Abstract
Locus control regions are regulatory elements that activate distant genes and typically consist of several DNase I hypersensitive sites coincident with clusters of transcription activator binding sites. To what extent nucleosomes and activators occupy these sites together or exclusively has not been extensively studied in vivo. We analyzed the chromatin structure of human β-globin locus control region hypersensitive sites in erythroid cells expressing embryonic and fetal globin genes. Nucleosomes were variably depleted at hypersensitive sites HS1-HS4 and at HS5 which flanks the 5′ of the locus. In lieu of nucleosomes, activators were differentially associated with these sites. Erythroid–specific GATA-1 resided at HS1, HS2 and HS4 but the NF-E2 hetero-dimer was limited to HS2 where nucleosomes were most severely depleted. Histones H3 and H4 were hyperacetylated and H3 was di-methylated at K4 across the LCR, however, the H3 K4 MLL methyltransferase component Ash2L and histone acetyltransferases CBP and p300 occupied essentially only HS2 and the NF-E2 motif in HS2 was required for Ash2L recruitment. Our results indicate that each hypersensitive site in the human β-globin LCR has distinct structural features and suggest that HS2 plays a pivotal role in LCR organization at embryonic and fetal stages of globin gene expression.
INTRODUCTION
Many gene families in mammals are expressed in a developmental stage and/or tissue-specific pattern under the influence of a locus control region (LCR) (1). LCRs are complex transcriptional enhancer elements located at a distance from target genes. Typically, LCRs comprise clustered DNase I hypersensitive sites (HSs) in a region of chromatin enriched for acetylation of H3 and H4 and di-methylation of H3 K4, histone modifications associated with active chromatin regions (2). The sequences within DNase I HSs contain binding motifs for transcriptional activators which are important for establishment of the distinct chromatin structure of the LCR and for LCR enhancer function (3–6). Specific LCR-binding activators may mediate chromatin modifying activity directly (7,8) or they may recruit or increase chromatin modifying complexes at an LCR, including SWI/SNF type complexes and histone acetyltransferases (HATs) (9–14).
It has long been observed that DNase I HSs denote regulatory elements in chromatin such as enhancers and promoters but the nature of these sites has been difficult to ascertain. The current view is that these sites may be nucleosome-free regions of DNA or they may contain ‘altered’ nucleosomes that do not strongly protect DNA from nuclease attack (15–17). One approach to distinguish these possibilities is to determine nucleosome occupancy by chromatin immunoprecipitation (ChIP) with antibodies to total histone H3. H3 occupancy data can be used to correct histone modification profiles and was used on a genome-wide scale to show that active gene promoters are depleted of nucleosomes (18). It is likely that nucleosome-free regions are also associated with enhancers/LCRs (19). Although it is a prediction that transcription activators will occupy these DNase I HSs in lieu of nucleosomes, this relationship has not been probed at high resolution.
The human β-globin locus contains an LCR that confers erythroid-specific enhancer activity to the globin genes which are expressed sequentially during development (20). The LCR consists of four HSs (HS1–HS4) that are located far upstream of the globin genes. The sites contain similar binding motifs for erythroid-specific and ubiquitous transcription activators and are generally considered to contribute to LCR enhancer activity collectively and to come into close physical contact in a ‘chromatin hub’ in erythroid cells (21). HS5, which flanks the 5′ end of the locus, is also part of the hub and displays certain characteristics of a chromatin insulator including binding of the insulator protein CTCF (22–25).
NF-E2 and GATA-1 are among the erythroid activators interacting at the β-globin LCR HSs that play critical roles in transcription activation of the globin genes (26). These factors interact differentially at the LCR HSs of the murine globin locus (27). Chromatin hub interactions bringing the LCR and actively transcribed globin genes into physical proximity in murine erythroid cells require GATA-1 (28). GATA-1 also participates in recruitment of CBP to murine LCR HS3 (9) and increases LCR recruitment of BRG1, a component of the SWI/SNF nucleosome remodeling complex (11). In human erythroid K562 cells that express the embryonic ε and fetal γ-globin genes but not the adult β-globin gene NF-E2 and GATA-1 play distinct roles in the enhancer activity of HS2 (4,29) and influence recruitment of CBP, p300 and SWI/SNF components to HS2 (10,12).
