Abstract
Constitutive and induced protein SUMOylation is involved in the regulation of a variety of cellular processes, such as regulation of gene expression and protein transport, and proceeds mainly in the nucleus of the cell. So far, several hundred SUMOylation targets have been identified, but presumably they represent only a part of the total of proteins which are regulated by SUMOylation. Here, we used the Ubc9 fusion-dependent SUMOylation system (UFDS) to screen for constitutive and induced SUMOylation of 46 randomly chosen proteins with proven or potential nuclear localization. Fourteen new UFDS-substrate proteins were identified of which eight could be demonstrated to be SUMOylated in a UFDS-independent manner in vivo. Of these, three were constitutively SUMOylated (FOS, CRSP9 and CDC37) while the remaining five substrates (CSNK2B, TAF10, HSF2BP, PSMC3 and DRG1) showed a stimulation-dependent SUMOylation induced by the MAP3 kinase MEKK1. Hence, UFDS is appropriate for the identification and characterization of constitutive and, more importantly, induced protein SUMOylation in vivo.
INTRODUCTION
SUMOylation is a protein conjugation process which leads to covalent modification of many proteins involved in transcriptional regulation, protein transport, chromosome segregation and signal transduction in a constitutive or even stimulation-dependent manner (1). The three mammalian SUMO isoforms (SUMO1–3) are expressed as precursor proteins and subsequently cleaved at their C-terminus by one of five SUMO-specific proteases (SENP1–3, SENP5 and SENP6) (1). A complex consisting of SAE1 and SAE2 (E1) (2–5) activates the maturated SUMO and transfers it to the conjugating enzyme Ubc9 (E2) (6,7). Its interaction with a substrate protein then induces the transfer of the SUMO moiety onto the substrate protein and the formation of the isopeptide bond (8). Most SUMOylation processes also seem to be assisted by SUMO ligases, which beside their different functions act as adapters that bind the conjugating enzyme Ubc9 and the substrate protein. Such SUMO ligases are the members of the PIAS (protein inhibitor of activated STAT; PIAS1, PIAS3, PIASxα, PIASxβ and PIASy) protein family (9), the Polycomb group protein Pc2 (10) and the RanBP2 (11,12), a protein of the nuclear pore. The amount of SUMOylated proteins in the cell is further regulated by the five SUMO-specific proteases, which display SUMO deconjugating activity as well (1). The number of identified SUMOylation substrates is steadily increasing especially through proteomic studies (13–17) but only for about 60 SUMOylation substrates the function of the SUMOylation is characterized in detail (18).
Recently, a Ubc9 fusion-directed SUMOylation (UFDS) system was developed that strongly increases the degree of SUMOylation of a specific substrate protein fused to Ubc9 in vivo. UFDS is efficient, selective for the in vivo SUMOylation sites and independent of SUMO ligases (19). This method should be well suited to screen for new SUMOylation substrates and, therefore, we applied UFDS for the identification of new in vivo SUMOylation substrates leading to 14 potential new substrate candidates. Verification of eight of these new substrates by UFDS independent methods revealed that the UFDS system is capable of identifying constitutive and induced SUMOylations.
MATERIALS AND METHODS
Plasmids
The destination vector (pCU-B) for the fusion of open reading frames to the N-terminus of Ubc9 was made by inserting the Gateway RfB recombination cassette (Invitrogen) into the filled-in EcoRI site of the pcDNA3-MCS-Ubc9. The destination vector for the fusion of open reading frames to the C-terminus of Ubc9 was made by amplifying the Ubc9 cDNA using the primers se-5′-CCTCGGATCCGTTATGTCGGGGATCGCCCTCAG-3′ and ase-5′-CCTCGAATTCTGAGGGGGCAAACTTCTTCG-3′ and cloning the PCR product into the BamHI/EcoRI sites of pcDNA3. Then, the Gateway-RfC.1 recombination cassette (Invitrogen) was cloned in the EcoRV site of the pcDNA3-Ubc9-MCS to obtain the destination vector pNU-C.1. The Ubc9-ORF/ORF-Ubc9 fusion protein expression vectors were obtained by recombination of the above described destination vectors with the ORF harboring entry plasmids (from the RZPD Deutsches Ressourcenzentrum für Genomforschung) with the Gateway recombination system (Invitrogen).
Cells and materials
HEK293 cells were cultured in Dulbecco's modified Eagle's medium with high glucose, complemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin. Antibodies against the following proteins or peptides were used: Ubc9 (H81, Santa Cruz) and GFP (B-2, Santa Cruz). Horseradish peroxidase-coupled secondary antibodies were from Santa Cruz.
