was detected after erythroid differentiation within a restricted area of the βmaj-globin gene coding region (exon 2, ~400 bp downstream of the transcription start site). Exon 3 (located 1 kb downstream of the transcription start site) also displays an increase in H3K4me3 during differentiation, although to a lesser extent than exon 2 (Figure 4). This overall pattern of H3K4me3 enrichment within the 5′ coding region is similar to that reported for other active genes (Bernstein et al., 2005; Schneider et al., 2004). Interestingly, during differentiation we did not observe an increase in H3K4me3 directly at the promoter or at the +80 bp region corresponding to exon 1. Instead, the strong increase in H3K4me3 starts after the first 80 bp within the transcribed region of the gene where Pol II is already switched to the elongation mode (Sims et al., 2004). This further suggests that H3K4me3 is linked to transcriptional elongation after promoter clearance. Finally, the LCR and the region between the LCR and the active β-maj globin gene is devoid of H3K4me3 before and after erythroid differentiation (Figure 4). Thus, even though MLL2 is present at the LCR and in the region between the LCR and the βmaj-globin gene (Figure 3B), it is not sufficient to methylate H3K4, suggesting that additional events are required to activate MLL2 methyltransferase activity at the β-maj globin gene. Alternatively, active demethylation of H3K4 could be taking place at the LCR and the intervening regions. Interestingly, large-scale mapping of histones modifications revealed a strong correlation between the distribution of H3K4me3 and H3K9ac/H3K14ac across the human chromosomes 21 and 22 (Bernstein et al., 2005). Furthermore, a cross talk between H3K4me3 and H3K9ac has previously been described (reviewed in (Margueron et al., 2005)). Thus, we examined the enrichment of the H3K9ac mark across the β-globin locus by ChIP during differentiation (Figure 4). We found that H3K9ac profile indeed parallels that of H3K4me3 across the locus, whereby the 5′ region of the βmaj-globin gene open reading frame becomes highly enriched for these modifications after differentiation (Figure 4). This suggests that MLL2 methyltransferase activity could be triggered by the H3K9ac mark when reaching the coding region of the βmaj-globin gene. Interestingly, it has recently been suggested that the repressive H3K9me3 mark is present within the transcribing βmaj-globin gene (Kim et al., 2006; Vakoc et al., 2005). However, H3K9ac and H3K9me3 modifications are mutually exclusive while H3K9me3 and H3K4me3 inhibit each other in vitro (Wang et al., 2001) and do not correlate on other genes in vivo (Vakoc et al., 2006). This suggests that the active (H3K4me3/H3K9ac) and repressive (H3K9me3) states exist in a dynamic equilibrium on the β-globin locus, and that a shift towards the active state may result in high level transcription of the β-globin gene during erythroid differentiation.
Figure 4. Enrichment of H3K4me3 and H3K9ac is limited to the 5′ region of the βmaj-globin gene coding area.
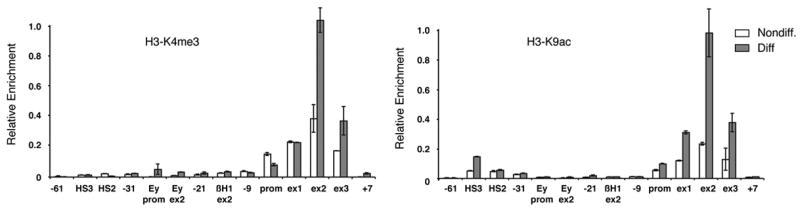
(A) Analysis of H3K4me3 at the β-globin locus during erythroid differentiation by ChIP.
(B) Analysis of H3K9ac at the β-globin locus during erythroid differentiation by ChIP.
TaqMan probes used to reveal the ChIP by real-time qPCR are the same as in Figure 3. ChIPs were performed in MEL cells before (Nondiff.) and after (Diff.) induction of erythroid differentiation. Average values of qPCR duplicates are expressed as a function of the highest enrichment on the locus with error bars corresponding to standard deviations. Each ChIP experiment was performed at least twice independently.
