Abstract
Estrogen receptor (ER) status is an important parameter in breast cancer management as ER-positive breast cancers have a better prognosis than ER-negative tumors. This difference comes essentially from the lower aggressiveness and invasiveness of ER-positive tumors. Here, we demonstrate, that IL-8 was clearly overexpressed in most ER-negative breast, ovary cell lines and breast tumor samples tested, whereas no significant IL-8 level could be detected in ER-positive breast or ovarian cell lines. We have also cloned human IL-8 from ER-negative MDA-MB-231 cells and we show that IL-8 produced by breast cancer cells is identical to monocyte-derived IL-8. Interestingly, the invasion potential of ER-negative breast cancer cells is associated at least in part with expression of interleukin-8 (IL-8), but not with IL-8 receptors levels. Moreover, IL-8 increases the invasiveness of ER-positive breast cancer cells by 2 fold, thus confirming the invasion-promoting role of IL-8. On the other hand, exogenous expression of estrogen receptors in ER-negative cells led to a decrease of IL-8 levels. In summary, our data show that IL-8 expression is negatively linked to ER-status of breast and ovarian cancer cells. We also support the idea that IL-8 expression is associated with a higher invasiveness potential of cancer cells in vitro, which suggests that IL-8 could be a novel marker of tumor aggressiveness.
Keywords: Amino Acid Sequence; Base Sequence; Breast Neoplasms; genetics; metabolism; pathology; Carcinogenicity Tests; Carcinoma, Ductal, Breast; genetics; metabolism; pathology; Cell Division; drug effects; genetics; Female; Gene Expression Regulation, Neoplastic; Humans; Interleukin-8; genetics; metabolism; pharmacology; Molecular Sequence Data; Neoplasm Invasiveness; Receptors, Estrogen; metabolism; Tumor Cells, Cultured
Introduction
Breast cancer is one of the most frequent malignant neoplasm occurring in women in developed countries (Pisani, 1992). Among the different prognostic factors, lack of estrogen receptor has been consistently associated with a poorer prognosis (Skoog et al., 1987). Two estrogen receptors (ERα and ERβ) have been identified (Green & Chambon, 1986; Mosselman et al., 1996). Most human breast cancers, at least initially, express ERα, and the presence of ERα is generally considered as an indication of hormone-dependence, even though only 60% of ERα-positive tumors will respond to adjuvant therapy with tamoxifen (McGuire, 1975). On the contrary, ERβ expression is low in most breast and ovarian cancers compared to normal tissues (Pujol et al., 1998; Roger et al., 2001). Due to the low levels of ERβ in breast and ovarian cancers, ER-positive status of samples of this study will refer to estrogen-responsive ERα-positive cells.
In the search of markers of cancer aggressiveness, interleukin-8 (IL-8) has been suspected to play an important role in breast cancer. Since metastasis represents the major remaining cause of mortality in human breast cancer, we sought to examine the possibility that increased metastatic potential (invasiveness) is associated with lack of ER and changes in IL-8 expression.
IL-8 is a 8,4 kDa protein belonging to the CXC subfamily of chemokines which is characterised by two essential cysteine residues, separated by a third intervening amino acid (Baggiolini et al., 1989; Matsushima & Oppenheim, 1989). There are two major forms of IL-8, that are the 72-amino acid monocyte-derived form, predominant in cultures of monocytes and macrophages, and the endothelial form which has five extra N-terminal amino acids, predominating in cultures of tissue cells such as endothelial cells and fibroblasts (Terui et al., 1998).
The double aims of the present study were to determine whether there are differences in IL-8 expression in human breast cancer cell lines with a particular attention to estrogen receptor levels, and to investigate the effect of ectopic expression of IL-8 on proliferation and invasion of breast carcinoma cell lines.
Results
IL-8 is considered as a potential metastatic factor in breast cancers (De Larco et al., 2001), however, no study has investigated the potential involvement of IL-8 expression in the aggressiveness of estrogen receptor (ER)-negative breast cancers. Therefore, we have conducted a study on a collection of breast cancer cell lines to determine whether IL-8 was correlated to ER-status and whether differences in interleukin-8 expression could reflect the invasiveness potential of ER-negative and ER-positive breast cancer cells.
We first checked IL-8 expression in two different model cell lines: MDA-MB-231 (ER-negative) and MCF-7 (ER-positive) cells. By performing a northern blot with an IL-8 probe, we observed a striking difference of IL-8 expression in MDA-MB-231 and MCF-7 cells. IL-8 was highly expressed in MDA-MB-231 cells and not detectable in MCF-7 cells (Fig. 1A). Moreover, E2 treatment did not affect IL-8 RNA levels in MCF-7 and MDA-MB-231 cells, but induced pS2 expression in MCF-7 cells, confirming the responsiveness of these cells (Fig. 1A). At the protein level, IL-8 was also restricted to MDA-MB-231 cells (Fig. 1B).
Fig. 1.
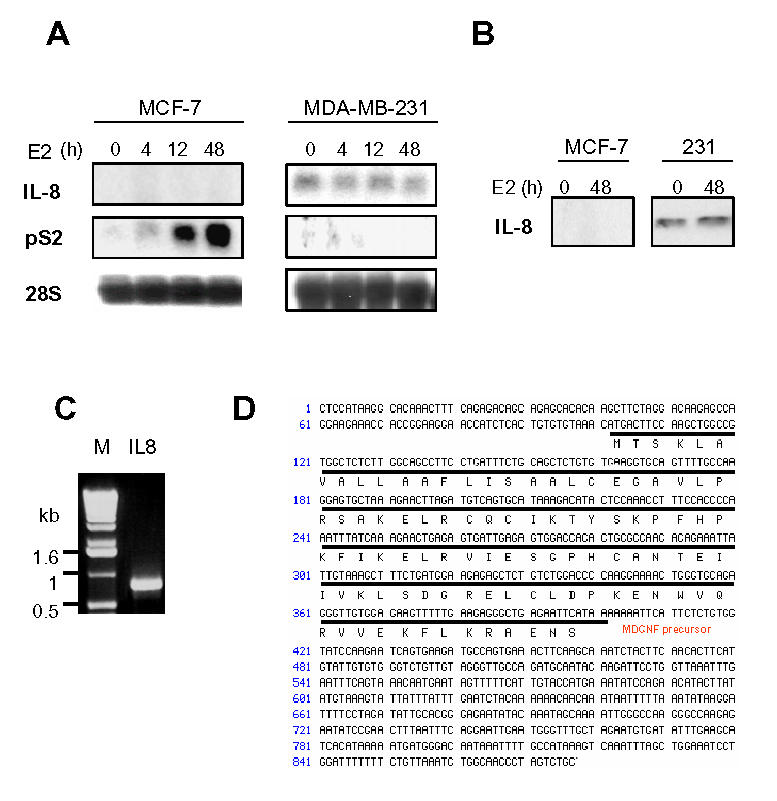
IL-8 is highly expressed in MDA-MB-231 cells. A. MDA-MB-231 or MCF-7 cells were treated with ethanol vehicle or E2 (10−9M) for 4, 12 or 48 h. RNA were used to for northern analysis using an IL-8 probe. B. IL-8 protein content was analyzed by western blot in MDA-MB-231 and MCF-7 cells treated with ethanol vehicle or E2 (10−9M) for 48h. C. IL-8 cDNA was amplified from a reverse transcribed reaction of MDA-MB-231 total RNA with IL-8 specific primers. The PCR product had an approximate size of 900 bp as compared to the 1kb marker (invitrogen) (M). D. After cloning, the PCR product was sequenced. The sequence is 100% homologous to the known IL-8 sequence.
