Abstract
Cell adhesion and migration are dynamic processes requiring the coordinated action of multiple signaling pathways, but the mechanisms underlying signal integration have remained elusive. Drosophila embryonic dorsal closure (DC) requires both integrin function and c-Jun amino-terminal kinase (JNK) signaling for opposed epithelial sheets to migrate, meet, and suture. Here, we show that PINCH, a protein required for integrin-dependent cell adhesion and actin–membrane anchorage, is present at the leading edge of these migrating epithelia and is required for DC. By analysis of native protein complexes, we identify RSU-1, a regulator of Ras signaling in mammalian cells, as a novel PINCH binding partner that contributes to PINCH stability. Mutation of the gene encoding RSU-1 results in wing blistering in Drosophila, demonstrating its role in integrin-dependent cell adhesion. Genetic interaction analyses reveal that both PINCH and RSU-1 antagonize JNK signaling during DC. Our results suggest that PINCH and RSU-1 contribute to the integration of JNK and integrin functions during Drosophila development.
Introduction
Adhesion and migration of epithelial sheets are critical for wound healing, organ integrity, and morphogenetic movements during development. Cellular circuits that orchestrate these processes require coordination of integrin function with multiple signaling pathways. Integrins are transmembrane heterodimeric receptors for ECM that convey information bi-directionally between the extracellular environment and intracellular signaling machinery (Bokel and Brown, 2002). Engagement of integrins leads to the concentration of tyrosine kinases and their substrates at focal adhesions, a type of adherens junction that acts as a signaling nexus, a tethering site for actin filaments, and a region for generation of traction force during cell migration.
During Drosophila embryogenesis, lateral epidermal sheets migrate to close a hole in the dorsal epidermis in the process of dorsal closure (DC). DC is executed through cytoskeletal rearrangements and cell shape changes with no accompanying cell division (Harden, 2002). Because many proteins involved in DC also function in epithelial migration in other organisms, DC has emerged as an ideal model system to dissect the mechanisms driving migration and fusion of epithelial sheets. During DC, structures related to focal adhesions are assembled at the leading edge (LE) of advancing lateral epithelial cells and integrins are concentrated at these sites (Reed et al., 2001; Harden, 2002). Moreover, genetic analysis has revealed that integrins are essential for normal DC (Brown, 1994; Stark et al., 1997). Based on the established roles of integrins in mammalian systems, these adhesion receptors could influence DC by supporting cell–substratum interactions, modulating signaling pathways, or both. One signaling cascade that is essential for successful execution of DC results in activation of c-Jun amino-terminal kinase (JNK). Fine tuning of JNK output is critical, as both attenuation and hyper-activation of JNK signaling result in a failure of DC. The formation of focal adhesion complexes at the apical borders of the LE cells during DC depends on proper modulation of the JNK cascade (Reed et al., 2001; Harden, 2002), highlighting the potential importance of crosstalk between integrin and JNK signaling.
Several cytoplasmic proteins that colocalize with integrins are known to be essential mediators of integrin function in mammalian systems (Zamir and Geiger, 2001). One of these, the LIM protein PINCH, interacts with the integrin-linked kinase (ILK) and is critical for adhesion and spreading of mammalian cells (Tu et al., 1999; Zhang et al., 2002). To elucidate the in vivo role and mechanism of action of PINCH, we undertook a genetic analysis of PINCH function. Drosophila PINCH is encoded by the steamer duck (stck) locus. Mosaic analysis has revealed a critical role for PINCH in integrin-dependent epithelial cell adhesion in the adult wing (Clark et al., 2003). Homozygous zygotic stck mutants die late in embryogenesis, exhibiting deficits in both muscle cell adhesion and actin–membrane anchorage (Clark et al., 2003). Involvement of PINCH in both integrin-mediated adhesion and actin–membrane linkages makes it an attractive candidate for coordination of integrin and JNK functions during DC.
Results and discussion
To determine if PINCH could contribute to DC, we examined its localization in stage 14 embryos. PINCH and β-PS integrin are colocalized in both the LE and the amnioserosa (Fig. 1 A), consistent with PINCH's established role as an integrin effector. The amnioserosa is an extraembryonic tissue present on the dorsal surface of the embryo. As it has been established that coordinated signaling between the amnioserosa and migrating epithelium is key to formation of LE focal complexes (Reed et al., 2001), PINCH could exert an effect in the LE epithelium, the amnioserosa, or both tissues. stck homozygous mutant embryos rescued with a PINCH:GFP transgene under the control of the endogenous PINCH promoter display PINCH-GFP at the LE of the advancing epithelial sheets. Within the LE, PINCH is precisely localized to areas of active phosphotyrosine signaling at triangular nodes corresponding to apical adherens junctions (Fig. 1 B, inset).
Figure 1.
