Abstract
Background
Fluctuations in external salinity force eukaryotic cells to respond by changes in the gene expression of proteins acting in protective biochemical processes, thus counteracting the changing osmotic pressure. The high-osmolarity glycerol (HOG) signaling pathway is essential for the efficient up-regulation of the osmoresponsive genes. In this study, the differential gene expression of the extremely halotolerant black yeast Hortaea werneckii was explored. Furthermore, the interaction of mitogen-activated protein kinase HwHog1 and RNA polymerase II with the chromatin in cells adapted to an extremely hypersaline environment was analyzed.
Results
A cDNA subtraction library was constructed for H. werneckii, adapted to moderate salinity or an extremely hypersaline environment of 4.5 M NaCl. An uncommon osmoresponsive set of 95 differentially expressed genes was identified. The majority of these had not previously been connected with the adaptation of salt-sensitive S. cerevisiae to hypersaline conditions. The transcriptional response in hypersaline-adapted and hypersaline-stressed cells showed that only a subset of the identified genes responded to acute salt-stress, whereas all were differentially expressed in adapted cells. Interaction with HwHog1 was shown for 36 of the 95 differentially expressed genes. The majority of the identified osmoresponsive and HwHog1-dependent genes in H. werneckii have not been previously reported as Hog1-dependent genes in the salt-sensitive S. cerevisiae. The study further demonstrated the co-occupancy of HwHog1 and RNA polymerase II on the chromatin of 17 up-regulated and 2 down-regulated genes in 4.5 M NaCl-adapted H. werneckii cells.
Conclusion
Extremely halotolerant H. werneckii represents a suitable and highly relevant organism to study cellular responses to environmental salinity. In comparison with the salt-sensitive S. cerevisiae, this yeast shows a different set of genes being expressed at high salt concentrations and interacting with HwHog1 MAP kinase, suggesting atypical processes deserving of further study.
Background
When a living organism is subjected to extreme environmental conditions for an extended period of time, an adaptive response may become crucial for its continued existence. The response of eukaryotic cells such as yeast to environmental stress involves complex changes in gene expression which subsequently lead to various metabolic responses to induce adaptation to the new conditions. Fluctuating external osmolarity, like changes in salt concentration, leads to altered transcription of many responsive genes in an effort to counteract the stress with the activity of their protein products. One of the earliest protective biochemical responses is the biosynthesis and accumulation of glycerol as an osmolyte via the activation of corresponding genes. The resulting glycerol accumulates in the cytosol and leads to increased internal osmolarity, thus restoring the osmotic gradient between the cells and their environment [1]. In the salt-sensitive yeast Saccharomyces cerevisiae, the hyperosmotic stress caused by 0.4 M NaCl leads to the transient transcriptional induction of more than 1500 genes, as a consequence of simultaneous action of the general stress response pathway together with the high-osmolarity glycerol (HOG) mitogen-activated protein kinase (MAPK) signaling pathway [2]. A functional HOG pathway is essential for the efficient up-regulation of the vast majority of genes in response to hyperosmotic conditions [3,4]. The terminal MAPK, Hog1, accumulates in the nucleus within minutes of exposure to high salt concentrations [5], whereupon it phosphorilates and activates the HOG-specific transcription factors Sko1 [6] and Smp1 [7], or recruits Hot1 [8] and the general stress-response transcriptional activators Msn1, Msn2 and Msn4 [9,10] to the promoters of osmoinducible genes. Findings that Hog1 could be an integral part of the upstream activation complex, targeting not only the activators but also components of the general transcription machinery, such as RNA polymerase II [11,12] together with Hog1-guided recruitment of Rpd3 histone deacetylase to the chromatin [13], have highlighted the additional level of complexity in the regulation of gene expression during hyperosmotic conditions.
To date, studies on hyperosmotic adaptation and salt tolerance in fungal species have been largely performed with the salt-sensitive model organism S. cerevisiae, for reasons of experimental convenience. However, the cellular machinery of S. cerevisiae is not adapted to the extreme hyperosmolar pressure caused by a salty environment with more than 1–2 M NaCl concentration. Therefore the specially adapted extremely halotolerant yeast-like fungus Hortaea werneckii represents a novel eukaryotic organism for studying cellular responses to extremely elevated environmental salinity. This naturally osmoadaptable species was first isolated from hypersaline crystallizer ponds in salterns [14], where NaCl concentration fluctuates from 0.5 M up to saturate solution levels (6 M). The increase in surrounding salt concentration is accompanied by increased intracellular glycerol accumulation in H. werneckii [15]. The glycerol accumulation suggested the activation of the HOG-like pathway. The MAPK HwHog1 was later identified, showing its highest activity at a concentration of 4.5 M NaCl [16]. Many cellular and physiological differences were observed between H. werneckii cells growing in the extremely hypersaline environment of 4.5 M NaCl and those growing in a moderate salinity of 3 M NaCl, which has been assigned as the optimal metabolic condition for H. werneckii [15,17-19]. Therefore the characterization of the differential transcriptional response and identification of HwHog1-target genes in H. werneckii when under extreme hypersaline conditions could provide new insight into eukaryotic saline-response genetics.
In this study we have identified a set of differentially expressed genes in hyperosmotically adapted H. werneckii. Expression profiles of the genes were determined for both hyperosmotically-adapted and hyperosmotically-stressed H. werneckii cells. The identified genes were tested for interaction with HwHog1 and RNA polymerase II. Here we report a new insight into the differential expression of osmoresponsive genes in an extremely halotolerant eukaryotic microorganism and suggest a role for these genes in adaptive metabolism.
Results and Discussion
Since H. werneckii occupies a different ecological niche than S. cerevisiae, it is not surprising that these yeasts perceive hyperosmotic environments differently. In contrast to S. cerevisiae, H. werneckii can actively grow in a wide range of external salt concentrations, which shows that the cells are also well adapted to an extremely hypersaline environment. Differentially expressed genes in H. werneckii cells grown in different salinities therefore represent the transcriptional response of the adapted cells, rather than the stress response. Previous work has shown differences in the activation of HwHog1 between the halotolerant H. werneckii and S. cerevisiae [16]. In the present work, we tried to address two questions: (i) what is the transcriptional response of H. werneckii adapted to a hypersaline environment compared with transcription under optimal growth conditions, and (ii) whether and how the major HOG pathway effector MAPK HwHog1 is associated with the expression of osmoresponsive genes in H. werneckii.
Bioinformatical characterization of the Hw4.5-3 subtracted library revealed over-representation of H. werneckii osmoresponsive genes in functional groups of metabolism and energy
It is a special challenge to identify a relatively large group of differentially-expressed genes that are specifically connected with the response to a certain environmental stimulus in an organism, without the availability of a sequenced genome. A suppression subtractive hybridization (SSH) cDNA library was prepared to study the differential expression of genes related to halotolerance in H. werneckii between cells growing in optimal salinity (3 M NaCl) and cells growing in extreme salinity (4.5 M NaCl). A mirror orientation selection (MOS) method [20] and differential cross-screening of the SSH-clones were employed after SSH to augment the subtracted library for differentially expressed genes. A total of 1088 colonies were subjected to highly stringent differential screening. According to this selection, 170 clones (15.6%) from the Hw4.5-3 subtracted library were strong candidates for differentially expressed genes. The nucleotide sequences were analyzed and their putative functions identified by searching BLASTX. As a result, a total of 95 (55.8%) unique singletons and assembled sequences were identified (see Table 1). Redundancy-check analysis (Table 2) revealed that the 10 most abundant ESTs occupied 35.8% of the Hw4.5-3 subtraction library. A comparison with the GenBank database revealed that 74 of the 95 cDNAs (77.8%) had a high degree of sequence similarity (E-value ≤ 2.00E-11) with genes from other organisms of fungal origin. From the remaining 24 sequences (25.3%), 11 sequences (11.6%) could be identified with a low degree of similarity and 13 sequences (13.7%) with weak or no similarity (E-value cut off at 1.00E-02) to genes from other organisms. The names for the identified sequences were assigned according to the Saccharomyces Genome Database [21] standard names of the closest orthologs from S. cerevisiae with the prefix Hw for Hortaea werneckii (Table 1). For unidentified genes, weakly similar genes or genes without known orthologs in S. cerevisiae, the names were assigned as SOL followed by a consecutive number in our arbitrary library of H. werneckii hyperosmolarity-induced expressed sequence tags (ESTs) [15]. The putative functions of the ESTs with E-values lower than 10-5 (Table 1) were categorized as suggested by the MIPS database FunCat functional annotation scheme based on S. cerevisiae classification [22,23]. For eight ESTs without a corresponding S. cerevisiae ortholog, a matched ortholog was found in other organisms and they were functionally classified by literature (Table 1, numbers in brackets). As can be seen in Table 3, general metabolism proteins (31.6% of total unique transcripts), followed by energy production (23.3%), were the most common categories among the 16 functional categories. Of 13 unclassified SOL genes, SOL10, SOL19, SOL20, SOL21, SOL23, SOL24, SOL26 and SOL29 showed very weak similarity with other S. cerevisiae genes as aligned by a Wu-BLAST2 search, whereas SOL18, SOL 22, SOL25, SOL27 and SOL28 were completely unrelated to known genes from databases. These transcripts might thus be specifically related to the adaptation capability of H. werneckii to extremely high salt concentrations. The most unexpected discovery among differentially expressed ESTs from the Hw4.5-3 subtracted library was the identification of the SOL13 gene, which shows the highest similarity with opsins from two other Dothideomycetes fungi, Phaeosphaeria nodorum and Leptosphaeria maculans, respectively. L. maculans rhodopsin represents the first proven case of a light-driven transmembraneproton translocation by retinal-binding protein from a fungal organism [24], suggesting that some fungal classes may use the rhodopsin-based bioenergetic pathway previously observed only in prokaryotes.