To determine the relevance of our observations, it was essential to ensure that IL-8 produced by MDA-MB-231 cells was functional. So, we cloned IL-8 present from MDA-MB-231 cells by performing RT-PCR with specific primers of human IL-8 cDNA. We isolated a fragment of IL-8 composed of the first 877 bp of the cDNA and containing the complete open reading frame of the protein (Fig. 1C). The sequencing revealed that IL-8 produced by MDA-MB-231 cells corresponds to the 72 aa monocyte form of IL-8 with a classical 27 aa signal peptide, which is the most active IL-8 form (Fig. 1D).
We have then analyzed whether the overexpression of IL-8 in MDA-MB-231 cells was a general difference between ER-negative and ER-positive breast cancer cells. We selected three negative and three positive breast cancer cell lines to evaluate IL-8 expression by northern blot (Fig. 2). Interestingly, none of the ER-positive cell lines expressed detectable levels of IL-8 RNA, whereas two ER-negative breast cancer lines (MDA-MB-231 and MDA-MB-436) were expressing high levels of IL-8 and SK-BR3 cells did not show significant levels of IL-8 (Fig. 2). In addition, estradiol did not modify IL-8 expression in ER-negative or ER-positive cancer cells, whereas it strongly induced pS2 expression (a marker of estrogen responsiveness) in ER-positive cells.
Fig. 2.
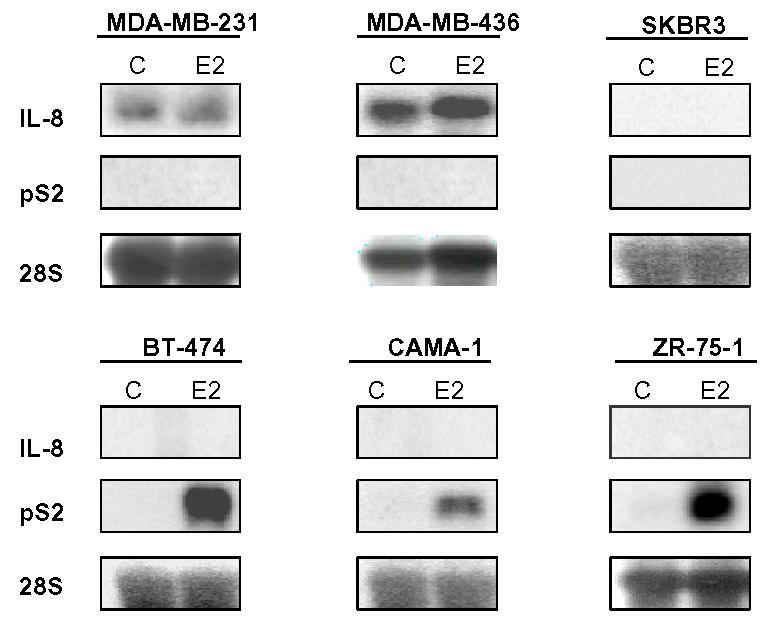
IL-8 RNA is differentially expressed in breast cancer cells. ER-negative (upper panel) or ER-positive (lower panel) breast cancer cells were cultured and treated or not with estradiol (E2: 10−9 M) for 48h. After extraction, 20 μg of total RNA from the different cells were used for Northern blot and hybridized with IL-8 or pS2 probes. Equal loading was checked with an RNA 28S probe. Data of a representative experiment are shown here.
As producing cells generally secrete IL-8, we then determined IL-8 secretion in a collection of breast and ovarian cancer cell lines, to assess whether this holds true for cancer cells in general. We analyzed IL-8 secretion by ER-negative and ER-positive breast cancer cells by ELISA (Fig. 3A, B). Interestingly, ER-negative breast cancer cells secreted variable amounts of IL-8 ranging from 0.1 to 336 ng IL-8/ml/48h/million cells. On the other hand, it was striking to observe that none of the ER-positive breast cancer cells tested secreted significant levels of IL-8. To evaluate whether this pattern of production was specific of breast cancer cells, we analyzed the IL-8 secretion of cervix carcinoma (Hela) and ovarian cancer cell lines (PEO14, BG1, PEO4) (Fig. 3C). There again, the same pattern was observed: no significant secretion of IL-8 by ER-positive cancer cells, and variable amounts of IL-8 secretion by ER-negative cancer cells. Very interestingly, the exogenous expression of ERα or ERβ in Hela cells led to decreased expression of IL-8 of about 50% in these cells (Fig. 3D) confirming that ER presence was negatively linked to IL-8 expression. This led us to determine whether IL-8 expression could be correlated to ERβ-status. We first checked the estrogen-responsiveness of our cell lines by transient transfection assay with an estrogen-responsive reporter construct (Fig. 3E). As expected, only ERα-positive cell lines (BT-474, CAMA-1, MCF-7, T47D, ZR-75-1, PEO4 and BG1) responded to estradiol. We then analysed the ERβ content of these cells by RT-PCR (Fig. 3F). We observed that ERβ expression was variable and low among the different cell lines, with the exception of MDA-MB-468 cells. Considering the unresponsiveness of these cells, it is likely that the expression of ERβ is either too low or leads to a mutated form of the protein. But the most important conclusion is that we could not see any correlation between ERβ and IL-8 levels.
Fig. 3.
