Abstract
Understanding the regulatory logic of a eukaryotic promoter requires the elucidation of the regulatory elements within that promoter. Current experimental or computational methods to discover regulatory motifs within a promoter can be labor intensive and may miss redundant, unprecedented or weakly activating elements. We have developed an unbiased combinatorial approach to rapidly identify new upstream activating sequences (UASs) in a promoter. This approach couples nonhomologous random recombination with an in vivo screen to efficiently identify UASs and does not rely on preconceived hypotheses about promoter regulation or on similarity to known activating sequences. We validated this method using the unfolded protein response (UPR) in yeast and were able to identify both known and potentially novel UASs involved in the UPR. One of the new UASs discovered using this approach implicates Crz1 as a possible activator of Hac1, a transcription factor involved in the UPR. This method has several advantages over existing methods for UAS discovery including its speed, potential generality, sensitivity and lack of false positives and negatives.
INTRODUCTION
The rapid increase in the number of sequenced genomes has augmented the need for efficient methods to define the regulatory logic of genes. Meeting this need requires the identification of each regulatory element within a promoter, including repressor-binding sites and upstream activation sequences (UASs), and the identification of the proteins that bind to these elements. These regulatory elements are typically short DNA sequences (4–10 bp) that are abundant in the genome but that only serve regulatory roles in a small subset of these occurrences (1).
The most common method researchers have used to identify new UASs in yeast places the promoter of interest upstream of a reporter gene, such as lacZ (2–6). Through a series of 5′-deletions, the promoter is truncated and the effects of each truncation on reporter gene expression is measured in the presence of a known inducer of that promoter. When induction is lost, it is assumed that a UAS has been deleted. Subsequently, higher resolution truncations of the putative UAS-containing region are used to define a minimal UAS. While this method has been successfully used to dissect dozens of promoters in yeast, it can be labor intensive and can miss UASs that are part of more complex promoter structures containing multiple UASs or repressors.
Recently, two new bioinformatic approaches have been developed to identify UASs: phylogenetic footprinting (7,8) and clustering of co-regulated genes (9,10). These methods are used to identify motifs that are conserved between related species or highly represented in the promoters of genes that are co-expressed under a given set of conditions. A limitation of these computational approaches is that regulatory regions in yeast are quite small compared with the size of a promoter (typically 600–1000 bp), leading to a low signal-to-noise ratio. As a result, these methods must apply strict selection criteria to yield only the most highly conserved sequences, resulting in a significant number of false negatives due to the degeneracy that occurs in many transcription-factor binding sites. Moreover, the uncertainty surrounding putative UASs identified using current computational methods requires that each predicted UAS be experimentally validated. This validation can be prone to a significant failure rate, since promoter regions can be highly conserved for reasons other than transcription-factor binding, including maintenance of chromatin structure and determination of long-range structure in the nucleus by specifying spatial organization of chromosomes (11). The creation of new, efficient experimental methods that not only generate but also simultaneously test hypotheses using a functional transcriptional readout would significantly enhance our ability to define the regulatory logic of promoters of interest.
Here we present a highly efficient method for the functional dissection of yeast promoters. This method involves the diversification of promoters using nonhomologous random recombination (NRR) (12), followed by the identification of functional promoter variants using an in vivo selection or high-throughput screen. We validated this method using the unfolded protein response (UPR), a transcriptional program activated in response to unfolded protein in the endoplasmic reticulum (ER). Using this approach, we rapidly identified known UASs in promoters that are upregulated during the UPR. In addition, this method also identified novel regions in these promoters that are sufficient for a transcriptional response in the presence of a UPR inducer. Our approach may be applicable to any yeast promoter whose activation or repression can be subjected to screening or selection.
MATERIALS AND METHODS
Yeast media and general materials
Media consisted of nitrogen base (Sigma), 2% dextrose and synthetic dropout supplements lacking uracil or uracil and tryptophan (Open Biosystems). All synthetic media was supplemented with 50 μg/ml myo-inositol (Sigma). X-Gal indicator plates were prepared as previously described (13), except that X-Gal was used at a final concentration of 80 μg/ml and adenine was added to a final concentration of 100 μg/ml. Saccharomyces cerevisiae yeast genomic DNA was purchased from Promega. Tunicamycin (Sigma) was stored as a 1000 × stock in dimethylformamide and used at a final concentration of 1 μg/ml.
Bacterial and yeast strains
Escherichia coli strain DH10B was purchased from Invitrogen. Yeast strains W303a (MAT a; ura3-1; leu2-3,-112; his3-11,-15; trp1-1; ade2-1; can 1-100) and CP263 (Δire1::TRP1 otherwise W303a) were kindly provided by Professor Peter Walter (University of California at San Francisco). Full open reading frame deletion of Crz1, replaced by the kanMX4 gene, was generated using a PCR-based deletion strategy in strain W303a (14,15). Gene disruption was confirmed by PCR and automated DNA sequencing.