Table 1.
Differentially expressed genes in H. werneckii adaptation to 3 M NaCl or 4.5 M NaCl. BLASTX matches of EST clones derived from the Hw4.5-3 subtraction library and their functional categorizations based on MIPS are collected. Similarities with probability <10-5 were regarded as being significant, and others as not determined (ND). Genes selected for the expression and chromatin immunoprecipitation analyses are printed in bold. aGenBank accession number. bNumbers indicate the functional groups (see Table 3). Numbers in brackets correspond to literature-based arbitrarily-assigned functional categories for ESTs with orthologs in organisms other than S. cerevisiae. cFold induction (fI, positive value) or fold repression (fR, negative value) in 4.5 M NaCl- vs. 3 M NaCl-adapted H. werneckii cells. Numbers indicated are mean values of three independent RT-PCR experiments, and the representative gels are shown in figures 1 and 2.
| GenBank.a | Gene | Characteristic or description | Functional categoryb | fI/f Rc |
| DQ822684 | HwAGP1 | Amino acid permease | 20. | 3.3 |
| DQ822624 | HwATP1 | Mitochondrial ATPase alpha-subunit | 2. 20. 34. | 1.8 |
| DQ822625 | HwATP2 | Mitochondrial ATPase beta-subunit | 2. 20. 34. 40. | 4.0 |
| DQ822626 | HwATP3 | Mitochondrial ATPase gamma-subunit | 2. 20. 34. | 2.3 |
| DQ822606 | HwBMH1 | 14-3-3 protein | 1. 10. 16. 30. 40. 43. | 1.8 |
| DQ822642 | HwCDC3 | Septin complex component | 10. 16. 34. 40. 42. 43. | -3.3 |
| DQ822620 | HwCIT1 | Mitochondrial citrate synthase | 1. 2. 42. | 4.6 |
| DQ822622 | HwCOB1 | Cytochrome b | 2. | 5.2 |
| DQ822621 | HwCOX1 | Cytochrome c oxidase subunit I | 2. 20. | 2.5 |
| DQ822687 | HwCYT1 | Cytochrome c1 | 2. 20. | 3.6 |
| DQ822643 | HwDBP2 | RNA helicase | 1. 11. | -4.2 |
| DQ822641 | HwECM33 | Extracellular matrix 33 protein | 10. 42. 43. | 2.0 |
| DQ822657 | HwEFT2 | Eukaryotic translation elongation factor 2 (eEF-2) | 12. | 1.9 |
| DQ822685 | HwELF1 | Transcription elongation factor | 10. 11. 40 | 7.6 |
| DQ822613 | HwERV25 | p24 component of the COPII-coated vesicles | 20. | 9.4 |
| DQ822619 | HwFAS1 | Mitochondrial acyl-carrier protein | 1. | 3.1 |
| DQ822689 | HwFRE7 | Ferric-chelate reductase-7 transmembrane component | 20. 34. | 10.5 |
| DQ822656 | HwFUN12 | Eukaryotic translation initiation factor 5B (eIF-5B) | 12. | 3.4 |
| DQ822655 | HwGAL10 | UDP-glucose 4-epimerase | 1. | ND |
| DQ822610 | HwGDH1 | NADP(+)-specific glutamate dehydrogenase | 1. 2. 16. | 7.5 |
| DQ822654 | HwGND2 | 6-phosphogluconate dehydrogenase | 1. 2. 16. | 4.1 |
| DQ822631 | HwGUT2 | FAD-dependent glycerol-3-phosphate dehydrogenase | 1. 2. 42. | 3.0 |
| DQ822645 | HwHHF1 | Histone H4 | 10. 11. 16. | ND |
| DQ822614 | HwHSP82 | Heat shock protein 90 | 1. 2. 10. 14. 32. 34. 43. | 5.1 |
| DQ822646 | HwHTA1 | Histone H2A | 10. 11. 16. | -2.6 |
| DQ858164 | HwIRE1 | Serine-threonine protein kinase and endoribonuclease | 1. 11. 14. 30. 32. | 2.1 |
| DQ822615 | HwKAR2 | Endoplasmic reticulum luminal chaperone | 1. 10. 14. 16. 20. 32. 41. 42. | 2.3 |
| DQ822627 | HwKGD2 | Dihydrolipoamide succinyltransferase | 1. 2. | 1.8 |
| DQ822629 | HwLSC2 | Mitochondrial succinyl-CoA ligase beta-chain | 1. 2. 16. | 2.7 |
| DQ822630 | HwMDH1 | Mitochondrial malate dehydrogenase | 1. 2. 42. | 3.4 |
| DQ858163 | HwMET14 | ATP adenosine-5'-phosphosulfate 3'-phosphotransferase | 1. | 3.4 |
| DQ822607 | HwMET17 | Cystein synthase | 1. | 8.2 |
| DQ822605 | HwMET6 | Cobalamin-independent methionine synthase | 1. | 3.2 |
| DQ822628 | HwMIR1 | Mitochondrial phosphate transport protein | 20. 34. | 8.4 |
| DQ822649 | HwNHP6A | High mobility group protein A | 10. 11. 40. 43. | 4.5 |
| DQ822623 | HwNUC1 | Mitochondrial nuclease | 1. 10. | 3.2 |
| DQ822635 | HwOPI3 | Unsaturated phospholipid methyltransferase | 1. | 9.7 |
| DQ822612 | HwPDI1 | Protein disulphide isomerase precursor | 14. 20. | 2.1 |
| DQ858158 | HwPGK1 | 3-phosphoglycerate kinase | 1. 2. 16. | 6.4 |
| DQ822634 | HwPMA2 | Plasma membrane proton-exporting ATPase | 2. 20. 34. | 7.9 |
| DQ822651 | HwPRY1 | Pathogenesis-related protein precursor | 34. | 7.5 |
| DQ822686 | HwPUF1 | Serine rich pumilio family RNA binding domain protein | 1. 10. 11. 16. | 4.6 |
| DQ822688 | HwRAD16 | SWI/SNF family DEAD/DEAH box helicase | 10. 32. | 4.9 |
| DQ822660 | HwRPL10 | 60S ribosomal protein 10 | 10. 12. 14. | ND |
| DQ822672 | HwRPL16A | 60S ribosomal protein 16A | 12. 16. | ND |
| DQ822664 | HwRPL22A | 60S ribosomal protein 22A | 12. | ND |
| DQ822670 | HwRPL2B | 60S ribosomal protein 2B | 12. | ND |
| DQ822671 | HwRPL3 | 60S ribosomal protein 3 | 12. | ND |
| DQ822669 | HwRPL6A | 60S ribosomal protein 6A | 12. 16. | 2.6 |
| DQ822661 | HwRPL7B | 60S ribosomal protein 7B | 12. | ND |
| DQ822617 | HwRPN2 | 26S proteasome regulatory particle subunit | 11. 40. 30. | 2.2 |
| DQ822673 | HwRPS10B | 40S ribosomal protein 10B | 12. | ND |
| DQ822666 | HwRPS12 | 40S ribosomal protein 12 | 12. | ND |
| DQ822665 | HwRPS15 | 40S ribosomal protein 15 | 12. 14. 20. | ND |
| DQ822667 | HwRPS16A | 40S ribosomal protein 16A | 12. | 7.6 |
| DQ822663 | HwRPS17A | 40S ribosomal protein 17A | 12. | ND |
| DQ822668 | HwRPS26B | 40S ribosomal protein 26B | 12. | ND |
| DQ822659 | HwRPS8A | 40S ribosomal protein 8A | 12. | ND |
| DQ822662 | HwRPS8B | 40S ribosomal protein 8B | 12. | ND |
| DQ822608 | HwSAM2 | S-adenosylmethionine synthetase | 1. | 2.8 |
| DQ822633 | HwSHY1 | Mitochondrial inner membrane protein chaperone | 2. 16. 32. | 2.5 |
| DQ858161 | HwSKN1 | Beta-1,6-glucan synthetase | 1. 2. 42. | 2.8 |
| DQ822611 | HwSSA4 | Heat shock protein 70 | 14. 16. 32. | 1.8 |
| AY731090 | HwSTT3 | Catalytic subunit of the oligosaccharyltransferase complex | 1. 14. | 2.8 |
| DQ822640 | HwSUN4 | Cell wall synthesis protein related to glucanases | 1. 40. 42. | -2.5 |
| DQ822676 | HwTDH1 | Glyceraldehyde-3-phosphate dehydrogenase | 1. 2. | 8.9 |
| DQ858159 | HwTEF1 | Eukaryotic elongation factor 1-alpha (EF-1A) | 12. | 2.1 |
