Exposure of DNA to free radicals and other electrophiles results in strand breaks, as well as modified nucleotides (lesions) within intact strands of the biopolymer. DNA lesions can be genotoxic and have been implicated in aging and diseases such as cancer.1-3 OxodG and Fapy•dG are examples of lesions produced in significant yields from dG when DNA is exposed to oxidative stress. It is well-known that OxodG gives rise to G→T transversions.1 Recent investigations suggest that Fapy•dG also has significant effects on repair and replication.4 For instance, OxodG and Fapy•dG result in high levels of G→T transversions when they are replicated in simian kidney cells.5 The former has been proposed as a biomarker when studying the effects of oxidative stress on DNA.2 In addition, it has also been suggested that the ratio of OxodG:Fapy•dG may be characteristic of the cancerous state of a cell.6 Currently, these lesions are detected and quantified via mass spectrometry following degradation of the DNA.7,8 However, there is some disagreement regarding how these measurements are made.9-11 We describe a reagent and accompanying fluorescent detection method that enables one to selectively quantify OxodG and Fapy•dG.
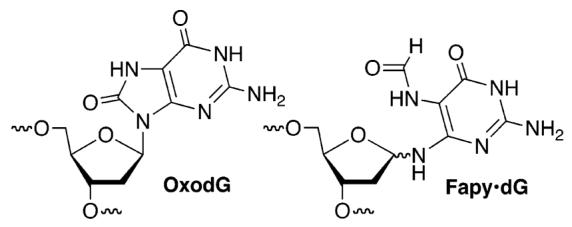
Selective DNA lesion detection is attractive because of its simplicity. One approach exploits a lesion's reactivity.12-14 The facile oxidation of OxodG and nucleophilic trapping of its oxidized product(s) by molecules such as spermine provided inspiration for developing a system for detecting it and Fapy•dG.15 OxodG (∼0.75 V, NHE) is more readily oxidized than the most readily oxidized native nucleotide, dG (1.29 V, NHE). The irreversible oxidation of 1 (1.08 V, NHE) was determined to be more favorable than that of dG, suggesting that using a similar oxidation/trapping approach for detecting Fapy•dG was plausible.16
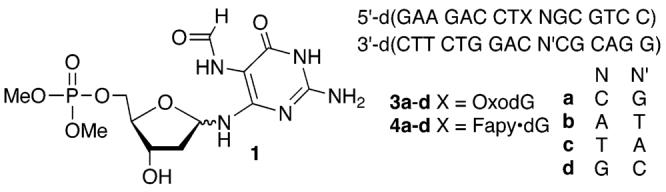
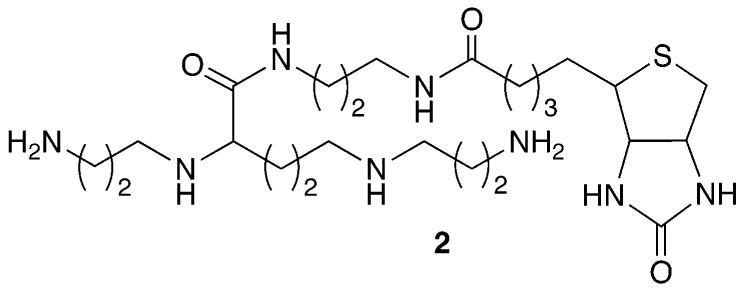
A carboxylated derivative of spermine was used as a precursor to 2.16,17 The spermine derivative (2) provides a means for tagging the oxidized species derived from OxodG and Fapy•dG with biotin, which is used for quantifying the lesions. Slower moving adducts were detected in good yield when duplex DNA (3 and 4) was reacted with Na2IrCl6 in the presence of 2 at 25 °C for 1 h (Figures 1 and 2).15 Minor amounts of a product that migrated even more slowly were also detected. Reaction of OxodG with spermine and other diamines under oxidative conditions is well-established.15 Confirmation that adducts corresponded to 1:1 adducts between 2 and oxidized OxodG or Fapy•dG was obtained using MALDI-TOF MS.16 Additional evidence that adduct formation occurred at Fapy•dG in 4 was obtained by ESI-MS following reaction of 1 and 2 under the same reaction conditions.16
Figure 1.
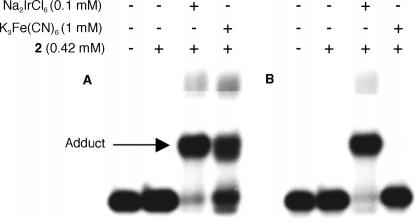
Reaction of 2 with DNA containing (A) OxodG (3b) or (B) Fapy•dG (4b) under oxidative conditions.
Figure 2.
