Abstract
CD4+ T cells from T cell receptor transgenic mice that have been vigorously primed to be interleukin (IL)-4 producers (TH2 cells) are capable of producing IL-4 even if restimulated in the absence of IL-4 and in the presence of IL-12. T cells vigorously primed in the absence of IL-4 (TH1 cells) fail to produce IL-4 even if restimulated under conditions that would cause a naive T cell to produce IL-4. In contrast, interferon γ (IFN-γ) production is highly cytokine-regulated. T cells primed in the presence of IL-4 develop into IFN-γ producers if IFN-γ is included in the priming culture and if the cells are challenged in the presence of IL-12, presumably reflecting the role of IFN-γ in inducing responsiveness to IL-12. Cells primed in the absence of IL-4 become highly responsive to IL-12 if IFN-γ is included in the priming culture, and these cells are excellent IFN-γ producers upon challenge; IL-12 considerably enhances their production of IFN-γ. If cells are primed in the absence of IL-4 and IFN-γ, they show very weak responsiveness to IL-12 as determined by STAT-4 activation. However, these cells acquire IL-12 responsiveness if cultured with IFN-γ for a period as short as 4 hr. Thereafter, they produce large amounts of IFN-γ upon challenge with antigen in the presence of IL-12. These results indicate that in primed CD4+ T cells, IL-4 production is largely cytokine-autonomous, whereas IFN-γ production is highly cytokine-regulated.
Naive CD4+ T cells produce interleukin (IL)-2 upon stimulation but can develop into cells that principally produce interferon γ (IFN-γ) or IL-4 and a set of related cytokines (1). In the mouse, this has often been referred to as TH1/TH2 polarization with the phenotype of TH1 cells being IFN-γ+/IL-4− and the phenotype of TH2 cells being IFN-γ−/IL-4+ (2). The development of naive cells into IL-4 producers requires the presence of IL-4 during the priming culture (3–6). In contrast, in the absence of IL-4, naive T cells develop into IFN-γ producers, a process that is enhanced by the presence of IL-12 and possibly IFN-γ (7–9).
The capacity of differentiated CD4+ T cells to alter their pattern of cytokine production has been a matter of interest. In contrast to long-term T cell lines in which cytokine-producing phenotypes show great stability (2), recently differentiated CD4+ T cells may have the capacity to alter the range of cytokines they produce (10, 11) and human CD4+ TH2 cells can acquire modest IFN-γ-producing capacity if cultured with IL-12 (12). Here we examine the issue of altered cytokine-producing phenotype in recently differentiated CD4+ T cells derived from mice transgenic (TG) for genes encoding T cell receptors specific for a cytochrome C peptide (88–104) in association with the class II molecule I-Ek (TG mice) (5). We show that after a vigorous in vitro priming regimen, the capacity (or lack thereof) of CD4+ T cells to produce IL-4 is a stable property and is not thereafter influenced by the cytokine environment, whereas the IFN-γ-producing capacity of such cells depends on the action of three distinct cytokines, IL-4, IFN-γ, and IL-12.
MATERIALS AND METHODS
Culture Medium.
cRPMI, RPMI 1640 medium (Biofluids, Rockville, MD) supplemented with 5% fetal bovine serum (Biofluids), 1 mM sodium pyruvate, 2 mM l-glutamine, 0.05 mM 2-mercaptoethanol, 100 units/ml penicillin, 100 μg/ml streptomycin, was used in cell cultures.
Cytokines and Antibodies.
Mouse IL-4 was obtained from a recombinant baculovirus prepared in our laboratory. Human IL-2 was a gift of Cetus. IFN-γ was purchased from Genzyme. IL-12 and goat anti-IL-12 were purchased from R & D Systems. Anti-IL-4 (11B11) (13) was prepared by Verax (Lebanon, NH). Anti-IFN-γ (XMG 1.2) (14), fluorescein isothiocyanate (FITC)-rat anti-mouse B220 (RA3–6B2) (15), FITC-rat anti-mouse I-Ak (AF6–120.1) (16), FITC-rat anti-mouse CD8 (53–6.7) (17), FITC anti-IL-4, biotinylated anti-IL-4, and FITC anti-IFN-γ were purchased from PharMingen. Streptavidin–horseradish peroxidase was obtained from Southern Biotechnology Associates. Rat anti-mouse Fcγ receptor antibody 2.4G2 (18) was used as 1:500 diluted ascites. Anti-STAT-4 antibodies were provided by J. Darnell (The Rockefeller University, New York) and used at 1 μl per reaction for supershift experiments.
Isolation of Naive CD4 T Cells, Antigen-Presenting Cells (APC), and Cell Priming Conditions.