Ash2L and MLL2 knockdowns lead to a decrease in H3K4me3 and β-globin transcription
It has been shown previously that knockdown (KnD) of various subunits of MLL complexes result in a 20% to 40 % decrease in transcription and a 40% to 60% reduction of H3K4me3 on some Hox genes (Dou et al., 2006). This mild transcriptional defect might be due to small amounts of proteins remaining after the KnD and/or to the possibility that MLL complexes might not be required for initiating transcription (Pavri et al., 2006) but instead for increasing the rate of transcriptional elongation (Margueron et al., 2005; Shilatifard, 2006). Because of the apparent divergence between Ash2L and MLL2 binding profiles with H3K4me3 enrichment at the β-globin locus, it was important to determine whether these proteins are involved in H3K4 trimethylation and β-globin transcription during erythroid differentiation. Thus, we used RNA interference (RNAi) to knockdown either Ash2L or MLL2 proteins in MEL cells, and analyzed the resulting effects on β-globin transcription and H3K4 trimethylation. Stable MEL clones expressing a doxycycline (Dox) inducible anti-Ash2L or -MLL2 small hairpin RNA (shRNA) constructs were derived and selected by western blot using anti-Ash2L and –MLL2 polyclonal Abs (Figure 5A). The parent and Ash2L and MLL2 KnD MEL clones were induced to differentiate for 3 days in the presence/absence of Dox. While the KnDs have no effect on cell growth (not shown), we observed a 30% decrease in the production of β-globin mRNA transcripts after erythroid differentiation following the Ash2L KnD and a 60% decrease after induction of the MLL2 KnD by Dox treatment (Figure 5B). These transcriptional defects were accompanied by a significant decrease in hemoglobin production (not shown). In contrast, Dox has no effect on β-globin transcription in the parent MEL cell line. Importantly, H3K4me3 ChIPs in the KnD cells also revealed a 20% and 30% decrease in H3K4me3 within the βmaj-globin coding region following Ash2L and MLL2 KnD, respectively (Figure 5C). These results show that Ash2L and MLL2 are important for β-globin transcription and H3K4 trimethylation during erythroid differentiation. Furthermore, the level of reduction of β-globin transcripts and H3K4me3 is comparable to that observed for certain Hox genes following KnD of MLL-associated proteins (Dou et al., 2006). Finally, to determine whether the activator NF-E2/p45 is also important for H3K4 trimethylation, we performed H3K4me3 ChIP experiments in the NF-E2/p45-null CB3 cell line and we observed that H3K4me3 on the βmaj-globin gene is increased upon re-expression of the NF-E2/p45 activator in the erythroid CB3 cell line (Figure 2C). These results provide direct evidence that NF-E2-mediated recruitment of the MLL2 complex at the β-globin locus is important for both H3K4 trimethylation and transcription at the β-globin locus during erythroid differentiation.
Figure 5. Ash2L and MLL2 are required for maximal levels of βmaj-globin transcription and H3K4 trimethylation during erythroid differentiation.

(A) Western blot analysis of the doxycyclin (Dox)-inducible knockdowns (KnDs) of Ash2L and MLL2 in differentiated MEL cells. Nuclear extracts were prepared and revealed using the Abs indicated on the right. SAP30 and TFIIHp89 are used as loading controls.
(B) Ash2L and MLL2 KnDs lead to a defect in transcription at the β-maj globin gene. Transcription at the βmaj-globin gene was assessed by real-time qRT-PCR before (Nondiff) and after (Diff) erythroid differentiation in Dox-induced (shaded bar) vs non-induced (white bar) MEL cells. The parent clone served as a negative control. βmaj-globin transcription relative to GAPDH is expressed as a percentage of the highest level of transcripts. Average values of RT-qPCR triplicates are indicated with error bars corresponding to standard deviations. Each experiment was performed at least 3 times independently.
(C) Ash2L and MLL2 KnDs lead to a defect in H3K4 trimethylation at the β-maj globin gene. H3K4me3 enrichment at the βmaj-globin gene was measured by chromatin immunoprecipitation (ChIP) after erythroid differentiation in Dox-induced (white bar) vs non-induced (shaded bar) MEL cells. TaqMan probes used to reveal the ChIPs by real-time qPCR are the same as in Figure 3. Average values of qPCR duplicates are expressed as a function of the highest enrichment on the locus with error bars corresponding to standard deviations. Each experiment was performed at least twice independently.
