Abstract
Defects in DNA replication fidelity lead to genomic instability. Gross chromosomal rearrangement (GCR), a type of genomic instability, is highly enhanced by various initial mutations affecting DNA replication. Frequent observations of GCRs in many cancers strongly argue the importance to maintain high fidelity of DNA replication to suppress carcinogenesis. Recent genome wide screens in Saccharomyces cerevisiae identified a new GCR suppressor gene, ELG1, enhanced level of genome instability gene 1. Its physical interaction with proliferating cell nuclear antigen (PCNA) and complex formation with Rfc2–5p proteins suggest that Elg1 functions to load/unload PCNA onto DNA during a certain DNA metabolism. High level of DNA damage accumulation and enhanced phenotypes with mutations in genes involved in cell cycle checkpoints, homologous recombination (HR), or chromatin assembly in the elg1 strain suggest that Elg1p-Rfc2–5p functions in a fundamental DNA metabolism to suppress genomic instability.
Keywords: Genomic instability, DNA replication, ELG1, RFC, PCNA
DNA is constantly challenged by intracellular as well as environmental stress. Inaccurate repair of DNA damage by such stress can result in genomic instability that can manifest as gross chromosomal rearrangements (GCRs). GCRs include translocations, deletions, inversions, amplifications, chromosome end-to-end fusions, and aneuploidy [1]. These genetic malformations often lead to cell death or carcinogenesis. Frequent observations of genomic instability in tumor cells support the argument for the importance of proper DNA repair to suppress carcinogenesis.
Multiple mutations in cancer normally exceed the expected number of mutations that cells are able to obtain with normal mutation rates. In order to generate multiple mutations, cells must develop specific initial mutations that can facilitate further mutagenesis [2]. These initial mutations, known as mutator mutations, have been suggested to facilitate multiple mutations in many oncogenes and tumor suppressor genes during carcinogenesis. Mutator mutations were first proposed from cases of the hereditary nonpolyposis colorectal cancer (HNPCC) and sporadic cancers where the mismatch-repair defects underlie cancer susceptibility [3]. Inherited cancer susceptibility syndromes including Ataxia Telangiectasia (AT), Nijmegen Breakage Syndrome (NBS), Bloom’s Syndrome (BS), Ataxia Telangiectasia-Like Disorder (ATLD), and familial breast cancer predisposition with a mutation in the BRCA1 or BRCA2 genes show increased numbers of chromosomal breaks and other GCRs suggesting that similar to HNPCC, certain mutator mutation could facilitate GCRs and carcinogenesis [1; 4]. Recent discoveries of multiple pathways involved in suppressing GCRs using yeast Saccharomyces cerevisiae and their evolutionary conserved mechanisms across species start to elucidate detail mechanisms that could lead to uncover clues for new therapeutic approach against tumorigenesis.
Genome stability is ensured by complex mechanisms that coordinate DNA replication, DNA repair, and chromosome segregation. GCRs are highly induced by various initial mutations affecting DNA replication. During DNA replication, DNA replication forks stall at damaged DNA or at naturally occurring sequences such as replication barriers. If not properly repaired, stalled DNA replication forks collapse and produce double strand breaks (DSBs) that are intermediates of GCRs. Extensive genetic interaction studies using various yeast GCR assays have identified eight pathways that suppress and six pathways that promote GCR in yeast (Table 1).
Table 1. Pathways that suppress or promote GCR formation in Saccharomyces cerevisiae.