Transfection, cell lysis and western blotting
Transfection of 50–80% confluent HEK293 cells was performed in 12-well plates using the Polyfect transfection reagent (Qiagen) according to the instructions of the supplier. Transfectants were grown for 24–48 h, then lysed in 150 µl gel loading buffer (80 mM Tris, pH 6.8, 2% SDS, 5% ß-ME, 0.01% bromphenol blue) and incubated for 10 min at 95°C. For western blot analysis the proteins were separated by SDS-PAGE, blotted on a PVDF membrane and developed with a specific primary antibody, an HRP conjugated secondary antibody, the ECL+ (Amersham) and the LAS-3000 imaging system (Fuji).
RESULTS
Identification of SUMOylation target proteins by UFDS
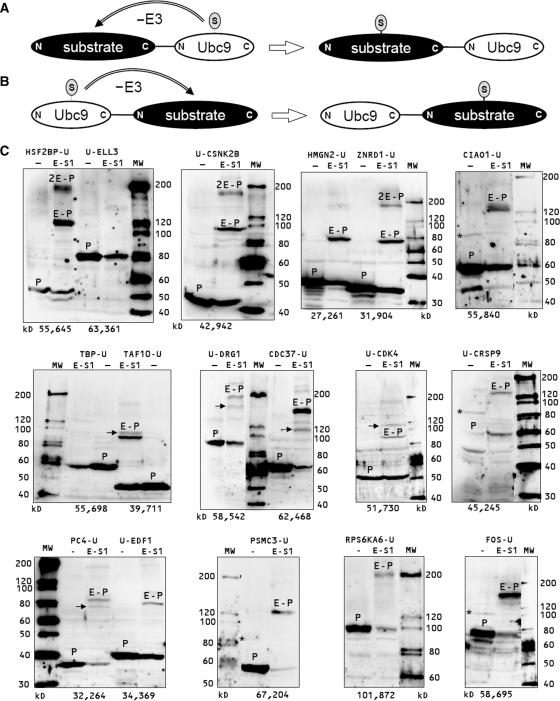
The analysis of protein functions necessitates the identification and characterization of post-translational modifications that are involved in their regulation. Since SUMOylation of most proteins is hardly detectable in vivo, we used UFDS to analyze 46 potential nuclear proteins (Table 1) for their SUMOylation. For an efficient generation of the expression plasmids for Ubc9 fusion proteins, we constructed Ubc9 fusion destination vectors that allowed the fusion of protein coding sequences to Ubc9 using the ‘Gateway’ cloning-by-recombination system (Invitrogen). We fused the coding sequences of the proteins (Table 1) to the N-terminus or the C-Terminus of Ubc9 (Figure 1A and B), to determine if Ubc9 can SUMOylate the fused protein in both structural arrangements. All Ubc9 fusion protein expression vectors were transfected alone or together with an EGFP-SUMO1 expression vector into HEK293 cells. Proteins of the transfectants were analyzed by immunoblotting using a Ubc9 antibody. Of the 46 fusion proteins analyzed, 37 were found to be expressed to different degrees, while 9 fusion proteins could not be detected. Of the expressed fusion proteins, 14 were strongly and 23 were not or only weakly SUMOylated (c.f. Figure 1C and Table 1). Nine of the strongly SUMOylated proteins were fusions to the C-terminus, while the five others were fusions to the N-terminus of Ubc9. This demonstrates that Ubc9 is able to SUMOylate the fused protein in both structural arrangements. Most of the strongly SUMOylated proteins we identified are involved in transcription (CIAO1, CRSP9, EDF1, FOS, HMGN2, HSF2BP, PC4, TAF10 and ZNRD1). The others are proteins involved in signal transduction by phosphorylation, such as the Casein kinase 2, beta polypeptide CSNK2B, the Ribosomal protein S6 kinase polypeptide 6 (RPS6KA6) and the p50CDC37, an Hsp90 chaperone protein kinase-targeting subunit. Furthermore, the proteasome 26S subunit 6A (PSMC3) and the developmentally regulated GTP binding protein 1 (DRG1) were identified.
Table 1.