Discussion
Recruitment, spreading and activation of the MLL2 H3K4 methyltransferase at the β-globin locus during erythroid differentiation
Our study provides evidence that a cell-specific DNA-bound transcriptional activator is required for initial targeting of the MLL2 methyltransferase complex to a specific gene locus and further methylation of H3K4 at this locus. First, we show that the hematopoietic activator NF-E2/p45 associates with the MLL2 methyltransferase complex in erythroid nuclear extracts, and that this complex is recruited to the β-globin locus during erythroid differentiation in a NF-E2 dependent manner. Recruitment of the Ash2L/MLL2 complex occurs at the β-globin LCR which contains cognate recognition sites for the activator and likely occurs independently of Pol II. Interestingly, we observed that while the Ash2L/MLL2 complex remains at the β-globin LCR during erythroid differentiation, the MLL2 subunit spreads across the β-globin locus, suggesting a mechanism by which a distal activator influences chromatin structure and transcription far downstream of its binding site (Figure 6). These results are supported by previous studies showing that Ash2L, RbBP5 and WDR5 can form a sub-complex in the absence of MLL (Dou et al., 2006; Steward et al., 2006) and by ChIP coupled with microarray experiments revealing that different subunits of MLL complexes bind separately to some genomic locations (Scacheri et al., 2006).
Figure 6. Model of recruitment, spreading and activation of the MLL2 methyltransferase complex on the β-globin locus during erythroid differentiation.
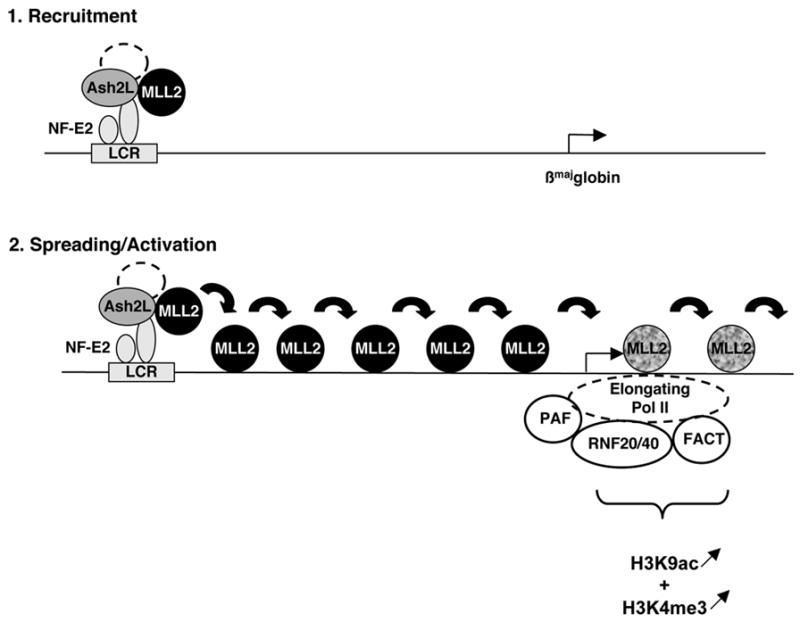
The MLL2 complex is initially guided to the β-globin Locus Control Region (LCR) via its interaction with the LCR-bound activator NF-E2. Binding of this H3K4 methyltransferase to the LCR does not however result in trimethylation of H3K4 in this region (presumably due to the histone code particular to this region of the locus). Then MLL2 detaches from its core components and spreads across 38 kb where it reaches the elongating Pol II and associated complexes. Interaction with basal transcriptional elongation machinery and/or the presence of specific histone modifications (H3K9ac and/or Ubiquitinated H2B) allows MLL2 to methylate H3K4. The inactive/active MLL2 protein is represented by a black/grey shading. H3K4me3 and H3K9ac are represented below the region where they are enriched on the β-globin locus and the direction of their variation during differentiation is figured by arrows.
