Abstract
Sequence-specific DNA detection is important in various biomedical applications such as gene expression profiling, disease diagnosis and treatment, drug discovery and forensic analysis. Here we report a gold nanoparticle-based method that allows DNA detection and quantification and is capable of single nucleotide polymorphism (SNP) discrimination. The precise quantification of single-stranded DNA is due to the formation of defined nanoparticle-DNA conjugate groupings in the presence of target/linker DNA. Conjugate groupings were characterized and quantified by gel electrophoresis. A linear correlation between the amount of target DNA and conjugate groupings was found. For SNP detection, single base mismatch discrimination was achieved for both the end- and center-base mismatch. The method described here may be useful for the development of a simple and quantitative DNA detection assay.
INTRODUCTION
There is a major need to develop fast, cheap and precise detection methodologies that detect DNA samples at extremely low concentration. This ability is critical to basic life sciences, medical diagnosis and treatment, pharmaceutical applications, identification of biological weapons, as well as forensic analysis (1–3). To fulfill this goal, the scientific community is striving to develop new methods and assays that are highly selective and sensitive. Optical/colormetric (4–6), fluorescent (7,8) and electrochemical (9–11) based methods have been reported for detection of DNA samples. Among these new methodologies, optical detection methods, which rely on the hybridization between target DNA and substrate modified with radioactive, fluorescent, chemiluminescent or nanoparticle tags, are of particular interest (12–14). The use of gold nanoparticles (nAu) as labeling tags receives most attention in recent years, due to their unique chemical and physical properties (15–17) that can be exploited in the development of highly sensitive detection assays (18,19). Although still in its infancy, the application of surface-functionalized nAu in sequence recognition has shown great promise in achieving high sensitivity that is difficult to achieve by conventional methods. Mirkin and co-workers have developed a series of nAu–based DNA detection methods, such as scanometric and bio-barcode assays, that reach attomolar and high zeptomolar sensitivity (2,19,20). Such sensitivity may allow the direct detection of genomic DNA and bypass the need of amplification that is usually done using polymerase chain reaction (PCR).
Besides sensitivity, quantification and selectivity are the other two important aspects for the evaluation of DNA biosensor devices. DNA quantification is critical for gene expression analysis, detection of DNA mutations or genetic defects, early stage diagnosis of critical illness such as HIV and cancers, and forensic applications (21–23). Furthermore, diagnosis of pathogenic and genetic diseases requires the device to have high selectivity that can discriminate single nucleotide mismatches (1,18). Single nucleotide polymorphisms (SNPs) are the most abundant form of genetic variation that occur once every 100–300 bases and there are greater than 3 million SNPs in the human genome (24). Identification of these SNPs and associate individual SNPs with specific diseases and pharmacological responses are clinically important for medical diagnostics, disease prevention and prognostics (25,26). These needs have driven intense efforts toward the development of new methodologies that enable quantitative, selective and cost-effective detection of SNP in DNA samples (19,27). Currently, real-time polymerase chain reaction (RT-PCR) is one of the most frequently used methods for DNA quantification and SNP discrimination in life science and clinical research. However RT-PCR is a time-consuming and labor-intense process, and its selectivity is not always satisfactory even with sophisticated optimizations (28,29). For commonly used DNA detection systems such as DNA chips, the selectivity and quantification are dependent on the dissociation properties of target DNA hybridized with capture strands immobilized on the chip (27). To achieve SNP discrimination, a stringent wash step has to be performed to remove mismatched DNA binding on the capture strands. However, the difference in binding affinity between a perfectly matched target DNA and one with a mismatched base is usually too small to achieve complete discrimination (19).
Previously, we have shown that gold nanoparticle–DNA (nAu–DNA) conjugates bearing definite number of short DNA (<20 bases) can be prepared by gel electrophoresis isolation followed by restriction endonuclease manipulation of the nAu–bound DNA (30). Simply loading short DNA onto the nAu directly followed by gel electrophoresis separation only yields a smear and not individual bands, which correspond to conjugates bearing definite number of DNA. This is because the mobility difference between conjugates bearing different number of short DNA is insignificant. Thus, we reported to first use gel electrophoresis to separate nAu bearing definite number of >50-base DNA strands. Subsequently restriction endonuclease can be used to cleave the long DNA to obtain the short DNA on nAu. In this study, we described a novel gold-nanoparticle (nAu)-based assay methodology that has reliable quantification ability and SNP discrimination selectivity. In this assay, two sets of specially designed nAu–DNA conjugates are fabricated via the gel electrophoresis and restriction endonuclease manipulation methods. These two sets of conjugates with definite number (1, 2, 3…) of short single-stranded DNA (ssDNA) probes are not directly complementary to each other. After mixing, these conjugates do not recognize and group each other until a target DNA that is complementary to both sets of conjugates is introduced. Only conjugate groupings with defined structure (dimer or trimer) can form due to definite number of DNA strands on each nAu. The resulting conjugate groupings are characterized and quantified by agarose gel electrophoresis. The size differentiation ability of gel electrophoresis allows strict discrimination between different conjugate structures (monomer, dimer and trimer) and enables precise quantification of target DNA samples. Furthermore, a strong discrimination between perfectly matched and single base mismatched DNA is achieved since only the perfectly matched target DNA allows the formation of conjugate groupings.