Proteins analyzed for SUMOylation by UFDS
| Gene | Protein | Ubc9 | SUMOyl |
|---|---|---|---|
| ARAF1 | A-Raf proto-oncogene serine/threonine-protein kinase | N | – |
| ATF3 | Cyclic AMP-dependent transcription factor ATF-3 | C | Weak |
| BACH1 | Transcription regulator protein BACH1 | C | – |
| BHLHB2 | Basic helix-loop-helix domain containing, class B, 2 | N | No |
| CDC37 | CDC37 cell division cycle 37 homolog (S cerevisiae) | N | Strong/verified |
| CDK4 | Cyclin-dependent kinase 4 | C | Weak |
| CDK5 | Cyclin-dependent kinase 5 | N | Weak |
| CDKN2D | Cyclin-dependent kinase inhibitor 2D (p19) | C | Weak |
| CDKN3 | Cyclin-dependent kinase inhibitor 3 | C | Weak |
| CIAO1 | WD40 protein Ciao1 | N | Yes |
| CINP | Cyclin-dependent kinase 2-interacting protein | N | Weak |
| CKS2 | CDC28 protein kinase regulatory subunit 2 | C | No |
| CRSP9 | Cofactor for Sp1 transcriptional activation subunit 9 | C | Yes/verified |
| CSK | C-src tyrosine kinase | C | – |
| CSNK2B | Casein kinase 2, beta polypeptide | C | Strong/double/verified |
| DRG1 | Developmentally regulated GTP binding protein 1 | C | Yes/verified |
| DSCR1 | Down syndrome critical region gene 1 | C | No |
| EDF1 | Endothelial differentiation-related factor 1 | C | Yes |
| ELL3 | Elongation factor RNA polymerase II-like 3 | N | No |
| FOS | Proto-oncogene protein c-fos | C | Strong/verified |
| HES1 | Hairy and enhancer of split 1, (Drosophila) | C | No |
| HMGN2 | High-mobility group nucleosomal binding domain 2 | N | Strong |
| HNF4G | Hepatocyte nuclear factor 4, gamma | C | – |
| HSF2BP | Heat shock transcription factor 2 binding protein | N | Strong/double/verified |
| MAP3K8 | Mitogen-activated protein kinase kinase kinase 8 | C | – |
| MAPK13 | Mitogen-activated protein kinase 13 | C | Weak |
| MYF6 | Myogenic factor 6 (herculin) | C | No |
| NEK6 | NIMA (never in mitosis gene a)-related kinase 6 | N | Weak |
| NFE2 | Nuclear factor (erythroid-derived 2), 45kDa | C | – |
| NFIL3 | Nuclear factor, interleukin 3 regulated | C | – |
| PC4 | Activated RNA polymerase II transcription cofactor 4 | N | Yes |
| POLR2C | Polymerase (RNA) II (DNA directed) polypeptide C | N | Weak |
| PSMC3 | Proteasome 26S subunit ATPase 3 | N | Strong/verified |
| PTTG1 | Pituitary tumor-transforming 1 | C | No |
| RARA | Retinoic acid receptor, alpha | N | No |
| RFXANK | DNA-binding protein RFXANK | C | No |
| RPL7 | Ribosomal protein L7 | N | No |
| RPS6KA6 | Ribosomal protein S6 kinase, 90kDa, polypeptide 6 | N | Strong |
| STK16 | Serine/threonine kinase 16 | C | No |
| STK17B | Serine/threonine kinase 17b (apoptosis-inducing) | C | – |
| TAF10 | Transcription initiation factor TFIID subunit 10 | N | Strong/verified |
| TBP | TATA box binding protein | N | Weak |
| TCF21 | Transcription factor 21 | C | Weak |
| VDRIP | Vitamin D receptor interacting protein | C | No |
| ZNF287 | Zinc finger protein 287 | N | – |
| ZNRD1 | Zinc ribbon domain containing, 1 | N | Strong/double |
The coding sequences of the listed genes were fused with the N-terminus (N) or C-terminus (C) to Ubc9.
SUMOyl = estimation of the SUMOylation by coexpressed EGFP-SUMO1, double indicates a clear double SUMOylation of the protein, verified indicates that SUMOylation was also shown without the Ubc9 fusion, - indicates that the fusion protein was not expressed. Examples for Western blots used for the estimations are shown in Figure 1C.
Figure 1.
Identification of new SUMOylation substrates using UFDS. The fusion of Ubc9 to the C-terminus (A) or the N-terminus (B) of a substrate protein induces the E3-ligase independent SUMOylation of the substrate protein. The expression plasmids for the Ubc9 (U) fusion proteins were expressed alone or together with EGFP-SUMO1 (E-S1) in HEK293 cells. After 24–48 h protein extracts of the transfectants were analyzed by immunoblotting using an Ubc9 antibody. Examples for strong mono-SUMOylated (HMGN2, TAF10, CDC37 and FOS), strongly di-SUMOylated (HSF2BP and CSNK2B and ZNRD1), weakly (CDK4) and non-SUMOylated (ELLE3) Ubc9 fusion proteins are shown (C, c.f. Table 1). The bands for the Ubc9 fusion proteins are marked with P, the EGFP-SUMO1 conjugated Ubc9-fusion proteins with E-P Bands for double EGFP-SUMO1 conjugated Ubc9-fusion proteins are marked with 2E-P, Ubc9-fusion proteins modified at alternative SUMOylation sites are marked by black arrows. Bands representing Ubc9-fusion proteins modified by endogenous SUMO (CIAO1, CRSP9 and FOS) are marked with black asterisks.