The molecular mechanism by which MLL2 detaches from its interacting partner Ash2L and spreads across 38 kb of the β-globin locus is currently unclear but could involve MLL2 self association through its Set domain (Rozovskaia et al., 2000). Alternatively, it could entail combinatorial interactions with nonmethylated CpG DNA (Ayton et al., 2004) and/or specifically modified histones and cofactors, reminiscent of the spreading of HP1 through interaction with methylated H3K9 (Hiragami and Festenstein, 2005). While MLL2 appears to spread across the β-globin locus, our data suggest that additional regulatory mechanisms are required to activate its H3K4 methyltransferase activity at particular gene locations. Indeed the H3K4me3 mark is restricted to a small region located around +400bp downstream of the βmaj-globin transcription start site suggesting that at the other regions, trimethylation at H3K4 is either inhibited or actively removed. Within the 5′ part of the βmaj-globin gene coding region, MLL2 enzymatic activity is likely enhanced by combinatorial modification of histones (i.e. H3K9ac (Figure 4)), phosphorylation of H3 (Rea et al., 2000), monoubiquitination of H2B (Dover et al., 2002; Sun and Allis, 2002)) together with events associated with the early elongating Pol II (Hughes et al., 2004; Milne et al., 2005a) and other interacting factors (i.e. the ubiquitin E3 ligases RNF20/40 and the E2 ubiquitin conjugating enzyme UbcH6 (Zhu et al., 2005b), the elongation complex PAF (Zhu et al., 2005a) and the histone chaperone FACT (Pavri et al., 2006) (see our Model on Figure 6). Based on these results, we propose that interaction with a DNA-bound activator is important initially for guiding MLL2 to a specific region of the genome containing one or several genes poised for activation. Subsequent spreading would then permit the inactive MLL2 methyltransferase to scan neighboring chromatin until reaching activation signals (e.g specific cofactors, phosphorylated pol II and/or acetylated H3K9) present only at the gene coding regions.
Is the “spreading” of MLL2 specific to the β-globin locus?
Recruitment of transcription factors to the β-globin LCR followed by long-range “transfer” to the β-globin genes is not without precedent. For example Pol II is recruited to the LCR (in a NF-E2/p45-independent manner) before being transported to the β-globin genes (in a NF-E2/p45-dependent manner) (Johnson et al., 2001). Communication between the LCR and the active β-globin genes is generally thought to occur via a “looping” mechanism whereby the LCR and the β-globin genes come into direct physical contact through their interacting proteins and the intervening DNA sequences are looped out (Dean, 2006; Li et al., 2006; Tolhuis et al., 2002). While this mechanism may be important for some instances of crosstalk between the LCR and the β-globin genes, our results suggest that transfer of the MLL2 protein follows a previously proposed “linking” mechanism whereby communication between the LCR and the β-globin genes occurs via spreading of proteins along the locus (Bulger and Groudine, 1999). Indeed, in differentiated erythroid cells, MLL2 binding clearly extends from the LCR to the coding region of the active βmaj globin gene, spanning regions of DNA that would generally be considered to be looped out in the “looping” model (Tolhuis et al., 2002). Thus, our results provide experimental evidence that spreading of a transcription factor indeed occurs between the LCR and the βmaj-globin gene, suggesting a mechanism other than looping for long-distance transfer of transcription factors.