B10.A mice were obtained from The Jackson Laboratory. B10.A TG mice were maintained in our animal facility.
TG lymph node and spleen cells were passed though a T cell enrichment column (R & D Systems). Enriched T cells were stained with FITC antibodies to the cell surface markers CD8, B220, and I-Ak. Labeled cells were negatively selected using sheep anti-FITC magnetic beads (Advanced Magnetics, Cambridge, MA) (19). Purified CD4+ cells were centrifuged for 20 min at 400 × g on a discontinuous 50%, 60%, and 70% Percoll (Pharmacia) gradient. Cells layering between 60% and 70% were collected and used for priming. APC were purified by treating B10.A splenocytes with anti-Thy-1.2 (HO13.4) antibody plus Low·Tox M rabbit complement 1:10 (Accurate Scientific, Westbury, NY) for 45 min at 37°C. Primary stimulation of TG cells was carried out by adding naive CD4+ T cells (105/ml) to irradiated APC (106/ml), 1 μM pigeon cytochrome C peptide 88–104 (prepared by the National Institute of Allergy and Infectious Diseases Biologic Resources Branch), and IL-2 (10 units/ml) for 5–7 days. For differentiation of TH1 cells, anti-IL-4 (10 μg/ml) and IL-12 (10 ng/ml) were also added to the culture; for the differentiation of TH2 cells, IL-4 (1,000 units/ml) was added. IL-12, IFN-γ (200 units/ml), anti-IL-12 (10 μg/ml), and anti-IFN-γ (10 μg/ml) were added in various combinations for priming where indicated. For some experiments, the priming culture was repeated two or three times.
Electrophoretic Mobility-Shift Analysis.
Primed cells were incubated overnight in cRPMI with IL-2. All cells were incubated for 2 hr at 37°C in RPMI medium 1640 without serum. Cells (5 × 106) were resuspended in 1 ml of cRPMI with or without IL-12 (50 ng/ml) for 15 min at room temperature and washed with 10 ml of ice-cold PBS/100 μM sodium vanadate. Cell pellets were resuspended in 20–40 μl of lysis buffer (0.5% Nonidet P-40/50 mM Tris, pH 8.0/10% glycerol/100 μM EDTA, pH 8.0/50 mM NaF/150 mM NaCl/100 μM Na3VO4/1 mM DTT/400 μM phenylmethylsulfonyl fluoride/1 μg/ml pepstatin A/1 μg/ml leupeptin/1 μg/ml aprotinin), and incubated on ice for 60 min with frequent vortexing. Lysates were centrifuged at 15,000 rpm for 15 min at 4°C. Supernatants were harvested and stored at −70°C. Protein concentrations were determined using the Bio-Rad protein assay reagent (Bio-Rad) with BSA as standard. For assays, 25 μg of cell lysate was incubated with 100 ng of 32P-labeled oligonucleotide in reaction buffer [40 mM KCl/1 mM MgCl2/0.1 mM EDTA/0.5 mM DTT/20 mM Hepes, pH 7.9/6% glycerol/1 mg/ml BSA/0.1 mg/ml poly(dI-dC)] for 15 min at room temperature. Reactants were separated by electrophoresis on a 4.5% polyacrylamide gel buffered with 0.22× TBE. Gels were dried and exposed directly to Kodak Biomax MR film (Sigma). A double-stranded oligonucleotide corresponding to an IFN-γ activation site (GAS)-like element found in the mouse FcγRI promoter (5′-gatcGCATGTTTCAAGGATTTGAGATGTATTTCCACAGAAAAGG) (20) was synthesized with a 5′-GATC overhang on each end (denoted by lowercase letters), and labeled with [32P]dCTP using Klenow DNA polymerase by standard techniques.
Intracellular Staining.
TH1 and TH2 cells were stimulated with IL-2 (10 units/ml), cytochrome C peptide (1 μM), and irradiated normal B10.A T-depleted spleen cells as APC in the presence of 2 μM monensin (Calbiochem). In some experiments, IL-12 or anti-IL-12 was added to the culture. After 6 hr of stimulation, cells were treated with 20 μg/ml of DNase 1 (Boehringer Mannheim) for 5 min at 37°C, washed with cold PBS, fixed with 4% paraformaldehyde for 5 min at 37°C, and washed with buffer containing 0.1% saponin and 0.1% BSA. Fixed cells were incubated with 2.4G2 for 5 min at 4°C and stained with labeled anti-cytokine antibodies for 30 min on ice (21). Samples were analyzed using a Becton Dickinson FACScan.
Lymphokine Measurements.
