Abstract
The definition of a typical sec-dependent bacterial signal peptide contains a positive charge at the N-terminus, thought to be required for membrane association. In this study the amino acid distribution of all Escherichia coli secretory proteins were analysed. This revealed that there was a statistically significant bias for lysine at the second codon position (P2), consistent with a role for the positive charge in secretion. Removal of the positively charged residue P2 in two different model systems revealed that a positive charge is not required for protein export. A well-characterized feature of large amino acids like lysine at P2 is inhibition of N-terminal methionine removal by methionyl amino-peptidase (MAP). Substitution of lysine at P2 for other large or small amino acids did not affect protein export. Analysis of codon usage revealed that there was a bias for the AAA lysine codon at P2, suggesting that a non-coding function for the AAA codon may be responsible for the strong bias for lysine at P2 of secretory signal sequences. We conclude that the selection for high translation initiation efficiency maybe the selective pressure that has led to codon and consequent amino acid usage at P2 of secretory proteins.
INTRODUCTION
Secretory proteins exported via the general secretory (sec) pathway are synthesized as precursors with an N-terminal signal peptide. This signal peptide is removed by leader peptidase I upon export to the periplasm [for a review of export, see (1)]. The signal peptide can be divided into three regions: a hydrophilic N-terminus often containing 1–3 positively charged residues, a hydrophobic core and a cleavage site for processing by the respective signal peptidase (2).
The role ascribed to the positively charged N-terminus of the signal peptide is to provide stable interactions with the negatively charged inner membrane phospholipids. This interaction is thought to be important for targeting the secretory protein translocase (1). The positive charge could also aid interaction with the export machinery, signal recognition particle (SRP) (3) and SecA (4). Studies by von Heijne (2,5) on the charge distribution of 39 and 32 prokaryotic signal sequences reported a net positive charge at the N-terminus of 1.7. Despite the abundance of positively charged residues at the N-terminus of signal peptides, studies have shown that removing the positive charges, such that the net charge at the N-terminus is 0, has differing effects on export. These include a reduced rate of export (6,7), lower rate of protein synthesis (8) and no discernable effect (9). If there is a net-negative charge, then export is impaired, with increased levels of unprocessed precursor (7–9). These results suggest that a net positive charge is not essential for export to occur, and raises the question of why the majority of secretory proteins have a positive charge at the N-terminus.
With the availability of the complete genome sequence of Escherichia coli (10), and the ability to sort secreted proteins from non-secreted proteins (11), distribution of charged residues was analysed in secretory proteins. The codon usage of the charged residues was further analysed and revealed preference for AAA at P2, implying that codon usage was a stronger selective pressure than a requirement for a positive charge.
MATERIALS AND METHODS
Molecular cloning techniques
All cloning was carried out in E. coli DH5α (F-(80dlacZ ▵ M15) (lacZYA-argF) U169 hsdR17 (r-m+) recA1 endA1 relA1 deoR). Kanamycin was used at 50 μg/ml. Primers were made by Pro-Oligo (Sigma, Lismore). All PCR reactions were carried out using Phusion Taq (Finnzyme). Ligations using T4 DNA ligase and digests were performed according to manufacturer's instructions (New England Biolabs). DNA sequencing was done using Big Dye Terminator method (Griffith University DNA Sequencing Facility).
Construction of pMalE::bla, pPhoA::bla and second position mutants
The wild-type bla gene and signal sequence mutants were generated by splice-overlap PCR and cloned into the multicloning site of pGBS19 [pUC19 with kanr replacing bla, (12)]. Briefly the pMalE::bla was made with splice-overlap PCR products generated from the 5′bla-MBP/3′MBPss and the 5′MBP-bla/bla_rev primer pairs, amplified with 5′bla-MBP and bla-rev primer pair. The pPhoA::bla construct was generated from the 5′phoA-bla/bla_rev and 5′bla-phoA/3′phoAss primer pairs, amplified with 5′bla-phoA and bla_rev primer pairs. Primers are listed in Table 1. The MBP signal sequence was amplified from the plasmid pMALp2e vector while the PhoA signal sequence was amplified from E. coli K-12 MG1655. For both constructs, the upstream primer (5′bla-MBP, 5′bla-phoA) incorporated 28 bp upstream of the bla start codon from pMALp2e, which contains a Shine Dalgarno sequence and an EcoRI site. The common downstream primer, bla_rev, incorporates a PstI site. The splice-overlap PCR product was digested with EcoRI and PstI and ligated into pGBS19. Prospective colonies were selected on LB-kanamycin plates and confirmed by sequencing using terminal primers. Amino acid changes were incorporated on oligonucleotides using pMalE::bla and pPhoA::bla as template for the PCR reaction. The PCR product was amplified using the mutagenic and bla-rev primer pair, digested with EcoRI and PstI and cloned into pGBS19. Prospective colonies were sequenced to check for the correct base changes.