Construction of the library plasmid
pJC104 is a two micron plasmid with a URA3 marker that contains a crippled CYC1 promoter upstream of lacZ and four copies of UPRE1 serving as a UAS (16). This plasmid was a generous gift from Professor Walter (16). A multiple cloning site was inserted in the large XhoI/BglII fragment of pJC104 using the phosphorylated primers 5′-GATCTGGTCACCTAGGTACCGCGGCCGGTAGCCCGGGTCGAC and 5′-TCGAGTCGACCCGGGCTACCGGCCGCGGTACCTAGGTGACCA to yield pProm.
Oligonucleotides for library construction
The following phosphorylated primers were used to amplify the promoters of KAR2, SIL1 and HAC1, respectively: K1 (5′-GTGGGAGTCAATCAAATCCC) and K2 (5′-GGTATGTTTGATACGCTTTTTCC); S1 (5′-CATCCAGGATCAAGTATATACC) and S2 (5′-TCTAAGTTTGCGTTCTTGGAAG); H1 (5′-AGAGCCACTATCATCGGC-GAC) and H2 (5′-AGTGGCGGTTGTTGTCGTAGG). Primers HP1 (5′-AGATCTGCATAGCAGCTGGCGCGCCATGGCGGCGCCGCCATGGCGCGCCAGCTGCTATGCAGATCT), HP2 (5′-GTCGACGCATAGCAGCTGGCGCGCCATGGCGGCGCCGCCATGGCGCGCCAGCTGCTATGCGTCGAC) and HP3 (5′-GGTGACCGCATAGCAGCTGGCGCGCCATGGCGGCGCCGCCATGGCGCGCCAGCTGCTATGCGGTCACC) contain BglII, SalI and BstEII restriction sites (underlined), respectively, for cloning into pProm. These primers also contain AscI and NcoI restriction sites (italicized) for removal of hairpins (see subsequently). Primers F1 (5′-CGCCAGCTGCTATGCAGATCT), F2 (5′-CGCCAGCTGCTATGCGTCGAC) and F3 (5′-CGCCAGCTGCTATGCGGTCACC) are used for the final PCR amplifications.
Construction of promoter libraries
Genomic DNA fragments from the KAR2, SIL1 and HAC1 promoters, −1 to −1000 relative to the start of translation, were amplified by PCR using the primers K1/K2, S1/S2 and H1/H2, respectively. NRR was performed on the resulting PCR product as previously described (12) using primers HP1 and HP2 for the KAR2 and HAC1 libraries. HP2 and HP3 were used for the SIL1 library. Fragments with a desired size range (see text) were purified by agarose gel electrophoresis after the hairpin ligation step. Recombined promoter fragments for the KAR2 and HAC1 libraries were digested with NcoI and AscI, then amplified with primers F1 and F2 and cloned into the large BglII/SalI fragment of pProm. Recombined fragments from the SIL1 library were also digested with NcoI and AscI, then amplified using F2 and F3, and cloned into the large BstEII/SalI fragment of pProm. All libraries were amplified in the E. coli strain DH10B.
Blue/white screening of promoter libraries
A promoter library (∼1 μg) was transformed into W303a using a standard lithium acetate protocol and selected on plates lacking uracil at 30°C. After 3 days of growth, the yeast colonies were replica plated onto X-Gal plates supplemented with tunicamycin and incubated at 30°C for 24–72 h. The bluest colonies were selected and grown individually in liquid media lacking uracil. Liquid cultures were then spotted onto X-Gal plates lacking uracil with and without tunicamycin to identify those cultures that specifically turned blue only in the presence of tunicamycin. The cells from an aliquot of each liquid culture were lysed using glass beads and a region of DNA encoding the promoter fragment was amplified using primers Seq1 (5′-GGAGACGCATTGGGTCAACAG) and Seq2 (5′-GTGTTTGCGTGTCTATAGAAG). These PCR products were then sequenced to identify promoter fragments. DNA sequences of the promoters from the blue colonies were then aligned to identify consensus regions.
Consensus region cloning
Phosphorylated, complementary oligonucleotides containing one, two or three copies of each consensus regions from the X-Gal screening were annealed and then individually cloned into the large BglII/SalI fragment of pProm for further analysis. For the KAR2 promoter, the consensus regions I–III are as follows: 5′-AATGTACACGTATC, 5′-TTTGAACACGTCAACAAC and 5′-TAGCCAAACGGACA, respectively. Extended consensus region III was 5′-TAGCCAAACGGACAGCTGTCCTCA. KAR2 consensus region IV was contained in pJC104. The SIL1 consensus region is 5′-GAAAAGGCCACGTAG. The HAC1 consensus regions I and II are as follows: 5′-GGCAAAGTGGCTCAGCAT and 5′-TGGTTTTGAACACCTTGTTCTCTTTTGT, respectively.
Quantitative β-galactosidase activity assay
Plasmids containing the consensus regions were then transformed in W303a, CP263 or the Crz1-deleteion yeast strains and assayed for β-galactosidase activity with or without tunicamycin in triplicate using o-nitrophenyl-β-galactopyranoside as previously described (17).