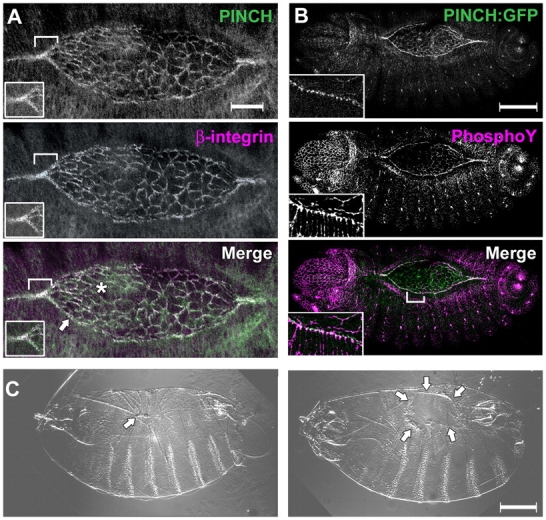
PINCH functions in DC and colocalizes with β-PS integrin and phosphotyrosine. (A) Confocal image of w1118 embryo shows PINCH (green), β-integrin (magenta), and their merge (white) highlighting the LE (arrow) and amnioserosa (asterisk). Inset shows a single z-slice and confirms colocalization. (B) Confocal image of P[w + GFP-PINCH] stck 18/l(3)097 embryo shows PINCH:GFP (green), phosphotyrosine (magenta), and their merge (white). Inset: magnification of the LE illustrating concentration of PINCH at apical junctions. (C) Cuticles of embryos carrying stck 17 loss-of-function allele both maternally and zygotically display puckers (left, arrow) and dorsal holes (right, arrows). Analogous stck 18 embryos have the same array of phenotypes. Control FLP-FRT embryos with wild-type PINCH are fully viable. Bars: 20 μm (A), 100 μm (B and C).
Zygotic stck mutants proceed normally through DC with complete lethality arising at the embryo-to-larval transition. When maternal PINCH contribution is eliminated, only 12% of cuticles have wild-type morphology. Dorsal puckers and dorsal holes (Fig. 1 C) characteristic of aberrant DC are observed at a 36 and 23% frequency, respectively (n = 180), indicating that maternally inherited PINCH is a key contributor to the process of DC. Moreover, in the absence of maternal PINCH, we also observe epithelial defects in ventral patterning and head involution (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200408090/DC1), indicating that PINCH may have additional functions in the developing embryo. Cuticles from embryos lacking both maternal and zygotic PINCH have the same array of phenotypes.
PINCH is composed of five LIM domains, each of which could serve as a protein-binding interface. The SH2-SH3 adaptor protein, Nck2, has been reported to interact with mammalian PINCH (Tu et al., 1998). This association is intriguing because the Drosophila homologue of Nck2, Dreadlocks, interacts directly with Misshapen (Msn), a MAP4K in the JNK signaling cascade (Ruan et al., 1999). As with other components of the JNK pathway, null mutations in msn result in embryonic lethality due to failure of DC. Although we were unable to detect direct binding of PINCH to Dreadlocks in Drosophila, we uncovered a link between PINCH's role in DC and the JNK cascade by testing for genetic interaction between stck and msn. Reduction of PINCH protein levels by introduction of a single copy of the loss-of-function allele, stck 18, into the msn 102 homozygous null background allows partial restoration of DC (P = 0.003; Fig. 2 A), suggesting that PINCH functions as a negative regulator of JNK signaling.
Figure 2.
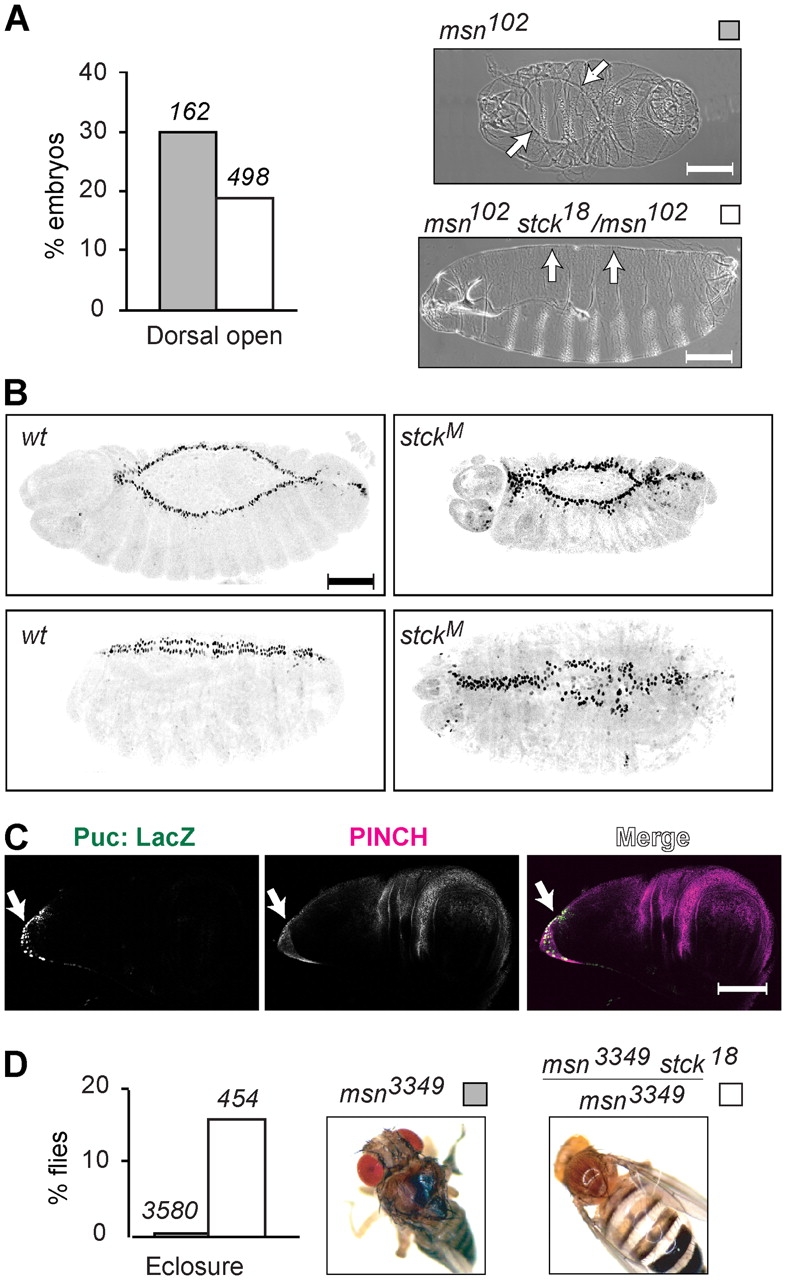