UFDS-independent verification of SUMOylation of target proteins
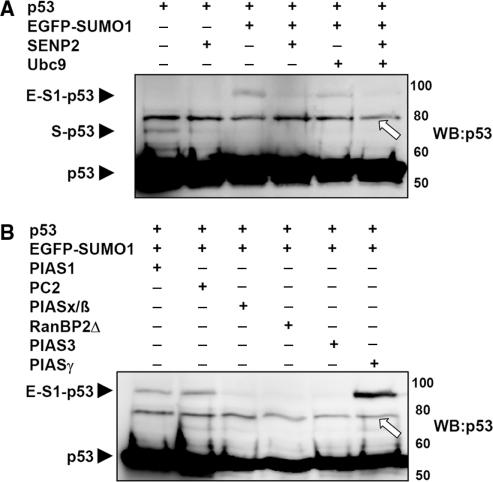
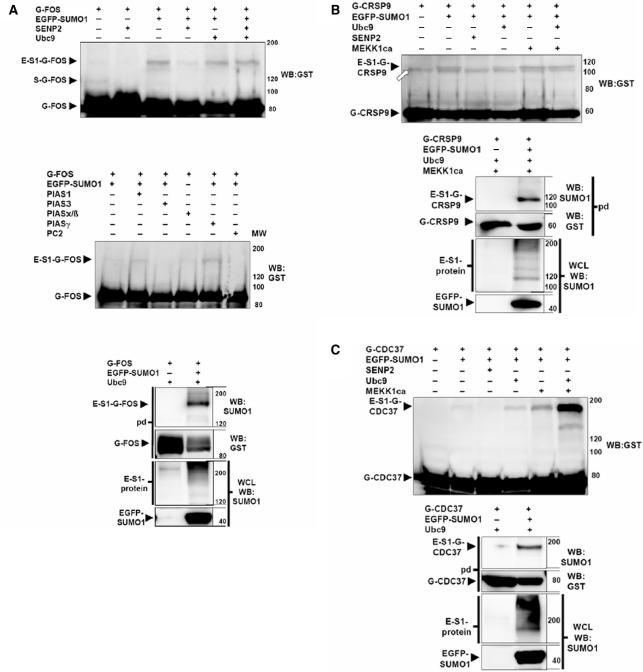
To verify the newly identified SUMOylation substrates, we analyzed their SUMOylation without the Ubc9 fusion. Because of the unavailability of suitable antibodies for most of the proteins, the SUMOylation was analyzed using GST-tagged proteins which can be pulled down with glutathion-S Sepharose and then be detected with a GST-antibody. Since SUMOylation of many proteins can hardly be detected without the presence of a specific SUMO ligase, we analyzed the potential SUMOylation target proteins also in the presence of the known SUMO ligases PC2, PIAS1/3/xß/γ and RanBP▵ which were ectopically co-expressed. As a further control, SENP2, one of the SUMO deconjugating enzymes, was coexpressed to demonstrate SUMOylation by its disappearance under these conditions. In vivo-SUMOylation was first tested for the known SUMOylation substrate p53 (21,22) and could be detected when coexpressed with EGFP-SUMO1 (Figure 2A). When SENP2 is coexpressed p53 SUMOylation is clearly diminished (Figure 2A). SUMOylation of p53 is enhanced by the SUMO ligase PIAS1 (23) and most strongly by PIASγ (24) (Figure 2B). Unexpectedly, in our hands PIASx/ß (25) did not enhance p53 SUMOylation (Figure 2B). But we found out that PC2 can enhance the p53 SUMOylation. Of the potential SUMOylation targets we could verify that FOS (Figure 3A) is SUMOylated when coexpressed with EGFP-SUMO1 and de-SUMOylated in the presence of SENP2. Furthermore its SUMOylation is enhanced by a coexpression of the SUMO ligases PIAS1 and PIASγ. CRSP9 (Figure 3B) and CDC37 (Figure 3C) are also SUMOylated when coexpressed with EGFP-SUMO1 and de-SUMOylated in the presence of SENP2, but there was no influence on their SUMOylation by coexpression of SUMO ligases (data not shown). For the remaining 11 SUMOylation substrates, we could not detect SUMOylation when coexpressed with EGFP-SUMO1 and Ubc9 (data not shown). Furthermore, none of the SUMO ligases PC2, PIAS1/3/xß/γ and RanBP▵ induced the SUMOylation of these proteins (data not shown), indicating the involvement of other specific SUMO ligases for these targets.
Figure 2.
p53 SUMOylation by Ubc9, SUMO-deconjugating enzyme (A) and SUMO ligases (B). (A and B) p53 was expressed alone or together with EGFP-SUMO1 and the indicated proteins in HEK293 cells. After 24 h, protein extracts of the transfectants were analyzed by immunoblotting using a p53 antibody (WB:p53). p53 and the p53 conjugated with endogenous SUMO (S) or with one EGFP-SUMO1 (E-S1) are indicated by black arrow heads. An unspecific band in A and B is marked by white arrows.