MATERIALS AND METHODS
Materials
Hydrogen tetrachloroaurate (III) trihydrate, trisodium citrate dihydrate, tannic acid, dithiothreitol (DTT), NaCl, MgCl2 and tris-borate-EDTA (TBE) buffer were purchased from Sigma-Aldrich. The synthetic DNA (modified with thiol linker at the 5′ end) was purchased from Integrated DNA Technologies (IDT). Restriction endonuclease EcoRV (500 000 U/ml) was obtained from New England Biolabs. Thirty percent acrylamide/Bis solution (29:1) and ethidium bromide were obtained from Bio-Rad Laboratories. Agarose was purchased from Cambrex. Dialysis tubing (Float-A-lyzer, MWCO: 3500) was obtained from Spectra/Por. Milli-Q water with resistance >18 MΩ/cm was used throughout the experiments.
Synthesis and characterization of nAu
nAu were synthesized by the reduction of hydrogen tetrachloroaurate (III) trihydrate by trisodium citrate dihydrate and tannic acid (31). In order to determine the size and size distribution of the resulting nAu, TEM characterization was performed on a Philips CM300 FEG system operating at 200 kV. At least 200 particles were sized from TEM micrographs via graphics software ‘Image-Pro Express’ (Media Cybernetics). The mean particle diameter was 10 nm and the size distribution was within 15% of the mean diameter.
Preparation of nAu–DNA conjugates
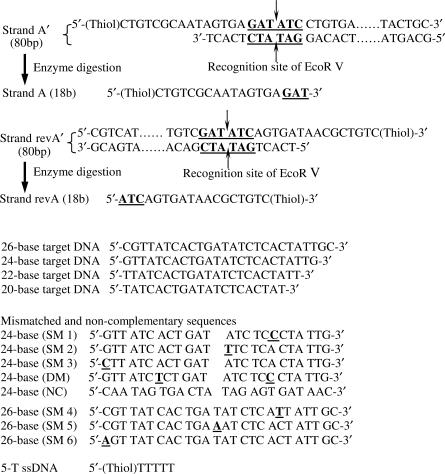
The sequences of DNA used in this study are shown in Figure 1 (complete sequences are shown in Figure S1, Supplementary Data). Two complementary single-stranded DNA (ssDNA), one modified with a thiol linker, was allowed to hybridize to form a double-stranded DNA (dsDNA). Two dsDNA, Strand A′ with a 5′ thiol group and Strand revA′ with a 3′ thiol group, were used to prepare two sets of conjugates (nAu–A′ and nAu–revA′) for conjugate grouping studies. The detailed procedure of these conjugates preparation can be found in our previous work (30). Briefly, 2 μM dsDNA (Strand A′ or revA′) was mixed with nAu for 2 h followed by 5-T ssDNA for surface passivation. Excess reagents were then removed by repeated washing and centrifuging the samples with 0.5X TBE.
Figure 1.
DNA sequences used in this study. For Strands A′ and revA′, the underlined sequences are the recognition site of EcoR V and the arrows indicate the enzyme cleavage site. For the mismatched DNA sequences, the underlined base is the mismatch. SM1/SM2/SM3/SM4/SM53/SM6 refer to single-base mismatched DNA, DM refers to double-base mismatched DNA and NC refers to non-complementary DNA.
Agarose gel electrophoresis isolation and extraction of nAu–DNA conjugates
Agarose gel electrophoresis (3% agarose at 5 V/cm, 0.5X TBE as running buffer) was carried out for 4 h to separate the conjugates bearing from one to three dsDNA molecules. As shown previously, discrete bands can easily be identified because of the wine-red color of nAu (30). Desired bands were extracted from the gel and the conjugates were then recovered by electrophoretic dialysis (32). After recovering from agarose gel, the conjugates were purified by repeated washing (with 50 mM tris buffer, pH 8.0) and centrifuging to ensure complete removal of EDTA which may deactivate the restriction endonuclease.
Enzyme manipulation of nAu–bound DNA and digestion efficiency
The restriction endonuclease digestion of nAu–bound DNA (Strands A′ and revA′ to Strands A and revA respectively) was performed by incubating the conjugates with 100 units of EcoRV at 37°C in a 200 μl reaction buffer for 20 h. After incubation, the enzyme was deactivated by adding 50 mM EDTA. The digested conjugates were analyzed by polyacrylamide gel electrophoresis to confirm the high digestion efficiency and high yield of nAu conjugated with definite number of DNA strands (Figure S2, Supplementary Data).