PINCH regulates JNK signaling. (A) Representative dorsal open msn 102 (dorsal view) and dorsal rescued msn 102stck18/msn102 (lateral view) embryos. Arrows mark the LE or dorsal suture. Graph shows percentage of dorsal open embryos observed in the sample size reported above each bar. (B) Inverted confocal images of stage 14 (top) and stage 15 (bottom) puc E69/+ embryos showing Puc:LacZ expression (black). Genetic backgrounds are wild type (left) or maternally stck deficient (right). (C) Confocal image of a puc E69 third instar larval wing disc showing Puc:LacZ (green), PINCH (magenta), and their merge (white). Arrow indicates the proximal stalk region. (D) Comparison of msn 3349 and rescued msn 3349stck18/msn3349 adult flies showing defective versus normal thorax. Graph reflects percent eclosure of the indicated genotype calculated from the sample size shown above each bar. Bars, 100 μM.
Puckered (Puc) is a JNK phosphatase whose expression is up-regulated in response to JNK activation, setting up a negative feedback loop (Martin-Blanco et al., 1998). During DC, JNK-regulated expression of a Puc-LacZ fusion reporter is restricted to the LE cells (Fig. 2 B). In embryos lacking maternal PINCH, expression of the Puc-LacZ fusion protein is disorganized and present in an expanded number of cells, including those beyond the LE border (Fig. 2 B). This phenotype is similar to Puc-LacZ expression observed in puc loss-of-function mutants and further supports a role for PINCH in the negative regulation of the JNK cascade.
Thorax closure is a post-embryonic developmental process with features common to DC, including migration of epithelial sheets and a dependence on JNK signaling. Within the wing disc, cells of the stalk region are functionally similar to LE cells during DC (Agnes et al., 1999; Martin-Blanco et al., 2000). These cells comprise the eventual fusion site for adjacent imaginal discs and are active in JNK signaling (Agnes et al., 1999). Spatially restricted JNK signaling in the stalk of wing disc can be visualized via a Puc-LacZ reporter, and PINCH expression overlaps with Puc-LacZ in this area of active JNK signaling (Fig. 2 C). Therefore, as in DC, PINCH is properly positioned to act as a regulator of the JNK cascade.
Although msn null mutations are embryonic lethal due to DC failure, flies homozygous for the hypomorphic allele msn 3349 are semi-viable and a large proportion of the eclosing adults have thorax closure defects (Fig. 2 D). These observations underscore the similarities between thorax closure and DC. In a stck 18 heterozygous background, a greater percentage of msn 3349 homozygotes are able to eclose (P < 0.0001; Fig. 2 D), supporting the hypothesis that PINCH is a negative regulator of the JNK pathway in both dorsal and thorax closure.