| DQ822658 | HwTIF1 | Ekcaryotic translation initiation factor 1-alpha (eIF-4A) | 12. | 5.0 |
| DQ822609 | HwTKL1 | Transketolase | 1. 2. | 5.2 |
| DQ822632 | HwTOM40 | Mitochondrial import receptor translocase | 14. 20. | 4.0 |
| DQ822644 | HwTPM1 | Tropomyosin | 16. 20. 34. 40. 42. 43. | ND |
| DQ822647 | HwTUB1 | Alpha-tubulin | 10. 14. 42. | ND |
| DQ822648 | HwTUB2 | Beta-tubulin | 10. 14. 42. | -1.8 |
| DQ822653 | HwUGP1 | UDP-glucose pyrophosphorylase | 1. 2. 14. | 2.8 |
| DQ822637 | HwVPS34 | Phosphatidylinositol 3-kinase homolog | 1. 14. 20. 30. | ND |
| DQ858157 | SOL10 | Weakly similar to H. sapiens cyclin-dependent kinase 6 | ND | ND |
| DQ822616 | SOL11 | Mannose-P-dolichol utilization defect 1 protein | (14. 16. 20.) | -1.5 |
| DQ822618 | SOL12 | Ubiquitin associated protein 2-like protein | (14. 16. 32.) | ND |
| DQ822636 | SOL13 | Opsin 1 | (2. 20.) | 2.1 |
| DQ822638 | SOL14 | Histidine-rich glycoprotein precursor | (32. 40.) | ND |
| DQ822639 | SOL15 | Acetyl xylan esterase | (1. 32. 34.) | 2.2 |
| DQ822650 | SOL16 | Senescence-associated protein | (10. 11. 40.) | 1.8 |
| DQ822652 | SOL17 | Feruloyl esterase | (1. 32. 34.) | 2.0 |
| DQ822674 | SOL18 | Hyperosmolarity-induced mRNA 18 | ND | 6.9 |
| DQ822675 | SOL19 | Weakly similar to yeast stress response transcription factor Crz1 | ND | -1.9 |
| DQ822676 | SOL20 | Weakly similar to yeast nitrosoguanidine resistance protein Sng1 | ND | 2.4 |
| DQ822677 | SOL21 | Weakly similar to yeast prespliceosomal RNA helicase Prp5 | ND | 7.2 |
| DQ822678 | SOL22 | Hyperosmolarity-induced mRNA 22 | ND | ND |
| DQ822679 | SOL23 | Weakly similar to yeast Hsp70 protein Ssz1 | ND | 5.5 |
| DQ822680 | SOL24 | Weakly similar to yeast t-SNARE protein Sso1 | ND | 3.8 |
| DQ822681 | SOL25 | Hyperosmolarity-induced mRNA 25 | ND | 5.5 |
| DQ822682 | SOL26 | Weakly similar to yeast transcriptional repressor Rgm1 | ND | 6.7 |
| DQ858160 | SOL27 | Hyperosmolarity-induced mRNA 27 | ND | 6.3 |
| DQ822683 | SOL28 | Hyperosmolarity-induced mRNA 28 | ND | 5.0 |
| DQ858162 | SOL29 | Weakly similar to yeast histone deacetylase complex subunit Pho23 | ND | ND |
Table 2.
Ten most frequent cDNAs in the Hw4.5-3 subtracted library. GenBank homologies, hit numbers and percentages of the most redundant cDNA clones are presented. aGenBank accession number of the most similar sequence identified by BLASTX alignment. bNumber of clones from Hw4.5-3 subtracted library assigned to the same GenBank accession number.
| BLAST hit Acc. No.a | Putative identity | Number of hits b | Percentage (%) |
| AAK94755 | Plasma membrane proton-exporting ATPase | 13 | 7.6 |
| XP750490 | Heat shock protein 70 | 9 | 5.3 |
| CAB38220 | Cytochrome c oxidase subunit I | 7 | 4.1 |
| XP658662 | Pathogenesis-related protein precursor | 6 | 3.5 |
| AAP57757 | Acetyl xylan esterase | 5 | 2.9 |
| EAS31689 | Unsaturated phospholipid methyltransferase | 5 | 2.9 |
| XP957152 | Cobalamin-independent methionine synthase | 5 | 2.9 |
| XP660489 | High mobility group protein A | 4 | 2.4 |
| CAA72382 | 14-3-3 protein | 4 | 2.4 |
| BAB33421 | Senescence-associated protein | 3 | 1.8 |
| Sum of top ten redundant clones | 60 | 35.8 | |
Table 3.
Distribution of differentially expressed genes from the Hw4.5-3 library by functional categories. Functional categorization was performed according to the MIPS database [22, 23] and annotated by the FunCat functional annotation scheme for systematic classification [22, 23]. Grouping of genes into indicated groups is shown as percent (%) of total identified EST from Hw4.5-3 library in descending order of abundance.
| Functional category | Description | % N = 95 |
| 1 | Metabolism | 31.6 |
| 2 | Energy | 23.3 |
| 12 | Protein synthesis | 21.1 |
| 20 | Cellular transport, transport facilitation and transport routes | 20.0 |
| 10 | Cell-cycle and DNA processing | 17.8 |
| 16 | Protein with binding function or cofactor requirement (structural or catalytic) | 16.7 |
| 14 | Protein fate | 14.4 |
| 34 | Interaction with the cellular environment | 13.3 |
| 42 | Biogenesis of cellular components | 12.2 |
| 40 | Cell fate | 11.1 |
| 32 | Cell rescue, defense and virulence | 11.1 |
| 11 | Transcription | 10.0 |
| 43 | Cell type differentiation | 6.7 |
| 30 | Cellular communication/signal transduction mechanism | 4.4 |
| 41 | Development (systemic) | 1.1 |
| ND | Not determined/Unknown function | 13.5 |
The majority of osmoresponsive genes in halotolerant H. werneckii are up-regulated in 4.5 M NaCl
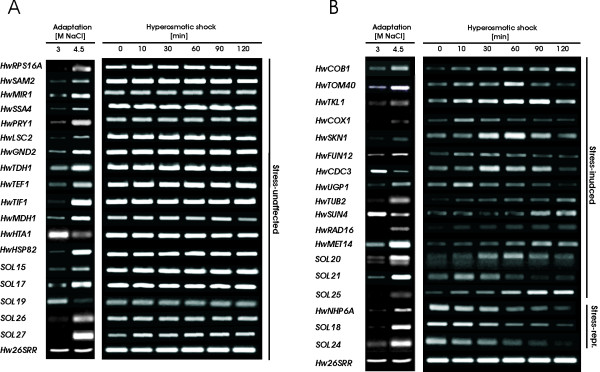
To confirm the differential expression and expression profiles of the ESTs obtained from the Hw4.5-3 library, a relative semi-quantitative RT-PCR was carried out. A total of 72 (75.8%) ESTs of interest, together with controls, were selected for further analyses. The levels of corresponding mRNAs in cells adapted to 3 M or 4.5 M NaCl were then analyzed. All of the 72 tested genes were confirmed as truly differentially expressed in H. werneckii adapted either to 3 M or 4.5 M NaCl. The magnitude of induction or repression of these genes is presented in Table 1, whereas the expression profiles are presented in Figures 1A, B and 2 (first columns). Fifty (69.4%) of the 72 genes were up-regulated 2- to 7-fold in 4.5 M NaCl. Besides HwGPD1A, used as a control (10.3-fold up-regulated), 10 genes (13.9%) were up-regulated more than 7-fold in 4.5 M NaCl compared with 3 M NaCl: HwELF1, HwERV25, HwFRE7, HwGDH1, HwOPI3, HwPMA2, HwPRY1, HwRPS16A, HwTDH1 and SOL21. The expression of these genes was virtually undetectable or barely detectable in cells from the 3 M NaCl medium. By contrast, only 6 (8.3%) of the genes studied by RT-PCR were preferentially induced in 3 M NaCl: HwCDC3, HwHTA1, HwSUN4, HwTUB2, SOL11 and SOL19. They were 1.5- to 3.3-fold down-regulated in 4.5 M NaCl. Of the 10 novel hyperosmolarity-induced SOL genes studied by RT-PCR, only SOL19 was down-regulated.