This model is likely to extend beyond β-globin gene regulation. For example, the developmentally regulated clustered Hox genes (Krumlauf, 1994) display several similarities with the β-globin genes. First, genome wide location analysis of MLL1 revealed that, in contrast to many active genes where MLL1 is restricted to a region immediately adjacent to transcription start sites, MLL1 binding to the HoxA cluster extends significantly in both directions from the transcription start site (at least 3.7 kb downstream and 2.2 kb upstream of individual Hox genes), and is distributed across the entire coding region of the genes (Guenther et al., 2005; Milne et al., 2005b). We also observe MLL2 spreading away from the LCR, across 38 kb into the βmaj-globin gene. Interestingly, in both the HoxA cluster (Guenther et al., 2005; Scacheri et al., 2006) and β-globin locus (Figure 3B), MLL1/MLL2 binding has been detected at genomic regions that are not enriched for H3K4me3. These observations suggest that the mechanism of recruitment and spreading of MLL2 that we have described at the β-globin locus might also be relevant to other developmentally-regulated clustered genes such as Hox genes. While similarities exist between the HoxA and the β-globin clusters, important differences remain between the two loci. Indeed, while the Hox A and B clusters contain broad domains of H3K4me2 regions covering multiple active genes (Bernstein et al., 2005), the β-globin locus displays a punctate pattern for both dimethyl ((Bulger et al., 2003) and data not shown) and trimethyl H3K4 (this study). These differences most likely arise from organizational differences between the two loci: the contiguous β-like globin genes are differentially expressed at specific developmental stages, while the HoxA clusters contain multiple active genes located in close proximity. Alternatively, these divergent H3K4 methylation patterns could arise from intrinsic differences between MLL1 and MLL2. However, we believe this is unlikely, as MLL1 and MLL2 methyltransferases display a high degree of similarity and interact with the same cofactors (Ruthenburg et al., 2007). Given these similarities, it is interesting to note that, while MLL−/− and MLL2−/− mice are both embryonic lethal (Ernst et al., 2004; Glaser et al., 2006; Yu et al., 1995), MLL+/− mice are severely anemic (Yu et al., 1995). It will be interesting to determine whether a similar erythroid phenotype is observed in the MLL2+/−mice.
Proteomics analysis of NF-E2/p45 interacting proteins has allowed us to identify a previously undefined mechanism by which the LCR regulates β-globin gene expression and chromatin structure at a distance during terminal erythroid differentiation. Continued analysis of protein complexes associated with the LCR should permit further insight into the mechanisms regulating developmental gene expression.
Experimental Procedures
Antibodies
Abs used for Western blot. From Santa Cruz Biotechnology, we used anti -NF-E2/p45 (sc-291), -CBP (sc-369), -MafK (sc-477), and -TFIIHp89 (sc-293). From Bethyl Laboratories, we used anti –MLL2/TRX2 (BL-835), -RbBP5 (BL-766) and menin (BL-342). From Upstate Biotechnology, we used anti-SAP30 (06-875). Polyclonal Abs against NF-E2/p45 (# 945) and Ash2L (# 1025) were generated in rabbits using purified full-length proteins. WDR5 and HP1γ Abs were gifts from W. Herr and R. Losson, respectively.
Abs used for IP and ChIP are the same except for Ash2L (# 907). From Upstate Biotechnology, we used anti-H3K9ac (07-352). From Abcam, we used anti- H3K4me3 (ab-8580). From Santa Cruz Biotechnology, we used the normal rabbit IgG (sc-2027).
Cell Culture, Immunoprecipitation and Quantitative Mass Spectrometry
MEL (clone 745) and CB3 cells were cultured in RPMI containing 10% serum, 1% glutamine and antibiotics. An expression vector encoding NF-E2/p45 (Kotkow and Orkin, 1995) was stably transfected into the CB3 cell line and clones expressing NF-E2/p45 were selected in 1mg/ml G418. CB3 and CB3+p45 clones were induced to differentiate along the erythroid lineage by adding 1.5% final DMSO for 3 days. MEL cells were induced to differentiate along the erythroid lineage by adding 2% final DMSO for 3 days and nuclear extracts were prepared as previously described (Brand et al., 2004). Anti-NF-E2/p45, -Ash2L and –MLL2 IPs were performed as described in (Brand et al., 2004) with the following modifications. Abs crosslinked to Dynabeads-protein A (Dynal) were used for IP. Elution of the proteins was performed at 37°C in 50% acetonitrile containing 0.1% TFA. Eluted proteins were labeled with the isotopically labeled ICAT reagents and identified and relatively quantified by mass spectrometry as described previously (Ranish et al., 2007). The probability of identification was determined for each protein using the PeptideProphet and ProteinProphet algorithms (Nesvizhskii et al., 2003).