Table 1.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| Splice overlap primers | |
| Bla-rev | TTGGCTGCAGATCAACCGGGGTAAATCAAT |
| 5′bla-MBP | CAGTGAATTCTATTGAAAAAGGAAGAGTAT GAAAATAAAAACAGGTGCACGCATCCT |
| 5′MBP-bla | GATGTTTTCCGCCTCGGCTCTCGCCCACCCA GAAACGCTGGTGAAAGT |
| 3′MBPss | GGCGAGAGCCGAGGCGGAAAACATC |
| 3′phoAss | GGCTTTTGTCACAGGGGTAAA |
| 5′bla_phoA_F | CAGTGAATTCTATTGAAAAAGGAAGAGTAT GAAACAAAGCACTATTGCACTGG |
| 5′phoA_bla | TTTACCCCTGTGACAAAAGCCCACCCAGAA ACGCTGGTG |
| Mutagenic primers | |
| MBP::K2G | CAGTGAATTCTATTGAAAAAGGAAGAGTAT GGGAATAAAAACAGGT |
| MBP::K2L | CAGTGAATTCTATTGAAAAAGGAAGAGTAT GCTGATAAAAACAGGT |
| MBP::K2N | CAGTGAATTCTATTGAAAAAGGAAGAGTAT GAATATAAAAACAGGT |
| MBP::K2A | CAGTGAATTCTATTGAAAAAGGAAGAGTAT GGCAATAAAAACAGGT |
| MBP::3N | CAGTGAATTCTATTGAAAAAGGAAGAGTATG AATATAAATACAGGTGCAAACATCCTCGC |
| PhoA::K2N | CAGTGAATTCTATTGAAAAAGGAAGAGTATG AATCAAAGCAC |
Determination of minimum inhibitory concentration (MIC) and protein expression
The MIC was determined using cultures grown overnight in LB-kanamycin and subcultured 1:100 the following morning. When cells were in mid-log phase, 106 cells were added to a microtitre plate for MIC determination, containing either freshly prepared ampicillin or kanamycin two-fold serially diluted. From the same culture 1 ml was spun down and resuspended in sample buffer for western analysis using a 1:10 000 dilution of a polyclonal β-lactamase antibody (Chemicon International, AB3738).
Bioinformatic analysis
Generation of data sets
At the outset of this study each ORF of E. coli K12 (MG 1655) complete genome sequence as annotated by genbank accession U00096.2 was sorted into three groups; secreted, non-secreted and uncertain (see supplementary Table 1). The proteins were sorted based on the result of signalP 3.0 analysis (http://www.cbs.dtu.dk/services/SignalP/) of their first 70aa. Sequences whose SignalP report featured 4 ‘Y's were classed secretory; if 4 ‘N's they were classed non-secretory; and sequences with reports featuring any other combination were grouped into uncertain category. This data set formed the basis of later analysis.
Determination of N-terminal methionine removal by MAP
The online TermiNator2 webserver (http://www.isv.cnrs-gif.fr/terminator2/index.html) was used to analyse the probability of formylated methionine (f-Met) removal from predicted secreted and non-secreted protein groups. The settings used were for ‘prokaryotic protein’, ‘Intrinsic, chromosome-encoded gene’ and ‘LPR cleaved: No’ for both the predicted secretory and non-secreted proteins. The results were tabulated in excel documents (data not shown) and analysed.