Figure 3.
Constitutive SUMOylation of newly identified substrate proteins without Ubc9 fusion. The expression plasmids for the GST fusion proteins G-FOS (A), G-CRSP9 (B), G-CDC37 (C) were transfected alone or together with expression plasmids for the indicated proteins in HEK293 cells. After 24 h, protein cell lysates were analyzed by immunoblotting using a GST antibody (WB: GST). The fusion proteins conjugated with endogenous SUMO (S) or with EGFP-SUMO1 (E-S1) are indicated by black arrow heads (A–C). For GST pull downs (pd) 24 h after transfection, the GST fusion proteins from the extracts of transfectants were purified on glutathione sepharose and analyzed in a western blot with a SUMO1 antibody (WB:SUMO1). Afterwards, the membranes were stripped and the fusion proteins were detected by western blot with a GST antibody (WB:GST). Additionally, the whole cell lysates (WCL) of the pull downs were analyzed for EGFP-SUMO1 expression by immunoblotting using a SUMO1 antibody (WB:SUMO1). The EGFP-SUMO1 protein is indicated by a black arrow head, the EGFP-SUMO1 conjugated proteins (E-S1-protein) are indicated by a black line. An unspecific band in B is indicated by a white arrow.
Verification of inducible SUMOylation
In view of the fact that proteomic studies have identified many stimulation-dependent SUMOylations of substrate proteins (20,26–28) and that phosphorylation-dependent SUMOylation sites have been recently described (29), we wondered whether UFDS had identified stimulation-dependent SUMOylations. To analyze a stimulation-dependent SUMOylation independent of UFDS, we coexpressed the candidate proteins with a constitutively active MEKK1 (MEKK1ca) (30). The constitutive SUMOylations of FOS (data not shown) and CRSP9 (Figure 3B) were not enhanced by the MEKK1ca coexpression, whereas the SUMOylation of CDC37 by EGFP-SUMO1 was enhanced by coexpressed MEKK1ca and was strongest when both MEKK1ca and Ubc9 were present (Figure 3C). For five other SUMOylation substrates, CSNK2B (Figure 4A), TAF10 (Figure 4B), HSF2BP (Figure 4C), PSMC3 (Figure 4D) and DRG1 (Figure 4E) we detected SUMOylation only when the various MAP-kinase cascades were activated by coexpression of MEKK1ca. Thus, UFDS had identified both target proteins for constitutive and induced SUMOylation. For the six remaining new SUMOylation substrate candidates, we could so far not unequivocally demonstrate SUMOylation in the absence of a fusion to Ubc9. This could be explained by the lack of expression of the specific SUMO ligases or the adequate stimulation in HEK 293 cells or by artificial SUMOylation in the Ubc9 fusion proteins. In spite of the large number of new SUMOylation substrates that were recently identified in proteomic studies, only one of the proteins identified in this study (FOS) was in parallel recognized as SUMOylation substrate (20). This clearly makes UFDS a powerful complementary approach for identifying in particular stimulation-dependent protein SUMOylation.
Figure 4.
Induced SUMOylation of substrate proteins without Ubc9 fusion. The expression plasmids for the GST fusion proteins G-CSNK2B (A), G-TAF10 (B), G-HSF2BP (C), G-PSMC3 (D) and G-DRG1 (E) were transfected alone or together with expression plasmids for the indicated proteins in HEK293 cells. After 24 h, protein extracts of the transfectants were analyzed by an immunoblot using a GST antibody (WB:GST). The fusion proteins conjugated with endogenous SUMO (S) or with EGFP-SUMO1 (E-S1) are indicated by black arrow heads (A–E). For GST pull downs (pd) 24 h after transfection, the GST fusion proteins from the extracts of transfectants were purified on glutathione sepharose and analyzed by a western blot with a SUMO1 antibody (WB:SUMO1). Afterwards, the membranes were stripped and the fusion proteins were detected by western blotting using a GST antibody (WB:GST). Additionally, the whole cell lysates (WCL) of the pull downs were analyzed for EGFP-SUMO1 expression by immunoblotting using a SUMO1 antibody (WB:SUMO1). The EGFP-SUMO1 protein is indicated by a black arrow head, the EGFP-SUMO1 conjugated proteins (E-S1-protein) are indicated by a black line. An unspecific band in A is marked by a white arrow.