Primed cells (106/ml) were stimulated with peptide (1 μM), irradiated APC (106/ml), and IL-2 (10 units/ml) in a 24-well plate. Supernatants were collected at 24 and 48 hr for IL-4 and IFN-γ measurements, respectively. IL-4 concentration was measured with the IL-4-dependent indicator cell line CT.4S (22) and with ELISA reagents from PharMingen. IFN-γ concentration was measured with a two-site ELISA (23, 24).
RESULTS
Phenotype Stability After in Vivo Residence.
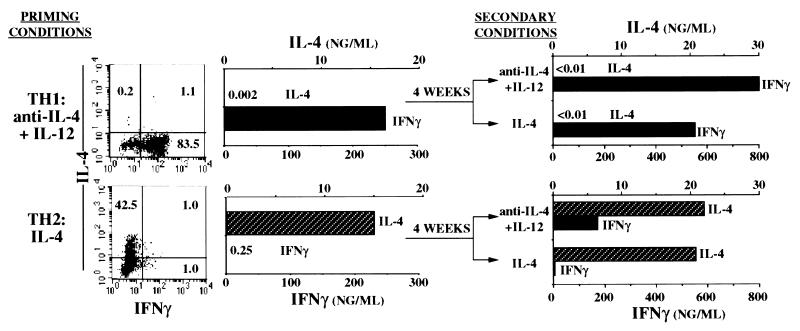
To examine the stability of cytokine-producing phenotypes of recently differentiated CD4+ T cells from TG mice, we purified small resting CD4+ T cells and subjected them to two rounds of in vitro priming. TH2 cells were prepared by stimulation with peptide, APC, IL-2, and IL-4, whereas TH1 cells were prepared by stimulation with peptide, APC, IL-2, and anti-IL-4 and IL-12. TH1 cells made large amounts of IFN-γ but little or no IL-4 in response to challenge with APC, peptide, and IL-2, whereas the TH2 cells displayed the opposite phenotype (Fig. 1). This is confirmed by the analysis of the frequency of cells that contained cytosolic IFN-γ and IL-4 upon challenge with peptide, IL-2, and APC. Primed cells were injected i.v. into non-TG B10.A mice. One month later, the TG (Vα11,Vβ3+) cells constituted 7–22% of spleen and lymph node CD4+ T cells. These cells were harvested, stimulated for 5 days with peptide, APC, IL-2, and IL-4 or anti-IL-4 and IL-12, and their capacity to produce IL-4 and IFN-γ upon challenge with APC, peptide, and IL-2 was measured. TH1 cells failed to produce IL-4 upon challenge whether restimulated in the presence of IL-4 or anti-IL-4 and IL-12 but did produce IFN-γ. TH2 cells restimulated in the presence of IL-4 or anti-IL-4 and IL-12 continued to produce IL-4 upon challenge. However, culture in the presence of anti-IL-4 and IL-12 resulted in a cell population that produced IFN-γ upon challenge, although in somewhat lower amounts that did cells initially primed without IL-4.
Figure 1.
Stability of cytokine phenotype of primed CD4+ T cells. CD4+ cells from B10.A TG mice were primed twice in vitro with cytochrome C peptide, APC, IL-2, and anti-IL-4 and IL-12 (TH1 cells) or IL-4 (TH2 cells). After the second priming culture, a portion of the cells were stimulated, and the number of cells containing cytosolic IL-4 and IFN-γ was determined and the secretion of the two cytokines was measured. The twice-primed cells (3 × 107) were also transferred into B10.A mice. Four weeks later, CD4+ T cells were purified from spleens and lymph nodes of the recipient mice and restimulated with peptide, APC, and IL-2, and with anti-IL-4 and IL-12 or IL-4. After 5 days, the cells (106) were challenged with peptide, APC, and IL-2. IL-4 and IFN-γ production were measured.