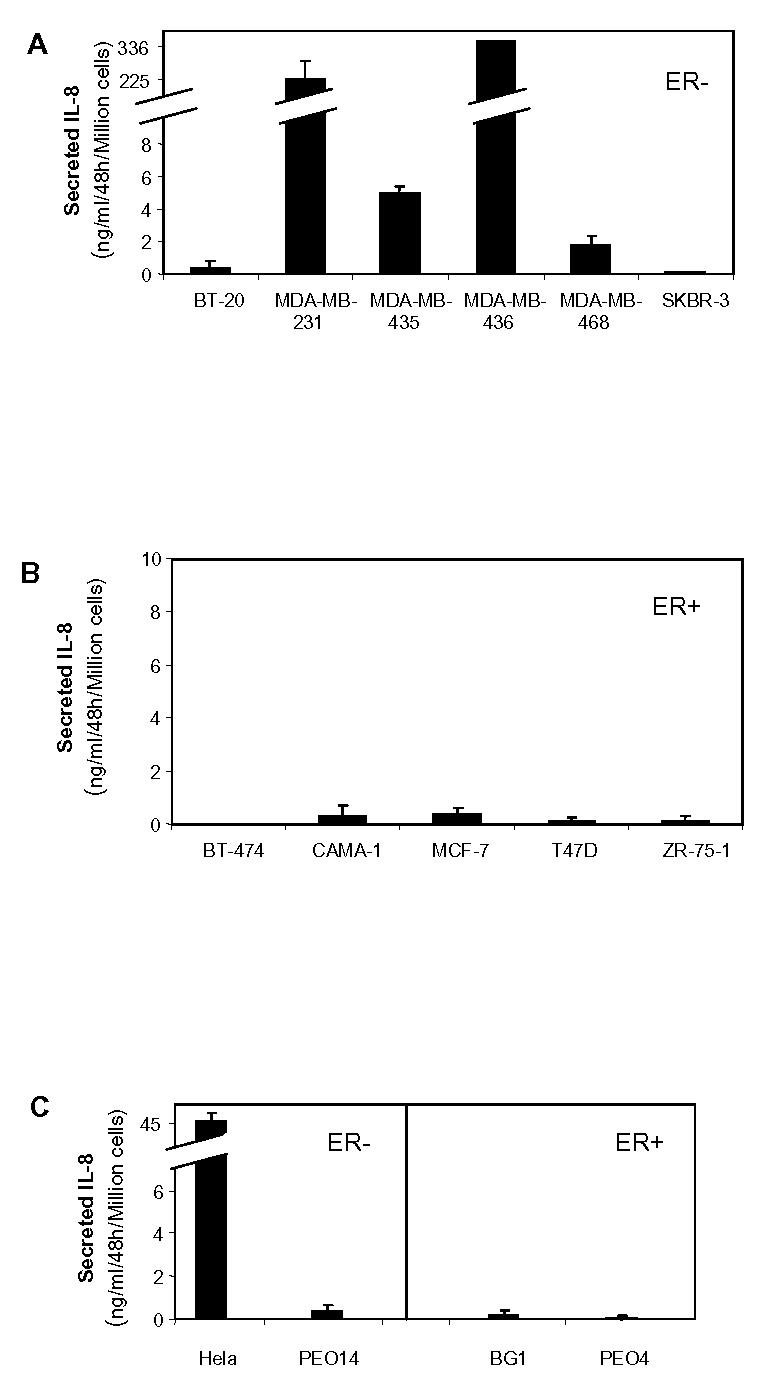
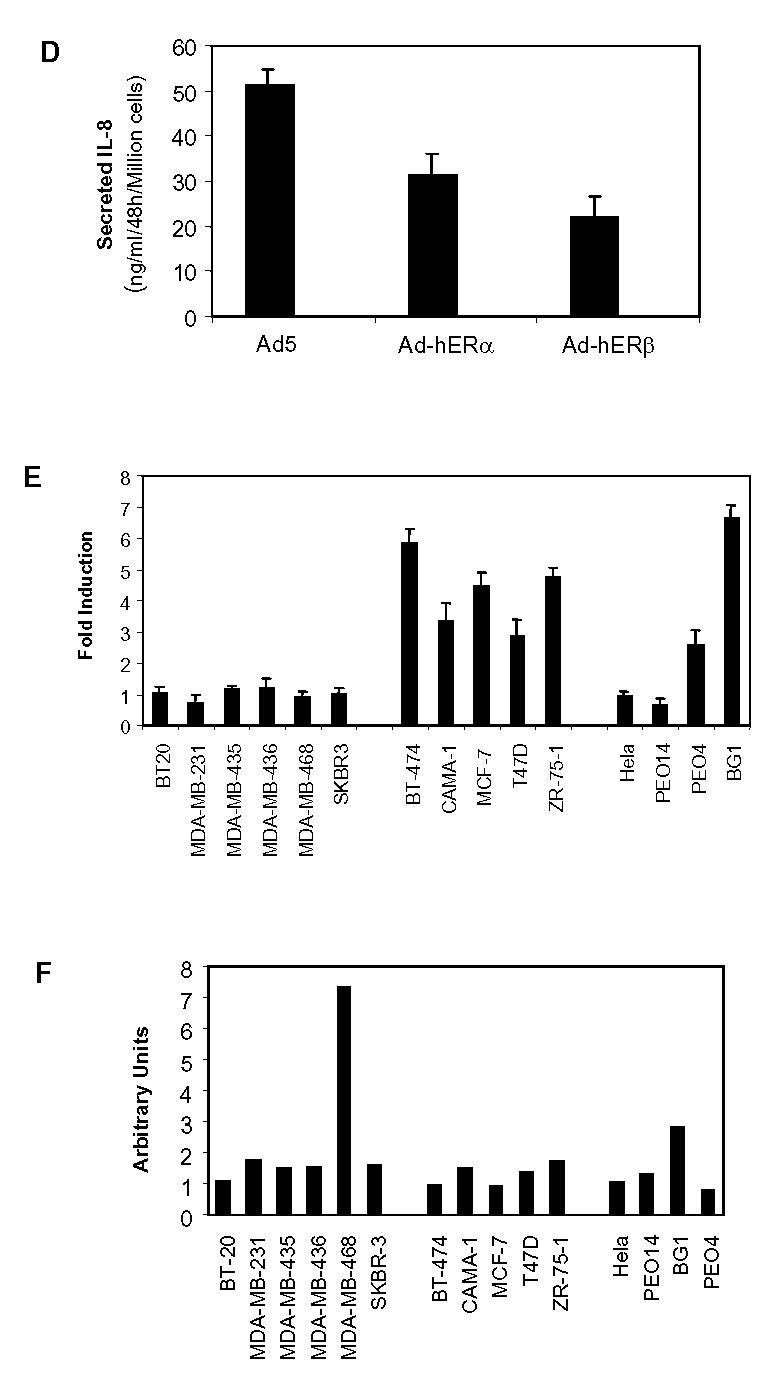
IL-8 is preferentially secreted by ER-negative cancer cells. A. ER-negative breast cancer cells. B. ER-positive breast cancer cells. C. ER-negative cervix carcinoma (Hela) or ovarian cells (PEO14) or ER-positive ovary cancer cells (BG1, PEO4). Cells were seeded in six-well plates in medium. After 48h of growth, 1.5 ml of medium was collected from each well to evaluate IL-8 levels by ELISA. Results are expressed as ng IL-8/ml/48h/million cells and represent the mean ± SD of three independent experiments. D. Hela cells were infected with Ad5, Ad-hERα or Ad-hERβ adenoviruses (see Materials and Methods) at MOI 100. After 48h of expression, the medium was collected and assayed for IL-8 content by Elisa. Results are expressed as ng IL-8/ml/48h/million cells and represent the mean ± SD of three independent experiments. E. Estrogen responsiveness of the different cell lines was determined by transient transfection with ERE2-TK-LUC reporter. Cells were grown for 24 h in the presence of control vehicle ethanol or 10−9 M E2. Results show fold induction of the reporter gene activity in the presence of E2 compared to the activity in the absence of ligand (n=3) after normalization for β-galactosidase activity. F. ERβ expression in the different cell lines was measured by semi-quantitative PCR as described in Materials and Methods section. The results are expressed in arbitrary units after normalization of ERβ expression to rS9 levels and represent the average of two experiments.
To extend our in vitro observations on differential expression of IL-8 in ER-negative versus ER-positive cancer cells, we have analyzed IL-8 expression in 14 breast tumor samples by RT-PCR. The ERα and ERβ-status was determined by RT-PCR (Fig. 4A) and by ELISA (Fig. 4B). First, we did not see by RT-PCR significant differences in terms of ERβ expression among the 14 samples, whereas the ERα-status was in total agreement with the Elisa data. Furthermore, at comparable amplification cycle number, ERβ expression was weaker than the one of ERα (see Materials and Methods). This confirms previous work from our laboratory, demonstrating that ERβ expression was weak in breast tumors and that ER-positive samples are indeed ERα-positive (Brouillet et al., 2001). We observed that most of ER-negative tumors (5/7) expressed detectable levels of IL-8 even though two of them did not contain significant levels of IL-8 (Fig. 4A, C). On the contrary, IL-8 was not detected in any of the ER-positive breast cancer tissues (Fig. 4A, C). When we evaluated the expression at the mRNA level of IL-8 between two groups of 7 ER-positive and ER-negative tumors, we found a significant difference between groups with a preferential expression of IL-8 only in the ER-negative tumors (p = 0.001). We also assessed PR levels in the samples, but no correlation could be seen between PR and IL-8 levels (Fig. 4B).