RESULTS
Rationale for the application of NRR to UAS identification
NRR (12) is a method to diversify nucleic acids that randomly rearranges any sequence using a fragment size defined by the researcher. NRR has been used to study protein topological requirements (18), sRNA mechanisms (19) and mRNA transport signal sequences [(20); Liu,J.M. and Liu,D.R., unpublished data). The first step in NRR involves the random digestion of a starting nucleic acid pool using DNase I (Figure 1). By modulating the digestion time, the average size of the digested fragments can be controlled. After blunting the pieces with T4 DNA polymerase, the DNA fragments are reassembled in the presence of DNA hairpins, allowing for the deletion, insertion and rearrangement of fragments. The average number of crossovers per recombinant is controlled by varying the relative molar ratio of hairpins to fragments.
Figure 1.
Generation of a nonhomologous random recombination library of a yeast promoter. The bottom figure represents the plasmid into which the library is cloned for blue/white screening.
To identify UASs within a promoter, we envisioned that an NRR-diversified promoter of interest could be cloned upstream of a crippled CYC1 core promoter that drives the expression of the lacZ gene (21) (Figure 1). The resulting library can be rapidly screened for transcriptional activation only in the presence of a signal known to induce transcription from the promoter of interest (13). Because the products of NRR are highly diversified in the size, order, spacing, redundancy and orientation of subsequences, the comparison of DNA sequences among promoter variants that maintain their ability to be induced should rapidly identify essential UASs. This approach requires virtually no knowledge of the promoter of interest other than a means of induction, and therefore could represent a general approach to the discovery of UASs.
NRR libraries are ideal for promoter analysis in this context for several reasons. First, NRR can use DNA fragments that approach the sizes of typical UASs (∼8–10 bp), facilitating the rapid identification of a small motif that is required for transcription. Second, complex combinations of necessary regulatory elements within a promoter can be identified using this approach because multiple fragments are allowed to recombine during the reassembly step. Third, precise spatial requirements, if any, between UASs or between a UAS and the start of transcription can be revealed because the spacing between fragments varies randomly within NRR-diversified libraries. Finally, discovery of a consensus region among the selected constructs allows for the rapid identification of a minimal UAS that is required for a response to a given stimulus because the inactive flanking regions of each UAS should be remaining highly diversified during the NRR and screening process.
Functional dissection of the KAR2 promoter
To validate the proposed method to identify UASs in promoters of interest, NRR coupled with high-throughput screening was applied to three promoters of genes involved in the UPR: KAR1, SIL1 and HAC1. Three UASs that specifically respond to unfolded protein in the ER have been previously identified as UPR elements 1, 2 and 3 (UPRE1, UPRE2 and UPRE3) (10,22,23).
The 1000-base KAR2 promoter contains one known UAS, UPRE1 (7 bp), that is responsive to the presence of unfolded protein in the ER (22–24). We subjected the promoter of KAR2 to NRR and screening to determine if UPRE1 could be quickly and accurately identified. The NRR-diversified KAR2 promoter library was cloned upstream of the crippled CYC1 promoter driving the expression of lacZ for blue/white screening in the presence of tunicamycin. The KAR2 promoter library contained DNA fragments that were an average of 31 bp in length. The library was transformed into yeast and replica plated onto X-Gal plates supplemented with tunicamycin to induce the UPR.
The 60 bluest colonies were isolated, then subjected to a secondary screen on X-Gal plates both with and without tunicamycin. The clones that showed robust transcriptional activation only in the presence of tunicamycin (15 out of 60 colonies) were classified as strong hits. All 60 clones were sequenced, and the promoter fragments were aligned to reveal any consensus regions among the selected promoters (Figure 2). Promoter fragments were grouped according to blue/white phenotype, with the strong hits at the top of Figure 2 (gray area), and then by sequence to show the consensus regions. This entire process, from initial amplification of the KAR2 promoter to the sequence alignment, required less than three weeks. Four consensus regions emerged from this analysis, including one consensus region present in most of the strong clones. Gratifyingly, this consensus region (region IV in Figure 1) contains the previously described UPRE1. Sequence alignment of the strongest clones yields a consensus region of 15 bp, which contains the 7 bp core of UPRE1 previously shown to be essential for ER stress-mediated activation of the KAR2 promoter (22). Since UPRE1 was the only UPRE previously described for the KAR2 promoter, we were surprised that three additional consensus regions emerged from the above analysis (Figure 3). Consensus region I contains UPRE2, a recently discovered UPRE, CACGTA, that is a putative binding site for Gcn4. Region II contains a very similar sequence that differs from UPRE2 by a single nucleotide, CACGTC. This sequence has not been specifically characterized as a Gcn4-binding site. Region III does not contain any previously reported UPREs though shares some similarity with UPRE1 (Figure 3).
Figure 2.