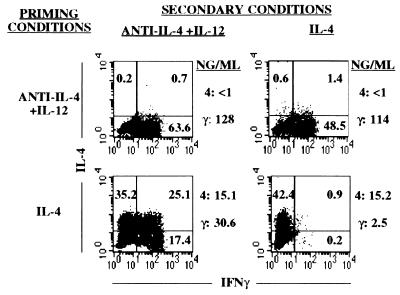
The cell population that was injected into B10.A mice was also immediately restimulated in vitro with antigen, APC, and either anti-IL-4 and IL-12 or IL-4. These cells behaved similarly to the cells that were “parked” in B10.A mice for 30 days (Fig. 2). When challenged at the end of the restimulation period, cells that had been initially subjected to two rounds of priming with anti-IL-4 and IL-12 failed to produce IL-4 whether restimulated in the presence or absence of IL-4. We have noted, as E. Murphy et al. have reported (11), that cells that had received only a single round of priming under TH1 conditions could acquire the capacity to produce modest amounts of IL-4 upon restimulation. The twice-primed TH1 cells, when restimulated with IL-4, contained somewhat fewer cells capable of producing IFN-γ than did such cells that had been restimulated with anti-IL-4 and IL-12. Strikingly, 42% of the cells that had been primed with IL-4 and restimulated with anti-IL-4 and IL-12 contained cytosolic IFN-γ upon challenge. Moreover, many of the IFN-γ-containing cells also contained IL-4. This result strongly argues that these were not simply cells that had failed to commit to IL-4 production in the first culture. If that were the case, they should not have acquired IL-4-producing activity in response to culture with anti-IL-4 and IL-12. Rather, this result indicates that these were TH2 cells that acquired IFN-γ-producing activity as a result of the conditions of the restimulation culture. The amounts of IL-4 and IFN-γ produced by these cultures are consistent with the results from the flow cytometric analysis of cytokine-containing cells.
Figure 2.
Stability of cytokine phenotype upon immediate restimulation. The cell populations primed twice in the experiment described in Fig. 1 were also immediately restimulated in vitro with peptide, APC, IL-2, and anti-IL-4 and IL-12 or IL-4. Seven days later, the cells were challenged with peptide, APC, and IL-2, and cytosolic cytokine content was determined by flow cytometry. Cytokine secretion was also measured. 4, IL-4; γ, IFN-γ.
Requirements for TH2 Cells to Produce IFN-γ.
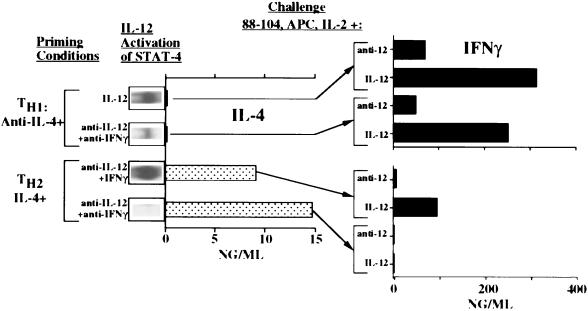
To analyze the requirements for the acquisition of IFN-γ-producing capacity by TH2 cells, we used two TH2-priming protocols in which we controlled the presence of IFN-γ and eliminated IL-12. T cells primed with IL-4, anti-IL-12, and anti-IFN-γ differed from cells primed with IL-4, anti-IL-12, and IFN-γ in certain striking ways. As anticipated, both produced IL-4 upon immediate challenge with peptide and APC. However, the cells primed in the presence of anti-IFN-γ failed to produce IFN-γ upon immediate challenge in the presence or absence of IL-12 (Fig. 3). Indeed, these cells appeared insensitive to IL-12 as shown by their failure to develop STAT-4-dependent DNA-binding activity when cultured with IL-12. In contrast, TH2 cells that had been primed in the presence of IFN-γ produced IFN-γ if challenged in the presence of IL-12 and showed striking activation of STAT-4 when incubated with IL-12 (Fig. 3). To verify that the IL-12-induced DNA-binding activity was due to STAT-4, anti-STAT-4 antibody was shown to “supershift” the complex in selected experiments.
Figure 3.
Presence of IFN-γ during priming determines responsiveness to IL-12. CD4+ TG T cells were primed with peptide, APC, IL-2, and either anti-IL-4 and IL-12 or anti-IL-4, anti-IL-12, and anti-IFN-γ (TH1 cells), or with IL-4 and either anti-IL-12 and IFN-γ or anti-IL-12 and anti-IFN-γ (TH2 cells). A portion of the cells were immediately challenged with or without IL-12; extracts were prepared, and STAT-4 binding to FcγRI GAS oligonucleotide sequence was determined as a measure of STAT-4 activation. The cells were also challenged with peptide, APC, and IL-2 (for IL-4 production) or with peptide, APC, IL-2, and either anti-IL-12 or IL-12 (for IFN-γ production).
When the intracellular cytokine content of TH2 cells primed in the presence of IFN-γ was analyzed, few contained IFN-γ if challenged in the presence of anti-IL-12 (5%); however, when challenged in the presence of IL-12, ≈65% of these cells produced IFN-γ (data not shown). Thus, TH2 cells primed in the presence of IFN-γ acquired IL-12-dependent IFN-γ production, whereas TH2 cells primed in the absence of IFN-γ were insensitive to IL-12 and failed to produce IFN-γ.