Formation and analysis of nAu–DNA conjugate groupings
Two sets of nAu, each carrying a single DNA probe (nAu–A or nAu–revA, 1.5 pmol of each) were mixed in a buffer containing 50 mM tris pH 8.0, 100 mM NaCl and 2 mM MgCl2 at a 1:1 molar ratio. Target/linker DNA molecules (matched and mismatched, Figure 1) were then added for hybridization with Strand A and rev A on the nAu surface in a tail-to-tail configuration (Figure 2). The ratio between the two sets of nAu–DNA conjugates and the target DNA (nAu–A: nAu–revA: target) varied from 1:1:0.1 to 1:1:1. The hybridization of target DNA was carried out by first heating the sample to 70°C for 2 min to ensure complete melting of the DNA strands and then the samples were slowly cooled down to 25°C at a rate of 0.2°C/min to form stable duplexes. After formation of conjugate groupings, agarose gel electrophoresis (3% agarose at 5 V/cm, 0.5X TBE as running buffer) was carried out at 4°C to characterize and quantify the conjugate groupings. Desired bands of conjugate groupings were visualized by a Syngene GeneGenius UV/white light gel documentation system or a Bio-Rad GS-800 calibrated densitometer. The grouping percentage of nAu–DNA conjugates was calculated by quantifying the relative optical intensity of the gel bands in the same lane using gel analysis software from manufacture, more specifically, by dividing the amount of conjugate groupings by the total amount of conjugates in the same lane (unbound conjugates + conjugate groupings). Each value reported was the average of three individual tests. After that the conjugate groupings were extracted from the gel and recovered by electrophoretic dialysis32 for TEM characterization.
Figure 2.
Schematic picture of the formation of nAu–DNA conjugate groupings.
RESULTS AND DISCUSSION
Formation of nAu–DNA conjugate dimers using target/linker DNA of different lengths
As reported previously, agarose gel electrophoresis was first used to isolate conjugates (nAu–A′ and nAu–revA′) bearing different number of DNA molecules (30). Subsequently the conjugates with definite number of DNA strands were exposed to endonuclease EcoRV digestion to cleave Strands A′ and revA′ (80 bp) into Strands A and revA (18 bases). The enzyme digestion efficiency of 10 nm nAu–bound DNA obtained from 12% PAGE (Figure S2, Supplementary Data) shows more than 90% digestion efficiency for both Strands A′ and revA′. The use of endonuclease to cleave nAu–bound DNA has been reported by several groups (33–36) including ourselves (30,37). Our high digestion efficiency here leads to short and homogeneous DNA on nAu which is critical for the subsequent quantitative DNA analysis.
Though nanoparticle assemblies has been studied extensively, most studies use nanoparticles with a high loading of DNA and lead to a coordinate effect from multiple DNA linkages. To avoid this scenario, nAu–A and nAu–revA conjugates with single DNA strands are used here. nAu–DNA conjugate dimer formed with various DNA targets/linkers (20 bases to 26 bases) was chosen as a model system. A tail-to-tail structure was selected to reduce the steric hindrance and allow maximum grouping efficiency (Figure 2). The conjugate dimers formed by nAu–A and nAu–revA grouping are shown in Figure 3. The stoichiometric ratio of nAu–A, nAu–revA, and target DNA used in hybridization is 1:1:1. Since Strand A and Strand revA are not complementary to each other, a linker is needed for the formation of conjugate dimers. Lane A in Figure 3 corresponds to nAu without Strand A or revA modification (control). Lanes B, C, D, E and F correspond to nAu–A + nAu–revA conjugates with no target DNA, with 26, 24, 22 and 20-base target DNA respectively. Using Lane B as a reference (contains only nAu–A and nAu–revA conjugate monomers), the bands on the bottom of Lanes C/D correspond to the unbound conjugate monomers (share the same mobility with the band in Lane B) and the bands on the top (with slower mobility) correspond to the conjugate dimers. The presence of only a single band in Lane B indicates that there is no nonspecific interaction between the nAu–A and nAu–revA. Therefore, the hybridization of complementary target DNA with these two conjugates, not nonspecific interaction, drives this conjugate dimer formation.
Figure 3.
Formation of nAu–DNA conjugate dimers with various lengths of DNA target. Lane A corresponds to nAu without Strand A or revA modification (control); Lane B corresponds to nAu–A + nAu–revA conjugates with no target; Lanes C, D, E and F correspond to nAu–A + nAu–revA conjugates with 26, 24, 22 and 20 bases of target DNA respectively.