Composition of selected UAS elements from the KAR2 promoter. The selected fragments (colored arrows) are compared to the native promoter to reveal consensus regions. Numbering across the top represents the nucleotide position in the native promoter relative to the start of translation. Arrow colors indicate the order of fragment assembly (5′-red-green-blue-purple-3′) and the arrow direction indicates a sense (pointing right) or antisense (pointing left) promoter fragment. The shaded gray encompasses clones with the strongest phenotype. Four consensus regions (I, II, III and IV) are indicated with a vertical line.
Figure 3.
Partial sequence alignments of clones from the NRR analysis of the KAR2 promoter to reveal consensus regions. For regions I, II and III, all clones with the conserved region were used for the alignments. For region IV, clones with a strong phenotype that also lacked other conserved regions were used for the alignment. Consensus regions are shown in red. For region III, an extended consensus region is highlighted in blue. Each consensus region is compared to a known UPRE, with the putative transcription-factor binding site underlined.
Of the four consensus regions, region III was the least abundant, occurring in only seven out of 60 clones (Figure 3). All seven clones contained a common sequence shown in red in Figure 3, but six of these clones also had a longer consensus region shown in blue. Since comparatively few clones defined the boundaries of this consensus region, both the minimal and extended consensus regions were used for further analysis. To test the functional significance of these consensus regions, fragments of the KAR2 promoter containing each consensus sequence, including both region III and the extended region III (Figure 3), were cloned upstream of the CYC1-lacZ construct and analyzed using quantitative β-galactosidase assays (Figure 4). Regions I, II and IV each showed significant lacZ activity only in the presence of tunicamycin. The minimal consensus region III did not show any activity in the presence or absence of tunicamycin, but the extended region III exhibited robust transcriptional induction in the presence of tunicamycin. This result reveals the necessity of carefully choosing the endpoints of a consensus region when only a small number of clones define the region.
Figure 4.
Quantitative β-galactosidase assays with the KAR2 consensus regions serving as UASs in the CYC1-lacZ construct. Multiple copies of regions I–IV, 3 ×, 2 ×, 3 × and 4 × respectively, were used in these constructs. Consensus region III and the extended region III (III +, Figure 3) were each analyzed. Assays were performed in wild-type (wt) and Δire1 strains in the presence and absence of tunicamycin. β-Galactosidase activity is shown in arbitrary units and error bars represent 1 SD.
Each of these consensus regions was also tested in a strain that lacks the gene for Ire1, the protein that initially senses unfolded protein in the ER (10). All four regions that showed tunicamycin-dependent lacZ activity in a wild-type strain had no activity in a Δire1 strain (Figure 4). Ire1 is essential for the interaction of the transcription factors Hac1 and Gcn4 with UPRE1 and UPRE2 (10). The Ire1 dependence of transcriptional activation mediated by consensus regions I, II, III (extended) and IV suggests a similar transcriptional pathway. Taken together, the above results demonstrate the ability of a coupled NRR/screening approach to identify both known and unknown upstream-activating regions within yeast promoters.
Functional dissection of the SIL1 promoter
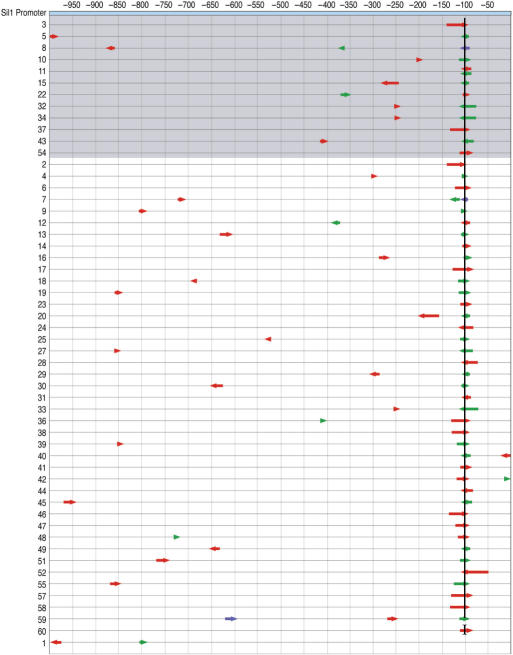
Sil1, a nucleotide exchange factor for Kar2 (25), is highly upregulated during the UPR (26). The promoter of SIL1 was analyzed because it has not been previously experimentally dissected, although it contains a sequencing matching UPRE2 (10). It is currently not known if addition transcriptional elements are also present in the SIL1 promoter. The promoter of SIL1 was subjected to NRR and cloned into the CYC1-lacZ construct. The resulting library had an average DNA fragment length of 27 bp. This NRR-diversified library was screened in the presence of tunicamycin and the 60 bluest colonies were isolated. Using the secondary screen, 12 of the 60 colonies showed robust lacZ transcription only in the presence of tunicamycin and these colonies were classified as strong hits (Figure 5, top). When all 60 colonies were subjected to sequence analysis, only one region was repeatedly selected (Figure 5). Alignment of clones showing a strong phenotype upon plating on media containing X-Gal in the presence and absence of tunicamycin revealed a 14-base consensus region containing UPRE2 (Figure 6). No other consensus regions emerged among the sequences.