DISCUSSION
Although hundreds of potential SUMOylation substrates have been described recently (1), many proteins were probably not accessible to this kind of analysis due to low expression rates or low amount of SUMOylation, which may result from a lack of expression of specific SUMO ligases or of an adequate stimulus. By using UFDS, out of 37 expressed proteins 14 were identified as SUMOylation substrates. Eight of these 14 potential SUMOylation substrates were also SUMOylated in vivo without fusion of Ubc9. While three of these are SUMOylated when coexpressed with EGFP-SUMO1 and one exhibited enhanced SUMOylation when coexpressed with known SUMO ligases, five proteins were only SUMOylated when constitutively activated MEKK1 was coexpressed. Therefore, constitutively active MEKK1 mimics various extracellular signal-dependent stimulations of the cell in parallel and serves as an ideal tool to analyze signal-regulated SUMOylation for candidate substrates. However, due to the broad effects of MEKK1ca, no information about the specific pathway involved in vivo can be obtained. Further work using specific stimuli and inhibitors of these pathways is needed to understand this signaling in detail.
The SUMOylation of many proteins is enhanced by different stress stimuli-like heat shock, ethanol, MG132, serum starvation (26–28) or serum stimulation (20). Recently, phosphorylation-dependent SUMOylation has been described where phosphorylation(s) C-terminal to the SUMOylation site are necessary (29) permitting signal-regulated SUMOylation. On the other hand, for the protein STAT1 we have recently shown that SUMOylation inhibits phosphorylation of a site in the vicinity of the SUMOylation (19). These findings demonstrate a full regulatory cross talk between phosphorylation and SUMOylation. Here, UFDS identified SUMOylation of substrate proteins in non-stimulated cells, whereas these substrates normally need a special stimulation to become SUMOylated. Therefore, UFDS can not only be used to identify constitutive SUMOylation, but also to identify the induced SUMOylation of proteins which are modified in vivo only in the presence of a specific, often unidentified SUMO ligase after a specific stimulation which may also alter subcellular localization or activity of this ligase. Furthermore, these results indicate that UFDS has the potency to study the function of stimulation-dependent SUMOylation without any stimulation of the cell, allowing the characterization of the function of one specific SUMOylated protein avoiding interference by parallel stimulation of other proteins.
The characteristics outlined above make UFDS eligible for the identification and analysis of unknown SUMOylation-dependent processes. This is demonstrated by the identification of 14 potential SUMOylation substrates of which only one (FOS) was described as substrate in parallel (20). The finding that UFDS is in vivo independent of both SUMO ligases (19) and specific stimuli favors that in case of the remaining six non-verified SUMOylation substrates expression of the specific SUMO ligase or a specific stimulation was not reached in HEK 293 cells. Of course, we can also not exclude that those were artificially SUMOylated by UFDS.
Interestingly, the majority of the 14 candidate proteins also represents interaction partners of other described SUMOylation substrates. CDC37 binds to the androgen receptor (32), Ciao1 is a binding partner of the Wilms Tumor suppressor protein WT1 (33), the Endothelial differentiation-related factor 1 (EDF1) binds to c-Jun (34) and Fos (20) and FOS binds to c-Jun as well(20). HSF2BP binds to the Heat shock transcription factor 2 (HSF2) (35) and PC4 interacts with p53 (36). TAF10 which we found to be strongly SUMOylated, is part of the TFIID complex that comprises the TATA box binding protein (TBP) and 13 TBP-associated factors (TAFs). Two proteins of this complex, TAF5 and TAF12, have also been reported to become SUMOylated (37). These data let us assume that the organization of several multiprotein complexes is assisted by the SUMOylation of more than one SUMOylatable proteins.
Nine of the fusion proteins were not expressed and others were strongly reduced in their expression by the coexpression of EGFP-SUMO1. Until now, most reports on SUMO conclude that SUMOylation is not involved in proteasomal protein degradation. We have not yet proved if some of the proteins we investigated are destabilized upon their Ubc9 fusion dependent SUMOylation, but UFDS should be an excellent tool for such investigations at least for proteins which are stated to be destabilized by SUMOylation (38).
Summarizing our data, it is evident that UFDS could be a valuable method for the identification of constitutive and stimulated SUMOylation and for the functional analysis of these kinds of protein SUMOylation leading to new insights on how SUMOylation regulates protein function.