With DNA targets of different lengths, the dimer grouping percentages of 26- and 24-base DNA targets are 55.6 and 55.3% respectively, even 1:1:1 ratio is used to allow complete grouping formation. Similar result was previously reported by Alivisatos and co-workers in which complete grouping of nAu–DNA conjugates could not be achieved even when a target with complementary sequence of 100-base was used (38,39). In our experiment, when the 20- and 22-base DNA targets are used, only a single band corresponding to conjugate monomers is unexpectedly found in the gel (Lanes E and F). With half of the DNA target hybridizes to each conjugate, reducing the target from 24 to 22 bases cuts only a single base in conjugate hybridization. This single base cut, however, results in total absence of dimer groupings. The modification of nAu with 5-T ssDNA as surface passivation in our experiment leads to a highly negatively charged surface. The strong electrostatic repulsion between both conjugates and perhaps target DNA may destabilize the hybridization of short sequences, and thus no dimer is formed. The critical length of DNA target for this particular system is found to be 24 bases.
Quantification of target DNA through the formation of nAu–DNA conjugate dimers
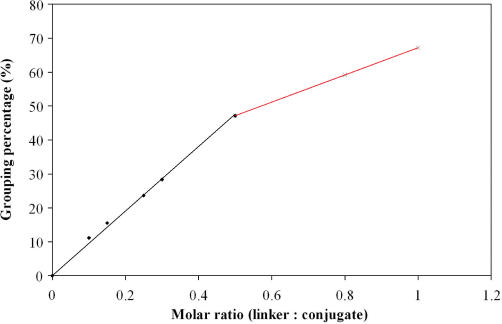
Our nAu–DNA conjugates carry only a single DNA probe and this allows quantitative analysis of target DNA that acts as a linker. This is because each conjugate dimer formation represents a hybridization event between a single complementary target DNA and the two nAu–DNA conjugates. By measuring the dimer formation, in principle, the amount of the target or linker DNA can then be quantified. In our experiments, equal molar of the nAu–A and nAu–revA conjugates was mixed with various ratios of 24-base target DNA (nAu–A: nAu–revA: target DNA ranges from 1:1:0.1 to 1:1:1). The target DNA amount was obtained by quantifying the percentage of conjugate dimers after agarose gel electrophoresis. In Figure 4, Lane J corresponds to the control where nAu was conjugated without Strand A/revA, and Lanes K is the mixture of nAu–A and nAu–revA without target DNA. Only one band corresponding to conjugate monomers is found in the gel, indicating no dimer formation. Lanes G, H, I, L, M, N and O are samples with various ratios of nAu–A and nAu–revA to target DNA, from 1:1:1 to 1:1:0.1 respectively. Two bands, which correspond to conjugate monomers and dimers, are found in the gel. The amount of the conjugate dimers increases with the increasing molar amount of target DNA. This trend is more clearly shown in Figure 5 after quantifying the dimer band intensity. When the molar ratio of the target DNA is between 0 and 0.5, the dimer grouping percentage is directly proportional to the target DNA ratio. Such linear relationship between the amount of target DNA and dimer formation has good potential for the development of new target quantification assay.
Figure 4.
Grouping percentage of nAu–DNA conjugates with various ratios of target DNA. Lanes G–O correspond to nAu–A: nAu–revA: target DNA ratio of 1:1:1, 1:1:0.8, 1:1:0.5, control (nAu without Strand A/revA conjugation), 1:1:0, 1:1:0.3, 1:1:0.25, 1:1:0.15 and 1:1:0.1, respectively. Gel picture shows the combined results from two experiments.
Figure 5.
Grouping percentage of nAu–DNA conjugates with different ratios of target DNA.
When the molar ratio goes beyond 1:1:0.5, both Figures 4 and 5 show that only minor increase in the grouping percentage (less than 20%) is found. For example, 1:1:0.8 and 1:1:1 can only result in 59 and 64% dimer formation respectively. Similar grouping percentage is also found when conjugates with two Strand revA (nAu–2x revA) were used. The top bands in Lanes P, Q and S in Figure 6 correspond to the conjugate trimers that have the lowest mobility among all the conjugates. The second bands from the top are conjugate dimers, and bands at the bottom are conjugate monomers. The maximum grouping percentage (dimers + trimers) of 66% is achieved when molar ratio of 2:1:2 is used. Excess target DNA, however, does not further improve the grouping percentage as the grouping percentage falls slightly to 60% when the ratio of target DNA increases to 2:1:5. Further drop in grouping is observed upon the introduction of large excess of target DNA, as the Lanes S and T (ratio 2:1:20 and 2:1:100) show fade bands of conjugate groupings with only 51 and 36% grouping percentage respectively. This decrease in grouping percentage can be attributed to the conjugate grouping inhibition by the excess target DNA. Since each nAu–DNA conjugate may hybridize with individual target DNA molecule separately, less conjugates are available for grouping and thus the dimer/trimer formation is impeded.
Figure 6.
Grouping percentage of nAu–DNA conjugates with different ratios of target DNA. Lanes P–T correspond to nAu–A and nAu–revA with target DNA at a ratio of 2:1:2 (nAu–A:nAu–2x revA:target DNA), 2:1:5, control (nAu without Strand A/revA conjugation), 2:1:20 and 2:1:100, respectively. Gel picture shows the combined results from two experiments.