Figure 1.
Expression profiles of HwHog-ChIP negative genes in adapted and stressed H. werneckii cells. RT-PCR was performed with RNA isolated from cells adapted to 3 M and 4.5 M NaCl (Adaptation) or cells exposed to hypersaline stress at indicated time points (Hyperosmotic stress). 26S rRNA (Hw26SRR) was used as an internal control for template normalization. Genes whose expression was not affected by salt-stress are presented in (A) and those that responded to salt stress are presented in (B).
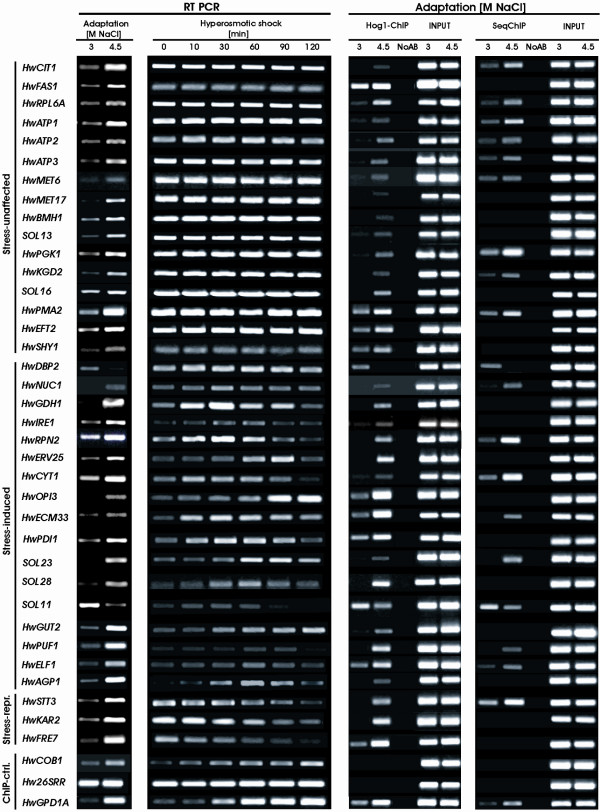
Figure 2.
Expression profiles of HwHog-ChIP positive genes in adapted and stressed H. werneckii cells and ChIP results in adapted cells. RT-PCR was performed with RNA isolated from adapted cells (Adaptation) or cells exposed to hypersaline stress (Hyperosmotic stress) as indicated. 26S rRNA (Hw26SRR) was used as an internal control for template normalization. The HwHog1-ChIP and sequential RNA polymerase II/HwHog1-ChIP (SeqChIP) results for cells adapted to 3 M NaCl and 4.5 M NaCl are presented in parallel. The negative control for nonspecific binding of antibodies in both ChIP experiments is represented in the "NoAB" line and the positive control for genomic DNA amplification from 100-fold diluted input samples is labeled as "INPUT". HwCOB1 and 26SRR genes were negative controls and HwGPD1A was the positive control for HwHog1 and RNA polymerase II cross-linking.
Expression of osmoresponsive genes is different in salt-adapted and salt-stressed H. werneckii cells
Induction of gene transcription has been recognized as a crucial mechanism for the adaptation of yeast cells to saline stress. Accurate whole genome expression analysis by microarray has been performed on S. cerevisiae at different salt concentrations (0.4 M–1.0 M NaCl) and different time points (10–90 min), resulting in different patterns of gene expression [2,25]. We asked if the salt concentration-dependent differences in the expression of H. werneckii osmoresponsive genes are characteristics of the adaptation phase only, or if these genes would also respond to acute hypersaline stress. To study the acute stress response, H. werneckii cells were exposed to hypersaline stress and the level of gene expression over the next 120 minutes was followed by RT-PCR. As shown in Figures 1A, B and 2 (second columns), time-dependent gene expression was observed only for half of the genes under study. Genes were nominated as "salt-stress responsive" if the mRNA level was induced or repressed at least 2-fold during the 120 min period following the salt stress. Whereas 38 (52.8%) genes were shown to be responsive to the acute hypersaline stress (see Table 4 for the list of the stress-responsive genes), the expression of 34 (47.3%) genes was not susceptible to the stress. As salt-stress responsive genes responded with different time courses of induction or repression, they were grouped into 21 early-induced genes and 11 delayed-induced genes if the 2-fold increase in mRNA level was observed within or after the first 30 min of salt stress, respectively; and 6 repressed genes, whose of mRNA level decreased within the first 10–30 min after salt stress. Those genes that were induced more than 5-fold during salt stress were HwECM33, HwRAD16, HwRPN2, HwSKN1, HwUGP1 and HwGPD1A as a control from the early-induced genes; and HwOPI3, HwSUN4, SOL23 and SOL28 from delayed-induced genes. RT-PCR examination of SOL20 repeatedly resulted in two PCR products (Figure 2B), suggesting a co-amplification of similar genes, both up-regulated in adapted cells (4.5 M NaCl), whereas it seems that only one of them responded to hypersaline stress. Among repressed genes,HwFRE7, HwNHP6A and SOL24 were more than 5-fold down-regulated after acute salt stress.
Table 4.
Induced and repressed genes in H. werneckii upon acute hypersaline stress. The stress-responsive genes were classified according to the kinetic of the response over the 120 min course of the experiment. Fold induction or repression is indicated as mean value of three independent RT-PCR experiments, and the representative gels are shown in figures 1 and 2.
| EARLY induced | DELAYED induced | REPRESSED | |||
| Gene | Fold induction | Gene | Fold induction | Gene | Fold repression |
| HwAGP1 | 5.2 | HwELF1 | 2.5 | HwFRE7 | 7.0 |
| HwCDC3 | 4.7 | HwERV25 | 3.0 | HwKAR2 | 4.4 |
| HwCOB1 | 3.1 | HwGUT2 | 4.0 | HwNHP6A | 5.2 |
| HwCOX1 | 2.8 | HwMET14 | 3.1 | HwSTT3 | 3.6 |
| HwCYT1 | 8.3 | HwOPI3 | 6.2 | SOL18 | 3.3 |
| HwECM33 | 5.5 | HwPUF1 | 2.7 | SOL24 | 5.5 |
| HwFUN12 | 2.4 | HwSUN4 | 5.7 | ||
| HwGDH1 | 4.2 | HwTOM40 | 3.4 | ||
| HwGPD1A | 6.3 | SOL23 | 5.3 | ||
| HwIRE1 | 4.7 | SOL25 | 4.4 | ||
| HwNUC1 | 3.0 | SOL28 | 5.6 | ||
| HwPDI1 | 3.9 | ||||
| HwRAD16 | 7.0 | ||||
| HwRPN2 | 5.6 | ||||
| HwSKN1 | 5.8 | ||||
| HwTKL1 | 3.9 | ||||
| HwTUB2 | 2.6 | ||||
| HwUGP1 | 7.5 | ||||
| SOL11 | 3.2 | ||||
| SOL20 | 2.8 | ||||
| SOL21 | 3.1 | ||||
These data demonstrate that in contrast to the response of the adapted cells, only a portion of the identified differentially expressed genes in H. werneckii transcriptionally responded to acute hypersaline stress as well. Interestingly, HwFRE7, HwKAR2, HwNHP6A, HwSTT3, SOL18 and SOL24 were up-regulated in cells adapted to extreme hypersaline conditions, but they were actually repressed after acute hypersaline stress. Additionally, HwCDC3, HwSUN4 and SOL11 were down-regulated in cells adapted to extreme hypersaline conditions, but induced during stress. This observation indicates an additional level of complexity in regulation of gene expression in halotolerant yeast when compared with the stress response of salt-sensitive S. cerevisiae.