RESULTS
Analysis of the charge distribution of secretory genes
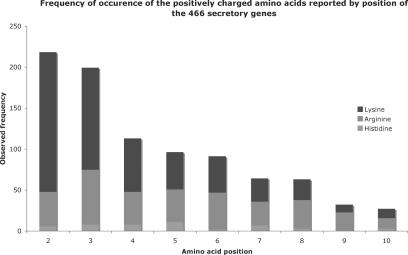
To study the distribution of positively charged residues in the signal peptide, first all protein-encoding genes (4153) from the E. coli K-12 (strain MG1655) were categorized as either secretory or non-secretory based on signalP (11) analysis of the first 70 amino acids (see Materials and Methods). This analysis generated 466 secreted and 3023 non-secreted genes with 664 not able to be classified in either group. To ensure that the entire N-region was included in the analysis, the distribution of positively charged amino acids was analysed for the first 10 amino acids from the secretory group (Figure 1). The net charge across this region was 1.84, compared to 0.4 for the non-secreted group. Approximately one-third of the net positive charge in the secretory group is due to lysine at P2 and P3.
Figure 1.
Stacked histogram showing the frequency of charged residues by position in the secretory genes of E. coli.
The extent of the bias for all charged amino acids was analysed using a χ2 test: ∑(observed−expected)2/(expected), with 19 degrees of freedom. The expected values for an amino acid were calculated using its frequency of occurrence at the respective position in all genes. This revealed that at P2, there was a massive bias for lysine (P = 1.72 × 10−24). At P3 both lysine and arginine were preferred (P = 2.71 × 10−10 × 10−9, P = 0.001, respectively), although arginine was clearly less than lysine (Table 2). This appears to be consistent with the reported key role for positively charged residues at the N-terminus of the signal peptide, but raises the question of why there is significant bias only for lysine, rather than any positively charged residues, at P2.
Table 2.
Observed and expected frequencies of amino acids at P2 and P3 in secretory group
| Amino acid | P2 | P3 | ||
|---|---|---|---|---|
| Observed | Expected | Observed | Expected | |
| Glycinea | 4 | 9 | 12 | 13 |
| Alaninea | 20 | 42 | 21 | 22 |
| Prolinea | 14 | 15 | 10 | 13 |
| Serinea | 35 | 73 | 25 | 29 |
| Threoninea | 24 | 41 | 30 | 40 |
| Valinea | 5 | 10 | 13 | 23 |
| Cysteine | 0 | 1 | 1 | 3 |
| Asparagine | 43 | 37 | 21 | 30 |
| Aspartic acid | 4 | 15 | 5 | 23 |
| Leucine | 16 | 29 | 32 | 36 |
| Isoleucine | 19 | 26 | 36 | 37 |
| Histadine | 6 | 6 | 8 | 12 |
| Glutamine | 12 | 19 | 10 | 31 |
| Glutamic acid | 9 | 20 | 2 | 27 |
| Phenylalanine | 21 | 15 | 14 | 15 |
| Methionine | 17 | 9 | 18 | 9 |
| Lysine | 170b | 67 | 124c | 55 |
| Tyrosine | 4 | 5 | 12 | 14 |
| Tryptophan | 1 | 1 | 5 | 6 |
| Arginine | 42 | 27 | 67d | 31 |
aamino acids that promote f-Met removal by MAP. The P-values are bP = 1.72 × 10−24, cP = 2.71 × 10−10, dP = 0.001, which were calculated using a χ2 test with 19 degrees of freedom. The expected values of amino acids at P2 and P3 were calculated by using their frequency of occurrence (f) at their respective position in all protein encoding E. coli genes (4153). For example, alanine occurs 372 times in all genes at P2. Expressed as an f-value this is: fala = 372/4153 = 0.0896. So the expected value of alanine in the secreted group is: 0.0896 × 466 = 41.74. The expected numbers shown in the table are the rounded to the nearest whole number.