For the grouping of nAu–DNA conjugates, the highest percentage is found to be ∼60–65%. This incomplete grouping can be attributed to the low number of Strand A/revA bound on nAu (1 or 2 strands per nAu), which may result in a much lower collision rate between the conjugates and target DNA. Thus extended incubation may be necessary to reach higher degree of hybridization. Furthermore, the relatively heavy nAu conjugates compared with the target DNA may further reduce the collision rate and contribute to the observed low hybridization percentage. Hybridization data in Supplementary Data (Figure S3 and Table S1) shows that without nAu, hybridization between Strands A, revA and target DNA is efficient and over 90%. This further suggests the possibility of nAu interference in the hybridization process. Further study is needed to obtain clearer understanding of DNA hybridization on nAu.
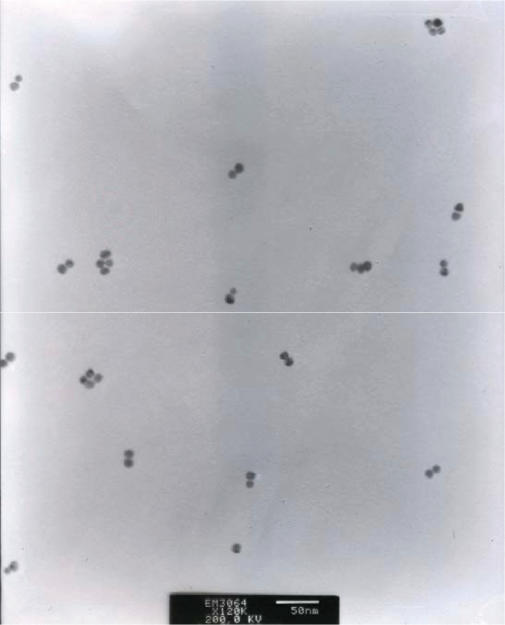
To give a direct connection between conjugates with different electrophoretic mobility and their actual structure, dimer groupings were recovered from the gel and visualized by TEM. Figure 7 shows the structure of the conjugate groupings from the dimer band extracted from Lane P. The large majority of the conjugates are participated in the same dimer grouping structure, indicating that each discrete gel band does contain a single type of grouping and not a mixture of several conjugate structures. A small number of monomers and high-order groupings were observed in the TEM image, and this may be due to the disruption during sample extraction from gels as well as during TEM preparation where solvent evaporation may result in particle aggregation.
Figure 7.
TEM image of nAu–DNA conjugate dimers.
SNP discrimination using nAu–DNA conjugate groupings
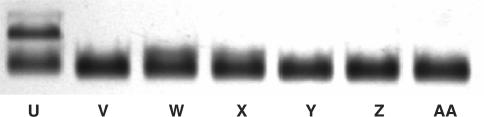
Conventional techniques for the detection of SNP using mass spectrometry or gel electrophoresis to discriminate DNA fragments (25,40) are time consuming and relatively costly. Chip-based detection methods using fluorescent dyes or nanoparticles as labels have become popular in recent years (19,24). These newly emerging methods are based on the melting temperature difference between perfectly matched and mismatched DNA duplex, and involve a stringent wash process with precise temperature control as well as skillful personnel and long analysis time. Furthermore, it may be difficult to discriminate DNA targets that exhibit insignificant melting temperature difference. Compared with the existing techniques mentioned above, our newly proposed method offers the advantage of straightforward single-base mismatch discrimination without the need of stringent wash (24). As shown in Figure 8, Lanes U-AA correspond to nAu–A and nAu–revA plus 24-base perfectly matched DNA (PM), single base matched DNA (SM1/SM2/SM3), double base matched DNA (DM), non-complementary DNA (NC) and no target DNA respectively. There is an obvious difference in Lane U where the PM case shows a top dimer band and a bottom monomer band. For the DM and NC cases (Lanes Y and Z), only a single band can be found in the gel, indicating that no dimer structure is formed. For SM1, 2 and 3 cases (Lanes V, W and X), only a single band corresponding to conjugate monomers is found, and no false positive signal is observed. The monomeric structure in these three lanes can be further confirmed by the one without target DNA (Lane AA), which shows exactly the same electrophoretic mobility. Even with end-mismatched SNP (Lane X), which introduces least interruption to the duplex DNA structure and is often difficult to discriminate, this method shows very good discrimination and no sign of conjugate dimers is observed in Lane X. The key strategy to achieve such high selectivity in SNP discrimination is to use unfavorable conditions for hybridization, so that only target DNA with a perfect match has a chance to hybridize with nAu–bound DNA. This unfavorable condition is most likely resulted from the highly negatively charged nAu surfaces (nAu–A) which repel the target DNA from hybridizing with the other conjugate (nAu–revA). As a result, even single base mismatched at the end position (SM3) can lead to sufficient destabilization to the dimers, and only perfectly matched duplex is energetically stable to form dimer grouping. To further confirm the critical role of nAu in SNP discrimination, the melting behavior of free dsDNA (PM, SM1, SM2 and SM3) in the absence of nAu was conducted using real-time PCR (Figure S4, Supplementary Data). The results show that the center-mismatched SM1 and SM2 show wider melting transition and lower peak value compared with PM. However, the end-mismatched SM3 shows almost the same melting transition and is hardly distinguishable compared with PM. This is not unexpected since end mismatching introduces least disturbance to the DNA duplex, the difference in melting behavior between PM and SM3 is too small to support an accurate discrimination. Due to the difficulty of detecting end-mismatched sequences, the location of SNP should be placed at the middle of the probe in the design of probe sequences to enhance discrimination ability.