It is noteworthy that the expression profile of H. werneckii adapted to 4.5 M NaCl is very different from the hypersaline stressed cells of S. cerevisiae. In comparison with the microarray studies performed with S. cerevisiae [2,25,26], using SSH-MOS we have identified only 18 gene orthologs that were clearly up-regulated in adapted or stressed H. werneckii and also in salt-stressed wild-type S. cerevisiae: HwBMH1, HwCIT1, HwGND2, HwGUT2, HwHSP82, HwKGD2, HwMDH1, HwOPI3, HwRPL16A, HwRPS10B, HwRPS15, HwRPS8A, HwSHY1, HwSSA4, HwTDH1, HwTIF1, HwTKL1, and HwUGP1. However, some discrepancies were observed between the compared microarray data in S. cerevisiae studies mentioned above. Among upregulated genes in salt-stressed S. cerevisiae, only HwGUT2, HwOPI3, HwTKL1 and HwUGP1 were also salt-stress responsive in H. werneckii. The mRNA levels of HwECM33, HwEFT2, HwFAS1, HwFUN12, HwGDH1, HwMET14, HwMET17, HwMET6, HwMIR1, HwNUC1, HwPMA2, HwPRY1 HwRPL6A, HwRPS16A, HwSAM2, HwSTT3, HwTEF1, HwTIF1 and HwTOM40 were induced in H. werneckii adapted to 4.5 M NaCl, whereas these levels diminished or remained unaffected in salt-stressed S. cerevisiae. Two genes, DBP2 and SUN4 were substantially downregulated in both, the hypersalinity-adapted H. werneckii and salt stressed S. cerevisiae. It was also found that GND2, GPD1 and SSA4 belong to a common environmental response (CER) genes in S. cerevisiae, which were affected not only by high salinity but various stresses such as heat, high or low pH, oxidative stress and sorbitol [27] and therefore might also present the CER response in H. werneckii. Interestingly, the induction of SSA4 was extremely stress-responsive in S. cerevisiae (47-fold induction), whereas it was only slightly affected by high salt concentration in H. werneckii (1.8-fold in 4.5 M NaCl).
One theory is that the main difference between salt-sensitive and halotolerant organisms in the expression of osmoresponsive genes relates to the inducibility and/or maintenance of the transcription level of protective genes. The perception threshold for the extracellular "hyper"-osmolarity in extremely halotolerant yeasts must be set at concentrations higher than salt-sensitive unicellular eukaryotes could even survive. In H. werneckii adapted to these extreme conditions, the protective response remains "on", meaning that there is a long-term up-regulation program of specific genes, and this up-regulation does not decrease over time.
HwHog1 associates with 36 novel osmoresponsive genes in chromatin of long-term adapted cells
The transcriptional induction or repression of approximately 500 genes in S. cerevisiae that are strongly responsive to salt stress was highly or fully dependent on the hyperosmolarity-responsive MAPK Hog1, indicating that the Hog1-mediated signaling pathway plays a key role in global gene regulation under saline stress conditions [2,28]. It has been shown that following exposure to salt stress, Hog1 is retained in the nucleus and becomes associated with the chromatin of target genes [12]. We approached the study of endogenous HwHog1 interaction with the chromatin regions of identified up-regulated genes in adapted H. werneckii cells by a chromatin immunoprecipitation (ChIP) PCR assay. Lacking information on promoter regions for the identified differentially-expressed genes in H. werneckii, a ChIP-coding region PCR amplification was performed. It has recently been shown that the activated Hog1 in S. cerevisiae is associated with elongating RNA polymerase II and is therefore recruited to the entire coding region of osmoinducible genes [29]. HwGPD1A was used as a positive control for the HwHog1-ChIP. As negative controls for the association of HwHog1 with DNA, the HwCOB1 gene encoded in mitochondrial DNA and the 26S rRNA gene (Hw26SRR) transcribed by RNA polymerase I were used.
The results of the HwHog1-DNA interactions determined by ChIP are shown in Figure 2, third column. As evidenced by PCR, the protein HwHog1 cross-linked with the coding region of the positive control HwGPD1A and 36 of the differentially expressed genes (50%), but not with the negative control genes HwCOB1 and Hw26SRR. As seen from the PCR product level, for 34 up-regulated genes the interaction with HwHog1 was stronger in cells adapted to 4.5 M NaCl. In contrast, for 2 down-regulated genes (HwDBP2 and SOL11), the HwHog1-ChIP signal was stronger in cells adapted to 3 M NaCl.
Genome-wide expression profiling studies using wild-type and hog1 mutant S. cerevisiae cells were performed to comparatively identify genes whose up-regulation of expression was dependent on Hog1. Of several hundred genes whose RNA levels were Hog1-dependent, a relatively small subset of approximately 40 high-osmolarity induced genes had a strong requirement for Hog1 for their induction [26,28]. Among them, only the UGP1 ortholog was induced in 4.5 M NaCl adapted and salt-stressed H. werneckii cells. Other yeast orthologs of HwHog1-ChIP positive genes in H. werneckii were reported for the first time in the present study in connection with MAPK Hog1. However, the HwHog1-ChIP did not confirm the HwHog1 interaction with the HwUGP1 gene in salt-adapted H. werneckii. The relative distribution of HwHog1-dependent genes was approximately equivalent among functional categories, except in the case of transcription, cellular transport, signal transduction mechanism and cell fate categories (MIPS categories 11, 20, 30 and 41, respectively), where the HwHog1-ChIP positive genes represented more than 70% of tested genes. Only 2 of 10 tested SOL genes (SOL23 and SOL28) with unknown functions (ND in table 1) were HwHog1-ChIP positive. Caution must be applied to the interpretation of genes which were HwHog1-ChIP negative, since many of them were salt-stress responsive. These genes could still be regulated by HwHog1. It is possible that during long-term adaptation the continuous interaction of HwHog1 with their genomic region is not obligatory, and thus HwHog1 was not cross-linked with the chromatin. Alternatively, HwHog1 could activate responsible factors more distal to the chromatin, thereby avoiding the cross-linking range.
HwHog1 and RNA polymerase II co-localization in coding regions of osmoresponsive genes is reflected by elevated levels of corresponding transcript in adapted cells
It has been previously shown that in the HOG response, the nuclear retention and chromatin association of Hog1 in S. cerevisiae depends on co-localization with general transcription machinery components [11,12]. We further asked whether HwHog1 cross-linking occurs with co-localization of RNA polymerase II in the case of HwHog1-dependent genes. A sequential HwHog1-ChIP analysis (SeqChIP) using primers specific for the genes identified as HwHog1-positive was therefore performed after the primary RNAPolII-ChIP. If the interaction of HwHog1 and RNAPolII existed within the same genomic region of HwHog1-positive genes, the PCR signal should be obtained in eluates of SeqChIP. As shown in Figure 2, fourth column, the co-localization of HwHog1 and RNA polymerase II existed in 17 cases out of 36 HwHog1-ChiP positive differentially expressed genes. Similarly, if the co-occupancy of HwHog1 and RNAPolII favored gene expression, then the relative ratio of amplified PCR products from SeqChIP eluates should reflect the relative ratio of RT-PCR results from the gene expression in adapted cells. As shown in Figure 2, the ratio of amplified PCR products in SeqChIP-positive genes does indeed reflect the ratio of mRNA levels observed by RT-PCR in both conditions of adaptation. Co-occupation of HwHog1 and RNA polymerase II in target genes resulted in an increased PCR signal in SeqChIP, with an accompanying increased level of corresponding transcript in RT-PCR analyses.
Taken together, these observations indicate a stimulating role for HwHog1 and RNA polymerase II co-localization on the efficiency of transcription of indicated genes, even in long-term high-salt adapted cells. Regarding the coding region positioning of primer pairs used in this study, we can say that in H. werneckii HwHog1 also associates with the elongating RNA polymerase II, as has recently been shown in S. cerevisiae [29]. In our study, H. werneckii cells used for chromatin immunoprecipitation analyses were completely adapted to extremely high environmental salt concentration by long-term growth in media containing either 3 M or 4.5 M NaCl. These results thus reflect HwHog1/RNAPolII-chromatin interactions, relevant for the extremely high saline conditions which until now could not be studied in salt-sensitive organisms. Moreover, to date this study is the first large-scale exploration of Hog1 interaction with target genes by chromatin immunoprecipitation in an organism with unavailable genomes. All similar studies have been performed using tagged and over-expressed proteins. Our study relays information based entirely on the cross-linking of endogenous HwHog1 and RNA polymerase II with their downstream targets on the chromatin, showing actual physiological interactions never studied before in eukaryotic cells adapted to such an extremely hyperosmotic condition.