Fig. 4.
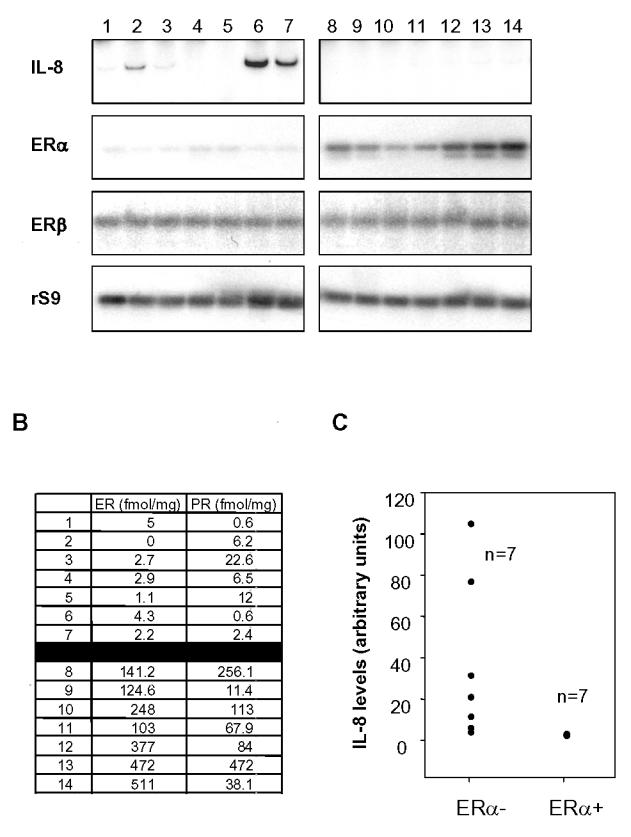
IL-8 is preferentially expressed in ER-negative breast tumors. A. ER-negative (left panel) or positive (right panel) breast tumors from 14 patients were analyzed by RT-PCR for ERα, ERβ and IL-8 expression. rS9 was used as a control of equal amplification between samples. 29 cycles were done for all markers, except ERβ (35 cycles). B. Estrogen receptor (ER) and progesterone receptor (PR) were quantified by ELISA in each tumor sample. Results are expressed in fmol/mg protein. C. Representation of IL-8 levels in the samples. IL-8 levels have been normalized to rS9 levels and the results are expressed in arbitrary units.
Our second objective was to evaluate whether IL-8 expression could be correlated to the higher invasiveness of ER-negative cancer cells. In vitro invasion test of the cells through a reconstituted basement membrane composed of matrigel was carried on in transwells. We observed that all ER-negative breast cancer cells except SKBR-3 (which do not express significant IL-8 levels) were highly invasive (Fig. 5A). On the contrary, ER-positive breast cancer cells displayed a low if any invasion ability (Fig. 5B). The same observation of a high invasion potential of ER-negative cancer cells was made for endometrium and ovary cancer cells, whereas ER-positive ovarian cancer cells displayed a low invasion ability, suggesting that it is a general phenomenon for cancer cells (Fig. 5C). When comparing IL-8 levels to their invasion potential between the different cell lines, the Spearman and Kendall test showed that the two variables were correlated (K = 0.5883 and Spa = 0.7293).
Fig. 5.
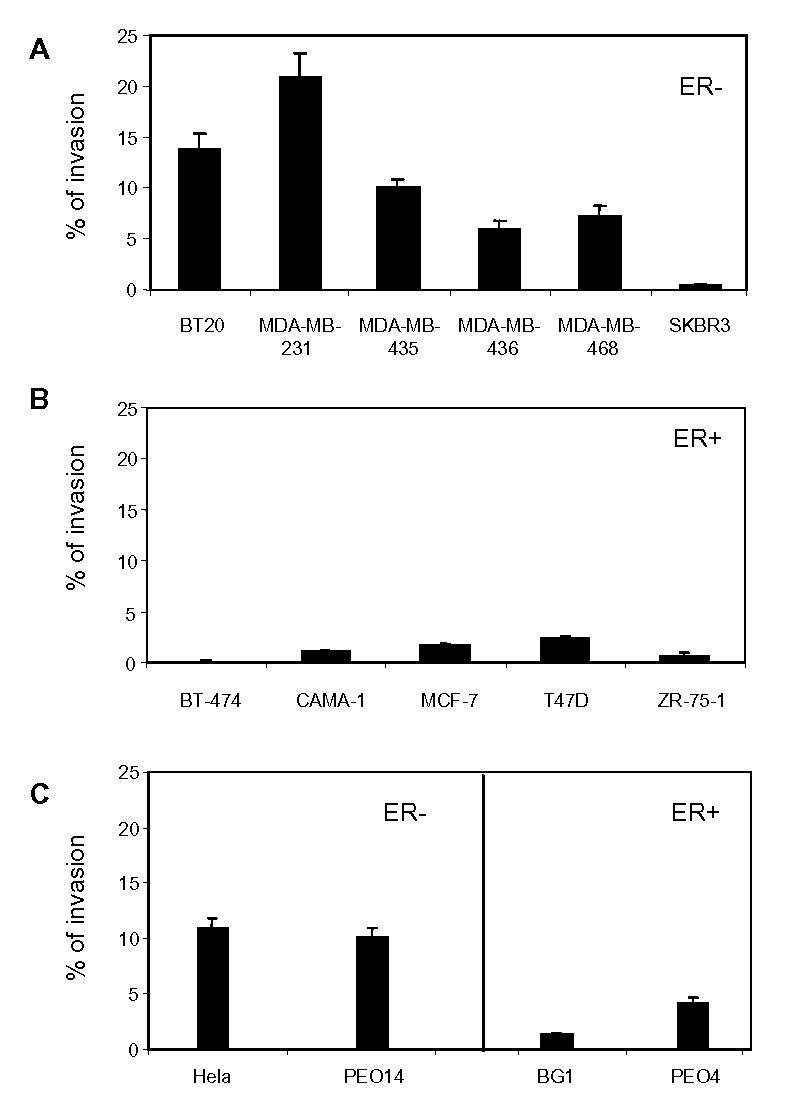
Invasiveness potential of cancer cells. Cells were plated on transwell or on control plates. After 24h of migration, the percentage of migrating cells was represented as the mean ± SD of three independent experiments. A. ER-negative breast cancer cells. B. ER-positive breast cancer cells. C. ER-negative cervix carcinoma or ovarian cells or ER-positive ovary cancer cells as in Fig. 3.