ACKNOWLEDGEMENTS
We thank A. Münster-Kühnel for the expression plasmids pEGFP-C1-PIASx/ß and the pGADT7-InhSTAT3 (PIAS3), F. Melchior for the pGST-RanBP▵FG, B. Valdez for the pCMV-mGBP (PIAS1), R. Bernards for the pRc/CMV-HA-Ubc9, B. Vogelstein for the pC53-SN3 and Michael Kracht for the pMEKK1ca. We also thank Alexey Kotlyarov and Edward Hitti for helpful discussions and Thomas Binz for critical reading of the manuscript. This work was supported by the Medizinische Hochschule Hannover HILF-program (to R. N.), and by BMBF program NGFN2 (PSR-S19T04, 01GR0413, to B.K.). Funding to pay the Open Access publication charges for this article was provided by Medizinische Hochschule Hannover, Institut für Physiologische Chemie.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hay RT. SUMO: a history of modification. Mol. Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Desterro JM, Rodriguez MS, Kemp GD, Hay RT. Identification of the enzyme required for activation of the small ubiquitin-like protein SUMO-1. J. Biol. Chem. 1999;274:10618–10624. doi: 10.1074/jbc.274.15.10618. [DOI] [PubMed] [Google Scholar]
- 3.Gong L, Li B, Millas S, Yeh ET. Molecular cloning and characterization of human AOS1 and UBA2, components of the sentrin-activating enzyme complex. FEBS Lett. 1999;448:185–189. doi: 10.1016/s0014-5793(99)00367-1. [DOI] [PubMed] [Google Scholar]
- 4.Johnson ES, Schwienhorst I, Dohmen RJ, Blobel G. The ubiquitin-like protein Smt3p is activated for conjugation to other proteins by an Aos1p/Uba2p heterodimer. EMBO J. 1997a;16:5509–5519. doi: 10.1093/emboj/16.18.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Okuma T, Honda R, Ichikawa G, Tsumagari N, Yasuda H. In vitro SUMO-1 modification requires two enzymatic steps, E1 and E2. Biochem. Biophys. Res. Commun. 1999;254:693–698. doi: 10.1006/bbrc.1998.9995. [DOI] [PubMed] [Google Scholar]
- 6.Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 7.Johnson ES, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 1997b;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez MS, Dargemont C, Hay RT. SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J. Biol. Chem. 2001;276:12654–12659. doi: 10.1074/jbc.M009476200. [DOI] [PubMed] [Google Scholar]
- 9.Hochstrasser M. SP-RING for SUMO: new functions bloom for a ubiquitin-like protein. Cell. 2001;107:5–8. doi: 10.1016/s0092-8674(01)00519-0. [DOI] [PubMed] [Google Scholar]
- 10.Kagey MH, Melhuish TA, Wotton D. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 11.Kirsh O, Seeler JS, Pichler A, Gast A, Muller S, Miska E, Mathieu M, Harel-Bellan A, Kouzarides T, et al. The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J. 2002;21:2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pichler A, Gast A, Seeler JS, Dejean A, Melchior F. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 2002;108:109–120. doi: 10.1016/s0092-8674(01)00633-x. [DOI] [PubMed] [Google Scholar]
- 13.Li T, Evdokimov E, Shen RF, Chao CC, Tekle E, Wang T, Stadtman ER, Yang DC, Chock PB. Sumoylation of heterogeneous nuclear ribonucleoproteins, zinc finger proteins, and nuclear pore complex proteins: a proteomic analysis. Proc. Natl Acad. Sci. USA. 2004;101:8551–8556. doi: 10.1073/pnas.0402889101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, Kwon SW, Anselmo A, Kaur K, White MA. Broad spectrum identification of cellular small ubiquitin-related modifier (SUMO) substrate proteins. J. Biol. Chem. 2004;279:20999–21002. doi: 10.1074/jbc.M401541200. [DOI] [PubMed] [Google Scholar]
- 15.Rosas-Acosta G, Russell WK, Deyrieux A, Russell DH, Wilson VG. A universal strategy for proteomic studies of SUMO and other ubiquitin-like modifiers. Mol. Cell Proteomics. 2005;4:56–72. doi: 10.1074/mcp.M400149-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gocke CB, Yu H, Kang J. Systematic identification and analysis of mammalian small ubiquitin-like modifier substrates. J. Biol. Chem. 2005;280:5004–5012. doi: 10.1074/jbc.M411718200. [DOI] [PubMed] [Google Scholar]
- 17.Vertegaal AC, Ogg SC, Jaffray E, Rodriguez MS, Hay RT, Andersen JS, Mann M, Lamond AI. A proteomic study of SUMO-2 target proteins. J. Biol. Chem. 2004;279:33791–33798. doi: 10.1074/jbc.M404201200. [DOI] [PubMed] [Google Scholar]
- 18.Dohmen RJ. SUMO protein modification. Biochim. Biophys. Acta. 2004;1695:113–131. doi: 10.1016/j.bbamcr.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 19.Jakobs A, Koehnke J, Himstedt F, Funk M, Korn B, Gaestel M, Niedenthal R. Ubc9 fusion-directed SUMOylation (UFDS): a method to analyze function of protein SUMOylation. Nat. Methods. 2007;4:245–250. doi: 10.1038/nmeth1006. [DOI] [PubMed] [Google Scholar]