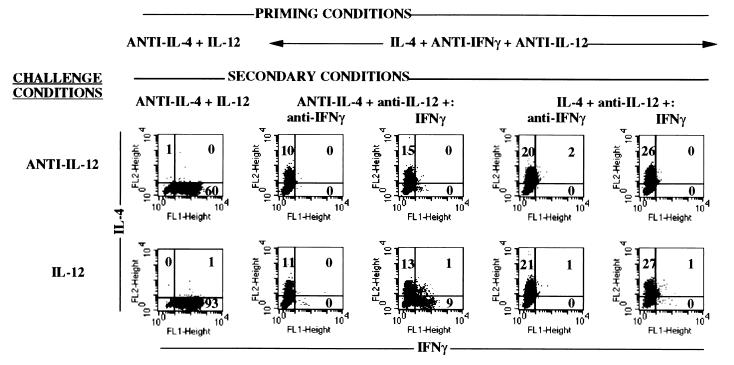
The IL-4-producing cells that had been primed with anti-IL-12 and anti-IFN-γ were recultured to examine whether and under what conditions they could acquire IFN-γ-producing activity. They were restimulated for 5 days in the absence of IL-12 with IL-4 or anti-IL-4 and with IFN-γ or anti-IFN-γ, and were then challenged with peptide, APC, and either IL-12 or anti-IL-12. Only cells restimulated with both IFN-γ and anti-IL-4 and challenged with IL-12 produced IFN-γ (Fig. 4). While only ≈9% of the cells produced IFN-γ, in contrast to ≈90% of the cells from cultures that had been subjected to two rounds of priming in anti-IL-4 plus IL-12, their production correlated with the ability of these cells to develop a STAT-4 electrophoretic mobility shift in response to IL-12 challenge. Thus, priming naive T cells in the presence of IL-4 and the absence of IFN-γ prevents the development of responsiveness to IL-12 and of the capacity to secrete IFN-γ upon challenge with antigen and IL-12. Such cells can acquire IL-12 responsiveness only if restimulated in both the presence of IFN-γ and the absence of IL-4. They retain IL-4-producing capacity under these various conditions.
Figure 4.
Requirements for TH2 cells primed in the absence of IFN-γ to develop IFN-γ-producing activity. TG CD4+ T cells were primed with peptide, APC, IL-2, IL-4, anti-IL-12, and anti-IFN-γ. The cells were then restimulated with peptide, APC, IL-2, anti-IL-12 and IL-4 or anti-IL-4 and IFN-γ or anti-IFN-γ. At the end of the restimulation culture, they were challenged with peptide, APC, and IL-12 or anti-IL-12. Cytosolic cytokine content was measured. As a control, cells that had been primed and restimulated in the presence of anti-IL-4 and IL-12 were also tested.
TH1 Cells: Control of IFN-γ-Producing Capacity and Responsiveness to IL-12.
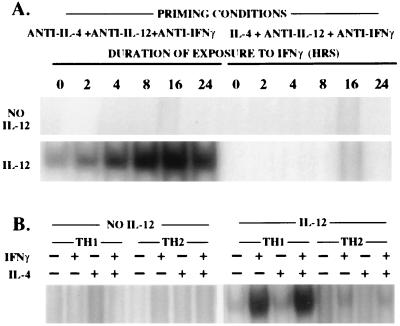
To examine the role of IFN-γ in priming TH1 cells for IFN-γ production, we stimulated naive TG CD4+ T cells in the presence of anti-IL-4, anti-IL-12, and anti-IFN-γ and compared those cells to TH1 cells primed in the presence of anti-IL-4 and IL-12. When challenged in the absence of IL-12, these cells made detectable and similar amounts of IFN-γ (Fig. 3). Addition of IL-12 to the challenge culture enhanced IFN-γ production by both cell populations approximately 3-fold. Interestingly, TH1 cells primed with anti-IL-12 and anti-IFN-γ showed a very weak induction of STAT-4 DNA-binding activity in response to IL-12, whereas TH1 cells primed in the presence of anti-IL-4 and IL-12 showed excellent IL-12 stimulation of STAT-4 DNA-binding activity (Fig. 3). This appeared somewhat at odds with the apparent similarity in sensitivity of the two cell populations to antigen challenge in the presence of IL-12. We reasoned that the IL-12-independent production of IFN-γ by the TH1 cells primed in the presence of anti-IFN-γ and anti-IL-12 might enhance IL-12 responsiveness in these cells. To test this, we prepared TH1 cells primed in the presence of anti-IL-4, anti-IL-12, and anti-IFN-γ, cultured them for 0–24 hr in IFN-γ, and then challenged them with IL-12. As few as 4 hr of culture with IFN-γ increased induction of STAT-4 DNA-binding activity in response to IL-12. TH2 cells primed in the absence of IFN-γ and IL-12 differ from TH1 cells prepared in the same way. Such TH2 cells acquire little or no ability to respond to IL-12 with STAT-4 DNA-binding activity in response to culture with IFN-γ for up to 24 hr (Fig. 5A). This difference does not appear to be due to the presence of IL-4 during the IFN-γ induction since the rapid acquisition of responsiveness by TH1 cells to IL-12 was not influenced by IL-4 (Fig. 5B). Furthermore, the addition of anti-IL-4 (to neutralize any endogenously produced IL-4) did not allow TH2 cells primed with anti-IFN-γ and anti-IL-12 to acquire responsiveness to IL-12. However, over longer culture periods with anti-IL-4 and IFN-γ, such TH2 cells do acquire IL-12 responsiveness, although it tends to be relatively weak (Fig. 4 and data not shown).