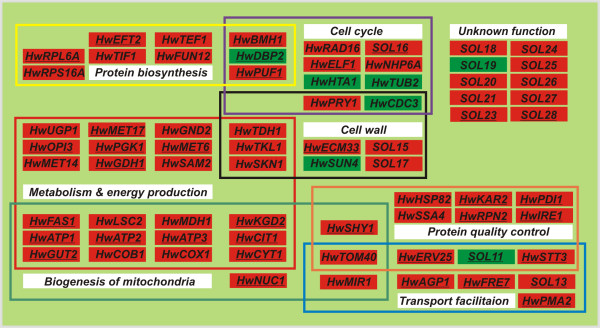
An integrative model of osmoresponsive gene action through functional modules in H. werneckii
The transcriptional behavior of some genes from our analysis correlated with known biochemical hyperosmotic stress responses in S. cerevisiae (for review see [4]). Nevertheless, important differences were observed between the two organisms, which presumably contribute to the remarkable capacity of H. werneckii for adaptation to extreme environments. We propose the following model of gene action through functional modules in H. werneckii adaptation to a hypersaline environment (Figure 3). During long-term adaptive growth in extreme salinity (4.5 M NaCl), there is a constant need to pump protons across the plasma membrane, which provides the electrochemical driving force for the Na+/H+-antiporters and P-type ATPases HwENA1/2 to exclude the intruding sodium cations [30] and to maintain the correct intracellular pH and plasma membrane potential. The 7.9-fold up-regulation of proton-exporting plasma membrane ATPase gene HwPMA2, which is also represented as the most frequent EST from the Hw4.5-3 subtracted library, indicates the importance of cytoplasmic pH regulation and proton gradient across the plasma membrane. We have observed a substantial decrease in growth media pH from pH 7.5 down to pH 2.5 during the log phase, which was more evident and faster with H. werneckii growing in 4.5 M NaCl than in 3 M NaCl (unpublished data). Therefore the cell has to increase its energy production machinery to supply the energetic demands of ion homeostasis. The functional category of energy supply was highly represented among up-regulated genes (Tables 1 and 3) in 4.5 M NaCl, including those coding for components of the electron transport chain (HwCOB1, HwCYT1, HwCOX1) and ATP production (HwATP1, HwATP2, HwATP3), and conveying the excessive cytosolic NADH into the mitochondrial respiratory chain (FAD-dependent glycerol-3-phospate dehydrogenase, HwGUT2). More than 10-fold up-regulation of HwFRE7 gene could indicate the increased requirements for the ferric-chelate reductase activity – namely, to satisfy the need for enhanced iron assimilation or reduction of oxidized iron-containing complexes under extremely hypersaline condition. Energy production metabolism seems to be additionally enhanced by the up-regulation of genes coding for the enzymes of the glycolytic pathway (HwTDH1, HwPGK1), the tricarboxylic acid cycle (HwCIT1, HwKGD2, HwLSC2, HwMDH1, HwGDH1), the pentose phosphate pathway (HwTKL1, HwGND2), and modulation of energy storage (HwUGP1, HwGAL10). More than 8-fold up-regulation of a specific mitochondrial inner-membrane Pi transporter gene, HwMIR1, further supported the idea of increased energy demands in H. werneckii growing at 4.5 M NaCl as compared to cells growing in a lower salinity, since most cellular ATP is produced within the mitochondria from ADP and Pi. Many of the Hw4.5-3 genes described so far, together with HwSUN4, HwTOM40, HwSHY1 and HwNUC1, code for mitochondrial enzymes, indicating the process of continuous adaptation to hypersaline conditions must be strongly supported by the biogenesis of mitochondria. Indeed, in the subcellular fractionation experiment we found that the quantity of isolated mitochondria was at least 2-fold higher in cells from 4.5 M NaCl, and that the succinate dehydrogenase activity per μg of mitochondria was 2.5 ± 0.5-fold higher compared with mitochondria from 3 M NaCl (data not shown). The biogenesis of mitochondria is coupled with elevated protein synthesis. Specific ribosomal protein isoforms (Table 1) and translation-regulating factors (HwFUN12, HwTIF1, HwEFT2, HwTEF1) are synthesized, which could be more effective in the translation of desired transcripts under unfavorable conditions and thereby increase the proliferative potential of the cell. The quantity of synthesized or damaged proteins could impair the ability of the cell to properly fold the resultant proteins; thus the components of protein quality control and the unfolded protein response (HwHSP82, HwSSA4, HwERV25, HwRPN2, HwSTT3, HwIRE1, HwKAR2) are constantly up-regulated. The increased protein synthesis is also coupled with up-regulation of genes coding for enzymes from amino acid biosynthetic pathways (HwMET6, HwMET14, HwMET17, HwSAM2, HwGDH1) and an amino acid permease (HwAGP1). From this point of view, the basic metabolic network is constitutively working to meet the demands of the increased energy expenditure needed to balance the ionic and the osmotic homeostasis. Because the environmental osmolarity of the natural habitat of H. werneckii can change, the plasma membrane has to be fluid enough, enabling the proper dynamics of signaling processes in a wide range of environmental salinities. Electron paramagnetic resonance spectroscopy measurements showed that the membranes of all halophilic fungi in extremely saline environments were more fluid than those of the salt-sensitive S. cerevisiae, which was in close agreement with the sterol-to-phospholipids ratio and fatty acid unsaturation level [19]. Plasma membrane fluidity could be supported by the biosynthesis of unsaturated or methylated lipids as a consequence of the 9.7-fold up-regulation of HwOPI3 and related genes (HwFAS1, HwSKN1). The interaction of cells with the cellular environment is mediated through the cell wall, which is highly implicated in the regulation of cellular turgor pressure. Two poorly described S. cerevisiae orthologs connected to cell wall integrity and architecture encoded by HwECM33 and HwSUN4 were identified as salt-responsive in H. werneckii. The finding that the major component of the septin complex HwCDC3 was strongly down-regulated, together with HwSUN4, could explain the substantially prolonged generation period of H. werneckii cells grown in 4.5 M NaCl compared to those grown in 3 M NaCl [15]. The role of interaction with the cellular environment is also assigned to SOL15 and SOL17, coding for acetyl-xylan esterase and feruloyl esterase orthologs, respectively, and to one of the most up-regulated genes at 4.5 M NaCl, the HwPRY1 gene, putatively coding for pathogenesis-related proteins in plants. The roles of both esterases in the automodulation of the cell wall dynamic as an adaptive response to high turgor pressure remains speculative.
Figure 3.
Model of gene interactions through functional modules responsible for the adaptive metabolism of H. werneckii in an extremely hypersaline environment. Interactions between functional modules are presented by overlapping frames. For visualization purposes, up-regulated genes are presented in red boxes and down-regulated genes are in green boxes. HwHog1-ChIP positive genes are underlined.
Conclusion
In our study, we have identified a set of 95 osmoresponsive genes in the extremely halotolerant black yeast H. werneckii adapted to a moderately saline environment of 3 M NaCl or an extremely saline environment of 4.5 M NaCl. Among them, more than half were related with general metabolism and energy. Thirteen unclassified SOL genes represent a specific transcriptional response unique to H. werneckii. A novel offset of 36 genes was shown as Hog1-dependent in long-term adaptation to extreme environments, previously not assigned as such in the salt-sensitive model organism S. cerevisiae. The combined data indicate important differences in the cellular processes of osmoadaptation between halotolerant and salt-sensitive yeasts. The novel set of osmoresponsive genes probably represents only a portion of actual differentially expressed genes in H. werneckii; however, we believe that valuable information was obtained concerning genes related to the hypersaline adaptation of extremely halotolerant eukaryotes.
Methods
Cell growth conditions
The H. werneckii strain MZKI B736 was obtained from culture collections of the Slovenian National Institute of Chemistry. Cells were grown at 28°C and 180 rpm in the defined YNB medium (0.17% (w/v) yeast nitrogen base, 0.08% (w/v) complete supplement mixture, 0.5% (w/v) ammonium sulphate, 2% (w/v) glucose in deionized water, pH 7.0), supplemented with the indicated NaCl concentration. Cells were harvested in the mid-exponential phase (OD600 nm 0.6–0.8) and frozen in liquid nitrogen. For hypersaline stress, H. werneckii cells were grown in YNB with 1 M NaCl to OD600 nm 0.8 and then transferred to the medium containing 4.5 M NaCl. Aliquots of the culture were removed before the stress was induced, and then at 10, 30, 60, 90 and 120 min after the stress. Cells were separated from the growth medium by fast filtration through a 0.45 μm-pore filters and then frozen in liquid nitrogen.