Figure 5.
Composition of selected UAS elements from the SIL1 promoter. The selected fragments (colored arrows) are compared to the native promoter to reveal consensus regions. Numbering across the top represents the nucleotide position in the promoter relative to the start of translation. Arrow colors indicate the order of fragment assembly (5′-red-green-blue-3′) and the arrow direction indicates a sense (pointing right) or antisense (pointing left) promoter fragment. The shaded gray encompasses clones with the strongest phenotype. One consensus region (I) is indicated with a vertical line.
Figure 6.
Analysis of the consensus region for the SIL1 promoter. (Left) A partial sequence alignment of the clones with the strong phenotype reveals a consensus region that contains UPRE2 with a putative Gcn4-binding site (underlined). (Right) Quantitative β-galactosidase assays with one copy or three copies of the SIL1 consensus region serving as a UAS in the CYC1-lacZ construct. Assays were performed in wild-type (wt) and Δire1 strains in the presence and absence of tunicamycin. β-Galactosidase activity is shown in arbitrary units and error bars represent 1 SD.
When this consensus region was cloned into the CYC1-lacZ construct, it was responsive to tunicamycin in a wild-type strain (Figure 6). As expected for UPRE2, this consensus region did not function as a UAS in a Δire1 strain (10). The regulatory logic of the SIL1 promoter appears to be quite simple; the lack of any other consensus regions suggests that UPRE2 is the sole determinant of transcription from SIL1 during the UPR. The 14-bp region containing UPRE2 was retained in 100% of our selected clones (Figure 6), indicating that some part of this sequence which includes eight bases upstream of UPRE2 may be important for a strong transcriptional response.
Functional dissection of the HAC1 promoter
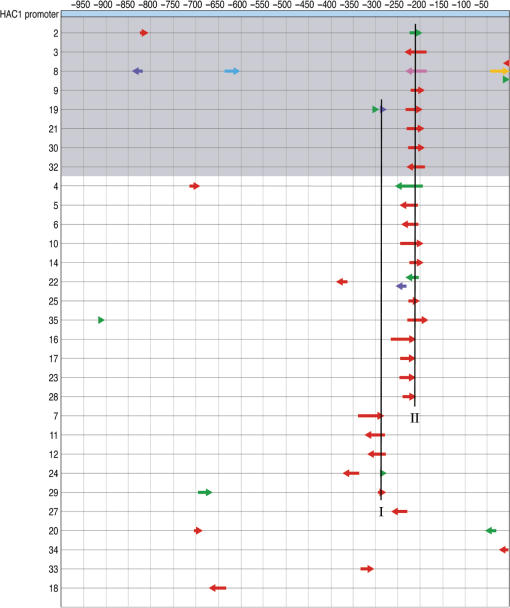
The promoter of HAC1, a key transcription factor involved in the UPR, was similarly analyzed by NRR and screening. A UPRE1-like sequence was recently identified in this promoter (27), and Hac1 has been shown to bind to this region in an autoregulatory mechanism that is IRE1-dependent. A second IRE1-independent regulatory mechanism for the HAC1 promoter has been observed during extensive ER stress, but the UAS responsible is unknown (28). To elucidate the various mechanisms of HAC1 transcriptional regulation, the promoter of HAC1 was subjected to NRR and cloned into the CYC1-lacZ construct. The resulting library had an average DNA fragment length of 33 bp. When this library was screened in a wild-type strain in the presence of tunicamycin, clones containing one of two consensus regions emerged (Figure 7). The DNA sequences of clones in groups I and II were aligned to reveal the identities of the minimal consensus regions (Figure 8).
Figure 7.
Composition of selected UAS elements from the HAC1 promoter tested in a wild-type strain. The selected fragments (colored arrows) are compared to the native promoter to reveal consensus regions. Numbering across the top represents the nucleotide position in the promoter relative to the start of translation. Arrow colors indicate the order of fragment assembly (5′-red-green-blue-purple-cyan-orange-3′) and the arrow direction indicates a sense (pointing right) or antisense (pointing left) promoter fragment. The shaded gray encompasses clones with the strong phenotype. Two consensus region (I and II) are indicated with a vertical lines.
Figure 8.
Partial sequence alignments of clones from the NRR analysis of the HAC1 promoter. (Top) Region I has a consensus sequence (red) common to all the clones and an extended consensus sequence (blue) shared by half of the clones. (Bottom) Wild-type region II contains the previously described UPRE1-like sequence and is shown with the KAR2 UPRE1 sequence for comparison. The putative transcription-factor binding site(s) in UPRE1 is underlined.