The next step was to analyze the mechanisms by which IL-8 affects invasion and proliferation of cancer cells. IL-8 is acting through specific receptors (CXCR1 and CXCR2). Thus, to estimate IL-8’s biological effects on cancer cells, it was important to check whether IL-8 receptors (CXCR1 and CXCR2) were present in the different cell lines. This was performed by RT-PCR to increase the sensitivity. CXCR1 receptors were expressed at very low levels (data not shown). On the contrary, CXCR2 was present in the different cell lines, with the exception of MCF-7 and MDA-MB-435 cells (Fig. 6A). No correlation could be seen between the ER-status and the levels of CXCR2, as most of ER-negative or positive cell lines express rather high levels of this receptor.
Fig. 6.
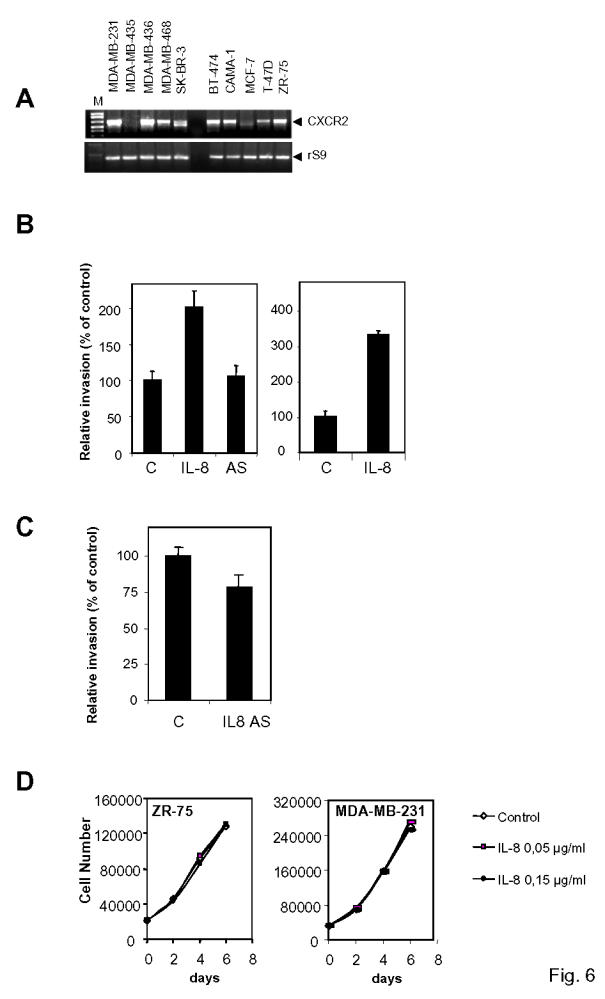
IL-8 expression is not correlated to IL-8 receptor levels and is promoting invasion but not proliferation. A. CXCR2 expression was monitored by RT-PCR with CXCR2 specific primers. A representative experiment is shown here. Equal reverse transcription and amplification was checked by performing a PCR with rS9 primers. B. Left panel. ZR-75-1 cells were transfected with the sense (IL-8) or anti-sense (IL-8 AS) version of IL-8 cDNA. 24h after transfection, cells were plated on transwell or on control plates. 24 h after the beginning of the invasion, migrating cells were evaluated by luciferase assay. The percentage of control migrating cells was set up to 100. Results are expressed as the percentage of control migrating cells and represent the mean ± SD of three experiments. Right panel. Same experiment but recombinant IL-8 (0.15 μg/ml) was added to the medium. C. Effect of anti-sense version of IL-8 on invasion in ER-negative MDA-MB-231 cells. D. IL-8 does not modify in vitro proliferation of breast cancer cells. The proliferation rate of ZR-75-1 and MDA-MB-231 cells was determined at days 2, 4, and 6 of treatment with vehicle, or IL-8 (0.05 μg/ml or 0.15 μg/ml). Results represent the mean of four determinations (SD < 15%).
To confirm IL-8 involvement in invasion, we performed in vitro invasion assays with ZR-75-1 cells, which had been transiently transfected with IL-8 sense or IL-8 anti-sense plasmids along with a luciferase reporter (Fig. 6B). This enabled us to follow only transfected cells. ZR-75-1 cells that had been transfected by IL-8 sense vector displayed a two fold enhanced invasion, whereas the anti-sense version had no effect, presumably due to the very low levels of IL-8 in these cells (Fig. 6B). On the other hand, addition of recombinant IL-8 to the medium also led to an increase of invasion of ZR-75-1 cells. These data confirm that IL-8 is playing a role in the invasion of cancer cells. Interestingly, transfection of IL-8 anti-sense construct in ER-negative MDA-MB-231 cells decreased by about 20% the invasiveness of the cells, confirming the pro-invasive role of IL-8 (Fig. 6C).
As IL-8 was affecting invasion, we were wondering if it could also modify the in vitro proliferation of breast cancer cells. We thus treated ZR-75-1 and MDA-MB-231 cells with two doses of IL-8 (Fig. 6D). IL-8 did not affect the proliferation of both type of cells, although these cells express IL-8 receptors (Fig. 6A), confirming that IL-8’s primary role was linked to invasion.
Discussion
Members of the α-chemokine family such as IL-8 have been considered as tumor cell products potentially involved in growth and progression of tumors. The goal of this study was to evaluate whether IL-8 was linked or not to ER-status and whether interleukin-8 could account for the higher aggressiveness of ER-negative breast cancer cells.
Cloning of IL-8 cDNA from MDA-MB-231 cells reveals that its sequence is totally identical to the 72 amino acids monocyte-derived form. This form is actually the most active among the different IL-8 isoforms and does not exhibit pro-apoptotic characteristics in leukemic cells compared to the 79 amino acids form (Terui et al., 1999).
More importantly, we show that IL-8 is overexpressed in ER-negative breast cancer cells. IL-8 expression is strongly different between ER-negative and ER-positive breast cancer cells. Not only IL-8 RNA, but also IL-8 secretion levels in the culture medium were extremely low in ER-positive breast cancer cell lines, whereas IL-8 expression was positive and variable in ER-negative breast cancer cells. The same difference of expression between ER-negative and ER-positive cells could be drawn from results obtained in ovarian cancer cells. ERβ expression was low and variable between the different cell lines, as we could only detect ERβ RNA by RT-PCR, but not ERβ protein (data not shown). There was no correlation between ERβ expression and IL-8 level, suggesting that ERα, the main estrogen receptor in ER-positive breast and ovarian cancer cells, was the receptor linked to IL-8 expression. Moreover, analyzis of tumor samples confirm this general pattern of high expression of IL-8 in ERα-negative cells and shows that there is a statistical difference between ERα-negative and ERα-positive breast tumors in terms of IL-8 expression.
IL-8 is produced by a wide panel of human cancer cells including colon (Brew et al., 1997), melanoma (Schadendorf et al., 1993), prostate (Greene et al., 1997), ovary (Lee et al., 2000; Xu & Fidler, 2000) or breast (Basolo et al., 1993; De Larco et al., 2001; Green et al., 1997; Youngs et al., 1997), but to our knowledge, this is the first demonstration of an inverse correlation between ERα-status and IL-8 expression. IL-8 is found not only in normal but also in cancerous breast (Basolo et al., 1993).
Some studies suggest that patients with recurrent prostate, breast or ovarian cancer exhibit higher levels of IL-8 in serum or peripherical blood leukocytes (Ivarsson et al., 1998; Merogi et al., 1997; Veltri et al., 1999) and in cancer tissues (Green et al., 1997). On the contrary, other studies show that IL-8 expression in breast tumors is identical between normal and cancer tissue (Speirs et al., 1996). Interestingly, IL-6 expression has been correlated with ER-status (Fontanini et al., 1999), which suggests that IL-6 and IL-8 are opposite markers.