Figure 8.
SNP discrimination using nAu–DNA conjugates and 24-base target DNA. Lanes U-AA correspond to, nAu–A and nAu–revA plus 24-base perfectly matched DNA (PM), single base matched DNA (SM1/SM2/SM3), double base matched DNA (DM), non-complementary DNA (NC) and no target DNA, respectively.
To check the SNP discrimination ability of our system with longer DNA, we used 26-base target DNA to test the same nAu–DNA conjugates. We found the discrimination is as good as that of the 24-base target. As shown in Figure 9, we mixed, in Lane BB, perfectly matched (PM) target with three other single mismatched sequences (SM4, SM5 and SM6) for conjugate dimer formation. The ratio between nAu–A, nAu–revA and PM/SM4/SM5/SM6 is 1:1:0.25/0.25/0.25/0.25. The result shows that ∼25% of dimer is formed, which is the amount of PM. Therefore the quantitative analysis of target DNA is not interfered by the presence of mismatched DNA. Using SM4, SM5 and SM6 alone, no dimer formation can be found in Lanes CC, DD and EE. SM6 has again an end-mismatched sequence and our nAu conjugate system is able to differentiate it. Therefore, we expect our system to be applied to even longer target DNA without jeopardizing the discrimination ability.
Figure 9.
SNP discrimination using nAu–DNA conjugates and 26-base target DNA. Lane BB corresponds to nAu–A and nAu–revA and 26-base perfectly matched DNA (PM) plus single base matched DNA SM4, SM5 and SM6 (ratio of nAu–A:nAu–revA:PM/SM4/SM5/SM6 = 1:1:0.25/0.25/0.25/0.25), and Lanes CC to EE correspond to nAu–A and nAu–revA plus SM4/SM5/SM6, respectively (ratio of nAu–A:nAu–revA:SM DNA = 1:1:1).
The data reported here show that the current conjugate grouping method reaches only 100 fmol range sensitivity, which is not as sensitive as other methods reported in the literature (2,19,20). Thus, the method is likely applicable for analyzing amplified DNA and not genomic DNA directly. Further work to improve the sensitivity is necessary for possible genomic DNA detection. Finally, a possible merit of the method reported here can be its ease of use. Most existing and highly sensitive methods require specialized readout platforms, such as surface plasmon resonance (SPR), fluorescent microarrays, and scanometry. Storhoff and co-workers reported a straightforward DNA detection method by allowing nAu–DNA conjugates to cross-link in the presence of target DNA in solution (41). The cross-linked nAu shows a red shift in the scattering spectrum and the result can be read even with naked eyes. Our method also requires fairly simple readout, which is a gel electrophoresis setup and is commonly found in biology and clinical labs.
CONCLUSIONS
In this study, we have demonstrated a novel nAu–based quantitative DNA assay method with SNP discrimination sensitivity. This method combines gel electrophoresis isolation and restriction endonuclease manipulation to produce precisely controlled nAu–DNA conjugates which allows quantitative analysis of DNA molecules based on the formation of conjugate groupings by target DNA linkage. A linear correlation between the amount of target DNA and conjugate groupings was obtained at lower target DNA concentration and can further be exploited for target quantification. For SNP study, single base mismatch discrimination is achieved for both the end- and center-mismatched cases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors would like to thank the National University of Singapore for providing the doctoral scholarship for W.J.Q. and the research funding (R-279-000-125-112 and R-279-000-205-112) for this work. Funding to pay the Open Access publication charges were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Patolsky F, Lichtenstein A, Willner I. Detection of single-base DNA mutations by enzyme-amplified electronic transduction. Nat. Biotechnol. 2001;19:253–257. doi: 10.1038/85704. [DOI] [PubMed] [Google Scholar]
- 2.Nam JM, Stoeva SI, Mirkin CA. Bio-bar-code-based DNA detection with PCR-like sensitivity. J. Am. Chem. Soc. 2004;126:5932–5933. doi: 10.1021/ja049384+. [DOI] [PubMed] [Google Scholar]