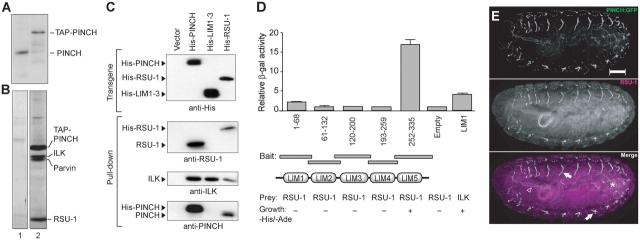
We purified Drosophila PINCH in complex with its binding partners using tandem affinity purification (TAP)–tagged PINCH (TAP-PINCH; Puig et al., 2001). stck homozygous mutant embryos rescued with a TAP:PINCH transgene driven by the endogenous stck promoter to wild-type levels (Fig. 3 A) afford material for purification of soluble, cytoplasmic TAP-PINCH complexes in the absence of endogenous PINCH protein. Three partners that copurified stoichiometrically with TAP-PINCH from embryos, as well as in complex with TAP-PINCH from cultured Drosophila S2R+ cells (Fig. 3 B), were identified by mass spectrometric analysis. Consistent with what is observed in mammalian cells (Tu et al., 1999) and our previous findings in Drosophila (Clark et al., 2003), ILK copurified with PINCH. The Drosophila homologue of the parvin/actopaxin family of proteins, CG32528, is also present in PINCH protein complexes. Parvin is known to bind ILK and actin in mammalian systems (Tu et al., 2001), but the isolated Parvin/ILK/PINCH complexes are the first to be described in Drosophila. Additionally, a novel 31-kD protein was identified as Drosophila CG9031. The CG9031 protein is 55% identical and 74% similar to human RSU-1, a leucine-rich repeat containing protein first identified as a suppressor of cell transformation by v-Ras (Cutler et al., 1992) and subsequently implicated in regulation of MAP kinase signaling, specifically the JNK and ERK cascades, when overexpressed in cultured cells (Masuelli and Cutler, 1996). Despite its potent ability to act as a tumor suppressor, little is known about the mechanism of action of RSU-1. Its partnership with the PINCH protein allows placement of RSU-1 in a molecular pathway that is linked to integrins.
Figure 3.
PINCH and RSU-1 physically interact and colocalize. (A) Quantitative anti-PINCH Western analysis of PINCH levels in wild-type flies (lane 1) versus TAP-PINCH–rescued flies (lane 2). (B) Silver-stained gel of purified proteins from S2R+ stably transfected with empty vector (lane 1) or pMT/TAP-PINCH (lane 2). (C) Western blots of Ni-NTA purified His-tagged complexes from S2 cells. RSU-1 interacts with full-length PINCH, but not LIM1–3. (D) Yeast two-hybrid reporter activity: growth on medium lacking histidine and adenine (+/−) and β-galactosidase activity relative to empty vector bait/RSU-1 prey. (E) Confocal images of P[w + GFP-PINCH] stck 18/l(3)097 stage 16 embryo shows that PINCH:GFP (green) and RSU-1 (magenta) colocalize (white) at muscle attachment sites (arrows) and in the pharynx (arrowhead) and gut (asterisk). Bar, 100 μm.
To assess the specificity and nature of the interaction between PINCH and RSU-1, domain-mapping studies were performed in cell culture and in yeast two-hybrid assays. Drosophila RSU-1 copurifies with full-length His-tagged PINCH, but not with a truncated His-tagged PINCH containing only LIM1–3 (Fig. 3 C), confirming the specificity of the interaction and suggesting LIM4 and/or 5 is the site of binding. ILK, which binds LIM1 of PINCH, copurifies with both full-length and the truncated LIM1–3 version of His-tagged PINCH (Fig. 3 C), serving as a positive control. Both PINCH and ILK are copurified with His-tagged RSU-1 (Fig. 3 C). Moreover, endogenous PINCH and RSU-1 can be coimmunoprecipitated (unpublished data). The site of RSU-1 binding to PINCH was further mapped using yeast two-hybrid analysis. Only cells expressing LIM5 bait/RSU-1 prey activated all three reporters (Fig. 3 D), indicating LIM5 is the site of RSU-1 binding. Consistent with the view that they interact in vivo, PINCH:GFP and RSU-1 are prominently colocalized at integrin-rich muscle attachment sites in Drosophila embryos (Fig. 3 E).