There are more proteins participating pathways listed here. Only proteins tested by GCR assay are listed. The “/” between Rad51 and Rad59 or Rad54 and Rhp54 means that both proteins are redundantly required for break induced replication. Detailed studies to identify pathways listed here are discussed in other reviews [1; 24].
| Pathways |
|
|---|---|
| GCR suppression | Proteins in pathways |
| 1. S phase checkpoints | |
| a. Replication checkpoint | Rfc5, Pol2, Dpb11, Drc1, Mec1, Ddc2, Dun1 |
| b. Intra-S checkpoint | Rad9, Rad17, Mec3, Ddc1, Mec1, Rad53, Dun1, yKu70, yKu80 |
| c. Intra-S checkpoint | Sgs1, Chk1, Mre11, Rad50, Xrs2, Tel1, yKu70, yKu80 |
| 2. Break induced replication | Rad52, Mre11, Rad50, Xrs2, Rad51/Rad59, Rad54/Rhp54 |
| 3. Telomerase inhibition | Pif1 |
| 4. Chromatin assembly | Cac1, Asf1 |
| 5. Chromosome fusion suppression | Tlc1, Est1, Est2, Est3, Cdc13, Mec1, Tel1 |
| 6. Mismatch repair pathway | Msh2, Sgs1 |
| 7. Detoxification of oxygen species | Tsa1, Alo1 |
| 8. Template switching | Rad18, Rad5, PCNA |
| GCR promoting | |
| 1. de novo telomere addition | Est1, Est2, Est3, Cdc13, Tlc1, Tel1, yKu70, yKu80 |
| 2. Non-homologous end joining (NHEJ) | Lig4, Lif1, yKu70, yKu80 |
| 3. Mitotic checkpoints & Mitotic exit network | Bub1, Bub3, Mad1, Mad2, Mad3 and Bub2 |
| 4. Endonuclease | Rad1, Rad10 |
| 5. PCNA sumoylation | Siz1, Srs2, PCNA |
| 6. Histone ubiquitination | Bre1, H2B |
1. A new member of RFC family (ELG1-RFC)
Processing of stalled replication forks are usually modulated by a homo-trimeric DNA sliding clamp called PCNA in eukaryotes. Differential post-translational modifications in PCNA have been documented as a regulatory mechanism for many different DNA metabolisms including GCR [5]. Replication factor C (RFC) complex is a heteropentameric complex that catalyzes the loading of PCNA [6]. There are four different RFC complexes (Fig. 1). All RFC complexes share a core complex consisting of four subunits (Rfc2p, Rfc3p, Rfc4p, and Rfc5p) and differ by a specific subunit. A RFC complex having the specific subunit Rfc1p loads PCNA onto DNA during general DNA replication [7]. The Rad24p (yeast homolog of human RAD17)-Rfc2–5p complex loads a PCNA-like hetero-trimeric clamp consisting of Rad17p, Ddc1p and Mec3p (yeast homologs of human RAD9, RAD1, and HUS1, respectively) onto damaged DNA for activation of DNA damage checkpoint [8]. The third RFC complex is Ctf18p-Rfc2–5p and unloads PCNA from DNA where sister chromatids are held by cohesion complex [9]. The fourth complex that was recently added to the RFC family contains Elg1p as a specific subunit [10; 11; 12]. Although the function of Elg1p-Rfc2–5p RFC is unclear, increases in GCR and prolonged S phase progression in the elg1 Δ strain suggest a function in genome stability [10; 11; 12; 13; 14]
Figure 1.