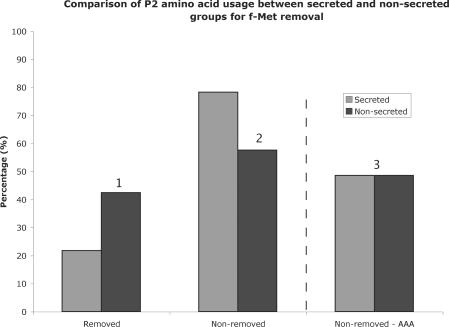
Other than a requirement for a positive charge, another possible factor constraining amino acid usage at P2 in secreted proteins could be the removal of the N-terminal methionine by methionyl amino-peptidase (MAP). All bacterial proteins start with a f-Met. The process of f-Met removal involves two steps, with the formyl group first removed by a deformylase (13), followed by excision of the N-terminal methionine by MAP. In general, amino acids at P2 that are small promote N-terminal methionine removal by MAP, whereas large amino acids prevent N-terminal methionine removal (14,15). If the N-terminus of the signal peptide is required to help initiate export by associating with the cytoplasmic membrane, then presumably removal of the N-terminal methionine by MAP, whose binding pocket recognizes the first two residues, would inhibit this process, albeit temporarily. This could form a selection pressure to use amino acids at P2 in secreted proteins that prevent N-terminal methionine removal. Using a program recently described in Frottin et al. (14), all secretory and non-secretory proteins were analysed for probability of N-terminal methionine removal (see Materials and Methods). In the secretory group, 78.33% (365/466) are predicted to retain the N-terminal methionine, compared to 57.62% (1742/3023) of non-secretory proteins (Figure 2). Assuming that there should be no difference between the two groups, this difference is significant (P < 10−5). However, the question still remains as to why only lysine, not arginine and histidine, is preferentially used in secreted proteins at P2.
Figure 2.
Graph showing the percentage of f-Met removal in secretory and non-secretory proteins using algorithm developed by Frottin et al. (14). The probability that the proportions are equivalent was calculated using a difference of proportions test. P(1 and 2) = < 10−5, P(3) = 0.999. The number in bracket corresponds to the number above three sections of the graph. The values on the right of the dotted line were with the lysine codon AAA removed from the analysis.
The lysine codon AAA is preferred at the second amino acid position in secreted proteins
Another factor that could affect amino acid composition at the second position is selection for codons that promote or hinder translation initiation efficiencies. Two independent studies have shown that codon usage at the second amino acid position can affect translation initiation efficiencies (16,17), which is thought to be the rate limiting step in translation (18,19). The strength of translation initiation correlates to the nucleotide composition, with adenosine content promoting high translation initiation efficiencies. As this factor is independent of amino acid properties but rather codon dependent, analysis on the codon usage at the second amino acid position in all groups was done using a χ2 test with 60 degrees of freedom (not including stop codons as possibilities). The expected values were calculated using the total frequency of codons in all E. coli genes at P2. The data were limited to amino acid families that occurred more than 30 times, as numbers below this are too small to do a χ2 test on. This analysis showed that only the lysine codon AAA, which is used 29.68% of the time in the secreted group at P2, occurred at levels significantly greater than expected (P = 1.7871 × 10−7) (Table 3). All other codons, including the other lysine codon AAG, occurred at frequencies that were expected (P = 0.99 for AAG). Extending the same analysis to P3, where there was a bias for the amino acids lysine and arginine, revealed no codon bias for any codon at that position. The bias for AAA at P2, and not for AAG, indicates that the selective pressure is for codon usage and not the amino acid at this position in secretory proteins.
Table 3.
Observed and expected codon usage at P2 for secretory group
| Codon | Amino Acid | Observed | Expected |
|---|---|---|---|
| UCU | Ser | 4 | 13 |
| UCC | Ser | 7 | 7 |
| UCA | Ser | 2 | 9 |
| UCG | Ser | 2 | 5 |
| AGU | Ser | 13 | 20 |
| AGC | Ser | 7 | 19 |
| AAU | Asn | 25 | 23 |
| AAC | Asn | 18 | 14 |
| AAA | Lys | 138a | 53 |
| AAG | Lys | 32 | 13 |
| CGU | Arg | 16 | 8 |
| CGC | Arg | 10 | 6 |
| CGA | Arg | 4 | 4 |
| CGG | Arg | 3 | 2 |
| AGA | Arg | 7 | 4 |
| AGG | Arg | 2 | 1 |
aP-value is 1.79 × 10−7 calculated using a χ2 test with 60 degrees of freedom. Expected values were calculated using the frequency of codon usage of all genes in the E. coli genome.