Consensus region II contains the previously described UPRE1-like sequence that has been shown to be sufficient for tunicamycin-induced transcription from the HAC1 promoter (27). The consensus sequence from region I is derived from only six clones, thus making it difficult to precisely identify the endpoints of the minimal region that contains a UAS (Figure 8). These six clones fall into two categories, one containing a minimal consensus region (red in Figure 8) and one containing an extended consensus region (red + blue in Figure 8). The consensus region I does not contain a DNA sequence that matches any of the known UPREs, suggesting that a novel transcription factor is likely binding to this region. A search for transcription factor-binding sites in this consensus region revealed only one known motif (29), a binding site for the transcription factor Crz1 (Figure 8) (30). Crz1 is a calcineurin-dependent transcription factor responsible for the transcriptional response to changes of Ca2+ concentration in the yeast calcium cell survival (CCS) pathway (31) and has not been previously implicated in the UPR.
To test if the consensus regions from the wild-type analysis can serve as UPREs, each region was cloned into the CYC1-lacZ construct and analyzed with quantitative β-galactosidase assays (Figure 9). These two regions were also tested in the Δire1 strain to determine if the Hac1/Gcn4 pathway is necessary for transcription. Region II showed robust transcriptional activation in the presence of tunicamycin in the wild-type strain but not in the Δire1 strain, consistent with the previously described analysis of this region in the HAC1 promoter (27). In contrast, both region I and the extended region I showed similar tunicamycin-induced transcription in both the wild-type and Δire1 strains, indicating that a transcription factor-binding site likely resides in the minimal consensus region, which contains the putative Crz1-binding site (Figure 9 and data not shown). Regions I and II were then tested in a Δcrz1 strain. While transcription from region II was unaffected by this deletion, all transcriptional induction was lost from region I, further implicating Crz1 as a factor responsible for transcriptional regulation from this region during the UPR.
Figure 9.
Quantitative β-galactosidase assays with one copy of the HAC1 consensus regions I and II each serving as a UAS in the CYC1-lacZ construct. Assays were performed in wild-type (wt), Δire1 and Δcrz1 strains in the presence and absence of tunicamycin. β-Galactosidase activity is shown in arbitrary units and error bars represent 1 SD.
DISCUSSION
Our results collectively suggest that NRR is well suited to the rapid and efficient identification of cis-regulatory elements from yeast promoters. All three sets of experiments described here including promoter library construction, in vivo screening and sequence analysis collectively required only 3 weeks in total and can easily be performed in parallel for several promoters. The ability to use fragment sizes that approach the sizes of the typical UAS elements allows for the rapid identification of consensus regions that are responsible for transcriptional activity under the chosen experimental conditions. In the three examples shown here, the fragment sizes were 10–50 bp (note that manipulation of double-stranded DNA fragments smaller than ∼ 10 bp can be difficult due to spontaneous fragment denaturation).
The ability of the NRR method to introduce crossovers between two or more functional regions in the libraries was particularly helpful in the analysis of the KAR2 promoter. Although most of the clones containing consensus regions I and II had a weak phenotype upon secondary screening, these weak consensus regions can still yield functional UASs since they can act additively to yield a strong phenotype as observed in clones 25 and 29 (Figure 2). For an optimum library, the average number of crossovers should match the number of regulatory elements that might act combinatorially in the promoter. While this number cannot be known a priori, ChIP-chip experiments suggest that more than one-third of yeast promoters are bound by more than one transcriptional regulator, indicating that ideal libraries would have an average number of crossovers > 1 (32).
Our findings further suggest that, unlike previously described bioinformatic approaches (10), the NRR method as applied to UPRE identification does not appear to suffer from a significant false-positive or false-negative rate. The method was able to find all four of the sequences that had previously been described as UASs: UPREs 1 and 2 from the KAR2 promoter, UPRE2 from the SIL1 promoter and the UPRE1-like sequence from the HAC1 promoter. Each consensus region revealed by the NRR analysis was identified every time it occurred in these three promoters, suggesting that the NRR method can cover candidate promoter fragment space exhaustively, at least above a certain threshold of activity determined by the screening method used. In addition, every consensus region that was tested as a UAS, when the boundaries were carefully chosen, was responsive to tunicamycin, indicating an absence of false positives from this analysis.
Current methods to experimentally dissect promoters usually take advantage of 5′-promoter truncation to identify regions that lead to a loss of inducible signal under the assay conditions. This method, however, can miss UASs that can function independently of and redundantly with downstream UASs, because truncation of a region containing an independent and redundant upstream UAS simply leads to a reduction in signal but not a loss of inducibility. For example, the deletion of a DNA sequence containing the consensus region I from the HAC1 NRR analysis (Figure 8) was previously shown to cause a 2-fold loss in signal, but no loss of inducibility because the UPRE1-like sequence was still present (27). As a result, this region was previously not implicated by researchers who were dissecting the HAC1 promoter (27). Since NRR can analyze DNA fragments independently and combinatorially, without any directional bias for UASs closer to the start of translation, this approach may be better suited than previous methods for the comprehensive characterization of yeast promoters.