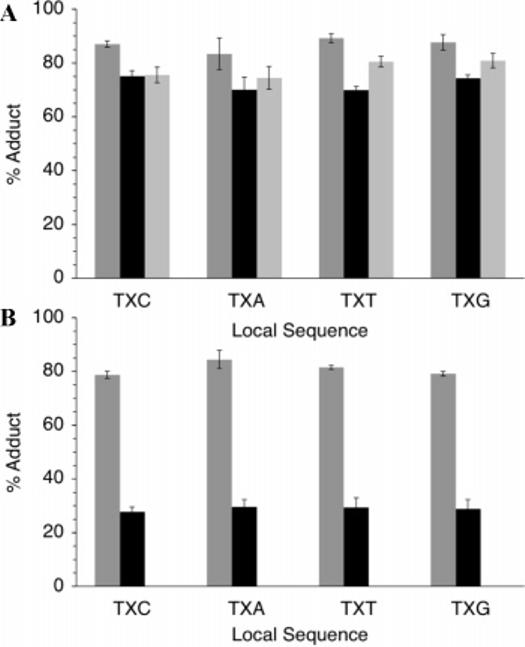
Average adduct yield with 2 in DNA as a function of sequence. (A) OxodG (3a–d). (B) Fapy•dG (4a–d). Dark gray, Na2IrCl6; black, Na2IrBr6; light gray, K3Fe(CN)6.
Although the yields of adducts were different for Fapy•dG and OxodG, for each lesion, they were independent of the flanking sequence (Figure 2). This was true even when OxodG was present in sequences that were expected to significantly alter its oxidation potential from that in 3a–d.16,18 In addition, no adducts were detected when duplexes containing OxodA or Fapy•dA were reacted with the strongest oxidant, Na2IrCl6, in the presence of 2 (data not shown). Moreover, no adducts were observed when DNA containing 5-hydroxy-2′-deoxyuridine or 5-hydroxy-2′-deoxycytidine, which have more favorable oxidation potentials than OxodG, were reacted with Na2IrCl6 and 2 (data not shown).19 Their lack of reactivity under the reaction conditions indicates that oxidation is a necessary but insufficient requirement for tagging.
In order to quantify the individual amounts of Fapy•dG and OxodG, other oxidants were explored in the hope of taking advantage of the lesions' differing oxidation potentials. Although both produced adducts with 2 in the presence of Na2IrBr6, only OxodG was trapped when K3Fe(CN)6 was used as the oxidant (Figures 1 and 2). The selective biotinylation of DNA containing the lesions as a function of oxidant indicated that individual amounts of OxodG and Fapy•dG in randomly damaged DNA could be determined using 2 as part of a fluorescence assay (Scheme 1). Amplex Red, which is oxidized to fluorescent resorufin by horseradish peroxidase in the presence of H2O2, was used to quantify adducts of 2.20 Experiments were carried out immediately after tagging by 2 because the adducts decompose with half-lives on the order of 12–15 h.15,16 The system was calibrated for application in a microtiter plate using a 287 nt duplex fragment of M13mp7(L2) prepared by PCR that contained biotin at the 5′-terminus of one strand.16 The fluorescent signal produced by resorufin depended linearly on the amount of biotinylated DNA as it varied 10-fold.
Scheme 1.

Fluorescence Detection of OxodG and Fapy•dG
The ELISA type method was used to measure the amounts of OxodG and Fapy•dG produced in aqueous solution by γ-radiolysis. The amount of biotinylated DNA was measured as a function of dose using Na2IrCl6 and K3Fe(CN)6 as oxidants (Figure 3). The adduct yield varied linearly with dose when either oxidant was used. The amounts of OxodG and Fapy•dG per dose (Gray) of radiation were determined (eqs 1 and 2) using the average yields of adduct formation established from experiments with 3 and 4 (Figure 2).16 Femtomoles of adduct(s) were readily detectable using this method. The data reveal that the ratio of Fapy•dG to OxodG formed in
| (1) |
| (2) |
aqueous solution by 137Cs equals 1.12. This is higher than the ratio reported when chromatin is irradiated but lower than when monocyte cells were subjected to γ-radiolysis.21,22 In summary, we have developed a simple and sensitive method for quantifying OxodG and Fapy•dG, two important DNA lesions. This tool will be widely accessible to researchers studying the effects of oxidative stress on DNA. In addition, the fluorescence assay should be generally applicable to the quantification of other lesions that can be selectively tagged with biotin.
Figure 3.
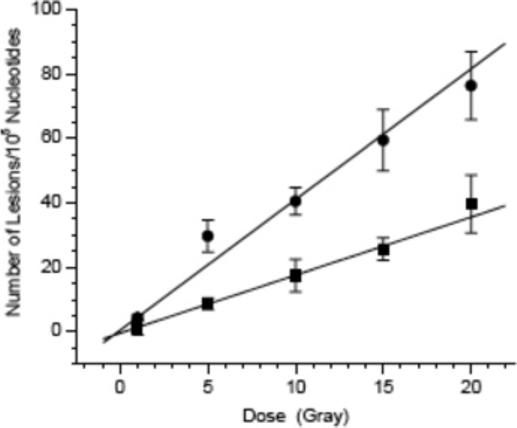
Average adduct yield with 2 in 287 nt duplex DNA (2.5 pmol) as a function of 137Cs dose. Circles, Na2IrCl6; squares, K3Fe(CN)6.