Bias for non-MAP residues entirely due to preference for AAA
To see if the statistically significant bias for non-MAP-residues at P2 in the secretory group could be attributed to AAA, all genes with AAA at P2 were removed from the analysis. This revealed that the two groups were now statistically equivalent (P = 0.99, Figure 2). This means that the bias for residues that do not promote N-terminal methionine removal at P2 seen in the secretory group is solely due to the bias for AAA at P2 in the secretory group. Removal of AAA at P2 reduces the net-positive charge at the N-terminus from 1.84 to 1.55. Therefore approximately one-sixth of the net-positive charge in secretory proteins is due to the AAA codon at P2. As AAA is the highest initiator of translation, and nearly all other codons are half as efficient (16,17), this might explain why only lysine, and in particular the codon AAA is preferred in secretory proteins.
Experimental verification that efficient translation initiation is required at P2 for secretory proteins
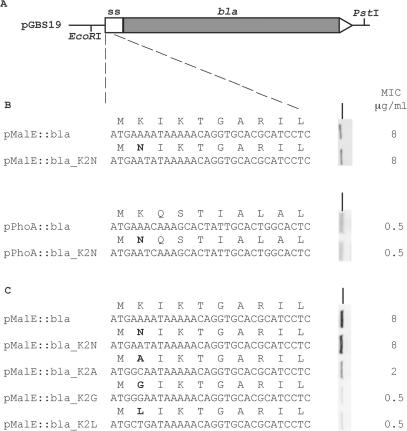
The above analysis raises the possibility that the bias for lysine in secreted proteins at the second amino acid position is due to the codon AAA to promote high translation initiation efficiencies, rather than a requirement for positive charge or to avoid N-terminal methionine removal. To investigate this experimentally, two model signal sequences with AAA at P2, maltose binding protein (MBP) and alkaline phosphatase (AP), were fused to the mature region of β-lactamase (bla) on pGBS19 generating the plasmids pMalE::Bla and pPhoA::Bla (Figure 3A). By creating fusions to β-lactamase, export to the periplasm can be measured by resistance levels to β-lactam antibiotics, as this only occurs when β-lactamase is correctly folded in the periplasm, a property that has been utilized in previous studies (20,21).
Figure 3.
(A) Schematic of the pMalE::bla and pPhoA::bla constructs used in this study. The cloning sites EcoRI and PstI are shown. The white box (to scale) represents the fusion of the signal sequence of malE and phoA to bla. For panels (B) and (C), the DNA and amino acid sequence of the first 10 amino acids are shown for all constructs used in this study. Changes to the amino acid code from the respective wt-sequences are shown in bold. To the right of each sequence is a vertical Western blot, showing the respective protein expression and the corresponding MIC values in DH5α. Panel B deals with the requirement for positive charge at P2, whereas panel C deals with the effect of N-terminal methionine removal on export. All MIC values for kanamycin were 8 mg/ml showing that plasmid copy number was constant for all constructs in this study. The black line indicates the 37.1 kDa molecular weight marker (Invitrogen, Cat. No. 10748-010). Key: ss: signal sequence.
For both fusion proteins, the lysine codon AAA was changed to the asparagine codon AAT (pMalE::Bla_K2N, pPhoA::Bla_K2N). The AAT codon is the second best initiator of translation after AAA as measured by two independent studies (16,17). This reduces the net-positive charge at the N-terminus from +1 to 0 for pPhoA::bla_K2N, and +3 to +2 for pMalE::bla_K2N. Analysis of expression as measured by MIC levels showed that the net loss of one positive charge in the MBP and AP signal peptide caused no change in ampicillin resistance levels relative to the unmodified fusion protein (Figure 3B). The Western analysis showed equivalent production of mature β-lactamase relative to the unmodified fusion protein, with no increase in precursor forms for both fusion proteins, indicating that there was no general defect in secretion caused by the reduction in net positive charge (Figure 3B). This indicates that a positive charge at the second amino acid position is not required for export, and that using a codon that initiates translation strongly has no discernable effect on export or expression.