The identification of a consensus sequence as a UAS does not automatically imply that it is physiologically relevant in the context of the original promoter from which it was discovered. For example, an identified consensus sequence may normally be sequestered in heterochromatin, inaccessible to the transcription factors that bind to it in the artificial context of the CYC1-lacZ assay. Also, a UAS could be controlled by negative regulatory elements that are lost in the analyses described above. However, while the UASs identified by the NRR method may not be relevant in the promoters from which they are derived, it is likely that they have a physiological role in other promoters. The evolution of DNA-binding specificity is under strong positive and negative selective pressures during evolution due to the importance of tight transcriptional regulation to cell survival (7,8). Therefore, interactions between a transcription factor and a DNA target in vivo that lead to a transcriptional readout are likely to be physiologically relevant in some cellular context. Identification of a new UAS allows researchers to identify a list of relevant promoters for which that UAS may be relevant. Each of these promoters can then be analyzed individually further to test for biological significance of the UAS.
A new UAS was also discovered in region I of the HAC1 promoter that responds to tunicamycin in an IRE1-independent manner. This UPRE contains a putative binding site for the transcriptional factor Crz1, a calcineurin-dependent transcription factor implicated in Ca2+ signaling (30), and its activity is dependent on Crz1. High salt, alkaline pH and cell wall damage lead to increased cytosolic levels Ca2+, activating calcineurin's phosphatase activity (31). Calcineurin dephosphorylates Crz1, leading to its nuclear localization and transcription of its target genes. Previous studies have shown that unfolded protein in the ER leads to an influx of Ca2+ at the plasma membrane in an IRE1- and HAC1-independent manner, leading to activation of Crz1 (31). The CCS pathway is not required for the initial UPR response, but may be required for a prolonged response to tunicamycin treatment (31). In addition to the canonical UPR, Walter and coworkers (28) have described a ‘super-UPR’ that requires at least two ER stress signals, such as inositol deprivation and tunicamycin treatment. Under these conditions, HAC1 transcription is upregulated in an IRE1-independent manner, suggesting the possible existence of a new transcription factor in targeting the HAC1 promoter. Our identification of a Crz1-dependent putative Crz1-binding site in the HAC1 promoter and the previous description of Crz1 activation in response to ER stress suggest a possible connection between the CCS pathway and the super-UPR. Cross-talk between these two pathways has not been previously reported, and additional experimental work is needed to explore this hypothesis in depth.
In conclusion, NRR coupled with screening is well suited to the dissection of the regulatory logic of yeast promoters, resulting in the identification of UASs with no significant false-positive or false-negative rate in the case of the UPR. From a single set of experiments that requires less than 3 weeks, fairly precise consensus regions can be rapidly defined that are close to the minimal requirements for a UAS that responds to the assay conditions. The speed and efficiency of this method to find UASs represent a significant advance over other current methods used to find UASs. Our initial efforts to validate the method described here have already yielded unexpected UASs that may play a role in the UPR or the super-UPR. Further work is needed to understand the biological role of these new UASs and to test the generality of this method in other promoters and organisms.
ACKNOWLEDGEMENTS
We are grateful to Professor Peter Walter at UCSF for providing plasmids and strains. We thank Jane Liu, Polina Kehayova and Courtney Yuen for technical assistance and helpful discussions. This work was supported by NSF CAREER Award MCB-0094128, NIH/NIGMS R01 GM065400 and the Howard Hughes Medical Institute. Funding to pay the Open Access publication charges for this article was provided by the Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.GuhaThakurta D. Computational identification of transcriptional regulatory elements in DNA sequence. Nucleic Acids Res. 2006;34:3585–3598. doi: 10.1093/nar/gkl372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srikanth CV, Vats P, Bourbouloux A, Delrot S, Bachhawat AK. Multiple cis-regulatory elements and the yeast sulphur regulatory network are required for the regulation of the yeast glutathione transporter, Hgt1p. Curr. Genet. 2005;47:345–358. doi: 10.1007/s00294-005-0571-7. [DOI] [PubMed] [Google Scholar]
- 3.Kim TS, Lee SB, Kang HS. Glucose repression of STA1 expression is mediated by the NRG1 and Sfl1 repressors and the Srb8-11 complex. Mol. Cell. Biol. 2004;24:7695–7706. doi: 10.1128/MCB.24.17.7695-7706.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shakoury-Elizeh M, Tiedeman J, Rashford J, Ferea T, Demeter J, Garcia E, Rolfes R, Brown PO, Botstein D, et al. Transcriptional remodeling in response to iron deprivation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:1233–1243. doi: 10.1091/mbc.E03-09-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braus GH, Grundmann O, Bruckner S, Mosch HU. Amino acid starvation and Gcn4p regulate adhesive growth and FLO11 gene expression in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:4272–4284. doi: 10.1091/mbc.E03-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brons JF, de Jong M, Valens M, Grivell LA, Bolotin-Fukuhara M, Blom J. Dissection of the promoter of the HAP4 gene in S-cerevisiae unveils a complex regulatory framework of transcriptional regulation. Yeast. 2002;19:923–932. doi: 10.1002/yea.886. [DOI] [PubMed] [Google Scholar]