Supplementary Material
Acknowledgment
We are grateful for financial support from the National Institutes of Health (CA-074954). We thank Professor Gerald Meyer, Mr. James Gardner, and Dr. Shanta Dhar for assistance with the cyclic voltammetry experiment.
Footnotes
Supporting Information Available: Procedures for synthesis and characterization of all molecules, and all other experiments. ESI-MS and MALDI-TOF MS from experiments described herein. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Neeley WL, Essigmann JM. Chem. Res. Toxicol. 2006;19:491–505. doi: 10.1021/tx0600043. [DOI] [PubMed] [Google Scholar]
- 2.Malins DC, Hellstrom KE, Anderson KM, Johnson PM, Vinson MA. Proc. Natl. Acad. Sci. U.S.A. 2002;99:5937–5941. doi: 10.1073/pnas.082111199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamiya H. Nucleic Acids Res. 2003;31:517–531. doi: 10.1093/nar/gkg137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Greenberg MM. Biochem. Soc. Trans. 2004;32:46–50. doi: 10.1042/bst0320046. [DOI] [PubMed] [Google Scholar]
- 5.Kalam MA, Haraguchi K, Chandani S, Loechler EL, Moriya M, Greenberg MM, Basu AK. Nucleic Acids Res. 2006;34:2305–2315. doi: 10.1093/nar/gkl099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malins DC, Johnson PM, Barker EA, Polissar NL, Wheeler TM, Anderson KM. Proc. Natl. Acad. Sci. U.S.A. 2003;100:5401–5406. doi: 10.1073/pnas.0931396100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dizdaroglu M, Jaruga P, Rodriguez H. Crit. Rev. Oxid. Stress Aging. 2003;1:165–189. [Google Scholar]
- 8.Cadet J, Douki T, Gasparutto D, Ravanat JL. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 9.Collins AR, Cadet J, Moeller L, Poulsen HE, Vina J. Arch. Biochem. Biophys. 2004;423:57–65. doi: 10.1016/j.abb.2003.12.022. [DOI] [PubMed] [Google Scholar]
- 10.Dizdaroglu M. Free Radical Res. 1998;29:551–563. doi: 10.1080/10715769800300591. [DOI] [PubMed] [Google Scholar]
- 11.Cadet J, D'Ham C, Douki T, Pouget JP, Ravanat JL, Sauvaigo S. Free Radical Res. 1998;29:541–550. doi: 10.1080/10715769800300581. [DOI] [PubMed] [Google Scholar]
- 12.Lhomme J, Constant JF, Demeunynck M. Biopolymers. 1999;52:65–83. doi: 10.1002/1097-0282(1999)52:2<65::AID-BIP1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 13.Sato K, Greenberg MM. J. Am. Chem. Soc. 2005;127:2806–2807. doi: 10.1021/ja0426185. [DOI] [PubMed] [Google Scholar]
- 14.Xue L, Greenberg MM. Angew. Chem., Int. Ed. 2007;46:561–564. doi: 10.1002/anie.200603454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hosford ME, Muller JG, Burrows CJ. J. Am. Chem. Soc. 2004;126:9540–9541. doi: 10.1021/ja047981q. [DOI] [PubMed] [Google Scholar]
- 16.See Supporting Information.
- 17.Huber M, Pelletier JG, Torossian K, Dionne P, Gamache I, Charest-Gaudreault R, Audette M, Poulin R. J. Biol. Chem. 1996;271:27556–27563. doi: 10.1074/jbc.271.44.27556. [DOI] [PubMed] [Google Scholar]
- 18.Hickerson RP, Prat F, Muller JG, Foote CS, Burrows CJ. J. Am. Chem. Soc. 1999;121:9423–9428. [Google Scholar]
- 19.Riviere J, Bergeron F, Tremblay S, Gasparutto D, Cadet J, Wagner JR. J. Am. Chem. Soc. 2004;126:6548–6549. doi: 10.1021/ja049438f. [DOI] [PubMed] [Google Scholar]
- 20.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. Anal. Biochem. 1997;253:162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 21.Gajewski E, Rao G, Nackerdien Z, Dizdaroglu M. Biochemistry. 1990;29:7876–7882. doi: 10.1021/bi00486a014. [DOI] [PubMed] [Google Scholar]
- 22.Pouget JP, Frelon S, Ravanat JL, Testard I, Odin F, Cadet J. Radiat. Res. 2002;157:589–595. doi: 10.1667/0033-7587(2002)157[0589:fomdbi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