Experimental evidence suggests no selective pressure for non-MAP residues at P2
Changing the P2 residue from lysine to asparagine does not show whether N-terminal methionine removal has any effect on protein export, as both amino acids are not removed by MAP. In order to investigate if N-terminal methionine removal has an effect on export, the second amino acid from the pMalE::Bla construct was changed to alanine (GCA), glycine (GGA) and leucine (CTG), generating the constructs pMalE::Bla_K2A, K2G and K2L. Both alanine and glycine are small amino acids that promote N-terminal methionine removal by rates of 97 and 92%, respectively, however leucine does not promote N-terminal methionine removal (14). All three codons though promote poor translation initiation rates compared to AAA (16,17). If N-terminal methionine removal has a deleterious effect on export, then one would expect the pMalE::Bla_K2L construct would have higher MIC values compared to K2A and K2G constructs. However, there was no accumulation of precursor seen from Western analysis on whole cell extracts for any second amino acid change (Figure 3C), indicating there is no defect in secretion by changing to residues that promote N-terminal methionine removal. When assayed for resistance to ampicillin, K2A had 4-fold and K2G and K2L had a 16-fold reduction in ampicillin MIC compared to MBP::bla. This correlates well with reduced translation efficiency reported in previous studies (16,17). Hence it is unlikely that N-terminal methionine removal is deleterious for secretion, and therefore unlikely to be a selecting force constraining codon choice at the second amino acid position.
DISCUSSION
In this study, the charge distribution of all E. coli sec dependent signal peptides was analysed. This revealed a massive bias for lysine at P2, which could be attributed to a bias for the lysine codon AAA. The preference for lysine codon AAA at P2 was experimentally shown not to be a requirement for a positive charge. Two studies have shown that AAA at P2 is the best initiator of translation (16,17). We propose that the selective pressure at P2 is for codons that promote faster translation initiation efficiencies. There does not appear to be any selective pressure at P2 for residues that do not promote N-terminal methionine removal.
Sequences rich in adenine nucleotides immediately downstream of the start codon have been shown to enhance gene expression, presumably by enhancing translation initiation (22). As lysine is encoded by AAA and AAG, it could be these factors that enhance choice of lysine at P2 and P3, not a requirement for a positive charge. This is supported by the ratio of lysine to arginine, which is 4.05:1 at P2 and drops at every position down to 0.41:1 by P9. If the requirement were simply for a positive charge, then one would not expect preferential usage of one basic amino acid over another. This enhances the idea that secretory proteins require higher translation initiation rates, but raises the question why that is necessary?
Signal peptides also contain the highest levels of non-optimal codons seen anywhere in the genome (23). Studies have shown that the insertion of consecutive non-optimal codons downstream of the start codon significantly lowers protein production compared to insertion of the same codons further downstream (24–26), due to ribosomes dissociating from the transcript prematurely (27). Hence for secretory proteins it is likely that ribosomes would disassociate prematurely due to the high levels of non-optimal codons in the signal sequence. Preferential use of AAA at P2, and the high use of adenine rich nucleotides at P3, which promotes rapid translation initiation, would help to counteract that effect, as subsequent ribosomes would quickly replace the previously dissociated ones. This would likely result in more ribosomes per transcript. Conversely factors that promote slow translation initiation would likely result in the spacing between ribosomes being greater, as one ribosome would be able to translate more codons before the next one commences translation.
Biasing codons to ensure rapid translation initiation could help recycle chaperones required for export. For example the molecular chaperone SecB delivers the presecretory protein to SecA, while SRP delivers the presecretory to FtsY (1). Both SecA and FtsY are inner membrane proteins. Once directed to these proteins, the chaperone is free to associate with a new nascent peptide. If ribosomes are close together on an mRNA transcript, due to increased translation initiation efficiencies, this could allow efficient binding to a new nascent peptide emerging from an upstream ribosome. This time factor may be important, as proteins must be in a loosely folded state to allow protein export (28,29). If it takes longer for the chaperone to find the next nascent peptide, this could mean the nascent peptide folds into a conformation incapable of export.