- 7.Cliften P, Sudarsanam P, Desikan A, Fulton L, Fulton B, Majors J, Waterston R, Cohen BA, Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- 8.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 9.McCarroll SA, Li H, Bargmann CI. Identification of transcriptional regulatory elements in chemosensory receptor genes by probabilistic segmentation. Curr. Biol. 2005;15:347–352. doi: 10.1016/j.cub.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 10.Patil CK, Li H, Walter P. Gcn4p and novel upstream activating sequences regulate targets of the unfolded protein response. PLoS Biol. 2004;2:1208–1223. doi: 10.1371/journal.pbio.0020246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adams MD. Conserved sequences and the evolution of gene regulatory signals. Curr. Opin. Genet. Dev. 2005;15:628–633. doi: 10.1016/j.gde.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 12.Bittker JA, Le BV, Liu DR. Nucleic acid evolution and minimization by nonhomologous random recombination. Nat. Biotechnol. 2002;20:1024–1029. doi: 10.1038/nbt736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chien CT, Bartel PL, Sternglanz R, Fields S. The 2-hybrid system – a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl Acad. Sci. USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or pcr-based gene disruptions in Saccharomyces-cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 15.Baudin A, Ozierkalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces-cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- 17.Pryciak PM, Hartwell LH. AKR1 encodes a candidate effector of the G beta gamma complex in the Saccharomyces cerevisiae pheromone response pathway and contributes to control of both cell shape and signal transduction. Mol. Cell. Biol. 1996;16:2614–2626. doi: 10.1128/mcb.16.6.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bittker JA, Le BV, Liu JM, Liu DR. Directed evolution of protein enzymes using nonhomologous random recombination. Proc. Natl Acad. Sci. USA. 2004;101:7011–7016. doi: 10.1073/pnas.0402202101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu JM, Bittker JA, Lonshteyn M, Liu DR. Functional dissection of sRNA translational regulators by nonhomologous random recombination and in vivo selection. Chem.Biol. 2005;12:757–767. doi: 10.1016/j.chembiol.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 20.Jambhekar A, McDermott K, Sorber K, Shepard KA, Vale RD, Takizawa PA, DeRisi JL. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2005;102:18005–18010. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guarente L, Ptashne M. Fusion of Escherichia-coli-lacz to the cytochrome-C gene of Saccharomyces-cerevisiae. Proc. Natl Acad. Sci. USA-Biol.l Sci. 1981;78:2199–2203. doi: 10.1073/pnas.78.4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T. Palindrome with spacer of one nucleotide is characteristic of the cis-acting unfolded protein response element in Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:9912–9920. doi: 10.1074/jbc.273.16.9912. [DOI] [PubMed] [Google Scholar]
- 23.Mori K, Sant A, Kohno K, Normington K, Gething MJ, Sambrook JF. A 22 Bp cis-acting element is necessary and sufficient for the induction of the yeast kar2 (Bip) gene by unfolded proteins. EMBO J. 1992;11:2583–2593. doi: 10.1002/j.1460-2075.1992.tb05323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohno K, Normington K, Sambrook J, Gething MJ, Mori K. The promoter region of the yeast kar2 (bip) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic-reticulum. Mol. Cell. Biol. 1993;13:877–890. doi: 10.1128/mcb.13.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kabani M, Beckerich JM, Gaillardin C. Slslp stimulates Sec63p-mediated activation of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol. Cell. Biol. 2000;20:6923–6934. doi: 10.1128/mcb.20.18.6923-6934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- 27.Ogawa N, Mori K. Autoregulation of the HAC1 gene is required for sustained activation of the yeast unfolded protein response. Genes Cells. 2004;9:95–104. doi: 10.1111/j.1365-2443.2004.00704.x. [DOI] [PubMed] [Google Scholar]
- 28.Leber JH, Bernales S, Walter P. IRE1-independent gain control of the unfolded protein response. PLoS Biol. 2004;2:1197–1207. doi: 10.1371/journal.pbio.0020235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teixeira MC, Monteiro P, Jain P, Tenreiro S, Fernandes AR, Mira NP, Alenquer M, Freitas AT, Oliveira AL, et al. The YEASTRACT database: a tool for the analysis of transcription regulatory associations in Saccharomyces cerevisiae. Nucleic Acids Res. 2006;34:D446–D451. doi: 10.1093/nar/gkj013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshimoto H, Saltsman K, Gasch AP, Li HX, Ogawa N, Botstein D, Brown PO, Cyert MS. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:31079–31088. doi: 10.1074/jbc.M202718200. [DOI] [PubMed] [Google Scholar]
- 31.Bonilla M, Nastase KK, Cunningham KW. Essential role of calcineurin in response to endoplasmic reticulum stress. EMBO J. 2002;21:2343–2353. doi: 10.1093/emboj/21.10.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]