- 3.Storhoff JJ, Marla SS, Bao P, Hagenow S, Mehta H, Lucas A, Garimella V, Patno T, Buckingham W, et al. Gold nanoparticle-based detection of genomic DNA targets on microarrays using a novel optical detection system. Biosens. Bioelectron. 2004;19:875–883. doi: 10.1016/j.bios.2003.08.014. [DOI] [PubMed] [Google Scholar]
- 4.Alivisatos AP, Johnsson KP, Peng XG, Wilson TE, Loweth CJ, Bruchez MP, Schultz PG. Organization of ‘nanocrystal molecules’ using DNA. Nature. 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 5.Mirkin CA, Letsinger RL, Mucic RC, Storhoff JJ. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature. 1996;382:607–609. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 6.Sonnichsen C, Reinhard BM, Liphard J, Alivisatos AP. A molecular ruler based on plasmon coupling of single gold and silver nanoparticles. Nat. Biotechnol. 2005;23:741–745. doi: 10.1038/nbt1100. [DOI] [PubMed] [Google Scholar]
- 7.Dubertret B, Calame M, Libchaber AJ. Single-mismatch detection using gold-quenched fluorescent oligonucleotides. Nat. Biotechnol. 2001;19:365–370. doi: 10.1038/86762. [DOI] [PubMed] [Google Scholar]
- 8.Piestert O, Barsch H, Buschmann V, Heinlein T, Knemeyer JP, Weston KD, Sauer M. A single-molecule sensitive DNA hairpin system based on intramolecular electron transfer. Nano Lett. 2003;3:979–982. [Google Scholar]
- 9.Bardea A, Patolsky F, Dagan A, Willner I. Sensing and amplification of oligonucleotide-DNA interactions by means of impedance spectroscopy: a route to a Tay-Sachs sensor. Chem. Commun. 1999;1:21–22. [Google Scholar]
- 10.Patolsky F, Katz E, Willner I. Amplified DNA detection by electrogenerated biochemiluminescence and by the catalyzed precipitation of an insoluble product on electrodes in the presence of the doxorubicin intercalator. Angew. Chem. Int. Edit. 2002;41:3398. doi: 10.1002/1521-3773(20020916)41:18<3398::AID-ANIE3398>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 11.Park SJ, Taton TA, Mirkin CA. Array-based electrical detection of DNA with nanoparticle probes. Science. 2002;295:1503–1506. doi: 10.1126/science.1067003. [DOI] [PubMed] [Google Scholar]
- 12.Maxwell DJ, Taylor JR, Nie SM. Self-assembled nanoparticle probes for recognition and detection of biomolecules. J. Am. Chem. Soc. 2002;124:9606–9612. doi: 10.1021/ja025814p. [DOI] [PubMed] [Google Scholar]
- 13.Li HX, Rothberg L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc. Natl Acad. Sci. USA. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill R, Willner I, Shweky I, Banin U. Fluorescence resonance energy transfer in CdSe/ZnS-DNA conjugates: Probing hybridization and DNA cleavage. J. Phys. Chem. B. 2005;109:23715–23719. doi: 10.1021/jp054874p. [DOI] [PubMed] [Google Scholar]
- 15.Lee J, Govorov AO, Dulka J, Kotov NA. Bioconjugates of CdTe nanowires and Au nanoparticles: Plasmon-exciton interactions, luminescence enhancement, and collective effects. Nano Lett. 2004;4:2323–2330. [Google Scholar]
- 16.Dulkeith E, Ringler M, Klar TA, Feldmann J, Javier AM, Parak WJ. Gold nanoparticles quench fluorescence by phase induced radiative rate suppression. Nano Lett. 2005;5:585–589. doi: 10.1021/nl0480969. [DOI] [PubMed] [Google Scholar]
- 17.Govorov AO, Bryant GW, Zhang W, Skeini T, Lee J, Kotov NA, Slocik JM, Naik RR. Exciton-plasmon interaction and hybrid excitons in semiconductor-metal nanoparticle assemblies. Nano Lett. 2006;6:984–994. [Google Scholar]
- 18.Storhoff JJ, Elghanian R, Mucic RC, Mirkin CA, Letsinger RL. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes. J. Am. Chem. Soc. 1998;120:1959–1964. [Google Scholar]
- 19.Taton TA, Mirkin CA, Letsinger RL. Scanometric DNA array detection with nanoparticle probes. Science. 2000;289:1757–1760. doi: 10.1126/science.289.5485.1757. [DOI] [PubMed] [Google Scholar]
- 20.Thaxton CS, Hill HD, Georganopoulou DG, Stoeva SI, Mirkin CA. A bio-bar-code assay based upon dithiothreitol-induced oligonucleotide release. Anal. Chem. 2005;77:8174–8178. doi: 10.1021/ac0514265. [DOI] [PubMed] [Google Scholar]