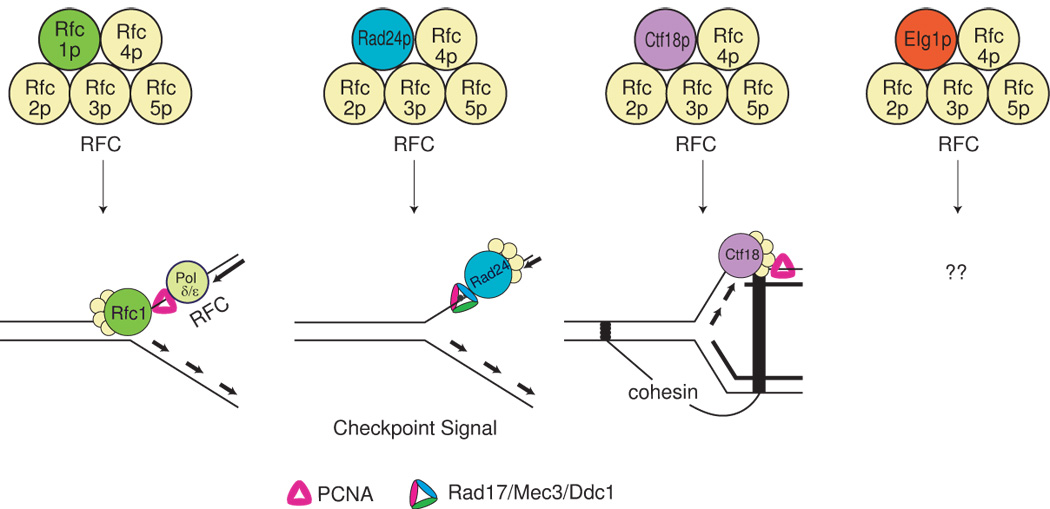
There are four different replication factor C (RFC) complexes. Each RFC is composed of four common subunits (Rfc2p, Rfc3p, Rfc4p, and Rfc5p) in addition to a specific subunit for each RFC. Rfc1p makes a general RFC complex with common subunits and function in general DNA replication for loading PCNA. Rad24p with common subunits functions specifically to load DNA damage checkpoint sensor Rad17p-Mec3p-Ddc1p to DNA damage. Ctf18p makes another RFC to unload PCNA where chromosome cohesion is established. The function of a RFC carrying Elg1p as a specific subunit is not clearly understood, yet.
2. ELG1 functions in DNA replication to suppress GCR
We identified Elg1p-Rfc2–5p as a new suppressor of GCR in Saccharomyces cerevisiae through a genome wide screen in identifying additional GCR suppression pathways [13]. Independently, Elg1p-Rfc2–5p was discovered as a suppressor of the Ty1 mediated recombination and a synthetic lethality mutation with holiday junction resolvase mutations, mus81 or mms4 [10; 11; 12].
In the absence of ELG1, spontaneous intra- or inter-allelic recombination frequencies are elevated [10]. In addition to enhanced GCR, high level of chromosomal loss was observed in the elg1 strain [11; 12]. Thus, Elg1p-Rfc2–5p functions to maintain genome stability during normal cell growth. The synthetic growth defect of the elg1 strain by the mutation of homologous recombination (HR) repair genes (rad51, rad52, rad54, rad55, rad57, and mre11) suggests that the lack of Elg1p-Rfc2–5p would create DSBs that HR proteins need to repair [12]. Although Elg1p’s function is not essential for viability in yeast, the replication fork could be stalled more frequently in the absence of Elg1p and ultimately generate DSBs. In this situation, cell cycle checkpoints could be activated and alternative repair machinery such as HR can be elicited. Strong synergistic increase of GCRs by the inactivation of cell cycle checkpoints in the elg1 deletion strain supports the notion that cell cycle checkpoints indeed suppress GCR from DNA damage initiated by the elg1 mutation [14]. However, hetero allelic recombination frequency after DNA damaging agent treatment was reduced in the elg1 strain [15].
3. Possible molecular functions of Elg1p-Rfc2–5p for maintaining genome stability
Elg1p interacts with PCNA and Rad27p (yeast homolog of human FEN1, FLAP endonuclease) that removes the flap structure at the lagging strand during DNA replication [12]. Therefore, Elg1p-Rfc2–5p may play an important role during DNA replication to protect the genome by loading or unloading a clamp similar to other RFCs’ roles. Despite negative in vitro PCNA loading or unloading PCNA by Elg1p-Rfc2–5p [9], the physical interaction of Elg1p with PCNA [12] strongly suggests Elg1p-Rfc2–5p as a PCNA loader or unloader. The PCNA loading/unloading activity of Elg1p-Rfc2–5p seems to overlap with other RFCs because synergistic sensitivities to DNA damaging agents were observed by inactivation of other RFCs with Elg1p-Rfc2–5p [11; 12]. However, Elg1p-Rfc2–5p seems to have a unique function because the elg1 mutation itself made cells sensitive to DNA damaging agents and grow slowly [12; 14]. Elg1p-Rfc2–5p could also function as a platform for polymerase switching in the post replication repair (PRR) pathway. When the DNA replication machinery encounters a DNA lesion that blocks further replication, a PRR pathway called translesion synthesis (TLS) is initiated [5]. TLS is largely dependent on the mono-ubiquitination of PCNA by Rad6p–Rad18p. The mono-ubiquitination of PCNA triggers the switch of replicative polymerase δ or ε to a specialized TLS polymerase such as polζ or polη that is extremely efficient in extending terminal mismatch or inserting A opposite to T-T dimers produced by UV irradiation, respectively [5]. In S phase, these TLS polymerases bypass DNA lesion and when DNA replication ends, several DNA repair pathways repair it. Co-localization of human ELG1 and human DNA polymerase η and the interaction between ELG1 and PCNA suggest that human ELG1-RFC could function in a PRR pathway (Banerjee, S., Sikdar, N. and Myung, K. unpublished data). PCNA is further poly-ubiquitinated by Ubc13p-Mms2p-Rad5p and directs another PRR pathway called template switching [16; 17]. The exact mechanism of template switching is not clearly understood. However, their shared role in GCR suppression by template switching and Elg1p, suggests that Elg1p could function in template switching.