This study, as well as others (6–9), has found that a positive charge is not required for protein export. Studies have found that a net negative charge is deleterious for export, resulting in increased amounts of unprocessed precursor (7–9). Other than the extreme bias for lysine at P2 and P3, which could be to promote high translation initiation frequencies, there was no bias for a positively charged residue at any other position. This raises the possibility that the overall selective force in the entire N-region of signal peptides is to avoid a net-negative charge. Supporting this is the fact that the negatively charged residues glutamic acid and aspartic acid occur on average 0.01 times from P2-P10 in secretory proteins, compared to 0.81 times for all other genes (data not shown). Given that most secretory proteins are exported to the membrane by SRP or SecB (30), there is no requirement for the positive charge to interact with the membrane to initiate export. Once at the membrane, a net-negative charge would interfere with insertion into the membrane, due to the negatively charged phospholipids. Hence the observation that sec dependent signal peptides contain a positive charge at the N-region could be due to selection for lysine at P2 and P3 to promote high translation initiation efficiencies, and an overall selection against a net-negative charge.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
Peter Power is supported by a Beit Memorial Research Fellowship. MPJ lab is supported by NHMRC program grant 284214. Funding to pay the Open Access publication charge was provided by the NHMRC program grant 284214.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fekkes P, Driessen AJ. Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.von Heijne G. Signal sequences. The limits of variation. J. Mol. Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein HD. A surprising function for SRP RNA? Nat. Struct. Biol. 2000;7:179–181. doi: 10.1038/73265. [DOI] [PubMed] [Google Scholar]
- 4.Akita M, Sasaki S, Matsuyama S, Mizushima S. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J. Biol. Chem. 1990;265:8164–8169. [PubMed] [Google Scholar]
- 5.von Heijne G. Analysis of the distribution of charged residues in the N-terminal region of signal sequences: implications for protein export in prokaryotic and eukaryotic cells. Embo. J. 1984;3:2315–2318. doi: 10.1002/j.1460-2075.1984.tb02132.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nesmeyanova MA, Karamyshev AL, Karamysheva ZN, Kalinin AE, Ksenzenko VN, Kajava AV. Positively charged lysine at the N-terminus of the signal peptide of the Escherichia coli alkaline phosphatase provides the secretion efficiency and is involved in the interaction with anionic phospholipids. FEBS Lett. 1997;403:203–207. doi: 10.1016/s0014-5793(97)00052-5. [DOI] [PubMed] [Google Scholar]
- 7.Puziss JW, Fikes JD, Bassford P.J., Jr. Analysis of mutational alterations in the hydrophilic segment of the maltose-binding protein signal peptide. J. Bacteriol. 1989;171:2303–2311. doi: 10.1128/jb.171.5.2303-2311.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Inouye S, Soberon X, Franceschini T, Nakamura K, Itakura K, Inouye M. Role of positive charge on the amino-terminal region of the signal peptide in protein secretion across the membrane. Proc. Natl Acad. Sci. USA. 1982;79:3438–3441. doi: 10.1073/pnas.79.11.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlasuk GP, Inouye S, Ito H, Itakura K, Inouye M. Effects of the complete removal of basic amino acid residues from the signal peptide on secretion of lipoprotein in Escherichia coli. J. Biol. Chem. 1983;258:7141–7148. [PubMed] [Google Scholar]
- 10.Blattner F, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. A neural network method for identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Int. J. Neural Syst. 1997;8:581–599. doi: 10.1142/s0129065797000537. [DOI] [PubMed] [Google Scholar]