Concerning IL-8 receptors, we observed that CXCR1 expression was extremely low in all lines tested, whereas most of the cells investigated showed a nice expression of CXCR2, without any correlation with ER-status. CXCR1 and CXCR2 which are encoded by two distinct genes (Sprenger et al., 1994a; Sprenger et al., 1994b) have been shown to be expressed in most cancer cells with no apparent correlation with the grade of the tumor (Miller et al., 1998; Muller et al., 2001). On the contrary, Müller et al. have recently shown that CXCR4 and CCR7, two other chemokine receptors which are involved in chemotactic and invasive responses, were highly expressed in breast cancer cells as well as malignant breast tumors and metastases (Muller et al., 2001). Our results suggest that IL-8 content but not CXCR1 or CXCR2 expression can be considered as markers of ER-negative breast cancers.
An important question was to estimate whether IL-8 expression could be correlated to the invasive potential of the ER-negative breast cancer cell lines. We demonstrated that most of the ER-negative breast or ovarian cancer cells exhibited a higher invasion rate compared to ER-positive cancer cells which migration was uniformly low, suggesting a general inverse correlation between the ER-status and the invasion potential of the cells. This correlation was not true for all cells, suggesting that other factors might modulate the invasive potential of each cell. This is in agreement with several reports describing that ER-positive breast cancer cells are generally less invasive than ER-negative breast cancer cells (Madsen & Briand, 1990; Sommers et al., 1994; Thompson et al., 1992). The lack of ER in human breast cancers is associated with a poorer prognosis (Skoog et al., 1987). In addition, exogenous expression of ER down-regulated IL-8 expression as shown in Hela cells. It is worthwhile to point out that we and others previously showed that introduction of ERα or ERβ in ER-negative breast cancer cells leads to a decreased invasiveness of the cells (Garcia et al., 1992; Lazennec et al., 2001). To date, no real explanation could be given to such a phenomenon. Our new data suggest that the decreased expression of IL-8 in cells transfected with ERα or ERβ could account at least in part to the reduced invasiveness of these cells.
Moreover, we also show that exogenous expression of IL-8 increases by two fold the invasion rate of ER-positive breast cancer cells, without affecting the in vitro proliferation rate of these cells, confirming the pro-invasive role of IL-8. When IL-8 is transfected in cancer cells, both tumor inhibition (Lee et al., 2000; Nakashima et al., 1996), and promotion (Inoue et al., 2000; Luca et al., 1997) have been observed in vivo depending on the cell type.
Our data show that IL-8 expression is negatively correlated to ER-status and is expressed preferentially in invasive cancer cells, which suggests a potential correlation between IL-8 expression and tumor progression/invasiveness. However, available studies remain preliminary to conclude on this question. Indeed, in xenograft mouse models, this issue remains controversial. Most of the studies show that IL-8 expression is correlated with tumorigenesis of prostate (Greene et al., 1997), ovary (Yoneda et al., 1998) and breast (De Larco et al., 2001) cancers. Other reports suggest that a high expression of IL-8 inhibits tumor progression of ovarian cancers (Lee et al., 2000) and has negligible effect on prostate cancer (Balbay et al., 1999) development in nude mice. One hypothesis for IL-8 reduction of ovarian tumor progression in nude mice could be an increased neutrophil infiltration of the cancer tissues, leading to the release of proteases or apoptotic agents able to kill cancer cells. This hypothesis could account for the discrepancies between these studies.
Whether IL-8 expression is correlated with worse prognosis in humans needs to be evaluated in breast cancer in patients. But, to date, no extensive study of large human breast tumor collection has been done and it appears that there is no clear correlation between the IL-8 expression and metastatic potential of tumor cells representing different clinical stages or with tumor prognosis.
Investigation of IL-8 involvement in cell proliferation, angiogenesis, migration or infiltration of host cells appears complex and will require combined consideration of data obtained from cell cultures, animal models and vast clinical studies.
Materials and Methods
Cell Culture
Cells were maintained in media recommended by ATCC supplemented with 10% fetal calf serum (FCS) and gentamycin. For transfection assays, cells were seeded on 6-well plates. To wean the cells off steroids, they were cultured in phenol red-free DMEM/F12 supplemented with 10% CDFCS (charcoal dextran-treated FCS) for 4 days.
Plasmids
CMV-LUC reporter (Promega, Madison, WI) was used as an internal control for invasion assays and corresponds to the luciferase gene under the control of the cytomegalovirus (CMV) promoter. pShuttle-IL-8 sense and pShuttle-IL-8 AS (anti-sense) contain the sequences 1-877 of human IL-8 cDNA cloned into pShuttle vector (Clontech, Palo Alto, CA). The luciferase reporter plasmid ERE2-TK-LUC used to monitor estrogen-responsiveness of our cells contains two copies of the consensus estrogen-responsive element (ERE) cloned upstream of the minimal herpes simplex virus thymidine kinase promoter. A CMV-GAL reporter was used as an internal control and corresponds to the β-galactosidase gene under the control of the cytomegalovirus (CMV) promoter.
Transient Transfection
3.105 cells were plated in 6-well plates in phenol red-free DMEM-F12 supplemented with 10% CDFCS 24 h before transfection. Transfections were performed by lipofection (lipofectamine, Invitrogen, Groningen, The Netherlands) using 2 μg of pShuttle, pShuttle-IL-8 sense or pShuttle-IL-8 AS, along with 2 μg of the internal reference reporter plasmid (CMV-LUC) per well. 36h after lipofection, cells were harvested and assayed for luciferase activity with Renilla Luciferase assay system (Promega, Madison, WI) on a LKB luminometer. For the determination of estrogen-responsiveness of the cells, 2 μg of ERE2-TK-LUC and 0.8 μg of internal control CMV-GAL per well were cotransfected by lipofection. β-galactosidase and luciferase were determined as previously described (Lazennec et al., 1997).
RNA Extraction and Reverse Transcriptase PCR
Total RNA was isolated with TRIzol reagent (Invitrogen) as described by the manufacturer. Reverse transcription was performed using random primers and Superscript II enzyme (Invitrogen). The PCR was performed with Platinium Taq polymerase (Invitrogen) and 1:40 of reverse transcription reaction. Cycles of 30 s at 94 °C, 1 min at 60 °C, and 1 min 30s at 72 °C were done 29 times (ERα, rS9, IL-8, CXCR2) or 35 times (ERβ). A tenth of each PCR was electrophoresed on agarose gel. For Northern blot analysis, 20 μg total RNA were electrophoresed and then hybridized with the different probes. Primers (s: sense, as: anti-sense) used were:
| CXCR2s: | TTGGCTCTTCTTCAGGGCACACTT |
| CXCR2as: | CAGGGGCACAATCTCGGCTCAC |
| ERα s: | AAAAGACCGAAGAGGAGGGAGAAT |
| ERα as | ATCCGGAACCGAGATGATGTAG |
| ERβs: | GGTCCATCGCCAGTTATCACATC |
| ERβas: | CAGCTCTTGCGCCGGTTTTTATC |
| IL8s: | AGGTGCAGTTTTGCCAAGGAG |
| IL8as: | GCAGACTAGGGTTGCCAGATTTA |
| RS9s: | CAGGCGCAGACGGTGGAAGC |
| RS9as: | CGCGAGCGTGGTGGATGGAC |
Northern Blot Analysis
Equal amounts of RNA (20 μg per lane) were loaded on a 1% agarose-formaldehyde gel. RNA was then transferred to nylon membranes. Membranes were then hybridized with the different probes at 65°C in hybridization solution (1 mM EDTA, 7% SDS, 0.5 M Na2HPO4, pH 7.2). A probe for human 28S was used to confirm equal loading of RNA in all the wells.