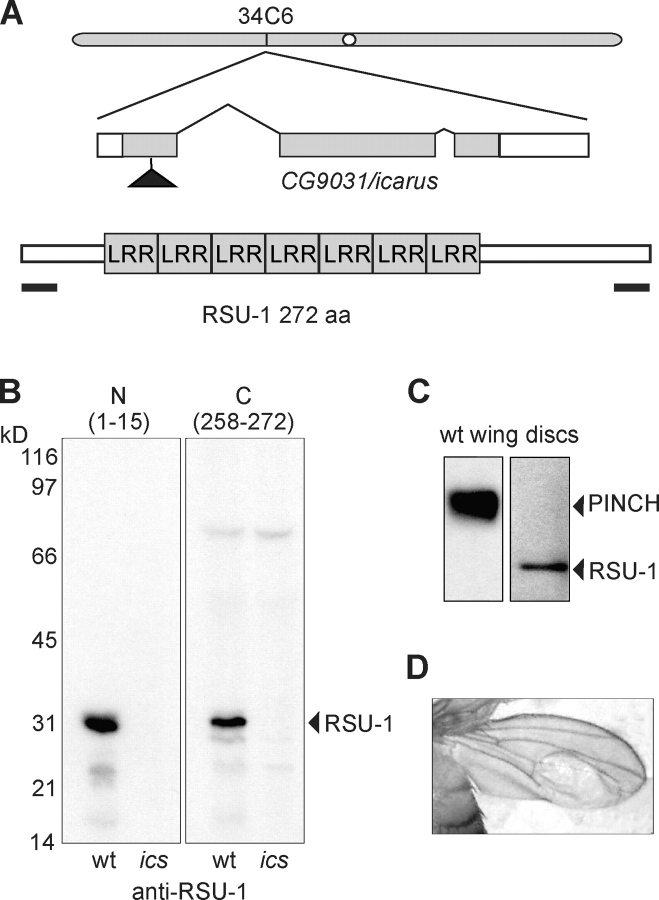
Drosophila RSU-1, which displays seven leucine-rich repeats with high sequence similarity to small GTPase regulators, is encoded by the CG9031 locus. We have characterized a P-element insertion allele that disrupts the RSU-1 coding sequence (Fig. 4 A). Flies homozygous for this mutation within CG9031 are viable and fertile (unpublished data), and lack RSU-1 protein as indicated by Western analysis with multiple anti-RSU-1 antibodies (Fig. 4 B). PINCH and RSU-1 are both expressed in larval wing discs (Fig. 4 C) and similar to stck wing clones (Clark et al., 2003), the mutation within CG9031 produces flies with wing blisters (Fig. 4 D) at 60% penetrance. These data are consistent with PINCH and RSU-1 acting in concert to support integrin-dependent adhesion. We have named the CG9031 gene icarus (ics) after the son of Daedalus who had unstable wings.
Figure 4.
ics encodes RSU-1. (A) The gene encoding RSU-1, located on chromosome 2 at cytological position 34C6, comprises three exons (coding regions shaded). Location of the peptide antigens used for antibody production (bars) and P-element insertion allele (triangle; Drosophila Gene Disruption Project, http://www.fruitfly.org/p_disrupt/index.html) are indicated. (B) Western blots of third instar larval extracts using antibodies directed against the amino or carboxy termini of RSU-1. (C) PINCH and RSU-1 are expressed in wing discs. (D) ics homozygotes display wing blisters.
Although elimination of RSU-1 function does not result in dorsal or thorax closure defects (unpublished data), we evaluated the role of RSU-1 in these processes by testing for genetic interactions between ics and msn. Similar to what occurs with reduction of stck dosage (Fig. 2 A), homozygous mutation of ics suppresses DC defects observed in msn 102 mutant embryos (P < 0.001; Fig. 5 A). Absence of RSU-1 also increases eclosure rates (P < 0.0001) of msn 3349 hypomorphs (Fig. 5 B) and completely suppresses the thorax defects present in msn 3349 animals (unpublished data), suggesting that like PINCH, RSU-1 can function as a negative regulator of JNK signaling. To confirm that the suppression of msn DC defects by ics mutation is mediated by the JNK signaling cascade, we eliminated RSU-1 in basket (bsk) embryos that lack zygotic JNK, the terminal kinase in this cascade. Homozygous ics mutation suppresses the DC defects of bsk 1 mutants (P < 0.001; Fig. 5 C), confirming that ics loss-of-function mutations affect DC by influencing the JNK cascade. Moreover, wing discs isolated from ics mutants display a 30% increase in active phospho-JNK relative to wild type (Fig. 5 D), providing direct biochemical confirmation that RSU-1 influences JNK activation state in vivo. Although we have not detected any localized accumulation of RSU-1 during DC (unpublished data), RSU-1 is readily detected by Western analysis in stage 13 embryos that are undergoing DC (Fig. 5 E, lane 1). Thus, the temporal pattern of RSU-1 expression is consistent with genetic results that highlight its role in regulation of JNK-dependent morphogenesis.
Figure 5.