Suppression subtractive hybridization, mirror orientation selection and differential screening of subtracted library
Total RNA from cells of H. werneckii was isolated using TRI Reagent (Sigma-Aldrich) according to the manufacturer's instructions from mid-exponential phase cells grown in YNB media with 3 M or 4.5 M NaCl. The poly(A)RNA was isolated using the Oligotex mRNA Mini Kit (Qiagen). 1 μg of poly(A)RNA from each sample was used for reverse transcription to perform the cDNA suppression subtractive hybridization using the Clontech PCR-select cDNA Subtraction Kit (BD Bioscience) and Advantage cDNA PCR Polymerase Mix (BD Bioscience) according to the manufacturer's protocols. cDNA from a sample of cells growing in YNB medium with 4.5 M NaCl was used as a tester, while a sample from the 3 M NaCl medium was used as a driver in forward subtraction (and vice versa for reverse subtraction). Mirror orientation selection was implemented as described by Rebrikov et. al. [20] with some modifications. Briefly, after subtractive hybridization, the primary PCR of 27 cycles was performed in 5 parallel tubes. The samples were then combined, diluted 500-fold and subsequently amplified by 12 additional PCR cycles using the same primer and conditions as described for the primary PCR. A secondary PCR was then performed as described in the manufacturer's protocol. 150 ng of the resultant subtracted cDNA samples were digested with 10 U of Cfr9I (Fermentas) for 1 h at 37°C in total volume of 20 μL. 1 μL of Cfr9I-digested cDNA was mixed with 1 μL of 4 × hybridization buffer (2 M NaCl, 200 mM HEPES pH 8.3, 0.8 mM EDTA), 2 μL of water, denatured at 98°C for 1.5 min, and then hybridized at 68°C for 4 h. Samples were then diluted with 200 μL of the dilution buffer (50 mM NaCl, 20 mM HEPES pH 8.3, 0.2 mM EDTA) and heated at 70°C for 7 min. 1 μL of diluted cDNA was taken for 20 μL tertiary PCR with 0.6 μM adapter-specific primer NP2Rs (5'-GGTCGCGGCCGAGGT-3') by the following temperature program: 2 min at 72°C for initial extension of 3'-ends, followed by 23 cycles with 7 s at 95°C, 20 s at 62°C, and 2 min at 72°C. 1.5 μL of PCR products was cloned into pGEM-T Easy Vector (Promega) and transformed in JM109 competent cells (Promega). After blue/white selection on LB-Ampicillin/IPTG/X-Gal plates, white colonies were picked and arrayed on LB-Ampicillin plates, including 2 blue colonies as negative hybridization controls for the differential screening. Two identical colony lifts were made from each plate onto nitrocellulose membranes (Sigma), soaked with denaturation solution (0.5 M NaOH, 1.5 M NaCl), neutralizing solution (1.5 M NaCl, 0.5 M Tris-HCl, pH 7.4) and finally with washing solution (2 × SSC, 0.5 M Tris-HCl, pH 7.4), and then fixed by baking for 1–2 hours at 80°C. Forward and reverse subtracted hybridization probes were prepared from SSH-MOS secondary PCR products digested with RsaI, Cfr9I and EaeI to remove and degrade adaptors. After clean-up, 100 ng of each probe was labeled with [γ-32P]dCTP. Colony lifts were pre-hybridized with hybridization solution (5 × SSC, 0.5% SDS, 5 × Denhard's reagent, 0.1% SDS, 150 μg/mL salmon sperm DNA) for 1 h at 72°C, and then hybridized overnight at 72°C with labeled probes. Membranes were then washed once with low-stringency solution (2 × SSC, 0.5% SDS), twice with high-stringency solution (0.2 × SSC, 0.5% SDS), each for 20 min at 68°C, and then exposed to BioMax MR film (Kodak) overnight. For the forward-subtracted cDNA library (Hw4.5-3), clones that hybridized only to the forward-subtracted probe but not to the reverse-subtracted probe and clones that hybridized to both subtracted probes with a difference in signal intensity >5-fold were assigned as truly differential and chosen for sequencing. Plasmid DNA from positive clones was isolated using the Wizard Plus Minipreps Purification System (Promega), sequenced and analyzed by BLAST algorithms.
Reverse transcription PCR
The total RNA was isolated as described above and treated with DNase I (Fermentas). 1 μg of RNA was used for 20 μl of reverse transcription reaction using Superscript III Reverse Transcriptase (Invitrogen, USA) and random hexamer-primers (Promega) according to the manufacturer's protocols. PCR with Gotaq DNA polymerase (Promega) was performed using 0.5 μl of cDNA in a 20 μl PCR reaction with 15 nmol of specific primers listed in Additional file 1. Thermal cycling was programmed for 23 cycles, each consisting of 30 sec at 94°C, 30 sec at 60°C and 30 sec at 72°C. The cycle number of 23 was empirically determined in the optimal linear range of amplification by measuring the concentration of PCR products after the 20th, 22nd and 24th cycles. PCR products were resolved in agarose gels, documented by MiniBis (DNR BioImaging Systems), and the band intensities quantified with the TotalLab gel analysis program (Nonlinear Dynamics).
Chromatin immunoprecipitation
Immunoprecipitation of cross-linked chromatin (ChIP) was performed as described by Hecht et al.[31] and Proft et al.[32], and sequential ChIP (SeqChIP) as described by Geisberg et al. [33] with some modifications. Briefly, cells of H. werneckii growing in YNB media with 3 M or 4.5 M NaCl were cross-linked at OD600 0.8 using formaldehyde in a final concentration of 1% for 15 min at room temperature. Cross-linking was stopped with glycin at a final concentration of 0.125 M for 5 min at room temperature. Cells were harvested, washed twice with ice-cold PBS, pelleted, frozen in liquid nitrogen and broken with a dismembrator. 2.5 g of powdered cells were re-suspended in 10 mL of ChIP-L buffer (1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.1) containing a cocktail of fungal protease inhibitors (Sigma) and mixed by inversion for 30 min on ice. The samples were sonicated for five 15-s pulses, resulting in DNA fragments with an average length of 500 bp, and then centrifuged for 15 min at 10.000 × g to remove insoluble debris. For each ChIP experiment, 100 μL of supernatant was frozen as an input and 1 mL of supernatant was diluted in 9 mL of ChIP-D buffer (0.01% SDS, 1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl, pH 8.1, 167 mM NaCl) with inhibitors. Chromatin solutions were pre-cleared with the salmon sperm DNA-precoated protein G-sepharose (Amersham) and then incubated with 5 μg of rabbit anti-Hog1 antibody (Santacruz) or mouse monoclonal anti-RNAPol II antibody 4H8 (Abcam) overnight at 8°C. 250 μL of precoated protein G-sepharose was added to each sample. Immunocomplexes were precipitated for 2 h at 8°C with shaking, pelleted 1 min at 1000 × g, and then pellets were washed with 2 mL of the following washing buffers: twice with ChIP-W1 (150 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1), once with ChIP-W2 (500 mM NaCl, 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl, pH 8.1), once with ChIP-W3 (250 mM LiCl, 1% Na-deoxycholate, 1 mM EDTA, pH 8.0) and twice with ChIP-W4 (10 mM Tris-HCl, 1 mM EDTA, pH 8.0). Immunocomplexes were eluted twice with 200 μL of ChIP-E buffer (1% SDS, 100 mM NaHCO3) for 10 min at 65°C and eluates were collected. For SeqChIP, the RNAPolII-ChIP eluates (400 μL) were diluted with 3.6 mL of ChIP-dilution buffer and incubated for 2 h at room temperate with 1 μg of rabbit anti-Hog1 antibodies, and then processed as described above. ChIP eluates and inputs were reverse cross-linked in 0.2 M NaCl for 5 h at 65°C, incubated with 20 μg of RNase for 30 min at 37°C, followed by treatment with 10 μg of proteinase K. DNA was purified using the Wizard PCR Clean-up Purification System (Promega) and eluted with 150 μL of water for Hog-ChIP samples or with 50 μL for SeqChIP samples. PCR with Gotaq DNA polymerase was performed using 1 μl of eluted DNA from immunoprecipitated samples or 1 μl of 100-fold diluted input in 20 μl PCR reaction with Gotaq DNA polymerase and 15 nmol of specific primers (Additional file 1). Thermal cycling was programmed for 30 cycles, each consisting of 30 sec at 94°C, 30 sec at 55°C and 60 sec at 72°C.