- 20.Bossis G, Malnou CE, Farras R, Andermarcher E, Hipskind R, Rodriguez M, Schmidt D, Muller S, Jariel-Encontre I, et al. Down-regulation of c-Fos/c-Jun AP-1 dimer activity by sumoylation. Mol. Cell. Biol. 2005;25:6964–6979. doi: 10.1128/MCB.25.16.6964-6979.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gostissa M, Hengstermann A, Fogal V, Sandy P, Schwarz SE, Scheffner M, Del Sal G. Activation of p53 by conjugation to the ubiquitin-like protein SUMO-1. EMBO J. 1999;18:6462–6471. doi: 10.1093/emboj/18.22.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez MS, Desterro JM, Lain S, Midgley CA, Lane DP, Hay RT. SUMO-1 modification activates the transcriptional response of p53. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahyo T, Nishida T, Yasuda H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell. 2001;8:713–718. doi: 10.1016/s1097-2765(01)00349-5. [DOI] [PubMed] [Google Scholar]
- 24.Bischof O, Schwamborn K, Martin N, Werner A, Sustmann C, Grosschedl R, Dejean A. The E3 SUMO ligase PIASy is a regulator of cellular senescence and apoptosis. Mol. Cell. 2006;22:783–794. doi: 10.1016/j.molcel.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 25.Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl Acad. Sci. USA. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitoh H, Hinchey J. Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J. Biol. Chem. 2000;275:6252–6258. doi: 10.1074/jbc.275.9.6252. [DOI] [PubMed] [Google Scholar]
- 27.Hietakangas V, Ahlskog JK, Jakobsson AM, Hellesuo M, Sahlberg NM, Holmberg CI, Mikhailov A, Palvimo JJ, Pirkkala L, et al. Phosphorylation of serine 303 is a prerequisite for the stress-inducible SUMO modification of heat shock factor 1. Mol. Cell. Biol. 2003;23:2953–2968. doi: 10.1128/MCB.23.8.2953-2968.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo D, Han J, Adam BL, Colburn NH, Wang MH, Dong Z, Eizirik DL, She JX, Wang CY. Proteomic analysis of SUMO4 substrates in HEK293 cells under serum starvation-induced stress. Biochem. Biophys. Res. Commun. 2005;337:1308–1318. doi: 10.1016/j.bbrc.2005.09.191. [DOI] [PubMed] [Google Scholar]
- 29.Hietakangas V, Anckar J, Blomster HA, Fujimoto M, Palvimo JJ, Nakai A, Sistonen L. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl Acad. Sci. USA. 2006;103:45–50. doi: 10.1073/pnas.0503698102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thiefes A, Wolf A, Doerrie A, Grassl GA, Matsumoto K, Autenrieth I, Bohn E, Sakurai H, Niedenthal R, et al. The Yersinia enterocolitica effector YopP inhibits host cell signalling by inactivating the protein kinase TAK1 in the IL-1 signalling pathway. EMBO Rep. 2006;7:838–844. doi: 10.1038/sj.embor.7400754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc. Natl Acad. Sci. USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smolen GA, Vassileva MT, Wells J, Matunis MJ, Haber DA. SUMO-1 modification of the Wilms' tumor suppressor WT1. Cancer Res. 2004;64:7846–7851. doi: 10.1158/0008-5472.CAN-04-1502. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt D, Muller S. Members of the PIAS family act as SUMO ligases for c-Jun and p53 and repress p53 activity. Proc. Natl Acad. Sci. USA. 2002;99:2872–2877. doi: 10.1073/pnas.052559499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hilgarth RS, Murphy LA, O’Connor CM, Clark JA, Park-Sarge OK, Sarge KD. Identification of Xenopus heat shock transcription factor-2: conserved role of sumoylation in regulating deoxyribonucleic acid-binding activity of heat shock transcription factor-2 proteins. Cell Stress Chaperones. 2004;9:214–220. doi: 10.1379/CSC-8R.1. Erratum in: Cell Stress Chaperones, 9, 397 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee S, Kumar BR, Kundu TK. General transcriptional coactivator PC4 activates p53 function. Mol. Cell. Biol. 2004;24:2052–2062. doi: 10.1128/MCB.24.5.2052-2062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boyer-Guittaut M, Birsoy K, Potel C, Elliott G, Jaffray E, Desterro JM, Hay RT, Oelgeschlager T. SUMO-1 modification of human transcription factor (TF) IID complex subunits: inhibition of TFIID promoter-binding activity through SUMO-1 modification of hsTAF5. J. Biol. Chem. 2005;280:9937–9945. doi: 10.1074/jbc.M414149200. [DOI] [PubMed] [Google Scholar]
- 37.Gresko E, Moller A, Roscic A, Schmitz ML. Covalent modification of human homeodomain interacting protein kinase 2 by SUMO-1 at lysine 25 affects its stability. Biochem. Biophys. Res. Commun. 2005;329:1293–1299. doi: 10.1016/j.bbrc.2005.02.113. [DOI] [PubMed] [Google Scholar]