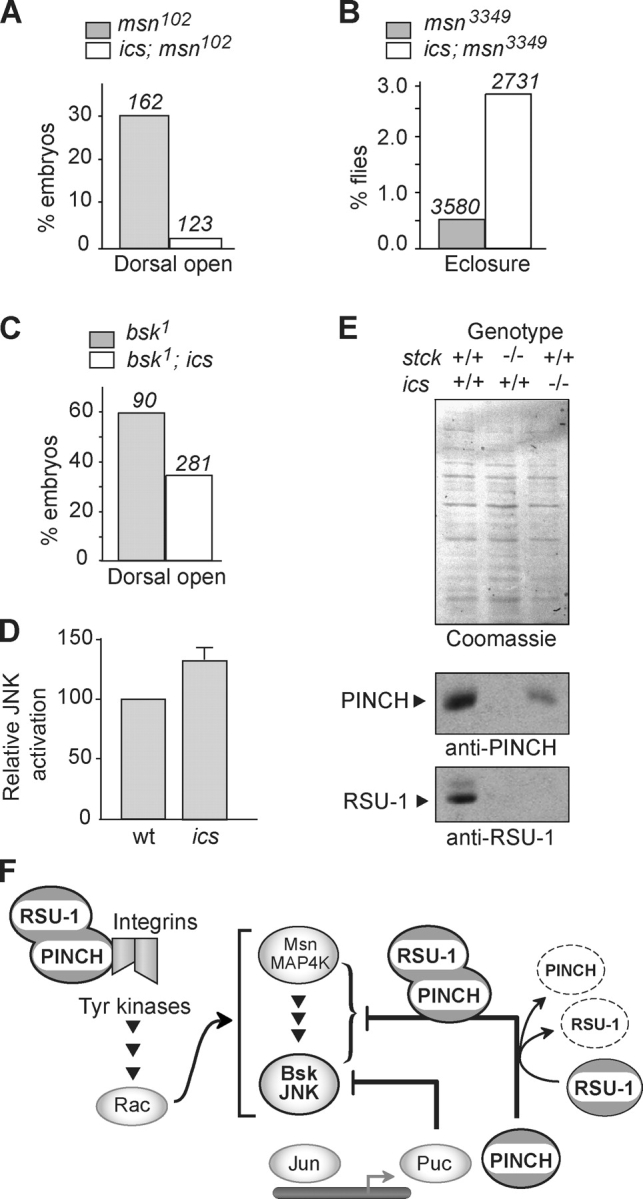
RSU-1 modulates JNK signaling and forms a stabilized complex with PINCH. (A) Comparison of DC defects in msn 102 versus ics; msn 102 embryos as in Fig. 2 A. (B) Comparison of percent eclosure in msn 3349 and ics; msn 3349 flies as in Fig. 2 D. (C) Comparison of DC defects in bsk 1 versus bsk 1 ; ics embryos. (D) Quantification from Western blots (n = 6) of phospho-JNK levels in third instar larval wing discs. Total JNK levels were unchanged (not depicted). (E) Quantitative anti-PINCH and anti-RSU-1 Western blots of w1118 (lane 1), stck germ line clone (lane 2), and ics (lane 3) stage 13 whole embryo lysates. Coomassie staining confirms equal loading. (F) Proposed model for PINCH action in JNK and integrin signaling.
Analysis of PINCH and RSU-1 levels in wild-type versus stck or ics mutant embryos provided insight into the physiological significance of their association. In embryos mutant for both maternal and zygotic stck, RSU-1 is dramatically reduced relative to wild-type levels (Fig. 5 E, lane 2). Likewise, in ics embryos, PINCH levels are also decreased (Fig. 5 E, lane 3). These observations suggest that PINCH and RSU-1 are reciprocally dependent on each other for maximal expression and/or stability. The mechanism for coordinate regulation of RSU-1 and PINCH remains to be determined. Because the phenotypes associated with loss of RSU-1 represent a subset of stck phenotypes, the processes disturbed in ics mutants may be exquisitely sensitive to PINCH levels. Alternately, RSU-1 may have functions that are independent of its role in PINCH stabilization.
Our data are consistent with a model (Fig. 5 F) in which PINCH could modulate JNK signaling in two distinct ways. First, PINCH is present at areas where JNK is active and antagonizes JNK signaling. This behavior is reminiscent of Drosophila Puc, a phosphatase regulator of the JNK cascade that establishes a negative feedback loop (Martin-Blanco et al., 1998). PINCH has no intrinsic catalytic activity, but might recruit proteins that could alter the availability or activity of JNK signaling components. Like Puc, PINCH expression is up-regulated in response to constitutive JNK signaling (Jasper et al., 2001). Availability of RSU-1 at these sites of active JNK signaling could independently regulate JNK signaling or modulate the effects of PINCH on JNK through regulation of PINCH stability. Second, PINCH and RSU-1 are required for integrin-dependent adhesion. PINCH has previously been shown to link integrins to the actin cytoskeleton via ILK and Parvin (Tu et al., 2001; Bokel and Brown, 2002; Clark et al., 2003), and these connections could influence both integrin-dependent adhesion and signaling. Integrin signaling, through a variety of tyrosine kinases and Rac, stimulates the JNK cascade (Harden, 2002); therefore, PINCH may also exert an influence on JNK signaling via integrin. Our findings illustrate that the cellular concentration of PINCH affects the level of RSU-1 and vice versa. Thus, modulation of the ratio of RSU-1 to PINCH could provide a mechanism to regulate JNK signaling during DC and thorax closure in Drosophila. We hypothesize that PINCH/RSU-1 complexes fine-tune and integrate the JNK and integrin signaling cascades required during morphogenesis, highlighting the potential role of integrin-associated apical junctional complexes as signal coordination points for epithelial morphogenesis.