Whole-cell extract preparation and Western blot
Cells extracts were prepared as described previously (Lazennec et al., 2001). 30 μg whole-cell extract proteins were subjected to SDS-PAGE followed by electrotransfer onto a nitrocellulose membrane. The blot was probed with anti-IL-8 antibody (MAB-208, R&D Systems, Minneapolis, MN) (1:1000) and then incubated with goat anti-mouse Gig horseradish peroxidase conjugated antibody (1 μg/ml). An ECL kit (Amersham Pharmacia Biotech, Arlington, IL) was used for detection.
IL-8 ELISA
IL-8 concentration in culture supernatants was determined by ELISA as recommended by the manufacturer. Briefly, samples were added to 96-well microtiter plates, which were coated with monoclonal anti-IL-8 antibody (MAB-208, 4 μg/ml, R&D Systems, Minneapolis, MN). After 2h, the wells were washed three times and biotinylated anti-IL-8 antibody (BAF-208, 20 ng/ml, R&D Systems) was added. After 2 h of incubation, the plates were washed three times, and streptavidin-HRP conjugate (RPN1231, Amersham-Pharmacia, Buckinghamshire, UK) supplied, and the plates incubated for 20 min. Plates were washed again and chromogen substrate (Sigma Fast OPD, Sigma, St Louis, MO) added. The plates were read at 450 nm.
Recombinant adenovirus construction, propagation and infection
The adenoviruses used in this study have been described previously (Lazennec et al., 2001). Briefly, the complete coding sequence of wild-type human ER (hERα) or hERβ cDNAs were subcloned in BamHI site of the pACsk12CMV5 shuttle vector. To obtain recombinant viruses, permissive HEK-293 cells (human embryonic kidney cells) were cotransfected with pACsk12CMV5 or the recombinant pACsk12CMV5-hER plasmid and with pJM17, which contains the remainder of the adenoviral genome. In vivo recombination of the plasmids generates infectious viral particles non-recombinant adenovirus Ad5 or recombinant adenovirus Ad-hERα and Ad-hERβ. Hela cells were infected overnight at a multiplicity of infection (MOI) of 100 with the different adenoviruses in DMEM/F12 10% CDFCS. The next day, the medium was changed and the cells were let to express IL-8 for 48h before collecting the medium.
Invasion through Matrigel, Renilla Luciferase Assay and MTT assay
Cells were grown overnight in 6-well culture plates in DMEM/F12 supplemented with 10% FCS. 2 μg of pShuttle IL-8 sense or pShuttle IL-8 AS, or 2 μg of control vector pShuttle, were co-transfected with 2 μg of CMV-LUC vector as mentioned above. Invasion assay as previously described using 12-well polycarbonate filter (12 μm pore size) Transwells (Corning-Costar, Corning, NY) (Platet & Garcia, 1998). After 24 h of migration, cells that had migrated to the lower side of the filter and cells present in the control plates were collected and luciferase activity was measured as described above. The percentage of Matrigel-invading cells was represented by the ratio of luciferase activity of invasive cells to luciferase activity of total cells.
The same type of experiments was also performed without transfection to determine the invasion potential of each cell line as described earlier (Lazennec et al., 2001).
Cell Proliferation Studies
Cells were maintained for 4 days in 10% CDFCS DMEM/F12 and then seeded at 20000 cells/well in 24-well dishes in 10% CDFCS DMEM/F12, complemented or not with recombinant human IL-8 (R&D Systems, Minneapolis, MN). After 2, 4, or 6 days, the total cell DNA was quantified by DABA (DiAmino Benzoic Acid) as described earlier (Lazennec et al., 2001).
Tumor material and tumor sample processing
The study included 14 primary breast invasive ductal carcinoma sent to the laboratory for routine analysis of ER and PR status. Frozen biopsy samples were ground to a fine powder in liquid nitrogen and subsequently homogenized in a 10mM Tris pH 7.4, 1.5 mM EDTA, 4.2 mM Thioglycerol, 5 mM Molybdic acid, using a Dounce homogenizer. The homogenate were spun at 100 000 g for 1 h at 4°C yielding 100 000g supernatants immediately used for ER and PR evaluation using the ABBOTT ER EIA Monoclonal Kit and the ABBOTT PR Monoclonal kit (ABBOTT, Rungis, France).
Statistical analysis
Statistical analysis. To compare IL-8 secretion levels or invasion potential between the different populations of cell lines, the Kruskal-Wallis one-way analysis of variance by ranks was used since the variable analysed was not Normally distributed, the groups being independent. To look for a correlation between IL-8 secretion levels and invasion potential of the cell lines, we have estimated the Kendall and Spearman rank correlation coefficients and have then tested them against the null value. The significance level of the P value was set at 0.05 for all tests.
Acknowledgments
We thank Dr. T. Maudelonde for critical reading of the manuscript and Pr. J.P. Daures for statistical analysis. We thank the Vector Core of the University Hospital of Nantes supported by the Association Française contre les Myopathies (AFM) for the production of Adenoviruses. This work was supported by grants from ARC (Association pour la Recherche contre le Cancer, Grant No. 4302), la Ligue Nationale contre le Cancer (Comite du Gard), INSERM, and CNRS.