Chromatin Immunoprecipitation
ChIP assays were performed as previously described (Brand et al., 2004) except that sonication was done using a Bioruptor (Diagenode). Real-time qPCR analysis was done on a ABI Prism 7500 detector using Taqman probes and primers (sequences available as Supplementary data). To calculate the enrichment of each protein to a particular target DNA, values obtained (using the standard curve method) for each target were divided by the amount of the corresponding target in the input fraction. Enrichments obtained from mock IPs performed in parallel using normal IgG were then subtracted from the enrichments values obtained using specific Abs. All the enrichments are expressed as a function of the highest enrichment obtained on the locus (set to 1).
Methyltransferase Assays
NF-E2/p45-, Mock- or MLL2- associated proteins were isolated on beads and incubated with 1 mg/ml purified core histones (Roche) or nucleosomes (prepared as described in (Brand et al., 1999)) for 1h at 30°C in the presence of 6.5 μM of [3H] s-adenosyl methionine in the EX50 buffer (10 mM Hepes K+ pH 7.6, 50 mM KCl, 10% Glycerol, 1.5 mM MgCl2, 0.5 mM EGTA, 1mM DTT, protease inhibitor cocktail). Proteins were eluted by adding ¼ volume of 4X loading dye buffer (100 mM Tris, pH6.8, 40% Glycerol, 8% SDS, 286 mM DTT, 0.4% bromophenol blue) and incubating at 95°C for 5 min. Proteins were separated by SDS-PAGE and electrotransferred to PVDF membrane before being revealed using Ponceau red and film exposure. Alternatively, eluted proteins were spotted onto phosphocellulose P81 squares and the radioactivity was counted in the presence of scintillation cocktail after washing the squares 4 times in 50mM Na2HPO4, pH9.0.
Ash2L and MLL2 knockdowns in MEL cells
Stable MEL cell lines stably expressing anti-Ash2L or anti-MLL2 shRNA in a doxycycline (Dox)-dependent manner were established as described in Supplemental Data. The knockdowns were induced by adding 5 μg/ml final of Dox in the culture medium for 3–4 days. Transcription of the βmaj-globin gene was assessed by RT-qPCR using Taqman probes (sequences available in Supplemental data) after RNA extraction and RNAse-free DNAse I digestion (Qiagen RNAeasy kit).
Supplementary Material
Acknowledgments
The authors would like to thank G. Blobel, E. Bresnick, A. Dean, B. Guyot and MC. Lorincz for critically reading the manuscript. We are also grateful to T. Sawado for the NF-E2/p45 protein, Y. Ben David for the CB3 cell line, S. Orkin for the mammalian NF-E2/p45 expression vector, M. Van de Vetering for the pTer vector, C. Guillouf for the pcDNA-6TR-EF1a vector, and W. Herr and R. Losson for Abs. Finally, we acknowledge the Fred Hutchinson Cancer Research Center Proteomics Facility for a subset of these experiments and useful advises. This project has been funded with grants to M.B. from the Human Frontier Science Program Organization and The Terry Fox Foundation (National Cancer Institute of Canada); to M.G. from the US National Institutes of Health (DK44746 and HL57620); to F.J.D. from the Canadian Institutes of Health Research; to R.A. from the National Heart, Lung, and Blood Institute (NIH) under contract No. N01-HV-28179; and to F.M. from the Ligue Nationale contre le Cancer (France) (équipe labellisée 2005).