Materials and methods
Fly genetics
PINCH:GFP and PINCH:TAP transgenics used standard methods. For msn 102 dorsal open rescue, msn 102/TM3, Sb flies were crossed to same and to msn 102 stck18/TM3, Sb, or ics; msn 102/TM3, Sb flies were crossed to same. For of the bsk 1 dorsal open rescue, bsk 1/CyO or bsk 1; ics/CyO flies were crossed to same. For rescue of msn 3349, msn 3349/TM3, Sb flies were crossed to same and to msn 3349 stck18/TM3, Sb, or ics; msn 3349/TM3, Sb flies were crossed to same. Embryos lacking maternal PINCH were generated using the FLP-FRT system (Chou and Perrimon, 1996).
Immunochemistry and microscopy
Rabbit polyclonal antisera were generated (Harlan Bioproducts) using antigens of the first and last 15 amino acids of Drosophila RSU-1. Antibodies used were rabbit anti-ACTIVE-JNK (Promega), anti-JNK (Chen et al., 2002), and anti-PINCH; mouse anti-ILK (BD Scientific), anti-phosphotyrosine 4G10 (Upstate Biotechnology), anti-penta His (QIAGEN), anti-β-PS integrin (CF6G11), and anti-LacZ (40-1a) (DSHB, University of Iowa, Iowa City, IA). Drosophila embryos and third instar larval wing discs were prepared as described previously (Clark et al., 2003). Confocal images were acquired at RT on an confocal microscope (model FV300; Olympus), using 20× 0.7 NA dry and 60× 1.4 NA oil immersion objectives, and were assembled using ImageJ and Adobe Photoshop 7.0. Third instar larval wing discs (n = 10–25) were homogenized and quantitatively immunoblotted for JNK and activated JNK as described previously (Chen et al., 2002).
DNA constructs
Plasmids for expression of tagged PINCH or RSU-1 were constructed by standard molecular biology techniques. See Fig. S2 for details (available at http://www.jcb.org/cgi/content/full/jcb.200408090/DC1).
PINCH complex purifications
10 g pCaspin-TAP–rescued stck embryos or 5 × 108 Drosophila S2R+ cells stably transfected with pMT/TAP-PINCH were washed and homogenized in lysis buffer (TBS, pH 7.9, plus 0.1% Triton X-100 and protease inhibitors) and 125,000 g soluble portion was used as described below. S2R+ cells were grown as recommended (Invitrogen) and lysed, and 30,000 g supernatant was batch-bound to 100 μl IgG Sepharose (Amersham Biosciences) prepared per manufacturer's recommendations and equilibrated in lysis buffer. After washing extensively with lysis buffer, proteins were eluted with a step gradient of 100 mM glycine from pH 5.0–2.75. Ni-NTA agarose (QIAGEN) purifications of His-tagged proteins used standard techniques.
Mass spectrometry
TAP-PINCH complexes were TCA precipitated and resuspended in Tris buffer, 8M urea, pH 8.6, reduced, and alkylated. Complexes were endoproteinase Lys-C digested (4 h), diluted to 2M urea, and digested with trypsin overnight (Washburn et al., 2001). Peptide mixtures were loaded onto a triphasic LC/LC column and analyzed as described previously (Cheeseman et al., 2002). Tandem mass spectra were analyzed using SEQUEST and the Drosophila sequence database with threshold values of 1.8 (+1), 2.8 (+2), and 3.5 (+3) (Washburn et al., 2001). Identities of specific bands were confirmed by sequence analysis.
Yeast two-hybrid
PINCH baits depicted in Fig. 3 C were constructed in pGBD-C1 (James et al., 1996). The full-length RSU-1 prey is cloned in pACT2. The yeast host strain, PJ69-2a, was transformed with bait and prey, and then reporter activities were assayed as described previously (James et al., 1996).
Online supplemental material
Fig. S1 shows pleitropic phenotypes of maternally deficient stck cuticles. Details of plasmid construction are provided in Fig. S2. Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200408090/DC1.
Acknowledgments
We thank A. Letsou, K. Bates, S. Noselli, M. Cutler, M. Cobb (GM56498), B. Seraphin and Cellzome, J. Bland, L. Pan, and D. Lim for their contributions.
This work was supported by the Huntsman Cancer Institute, National Institutes of Health (R01 GM50877 to M.C. Beckerle, RR11823-08 to J.R. Yates, and T32 GM07464-27 to M.A. Smith), and the Cancer Center Support grant P30CA42014 for shared resources.
J.L. Kadrmas and M.A. Smith contributed equally to this paper.
Abbreviations used in this paper: bsk, basket; DC, dorsal closure; ics, icarus; ILK, integrin-linked kinase; JNK, c-Jun amino-terminal kinase; LE, leading edge; Msn, Misshapen; Puc, Puckered; stck, steamer duck; TAP, tandem affinity purification.