BIBLIOGRAPHY
- Baggiolini M, Walz A, Kunkel SL. J Clin Invest. 1989;84:1045–9. doi: 10.1172/JCI114265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbay MD, Pettaway CA, Kuniyasu H, Inoue K, Ramirez E, Li E, Fidler IJ, Dinney CP. Clin Cancer Res. 1999;5:783–9. [PubMed] [Google Scholar]
- Basolo F, Conaldi PG, Fiore L, Calvo S, Toniolo A. Int J Cancer. 1993;55:926–30. doi: 10.1002/ijc.2910550609. [DOI] [PubMed] [Google Scholar]
- Brew R, Erikson JS, West DC, Flanagan BF, Christmas SE. Biochem Soc Trans. 1997;25:264S. doi: 10.1042/bst025264s. [DOI] [PubMed] [Google Scholar]
- Brouillet JP, Dujardin MA, Chalbos D, Rey JM, Grenier J, Lamy PJ, Maudelonde T, Pujol P. Int J Cancer. 2001;95:205–8. doi: 10.1002/1097-0215(20010720)95:4<205::aid-ijc1035>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- De Larco JE, Wuertz BR, Rosner KA, Erickson SA, Gamache DE, Manivel JC, Furcht LT. Am J Pathol. 2001;158:639–46. doi: 10.1016/S0002-9440(10)64005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanini G, Campani D, Roncella M, Cecchetti D, Calvo S, Toniolo A, Basolo F. Br J Cancer. 1999;80:579–84. doi: 10.1038/sj.bjc.6690394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia M, Derocq D, Freiss G, Rochefort H. Proc Natl Acad Sci U S A. 1992;89:11538–42. doi: 10.1073/pnas.89.23.11538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green AR, Green VL, White MC, Speirs V. Int J Cancer. 1997;72:937–41. doi: 10.1002/(sici)1097-0215(19970917)72:6<937::aid-ijc3>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Green S, Chambon P. Nature. 1986;324:615–7. doi: 10.1038/324615a0. [DOI] [PubMed] [Google Scholar]
- Greene GF, Kitadai Y, Pettaway CA, von Eschenbach AC, Bucana CD, Fidler IJ. Am J Pathol. 1997;150:1571–82. [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Slaton JW, Eve BY, Kim SJ, Perrotte P, Balbay MD, Yano S, Bar-Eli M, Radinsky R, Pettaway CA, Dinney CP. Clin Cancer Res. 2000;6:2104–19. [PubMed] [Google Scholar]
- Ivarsson K, Runesson E, Sundfeldt K, Haeger M, Hedin L, Janson PO, Brannstrom M. Gynecol Oncol. 1998;71:420–3. doi: 10.1006/gyno.1998.5198. [DOI] [PubMed] [Google Scholar]
- Lazennec G, Bresson D, Lucas A, Chauveau C, Vignon F. Endocrinology. 2001;142:4120–30. doi: 10.1210/endo.142.9.8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazennec G, Kern L, Valotaire Y, Salbert G. Mol Cell Biol. 1997;17:5053–66. doi: 10.1128/mcb.17.9.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LF, Hellendall RP, Wang Y, Haskill JS, Mukaida N, Matsushima K, Ting JP. J Immunol. 2000;164:2769–75. doi: 10.4049/jimmunol.164.5.2769. [DOI] [PubMed] [Google Scholar]
- Luca M, Huang S, Gershenwald JE, Singh RK, Reich R, Bar-Eli M. Am J Pathol. 1997;151:1105–13. [PMC free article] [PubMed] [Google Scholar]
- Madsen MW, Briand P. Eur J Cancer. 1990;26:793–7. doi: 10.1016/0277-5379(90)90154-l. [DOI] [PubMed] [Google Scholar]
- Matsushima K, Oppenheim JJ. Cytokine. 1989;1:2–13. doi: 10.1016/1043-4666(89)91043-0. [DOI] [PubMed] [Google Scholar]
- McGuire WL. Cancer. 1975;36:638–44. doi: 10.1002/1097-0142(197508)36:2+<638::aid-cncr2820360805>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- Merogi AJ, Marrogi AJ, Ramesh R, Robinson WR, Fermin CD, Freeman SM. Hum Pathol. 1997;28:321–31. doi: 10.1016/s0046-8177(97)90131-3. [DOI] [PubMed] [Google Scholar]
- Miller LJ, Kurtzman SH, Wang Y, Anderson KH, Lindquist RR, Kreutzer DL. Anticancer Res. 1998;18:77–81. [PubMed] [Google Scholar]
- Mosselman S, Polman J, Dijkema R. FEBS Lett. 1996;392:49–53. doi: 10.1016/0014-5793(96)00782-x. [DOI] [PubMed] [Google Scholar]
- Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, Barrera JL, Mohar A, Verastegui E, Zlotnik A. Nature. 2001;410:50–6. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- Nakashima E, Oya A, Kubota Y, Kanada N, Matsushita R, Takeda K, Ichimura F, Kuno K, Mukaida N, Hirose K, Nakanishi I, Ujiie T, Matsushima K. Pharm Res. 1996;13:1896–901. doi: 10.1023/a:1016057830271. [DOI] [PubMed] [Google Scholar]
- Pisani P. J Environ Pathol Toxicol Oncol. 1992;11:313–6. [PubMed] [Google Scholar]
- Platet N, Garcia M. Invasion Metastasis. 1998;18:198–208. doi: 10.1159/000024513. [DOI] [PubMed] [Google Scholar]
- Pujol P, Rey JM, Nirde P, Roger P, Gastaldi M, Laffargue F, Rochefort H, Maudelonde T. Cancer Res. 1998;58:5367–5373. [PubMed] [Google Scholar]
- Roger P, Sahla ME, Makela S, Gustafsson JA, Baldet P, Rochefort H. Cancer Res. 2001;61:2537–2541. [PubMed] [Google Scholar]
- Schadendorf D, Moller A, Algermissen B, Worm M, Sticherling M, Czarnetzki BM. J Immunol. 1993;151:2667–75. [PubMed] [Google Scholar]
- Skoog L, Humla S, Axelsson M, Frost M, Norman A, Nordenskjold B, Wallgren A. Acta Oncol. 1987;26:95–100. doi: 10.3109/02841868709091747. [DOI] [PubMed] [Google Scholar]
- Sommers CL, Byers SW, Thompson EW, Torri JA, Gelmann EP. Breast Cancer Res Treat. 1994;31:325–35. doi: 10.1007/BF00666165. [DOI] [PubMed] [Google Scholar]
- Speirs V, Green AR, White MC. Int J Cancer. 1996;66:551–6. doi: 10.1002/(SICI)1097-0215(19960516)66:4<551::AID-IJC21>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Sprenger H, Lloyd AR, Lautens LL, Bonner TI, Kelvin DJ. J Biol Chem. 1994a;269:11065–72. [PubMed] [Google Scholar]
- Sprenger H, Lloyd AR, Meyer RG, Johnston JA, Kelvin DJ. J Immunol. 1994b;153:2524–32. [PubMed] [Google Scholar]
- Terui Y, Ikeda M, Tomizuka H, Kasahara T, Ohtsuki T, Uwai M, Mori M, Itoh T, Tanaka M, Yamada M, Shimamura S, Ishizaka Y, Ikeda K, Ozawa K, Miura Y, Hatake K. Blood. 1998;92:2672–80. [PubMed] [Google Scholar]
- Terui Y, Tomizuka H, Mishima Y, Ikeda M, Kasahara T, Uwai M, Mori M, Itoh T, Tanaka M, Yamada M, Shimamura S, Ishizaka Y, Ozawa K, Hatake K. Cancer Res. 1999;59:5651–5. [PubMed] [Google Scholar]
- Thompson EW, Paik S, Brunner N, Sommers CL, Zugmaier G, Clarke R, Shima TB, Torri J, Donahue S, Lippman ME, et al. J Cell Physiol. 1992;150:534–44. doi: 10.1002/jcp.1041500314. [DOI] [PubMed] [Google Scholar]
- Veltri RW, Miller MC, Zhao G, Ng A, Marley GM, Wright GL, Jr, Vessella RL, Ralph D. Urology. 1999;53:139–47. doi: 10.1016/s0090-4295(98)00455-5. [DOI] [PubMed] [Google Scholar]
- Xu L, Fidler IJ. Oncol Res. 2000;12:97–106. doi: 10.3727/096504001108747567. [DOI] [PubMed] [Google Scholar]
- Yoneda J, Kuniyasu H, Crispens MA, Price JE, Bucana CD, Fidler IJ. J Natl Cancer Inst. 1998;90:447–54. doi: 10.1093/jnci/90.6.447. [DOI] [PubMed] [Google Scholar]
- Youngs SJ, Ali SA, Taub DD, Rees RC. Int J Cancer. 1997;71:257–66. doi: 10.1002/(sici)1097-0215(19970410)71:2<257::aid-ijc22>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]