Chromatin assembly is a fundamental biological process that is essential for the replication and maintenance of the eukaryotic genome [18]. In dividing cells, newly synthesized DNA is rapidly assembled into chromatin and packaged into nucleosomes that form the fundamental repeating subunits of chromatin. The nucleosome assembly is initiated by the deposition of a tetramer composed of two of each histone H3 and H4 to DNA. Subsequently, another tetramer comprised of two of each histone H2A and H2B is recruited to complete the nucleosome assembly. The synthetic lethality of the elg1 mutation with the htb1 or bre1/bre5 mutations that creates defects in histone levels or histone modification suggests a function for Elg1p in chromatin assembly through PCNA interaction [19]. Disruption of proper chromatin assembly by the inactivation of chromatin assembly factor 1 (CAF1) blocks DNA synthesis and induces DNA damage that activates checkpoints [20]. Intriguingly, the inactivation of each subunit of CAF1 including Cac1p, Cac2p, and Cac3p increased GCRs [21] and Cac1p physically interacts with PCNA [22]. Therefore, it is possible that Elg1p-Rfc2–5p could function in chromatin assembly by assisting proper chromosome condensation by PCNA loading or unloading.
4. Concluding Remarks
Elg1p is conserved throughout eukaryotic evolution with clear orthologues in C.elegans, Zebra fish, mouse, Rat and human. Mammalian ELG1 seems to have similar functions. Human ELG1 gene is mapped to Chr17q11.2 loci, proximal to Neurofibromatosis (NF) 1 gene. Intriguingly, the human ELG1 is often deleted in NF patients who are prone to cancer development [23]. It is possible that these cancer prone NF patients develop cancer as a result of ELG1 defect. High induction of GCRs and, faulty DNA replication presumably caused by stalled replication fork and abnormal HR in the elg1 strain suggest that Elg1 has a fundamental role of Elg1 to in maintaining genomic stability.