References
- Andrews NC. The NF-E2 transcription factor. Int J Biochem Cell Biol. 1998;30:429–432. doi: 10.1016/s1357-2725(97)00135-0. [DOI] [PubMed] [Google Scholar]
- Ayton PM, Chen EH, Cleary ML. Binding to nonmethylated CpG DNA is essential for target recognition, transactivation, and myeloid transformation by an MLL oncoprotein. Mol Cell Biol. 2004;24:10470–10478. doi: 10.1128/MCB.24.23.10470-10478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. 2005 doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Genomic maps and comparative analysis of histone modifications in human and mouse. Cell . 120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabtree GR, Aebersold R, Groudine M. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol. 2004;11:73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- Brand M, Yamamoto K, Staub A, Tora L. Identification of TATA-binding protein-free TAFII-containing complex subunits suggests a role in nucleosome acetylation and signal transduction. J Biol Chem. 1999;274:18285–18289. doi: 10.1074/jbc.274.26.18285. [DOI] [PubMed] [Google Scholar]
- Bulger M, Groudine M. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- Bulger M, Schubeler D, Bender MA, Hamilton J, Farrell CM, Hardison RC, Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse beta-globin locus. Mol Cell Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean A. On a chromosome far, far away: LCRs and gene expression. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004;6:437–443. doi: 10.1016/s1534-5807(04)00061-9. [DOI] [PubMed] [Google Scholar]
- Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ. MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol. 2001;21:2249–2258. doi: 10.1128/MCB.21.7.2249-2258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzGerald KT, Diaz MO. MLL2: A new mammalian member of the trx/MLL family of genes. Genomics. 1999;59:187–192. doi: 10.1006/geno.1999.5860. [DOI] [PubMed] [Google Scholar]
- Forsberg EC, Downs KM, Bresnick EH. Direct interaction of NF-E2 with hypersensitive site 2 of the beta-globin locus control region in living cells. Blood. 2000;96:334–339. [PubMed] [Google Scholar]
- Glaser S, Schaft J, Lubitz S, Vintersten K, van der Hoeven F, Tufteland KR, Aasland R, Anastassiadis K, Ang SL, Stewart AF. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development. 2006;133:1423–1432. doi: 10.1242/dev.02302. [DOI] [PubMed] [Google Scholar]
- Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther MG, Jenner RG, Chevalier B, Nakamura T, Croce CM, Canaani E, Young RA. Global and Hox-specific roles for the MLL1 methyltransferase. Proc Natl Acad Sci U S A. 2005;102:8603–8608. doi: 10.1073/pnas.0503072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R, Slightom JL, Gumucio DL, Goodman M, Stojanovic N, Miller W. Locus control regions of mammalian beta-globin gene clusters: combining phylogenetic analyses and experimental results to gain functional insights. Gene. 1997;205:73–94. doi: 10.1016/s0378-1119(97)00474-5. [DOI] [PubMed] [Google Scholar]
- Hiragami K, Festenstein R. Heterochromatin protein 1: a pervasive controlling influence. Cell Mol Life Sci. 2005;62:2711–2726. doi: 10.1007/s00018-005-5287-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- Hung HL, Kim AY, Hong W, Rakowski C, Blobel GA. Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J Biol Chem. 2001;276:10715–10721. doi: 10.1074/jbc.M007846200. [DOI] [PubMed] [Google Scholar]
- Johnson KD, Christensen HM, Zhao B, Bresnick EH. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- Kiekhaefer CM, Boyer ME, Johnson KD, Bresnick EH. A WW domain-binding motif within the activation domain of the hematopoietic transcription factor NF-E2 is essential for establishment of a tissue-specific histone modification pattern. J Biol Chem. 2004;279:7456–7461. doi: 10.1074/jbc.M309750200. [DOI] [PubMed] [Google Scholar]
- Kiekhaefer CM, Grass JA, Johnson KD, Boyer ME, Bresnick EH. Hematopoietic-specific activators establish an overlapping pattern of histone acetylation and methylation within a mammalian chromatin domain. Proc Natl Acad Sci U S A. 2002;99:14309–14314. doi: 10.1073/pnas.212389499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim A, Kiefer CM, Dean A. Distinctive signatures of histone methylation in transcribed coding and non-coding human {beta}-globin sequences. Mol Cell Biol. 2006 doi: 10.1128/MCB.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotkow KJ, Orkin SH. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol Cell Biol. 1995;15:4640–4647. doi: 10.1128/mcb.15.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. doi: 10.1016/0092-8674(94)90290-9. [DOI] [PubMed] [Google Scholar]