Figure 5.
IFN-γ causes rapid induction of IL-12 responsiveness in TH1 cells primed in the absence of IFN-γ. TG CD4+ T cells were primed with peptide, APC, IL-2, anti-IL-12, anti-IFN-γ, and either IL-4 (TH2) or anti-IL-4 (TH1). (A) Cells were incubated for a total of 24 hr. IFN-γ (20 units/ml) was added at varying times as indicated. (B) Cells were cultured for 24 hr with IFN-γ (+) or anti-IFN-γ (−) and with IL-4 (+) or anti-IL-4 (−). Cells were then challenged with or without IL-12, and the capacity of the extracts to bind Fcγ RI GAS oligonucleotide sequence was determined as a measure of STAT-4 activity.
DISCUSSION
Production of IFN-γ by primed T cells is highly dependent upon the cytokine environment. IL-12, IFN-γ, and IL-4 each play an important role. We summarize their roles as follows. IFN-γ induces responsiveness to IL-12 as judged by activation of STAT-4. It has been shown that, in the mouse, IFN-γ induces the β2-chain of the IL-12 receptor (Szabo, S. J., Dighe, A. S., Giebler, U. and Murphy, K. M., unpublished work), presumably explaining its induction of IL-12 responsiveness. IL-4 acts under certain circumstances to prevent the acquisition of IL-12 responsiveness, although it does not prevent the rapid induction of such responsiveness by IFN-γ in some primed TH1 cells. IL-12 activates STAT-4 (in IL-12-responsive cells) (25, 26) and plays a critical role in the induction of IFN-γ production. In contrast, once CD4+ T cells have been adequately primed, IL-4-producing capacity is not strikingly affected by the cytokine environment.
Our data reaffirm the important role of IL-4 in determining the capacity of naive T cells to develop into IL-4-producing cells. Recent work has demonstrated that the capacity of naive T cells to develop into IL-4 producers in response to stimulation by IL-4 is STAT-6-dependent (27, 28), suggesting that an immediate-early STAT-6-dependent gene is critical in the determination of the differentiation of the cell. Recently, c-maf has been shown to be a tissue-specific transcription factor that distinguishes TH2 from TH1 cells (29, 30). Whether c-maf is the factor that is directly induced by STAT-6 and begins the process of differentiation into a TH2 cell or whether it itself is induced as a result of the action of an earlier IL-4-dependent gene still remains to be established.
The induction of the capacity to produce IFN-γ is a very highly regulated process. Priming in the presence of IL-4 and the absence of IFN-γ results in cells that are unresponsive to IL-12 and that fail to promptly respond to IFN-γ with the acquisition of IL-12 responsiveness, as judged by activation of STAT-4 in response to IL-12, although they can develop such responsiveness over a longer period of culture with anti-IL-4 and IFN-γ. The presence of IL-12 during priming may enhance the acquisition of IL-12 responsiveness and IFN-γ-producing activity, possibly by causing the production of IFN-γ, which up-regulates IL-12 responsiveness, causing more IFN-γ production and further up-regulating IL-12 responsiveness.
Although the role of IFN-γ in controlling IL-12 responsiveness and IFN-γ-production is likely to be through induction of expression of IL-12 receptors, it is not clear how IL-4 blocks IFN-γ production. It does not block the rapid IFN-γ-induced acquisition of IL-12 responsiveness by cells that have been primed in the absence of IL-4, nor is it required for the failure of TH2 cells primed in the absence of IL-12 and IFN-γ to acquire such responsiveness. It is interesting that TH1 cells, primed with anti-IL-4 and anti-IFN-γ, are clearly responsive to IFN-γ, indicating that lack of responsiveness to IFN-γ is not a general feature of TH1 cells as has previously been proposed (31).