- 12.Spratt BG, Hedge PJ, te Heesen S, Edelman A, Broome-Smith JK. Kanamycin-resistant vectors that are analogues of plasmids pUC8, pUC9, pEMBL8 and pEMBL9. Gene. 1986;41:337–342. doi: 10.1016/0378-1119(86)90117-4. [DOI] [PubMed] [Google Scholar]
- 13.Pine MJ. Kinetics of maturation of the amino termini of the cell proteins of Escherichia coli. Biochim. Biophys. Acta. 1969;174:359–372. doi: 10.1016/0005-2787(69)90261-5. [DOI] [PubMed] [Google Scholar]
- 14.Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. The proteomics of N-terminal methionine cleavage. Mol. Cell Proteomics. 2006;5:2336–2349. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Ben-Bassat A, Bauer K, Chang SY, Myambo K, Boosman A, Chang S. Processing of the initiation methionine from proteins: properties of the Escherichia coli methionine aminopeptidase and its gene structure. J. Bacteriol. 1987;169:751–757. doi: 10.1128/jb.169.2.751-757.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Looman AC, Bodlaender J, Comstock LJ, Eaton D, Jhurani P, de Boer HA, van Knippenberg PH. Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. Embo J. 1987;6:2489–2492. doi: 10.1002/j.1460-2075.1987.tb02530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stenstrom CM, Jin H, Major LL, Tate WP, Isaksson LA. Codon bias at the 3′-side of the initiation codon is correlated with translation initiation efficiency in Escherichia coli. Gene. 2001;263:273–284. doi: 10.1016/s0378-1119(00)00550-3. [DOI] [PubMed] [Google Scholar]
- 18.Liljenstrom H, von Heijne G. Translation rate modification by preferential codon usage: intragenic position effects. J. Theor. Biol. 1987;124:43–55. doi: 10.1016/s0022-5193(87)80251-5. [DOI] [PubMed] [Google Scholar]
- 19.Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 1984;180:549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- 20.Kadonaga JT, Pluckthun A, Knowles JR. Signal sequence mutants of beta-lactamase. J. Biol. Chem. 1985;260:16192–16199. [PubMed] [Google Scholar]
- 21.Pluckthun A, Pfitzinger I. Membrane-bound beta-lactamase forms in Escherichia coli. J. Biol. Chem. 1988;263:14315–14322. [PubMed] [Google Scholar]
- 22.Chen H, Pomeroy-Cloney L, Bjerknes M, Tam J, Jay E. The influence of adenine-rich motifs in the 3' portion of the ribosome binding site on human IFN-gamma gene expression in Escherichia coli. J. Mol. Biol. 1994;240:20–27. doi: 10.1006/jmbi.1994.1414. [DOI] [PubMed] [Google Scholar]
- 23.Power PM, Jones RA, Beacham IR, Bucholtz C, Jennings MP. Whole genome analysis reveals a high incidence of non-optimal codons in secretory signal sequences of Escherichia coli. Biochem. Biophys. Res. Commun. 2004;322:1038–1044. doi: 10.1016/j.bbrc.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 24.Chen GF, Inouye M. Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res. 1990;18:1465–1473. doi: 10.1093/nar/18.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldman E, Rosenberg AH, Zubay G, Studier FW. Consecutive low-usage leucine codons block translation only when near the 5′ end of a message in Escherichia coli. J. Mol. Biol. 1995;245:467–473. doi: 10.1006/jmbi.1994.0038. [DOI] [PubMed] [Google Scholar]
- 26.Rosenberg AH, Goldman E, Dunn JJ, Studier FW, Zubay G. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J. Bacteriol. 1993;175:716–722. doi: 10.1128/jb.175.3.716-722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heurgue-Hamard V, Dincbas V, Buckingham RH, Ehrenberg M. Origins of minigene-dependent growth inhibition in bacterial cells. Embo J. 2000;19:2701–2709. doi: 10.1093/emboj/19.11.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verner K, Schatz G. Protein translocation across membranes. Science. 1988;241:1307–1313. doi: 10.1126/science.2842866. [DOI] [PubMed] [Google Scholar]
- 29.Pugsley AP. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Driessen AJ, Manting EH, van der Does C. The structural basis of protein targeting and translocation in bacteria. Nat. Struct. Biol. 2001;8:492–498. doi: 10.1038/88549. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.