Acknowledgement
We apologize to researchers whose studies could not be discussed or cited because of space limitation. We thank Drs. Cristina Strong and Kyoo-young Lee for comments on the manuscript; J. Fekecs (NHGRI) for figure preparation. K.M. especially thanks E. Cho. This work was supported by the intramural research program of the NHGRI, NIH (to K.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- 2.Loeb LA, Loeb KR, Anderson JP. Multiple mutations and cancer. Proc Natl Acad Sci U S A. 2003;100:776–781. doi: 10.1073/pnas.0334858100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolodner R. Biochemistry and genetics of eukaryotic mismatch repair. Genes Dev. 1996;10:1433–1442. doi: 10.1101/gad.10.12.1433. [DOI] [PubMed] [Google Scholar]
- 4.Khanna KK, Jackson SP. DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet. 2001;27:247–254. doi: 10.1038/85798. [DOI] [PubMed] [Google Scholar]
- 5.Moldovan GL, Pfander B, Jentsch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Bylund GO, Majka J, Burgers PM. Overproduction and purification of RFC-related clamp loaders and PCNA-related clamps from Saccharomyces cerevisiae. Methods Enzymol. 2006;409:1–11. doi: 10.1016/S0076-6879(05)09001-4. [DOI] [PubMed] [Google Scholar]
- 7.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 8.Lindsey-Boltz LA, Bermudez VP, Hurwitz J, Sancar A. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc Natl Acad Sci U S A. 2001;98:11236–11241. doi: 10.1073/pnas.201373498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bylund GO, Burgers PM. Replication protein A-directed unloading of PCNA by the Ctf18 cohesion establishment complex. Mol Cell Biol. 2005;25:5445–5455. doi: 10.1128/MCB.25.13.5445-5455.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-Aroya S, Koren A, Liefshitz B, Steinlauf R, Kupiec M. ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc Natl Acad Sci U S A. 2003;100:9906–9911. doi: 10.1073/pnas.1633757100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bellaoui M, Chang M, Ou J, Xu H, Boone C, Brown GW. Elg1 forms an alternative RFC complex important for DNA replication and genome integrity. Embo J. 2003;22:4304–4313. doi: 10.1093/emboj/cdg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanellis P, Agyei R, Durocher D. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr Biol. 2003;13:1583–1595. doi: 10.1016/s0960-9822(03)00578-5. [DOI] [PubMed] [Google Scholar]
- 13.Smith S, Hwang JY, Banerjee S, Majeed A, Gupta A, Myung K. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2004;101:9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee S, Myung K. Increased genome instability and telomere length in the elg1-deficient Saccharomyces cerevisiae mutant are regulated by S-phase checkpoints. Eukaryot Cell. 2004;3:1557–1566. doi: 10.1128/EC.3.6.1557-1566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ogiwara H, Ui A, Enomoto T, Seki M. Role of Elg1 protein in double strand break repair. Nucleic Acids Res. 2007;35:353–362. doi: 10.1093/nar/gkl1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence C. The RAD6 DNA repair pathway in Saccharomyces cerevisiae: what does it do, and how does it do it? Bioessays. 1994;16:253–258. doi: 10.1002/bies.950160408. [DOI] [PubMed] [Google Scholar]
- 17.Smirnova M, Klein HL. Role of the error-free damage bypass postreplication repair pathway in the maintenance of genomic stability. Mutat Res. 2003;532:117–135. doi: 10.1016/j.mrfmmm.2003.08.026. [DOI] [PubMed] [Google Scholar]
- 18.Kaufman PD, Botchan MR. Assembly of nucleosomes: do multiple assembly factors mean multiple mechanisms? Curr Opin Genet Dev. 1994;4:229–235. doi: 10.1016/s0959-437x(05)80049-8. [DOI] [PubMed] [Google Scholar]
- 19.Aroya SB, Kupiec M. The Elg1 replication factor C-like complex: a novel guardian of genome stability. DNA Repair (Amst) 2005;4:409–417. doi: 10.1016/j.dnarep.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 21.Myung K, Pennaneach V, Kats ES, Kolodner RD. Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci U S A. 2003;100:6640–6645. doi: 10.1073/pnas.1232239100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krawitz DC, Kama T, Kaufman PD. Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol Cell Biol. 2002;22:614–625. doi: 10.1128/MCB.22.2.614-625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenne DE, Tinschert S, Dorschner MO, Hameister H, Stephens K, Kehrer-Sawatzki H. Complete physical map and gene content of the human NF1 tumor suppressor region in human and mouse. Genes Chromosomes Cancer. 2003;37:111–120. doi: 10.1002/gcc.10206. [DOI] [PubMed] [Google Scholar]
- 24.Motegi A, Myung K. Measuring the rate of gross chromosomal rearrangements in Saccharomyces cerevisiae: A practical approach to study genomic rearrangements observed in cancer. Methods. 2007;41:168–176. doi: 10.1016/j.ymeth.2006.07.025. [DOI] [PubMed] [Google Scholar]