Although both TH1 and TH2 cells can produce IFN-γ, depending on the conditions of restimulation and challenge, there are interesting differences between them. TH1 cells (i.e., cells primed in the presence of anti-IL-4) can produce IFN-γ without the presence of IL-12 in either the priming or the challenge cultures, although IL-12 clearly augments IFN-γ production. In contrast, cells primed in the presence of IL-4 and IFN-γ produce substantial amounts of IFN-γ, but this production appears to completely depend on IL-12. Furthermore, priming in the presence of IL-4, and the absence of IFN-γ and IL-12, generates cells that acquire IFN-γ-producing capacity only if IL-4 is omitted and IFN-γ present in subsequent cultures. The failure of IL-4 to block acquisition of IFN-γ-producing capacity in the presence of IFN-γ in primary cultures, although it prevents TH2 cells, primed in the absence of IFN-γ, from acquiring IL-12 responsiveness in secondary cultures, may be related to the relatively slow pace at which IL-4 establishes a TH2 phenotype in primary responses (32).
Once cells have acquired the capacity to produce IL-4, they continue to do so largely independently of the cytokine environment, and, similarly, cells that have been vigorously primed in the absence of IL-4 fail to acquire IL-4-producing activity even if restimulated in the presence of IL-4. We conclude that IL-4 production by differentiated cells is largely cytokine-autonomous, whereas IFN-γ production is highly cytokine-regulated.
These results lead us to reexamine the qualities that characterize TH1 and TH2 cells. In most investigators’ views, the production of IL-4 by TH2 cells and the production of IFN-γ by TH1 cells have been defining properties. Our results forcefully argue that TH2 cells can develop the capacity to produce IFN-γ, often in amounts quite comparable to conventional TH1 cells. In contrast, vigorously primed TH1 cells fail to develop into IL-4 producers even if restimulated in the presence of IL-4. A more appropriate basis on which to distinguish primed CD4+ cells is that TH2 cells can produce IL-4, whereas TH1 cells cannot. IFN-γ-producing capacity is a property either cell type can express, depending upon its recent cytokine experience.
Acknowledgments
We thank Dr. Ken Murphy for discussions regarding the role of IFN-γ in inducing responsiveness to IL-12 and for providing us with a preprint of an unpublished manuscript. We acknowledge Cynthia Watson for providing the recombinant mouse IL-4 used in these studies, Dr. Calman Prussin for helpful suggestions on intracellular staining protocols, Pat Dearst for performing intravenous injections, and Jeff Nyswaner and the Animal Care Staff at the National Institute of Allergy and Infectious Diseases for excellent technical assistance. H.H. is a Fellow of the Leukemia Society of America. J.R. is a Fellow of the Cancer Research Institute.
ABBREVIATIONS
- IL
interleukin
- IFN-γ
interferon γ
- FITC
fluorescein isothiocyanate
- TG
transgenic
- APC
antigen-presenting cell(s)
References
- 1.Paul W E, Seder R A. Cell. 1994;76:241–251. doi: 10.1016/0092-8674(94)90332-8. [DOI] [PubMed] [Google Scholar]
- 2.Mosmann T R, Cherwinski H, Bond M W, Giedlin M A, Coffman R L. J Immunol. 1986;136:2348–2357. [PubMed] [Google Scholar]
- 3.Le Gros G, Ben-Sasson S Z, Seder R, Finkelman F D, Paul W E. J Exp Med. 1990;172:921–929. doi: 10.1084/jem.172.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swain S L, Weinberg A D, English M, Huston G. J Immunol. 1990;145:3796–3806. [PubMed] [Google Scholar]
- 5.Seder R A, Paul W E, Davis M M, Fazekas de St, Groth B. J Exp Med. 1992;176:1091–1098. doi: 10.1084/jem.176.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh C S, Heimberger A B, Gold J S, O’Garra A, Murphy K M. Proc Natl Acad Sci USA. 1992;89:6065–6069. doi: 10.1073/pnas.89.13.6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsieh C S, Macatonia S E, Tripp C S, Wolf S F, O’Garra A, Murphy K M. Science. 1993;260:547–549. doi: 10.1126/science.8097338. [DOI] [PubMed] [Google Scholar]