Authors' contributions
TV designed the study, carried out the molecular experiments and bioinformatics study, and drafted the manuscript. AP conceived of the study, participated in its design and coordination, and helped to draft the manuscript. Both authors read and approved the final manuscript.
Supplementary Material
List of primers used in this study.
Acknowledgments
Acknowledgements
We would like to thank Prof. S. A. Lukyanov from Evrogen for useful suggestions in MOS technique and Prof. M. B. Peterlin from UCSF for his support in ChIP methodology training. This work was supported in part by research grant P1-0170 and in part by a Young Researcher Fellowship from the Slovenian Research Agency.
Contributor Information
Tomaž Vaupotič, Email: tomaz.vaupotic@mf.uni-lj.si.
Ana Plemenitaš, Email: ana.plemenitas@mf.uni-lj.si.
References
- Gustin MC, Albertyn J, Alexander M, Davenport K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1998;62:1264–1300. doi: 10.1128/mmbr.62.4.1264-1300.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posas F, Chambers JR, Heyman JA, Hoeffler JP, de Nadal E, Arino J. The transcriptional response of yeast to saline stress. J Biol Chem. 2000;275:17249–17255. doi: 10.1074/jbc.M910016199. [DOI] [PubMed] [Google Scholar]
- de Nadal E, Alepuz PM, Posas F. Dealing with osmostress through MAP kinase activation. EMBO Rep. 2002;3:735–740. doi: 10.1093/embo-reports/kvf158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohmann S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev. 2002;66:300–372. doi: 10.1128/MMBR.66.2.300-372.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser V, Ruis H, Ammerer G. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol Biol Cell. 1999;10:1147–1161. doi: 10.1091/mbc.10.4.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Pascual-Ahuir A, de Nadal E, Arino J, Serrano R, Posas F. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. Embo J. 2001;20:1123–1133. doi: 10.1093/emboj/20.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nadal E, Casadome L, Posas F. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol Cell Biol. 2003;23:229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Reiser V, Gartner U, Thevelein JM, Hohmann S, Ammerer G, Ruis H. Osmotic stress-induced gene expression in Saccharomyces cerevisiae requires Msn1p and the novel nuclear factor Hot1p. Mol Cell Biol. 1999;19:5474–5485. doi: 10.1128/mcb.19.8.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Pastor MT, Marchler G, Schuller C, Marchler-Bauer A, Ruis H, Estruch F. The Saccharomyces cerevisiae zinc finger proteins Msn2p and Msn4p are required for transcriptional induction through the stress response element (STRE) Embo J. 1996;15:2227–2235. [PMC free article] [PubMed] [Google Scholar]
- Rep M, Albertyn J, Thevelein JM, Prior BA, Hohmann S. Different signalling pathways contribute to the control of GPD1 gene expression by osmotic stress in Saccharomyces cerevisiae. Microbiology. 1999;145 ( Pt 3):715–727. doi: 10.1099/13500872-145-3-715. [DOI] [PubMed] [Google Scholar]
- Alepuz PM, de Nadal E, Zapater M, Ammerer G, Posas F. Osmostress-induced transcription by Hot1 depends on a Hog1-mediated recruitment of the RNA Pol II. Embo J. 2003;22:2433–2442. doi: 10.1093/emboj/cdg243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alepuz PM, Jovanovic A, Reiser V, Ammerer G. Stress-induced map kinase Hog1 is part of transcription activation complexes. Mol Cell. 2001;7:767–777. doi: 10.1016/S1097-2765(01)00221-0. [DOI] [PubMed] [Google Scholar]
- De Nadal E, Zapater M, Alepuz PM, Sumoy L, Mas G, Posas F. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature. 2004;427:370–374. doi: 10.1038/nature02258. [DOI] [PubMed] [Google Scholar]
- Gunde-Cimerman N., Zalar, P., de Hoog, G. S., Plemenitaš, A. Hypersaline waters in salterns: natural ecological niches for halophilic black yeasts. FEMS microbiol ecol. 2000;32:235–340. doi: 10.1111/j.1574-6941.2000.tb00716.x. [DOI] [PubMed] [Google Scholar]
- Petrovic U, Gunde-Cimerman N, Plemenitas A. Cellular responses to environmental salinity in the halophilic black yeast Hortaea werneckii. Mol Microbiol. 2002;45:665–672. doi: 10.1046/j.1365-2958.2002.03021.x. [DOI] [PubMed] [Google Scholar]
- Turk M, Plemenitas A. The HOG pathway in the halophilic black yeast Hortaea werneckii: isolation of the HOG1 homolog gene and activation of HwHog1p. FEMS Microbiol Lett. 2002;216:193–199. doi: 10.1111/j.1574-6968.2002.tb11435.x. [DOI] [PubMed] [Google Scholar]
- Kogej T, Ramos J, Plemenitas A, Gunde-Cimerman N. The halophilic fungus Hortaea werneckii and the halotolerant fungus Aureobasidium pullulans maintain low intracellular cation concentrations in hypersaline environments. Appl Environ Microbiol. 2005;71:6600–6605. doi: 10.1128/AEM.71.11.6600-6605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovic U, Gunde-Cimerman N, Plemenitas A. Salt stress affects sterol biosynthesis in the halophilic black yeast Hortaea werneckii. FEMS Microbiol Lett. 1999;180:325–330. doi: 10.1111/j.1574-6968.1999.tb08813.x. [DOI] [PubMed] [Google Scholar]
- Turk M, Mejanelle L, Sentjurc M, Grimalt JO, Gunde-Cimerman N, Plemenitas A. Salt-induced changes in lipid composition and membrane fluidity of halophilic yeast-like melanized fungi. Extremophiles. 2004;8:53–61. doi: 10.1007/s00792-003-0360-5. [DOI] [PubMed] [Google Scholar]
- Rebrikov DV, Britanova OV, Gurskaya NG, Lukyanov KA, Tarabykin VS, Lukyanov SA. Mirror orientation selection (MOS): a method for eliminating false positive clones from libraries generated by suppression subtractive hybridization. Nucleic Acids Res. 2000;28:E90. doi: 10.1093/nar/28.20.e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccharomyces Genome Database . http://www.yeastgenome.org/; [Google Scholar]
- Mewes HW, Frishman D, Guldener U, Mannhaupt G, Mayer K, Mokrejs M, Morgenstern B, Munsterkotter M, Rudd S, Weil B. MIPS: a database for genomes and protein sequences. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, Mewes HW. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004;32:5539–5545. doi: 10.1093/nar/gkh894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waschuk SA, Bezerra AG, Jr., Shi L, Brown LS. Leptosphaeria rhodopsin: bacteriorhodopsin-like proton pump from a eukaryote. Proc Natl Acad Sci U S A. 2005;102:6879–6883. doi: 10.1073/pnas.0409659102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yale J, Bohnert HJ. Transcript expression in Saccharomyces cerevisiae at high salinity. J Biol Chem. 2001;276:15996–16007. doi: 10.1074/jbc.M008209200. [DOI] [PubMed] [Google Scholar]
- Rep M, Krantz M, Thevelein JM, Hohmann S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J Biol Chem. 2000;275:8290–8300. doi: 10.1074/jbc.275.12.8290. [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke SM, Herskowitz I. Unique and redundant roles for HOG MAPK pathway components as revealed by whole-genome expression analysis. Mol Biol Cell. 2004;15:532–542. doi: 10.1091/mbc.E03-07-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, Posas F. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell. 2006;23:241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- Gorjan A, Plemenitas A. Identification and characterization of ENA ATPases HwENA1 and HwENA2 from the halophilic black yeast Hortaea werneckii. FEMS Microbiol Lett. 2006;265:41–50. doi: 10.1111/j.1574-6968.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- Hecht A, Grunstein M. Mapping DNA interaction sites of chromosomal proteins using immunoprecipitation and polymerase chain reaction. Methods Enzymol. 1999;304:399–414. doi: 10.1016/s0076-6879(99)04024-0. [DOI] [PubMed] [Google Scholar]
- Proft M, Struhl K. Hog1 kinase converts the Sko1-Cyc8-Tup1 repressor complex into an activator that recruits SAGA and SWI/SNF in response to osmotic stress. Mol Cell. 2002;9:1307–1317. doi: 10.1016/S1097-2765(02)00557-9. [DOI] [PubMed] [Google Scholar]
- Geisberg JV, Struhl K. Quantitative sequential chromatin immunoprecipitation, a method for analyzing co-occupancy of proteins at genomic regions in vivo. Nucleic Acids Res. 2004;32:e151. doi: 10.1093/nar/gnh148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers used in this study.