- Li Q, Barkess G, Qian H. Chromatin looping and the probability of transcription. Trends Genet. 2006;22:197–202. doi: 10.1016/j.tig.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Lu SJ, Rowan S, Bani MR, Ben-David Y. Retroviral integration within the Fli-2 locus results in inactivation of the erythroid transcription factor NF-E2 in Friend erythroleukemias: evidence that NF-E2 is essential for globin expression. Proc Natl Acad Sci U S A. 1994;91:8398–8402. doi: 10.1073/pnas.91.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- Milne TA, Dou Y, Martin ME, Brock HW, Roeder RG, Hess JL. MLL associates specifically with a subset of transcriptionally active target genes. Proc Natl Acad Sci U S A. 2005a;102:14765–14770. doi: 10.1073/pnas.0503630102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Martin ME, Brock HW, Slany RK, Hess JL. Leukemogenic MLL fusion proteins bind across a broad region of the Hox a9 locus, promoting transcription and multiple histone modifications. Cancer Res. 2005b;65:11367–11374. doi: 10.1158/0008-5472.CAN-05-1041. [DOI] [PubMed] [Google Scholar]
- Mo R, Rao SM, Zhu YJ. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J Biol Chem. 2006;281:15714–15720. doi: 10.1074/jbc.M513245200. [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Popovic R, Zeleznik-Le NJ. MLL: how complex does it get? J Cell Biochem. 2005;95:234–242. doi: 10.1002/jcb.20430. [DOI] [PubMed] [Google Scholar]
- Ranish JA, Brand M, Aebersold R. Quantitative proteomics by mass spectrometry. Vol. 359. Clifton, N.J., Totowa, N.J.: Humana Press; 2007. [Google Scholar]
- Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases [see comments] Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Rozovskaia T, Rozenblatt-Rosen O, Sedkov Y, Burakov D, Yano T, Nakamura T, Petruck S, Ben-Simchon L, Croce CM, Mazo A, Canaani E. Self-association of the SET domains of human ALL-1 and of Drosophila TRITHORAX and ASH1 proteins. Oncogene. 2000;19:351–357. doi: 10.1038/sj.onc.1203307. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of Lysine 4 on Histone H3: Intricacy of Writing and Reading a Single Epigenetic Mark. Molecular Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Sawado T, Igarashi K, Groudine M. Activation of beta-major globin gene transcription is associated with recruitment of NF-E2 to the beta-globin LCR and gene promoter. Proc Natl Acad Sci U S A. 2001;98:10226–10231. doi: 10.1073/pnas.181344198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scacheri PC, Davis S, Odom DT, Crawford GE, Perkins S, Halawi MJ, Agarwal SK, Marx SJ, Spiegel AM, Meltzer PS, Collins FS. Genome-wide analysis of menin binding provides insights into MEN1 tumorigenesis. PLoS Genet. 2006;2:e51. doi: 10.1371/journal.pgen.0020051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. CHROMATIN MODIFICATIONS BY METHYLATION AND UBIQUITINATION: Implications in the Regulation of Gene Expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Sierra J, Yoshida T, Joazeiro CA, Jones KA. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 2006;20:586–600. doi: 10.1101/gad.1385806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–2468. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- Slany RK. Chromatin control of gene expression: mixed-lineage leukemia methyltransferase SETs the stage for transcription. Proc Natl Acad Sci U S A. 2005;102:14481–14482. doi: 10.1073/pnas.0507401102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward MM, Lee JS, O’Donovan A, Wyatt M, Bernstein BE, Shilatifard A. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol. 2006 doi: 10.1038/nsmb1131. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Sachdeva MM, Wang H, Blobel GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cao R, Xia L, Erdjument-Bromage H, Borchers C, Tempst P, Zhang Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell. 2001;8:1207–1217. doi: 10.1016/s1097-2765(01)00405-1. [DOI] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol. 2004;24:5639–5649. doi: 10.1128/MCB.24.13.5639-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu BD, Hess JL, Horning SE, Brown GA, Korsmeyer SJ. Altered Hox expression and segmental identity in Mll-mutant mice. Nature. 1995;378:505–508. doi: 10.1038/378505a0. [DOI] [PubMed] [Google Scholar]
- Zhu B, Mandal SS, Pham AD, Zheng Y, Erdjument-Bromage H, Batra SK, Tempst P, Reinberg D. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 2005a;19:1668–1673. doi: 10.1101/gad.1292105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol Cell. 2005b;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