- 8.Seder R A, Gazzinelli R, Sher A, Paul W E. Proc Natl Acad Sci USA. 1993;90:10188–10192. doi: 10.1073/pnas.90.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitt E, Hoehn P, Huels C, Goedert S, Palm N, Rüde E, Germann T. Eur J Immunol. 1994;24:792–798. doi: 10.1002/eji.1830240403. [DOI] [PubMed] [Google Scholar]
- 10.Perez V L, Lederer J A, Lichtman A H, Abbas A K. Int Immunol. 1995;7:869–875. doi: 10.1093/intimm/7.5.869. [DOI] [PubMed] [Google Scholar]
- 11.Murphy E, Shibuya K, Hosken N, Openshaw P, Maino V, Davis K, Murphy K, O’Garra A. J Exp Med. 1996;183:901–913. doi: 10.1084/jem.183.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manetti R, Gerosa F, Giudizi M G, Biagiotti R, Parronchi P, Piccinni M P, Sampognaro S, Maggi E, Romagnani S, Trinchieri G. J Exp Med. 1994;179:1273–1283. doi: 10.1084/jem.179.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohara J, Paul W E. Nature (London) 1985;315:333–336. doi: 10.1038/315333a0. [DOI] [PubMed] [Google Scholar]
- 14.Cherwinski H M, Schumacher J H, Brown K D, Mosmann T R. J Exp Med. 1987;166:1229–1244. doi: 10.1084/jem.166.5.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coffman R L. Immunol Rev. 1982;69:5–23. doi: 10.1111/j.1600-065x.1983.tb00446.x. [DOI] [PubMed] [Google Scholar]
- 16.Oi V T, Jones P P, Goding J W, Herzenberg L A, Herzenberg L A. Curr Top Microbiol Immunol. 1978;81:115–120. doi: 10.1007/978-3-642-67448-8_18. [DOI] [PubMed] [Google Scholar]
- 17.Ledbetter J A, Herzenberg L A. Immunol Rev. 1979;47:63–90. doi: 10.1111/j.1600-065x.1979.tb00289.x. [DOI] [PubMed] [Google Scholar]
- 18.Unkeless J C. J Exp Med. 1979;150:580–596. doi: 10.1084/jem.150.3.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshimoto T, Bendelac A, Hu-Li J, Paul W E. Proc Natl Acad Sci USA. 1995;92:11931–11934. doi: 10.1073/pnas.92.25.11931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pearse R N, Feinman R, Shuai K, Darnell J J, Ravetch J V. Proc Natl Acad Sci USA. 1993;90:4314–4318. doi: 10.1073/pnas.90.9.4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elson L H, Nutman T B, Metcalfe D D, Prussin C. J Immunol. 1995;154:4294–4301. [PubMed] [Google Scholar]
- 22.Hu-Li J, Ohara J, Watson C, Tsang W, Paul W E. J Immunol. 1989;142:800–807. [PubMed] [Google Scholar]
- 23.Curry R C, Kiener P A, Spitalny G L. J Immunol Methods. 1987;104:137–142. doi: 10.1016/0022-1759(87)90497-2. [DOI] [PubMed] [Google Scholar]
- 24.Mosmann T R, Fong T A. J Immunol Methods. 1989;116:151–158. doi: 10.1016/0022-1759(89)90198-1. [DOI] [PubMed] [Google Scholar]
- 25.Jacobson N G, Szabo S J, Weber N R, Zhong Z, Schreiber R D, Darnell J J, Murphy K M. J Exp Med. 1995;181:1755–1762. doi: 10.1084/jem.181.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bacon C M, Petricoin E R, Ortaldo J R, Rees R C, Larner A C, Johnston J A, O’Shea J J. Proc Natl Acad Sci USA. 1995;92:7307–7311. doi: 10.1073/pnas.92.16.7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shimoda K, van Deursen J, Sangster M Y, Sarawar S R, Carson R T, Tripp R A, Chu C, Quelle F W, Nosaka T, Vignali D A, Doherty P C, Grosveld G, Paul W E, Ihle J N. Nature (London) 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 28.Kaplan M H, Schindler U, Smiley S T, Grusby M J. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 29.Ho I C, Hodge M R, Rooney J W, Glimcher L H. Cell. 1996;85:973–983. doi: 10.1016/s0092-8674(00)81299-4. [DOI] [PubMed] [Google Scholar]
- 30.Hodge M R, Chun H J, Rengarajan J, Alt A, Lueberson R, Glimcher L H. Science. 1996;274:1903–1905. doi: 10.1126/science.274.5294.1903. [DOI] [PubMed] [Google Scholar]
- 31.Pernis A, Gupta S, Gollob K J, Garfein E, Coffman R L, Schindler C, Rothman P. Science. 1995;269:245–247. doi: 10.1126/science.7618088. [DOI] [PubMed] [Google Scholar]
- 32.Seder R A, Le Gros G, Ben-Sasson S Z, Urban J J, Finkelman F D, Paul W E. Eur J Immunol. 1991;21:1241–1247. doi: 10.1002/eji.1830210522. [DOI] [PubMed] [Google Scholar]