References
- Agnes, F., M. Suzanne, and S. Noselli. 1999. The Drosophila JNK pathway controls the morphogenesis of imaginal discs during metamorphosis. Development. 126:5453–5462. [DOI] [PubMed] [Google Scholar]
- Bokel, C., and N.H. Brown. 2002. Integrins in development: moving on, responding to, and sticking to the extracellular matrix. Dev. Cell. 3:311–321. [DOI] [PubMed] [Google Scholar]
- Brown, N.H. 1994. Null mutations in the αPS2 and βPS integrin subunit genes have distinct phenotypes. Development. 120:1221–1231. [DOI] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Anderson, M. Jwa, E.M. Green, J. Kang, J.R. Yates III, C.S. Chan, D.G. Drubin, and G. Barnes. 2002. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172. [DOI] [PubMed] [Google Scholar]
- Chen, W., M.A. White, and M.H. Cobb. 2002. Stimulus-specific requirements for MAP3 kinases in activating the JNK pathway. J. Biol. Chem. 277:49105–49110. [DOI] [PubMed] [Google Scholar]
- Chou, T.B., and N. Perrimon. 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 144:1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, K.A., M. McGrail, and M.C. Beckerle. 2003. Analysis of PINCH function in Drosophila demonstrates its requirement in integrin-dependent cellular processes. Development. 130:2611–2621. [DOI] [PubMed] [Google Scholar]
- Cutler, M.L., R.H. Bassin, L. Zanoni, and N. Talbot. 1992. Isolation of rsp-1, a novel cDNA capable of suppressing v-Ras transformation. Mol. Cell. Biol. 12:3750–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harden, N. 2002. Signaling pathways directing the movement and fusion of epithelial sheets: lessons from dorsal closure in Drosophila. Differentiation. 70:181–203. [DOI] [PubMed] [Google Scholar]
- James, P., J. Halladay, and E.A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 144:1425–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper, H., V. Benes, C. Schwager, S. Sauer, S. Clauder-Munster, W. Ansorge, and D. Bohmann. 2001. The genomic response of the Drosophila embryo to JNK signaling. Dev. Cell. 1:579–586. [DOI] [PubMed] [Google Scholar]
- Martin-Blanco, E., A. Gampel, J. Ring, K. Virdee, N. Kirov, A.M. Tolkovsky, and A. Martinez-Arias. 1998. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila. Genes Dev. 12:557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Blanco, E., J.C. Pastor-Pareja, and A. Garcia-Bellido. 2000. JNK and decapentaplegic signaling control adhesiveness and cytoskeleton dynamics during thorax closure in Drosophila. Proc. Natl. Acad. Sci. USA. 97:7888–7893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuelli, L., and M.L. Cutler. 1996. Increased expression of the Ras suppressor Rsu-1 enhances Erk-2 activation and inhibits Jun kinase activation. Mol. Cell. Biol. 16:5466–5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig, O., F. Caspary, G. Rigaut, B. Rutz, E. Bouveret, E. Bragado-Nilsson, M. Wilm, and B. Seraphin. 2001. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 24:218–229. [DOI] [PubMed] [Google Scholar]
- Reed, B.H., R. Wilk, and H.D. Lipshitz. 2001. Downregulation of Jun kinase signaling in the amnioserosa is essential for dorsal closure of the Drosophila embryo. Curr. Biol. 11:1098–1108. [DOI] [PubMed] [Google Scholar]
- Ruan, W., P. Pang, and Y. Rao. 1999. The SH2/SH3 adaptor protein dock interacts with the Ste20-like kinase misshapen in controlling growth cone motility. Neuron. 24:595–605. [DOI] [PubMed] [Google Scholar]
- Stark, K.A., G.H. Yee, C.E. Roote, E.L. Williams, S. Zusman, and R.O. Hynes. 1997. A novel α integrin subunit associates with βPS and functions in tissue morphogenesis and movement during Drosophila development. Development. 124:4583–4594. [DOI] [PubMed] [Google Scholar]
- Tu, Y., F. Li, and C. Wu. 1998. Nck-2, a novel Src homology2/3-containing adaptor protein that interacts with the LIM-only protein PINCH and components of growth factor receptor kinase-signaling pathways. Mol. Biol. Cell. 9:3367–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Y., F. Li, S. Goicoechea, and C. Wu. 1999. The LIM-only protein PINCH directly interacts with integrin-linked kinase and is recruited to integrin-rich sites in spreading cells. Mol. Cell. Biol. 19:2425–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu, Y., Y. Huang, Y. Zhang, Y. Hua, and C. Wu. 2001. A new focal adhesion protein that interacts with integrin-linked kinase and regulates cell adhesion and spreading. J. Cell Biol. 153:585–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn, M.P., D. Wolters, and J.R. Yates III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242–247. [DOI] [PubMed] [Google Scholar]
- Zamir, E., and B. Geiger. 2001. Molecular complexity and dynamics of cell-matrix adhesions. J. Cell Sci. 114:3583–3590. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., L. Guo, K. Chen, and C. Wu. 2002. A critical role of the PINCH-integrin-linked kinase interaction in the regulation of cell shape change and migration. J. Biol. Chem. 277:318–326. [DOI] [PubMed] [Google Scholar]