- 21.Nicklas JA, Buel E. Quantification of DNA in forensic samples. Anal. Bioanal. Chem. 2003;376:1160–1167. doi: 10.1007/s00216-003-1924-z. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt Y. Performance characteristics of quantification assays for human immunodeficiency virus type 1 RNA. J. Clin. Virol. 2001;20:31–33. doi: 10.1016/s1386-6532(00)00152-9. [DOI] [PubMed] [Google Scholar]
- 23.Gibson NJ. The use of real-time PCR methods in DNA sequence variation analysis. Clin. Chim. Acta. 2006;363:32–47. doi: 10.1016/j.cccn.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 24.Bao YP, Huber M, Wei TF, Marla SS, Storhoff JJ, Muller UR. SNP identification in unamplified human genomic DNA with gold nanoparticle probes. Nucleic Acids Res. 2005;33:e15. doi: 10.1093/nar/gni017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li J, Chu X, Liu Y, Jiang J.-H, He Z, Zhang Z, Shen G, Yu R.-Q. A colorimetric method for point mutation detection using high-fidelity DNA ligase. Nucleic Acids Res. 2005;33:e168. doi: 10.1093/nar/gni163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato K, Hosokawa K, Maeda M. Non-cross-linking gold nanoparticle aggregation as a detection method for single-base substitutions. Nucleic Acids Res. 2005;33:e4. doi: 10.1093/nar/gni007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niemeyer CM, Mirkin CA. Nanobiotechnology: Concepts, Applications and Perspectives. Weinheim: John Wiley & Sons; 2004. [Google Scholar]
- 28.Rebrikov DV, Trofimov DY. Real-time PCR: a review of approaches to data analysis. Appl. Biochem. Microbiol. 2006;42:455–463. [PubMed] [Google Scholar]
- 29.Li HK, Huang JH, Lv JH, An HJ, Zhang XD, Zhang ZZ, Fan CH, Hu J. Nanoparticle PCR: Nanogold-assisted PCR with enhanced specificity. Angew. Chem.-Int. Edit. 2005;44:5100–5103. doi: 10.1002/anie.200500403. [DOI] [PubMed] [Google Scholar]
- 30.Qin WJ, Yung LYL. Nanoparticle-DNA conjugates bearing a specific number of short DNA strands by enzymatic manipulation of nanoparticle-bound DNA. Langmuir. 2005;21:11330–11334. doi: 10.1021/la051630n. [DOI] [PubMed] [Google Scholar]
- 31.Handley DA. Colloidal Gold: Principles Methods and Applications. New York: Academic Press; 1989. [Google Scholar]
- 32.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd. NY: Cold Spring Harbor Laboratory Press; 2001. edn. [Google Scholar]
- 33.He L, Musick MD, Nicewarner SR, Salinas FG, Benkovic SJ, Natan MJ, Keating CD. Colloidal Au-enhanced surface plasmon resonance for ultrasensitive detection of DNA hybridization. J. Am. Chem. Soc. 2000;122:9071–9077. [Google Scholar]
- 34.Yun CS, Khitrov GA, Vergona DE, Reich NO, Strouse GF. Enzymatic manipulation of DNA - nanomaterial constructs. J. Am. Chem. Soc. 2002;124:7644–7645. doi: 10.1021/ja025971o. [DOI] [PubMed] [Google Scholar]
- 35.Kanaras AG, Wang ZX, Bates AD, Cosstick R, Brust M. Towards multistep nanostructure synthesis: programmed enzymatic self-assembly of DNA/gold systems. Angew. Chem. Int. Edit. 2003;42:191–194. doi: 10.1002/anie.200390075. [DOI] [PubMed] [Google Scholar]
- 36.Wang ZX, Kanaras AG, Bates AD, Cosstick R, Brust M. Enzymatic DNA processing on gold nanoparticles. J. Mater. Chem. 2004;14:578–580. [Google Scholar]
- 37.Qin WJ, Yung LYL. Efficient manipulation of nanoparticle-bound DNA via restriction endonuclease. Biomacromolecules. 2006;7:3047–3051. doi: 10.1021/bm060517o. [DOI] [PubMed] [Google Scholar]
- 38.Zanchet D, Micheel CM, Parak WJ, Gerion D, Williams SC, Alivisatos AP. Electrophoretic and structural studies of DNA-directed Au nanoparticle groupings. J. Phys. Chem. B. 2002;106:11758–11763. [Google Scholar]
- 39.Claridge SA, Goh SL, Frechet JMJ, Williams SC, Micheel CM, Alivisatos AP. Directed assembly of discrete gold nanoparticle groupings using branched DNA scaffolds. Chem. Mater. 2005;17:1628–1635. [Google Scholar]
- 40.Ross P, Hall L, Smirnov I, Haff L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat. Biotechnol. 1998;16:1347–1351. doi: 10.1038/4328. [DOI] [PubMed] [Google Scholar]
- 41.Storhoff JJ, Lucas AD, Garimella V, Bao YP, Muller UR. Homogeneous detection of unamplified genomic DNA sequences based on colorimetric scatter of gold nanoparticle probes. Nat. Biotechnol. 2004;22:883–887. doi: 10.1038/nbt977. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.