We previously observed varying levels of DNase I hypersensitivity among the β-globin LCR HSs in human K562 cells using a quantitative approach (25). Here we probed the relationship between nucleosome occupancy, activator and co-activator binding and histone modifications at the LCR HSs in a representative population of native mono and di-nucleosomes prepared from K562 cell nuclei. Sequences corresponding to HS1, HS2 and HS4 were highly depleted of nucleosomes. HS5 was less depleted and at HS3 nucleosomes were not lost. Histones remaining at HSs were hyperacetylated on H3 and H4 to varying degrees that did not always correlate with the extent of nucleosome loss. GATA-1 occupied HS1, HS2 and HS4, while NF-E2 was strongly detected only at HS2. The co-activators CBP/p300 and Ash2L occupied HS2 and NF-E2 interaction there was required for Ash2L recruitment. Interestingly, even though HS5 contains motifs for GATA-1 and NF-E2, these factors were not present at HS5 which was otherwise occupied by factors associated with insulator activity. These studies show that different HSs of the β-globin LCR in K562 cells have distinct chromatin structural attributes and factor occupancy which may reflect the organization of the complete LCR.
MATERIALS AND METHODS
Cell culture
K562 cells were grown in RPMI 1640 medium containing 10% FBS and harvested at confluence of 4–6 × 105 per ml for all experiments. HeLa cells were cultured in DMEM with 10% FBS. K562 cell clones carrying stably maintained episomes that contain HS2 linked to a complete ε-globin gene have been described (4). The tandem NF-E2 or GATA-1 binding motifs of HS2 were mutated by site-directed mutagenesis in episomes.
MNase sensitivity assay
Nuclei were prepared from 5 × 107 K562 or HeLa cells, digested with different MNase concentrations (0.0025 units/ul, 0.01 units/ul and 0.04 units/ul) and combined (30). Soluble chromatin was fractionated on a sucrose gradient (5–30%). DNA purified from fractions containing mainly mono- and di-nucleosomes was used for further study. To determine MNase sensitivity, 1 ng of purified DNA was compared with 1 ng of total genomic DNA (briefly digested with EcoRI) by real-time quantitative PCR using the comparative Ct method (4). In this analysis, nuclease sensitive sequences will be depleted compared to the total genomic sample.
Chromatin immunoprecipitation (ChIP)
Histone modifications were analyzed using purified nucleosomes without formaldehyde cross-linkage (31). Nuclei from 5 × 107 K562 cells were digested with different concentrations of MNase as for the sensitivity assay. Soluble chromatin was fractionated on a sucrose gradient (5–30%). Mono- and di-nucleosome were pooled and reacted with antibodies after pre-clearing with protein A agarose. Immunoprecipitated nucleosome-protein A agarose complexes were washed and DNA was eluted as described (31).
ChIP assays for transcription factors CBP, p300 and Ash2L were carried out using cross-linked chromatin (32). Briefly, 2 × 107 K562 cells were incubated in growth medium containing 1% formaldehyde for 10 min at 25°C, and then the cross-linking reaction was quenched by adding glycine to 0.125 M. Chromatin of primarily mono-nucleosome size was prepared by MNase digestion and sonication, and reacted with antibodies after pre-clearing with protein A or A/G agarose. Immunoprecipitated chromatin was bound to protein A or A/G agarose bead, washed and eluted as described (32).
Antibodies
Antibodies for NF-E2, GATA-1, CBP and p300 and normal rabbit IgG were purchased from Santa Cruz Biotechnology, Santa Cruz, CA, USA. The antibody for Ash2L was prepared by Marjorie Brand. Anti-di-acetylated (K9, K14) histone H3, anti-tetra-acetylated (K4, K8, K12, K16) histone H4, and anti-di-methylated H3 (K4) were obtained from Upstate Biotechnology, Lake Placid, NY, USA.
Real-time PCR analysis, primers and TaqMan probes
Purified DNA was analyzed by quantitative real-time PCR (ABI Prism 7900) using TaqMan probes and primers (Primer Express 1.0, PE Applied Biosystems). Real-time PCR was carried out with 200 nmol of TaqMan probes and 900 nmol of primers in a 25 µl reaction volume. Data were collected at the threshold where amplification was linear. In cross-linked ChIP assays, the relative enrichment for each primer pair was determined by comparing the amount of target sequence in 1.25% of immunoprecipitated DNA to the amount of target sequence in 0.1% of input DNA. In ChIP assays for histone modifications using nucleosomes, the fold enrichment was determined by comparing equal amounts (1 ng) of immunoprecipitated DNA and input DNA. Sequences of primers and TaqMan probes have been described (25).
RESULTS
Nucleosome loss in LCR DNase I HSs
To investigate the presence or absence of nucleosomes in the LCR directly, we digested non-cross-linked nuclei with graded concentrations of MNase and pooled and purified the digests to produce a complete, representative pool of native nucleosomes (30). At the lowest concentrations, MNase will attack linker regions between nucleosomes and sequences not protected by a nucleosome such as sites of extensive factor occupancy. At successively higher concentrations, DNA associated with nucleosomes of varying stability will eventually be digested. Nuclei from K562 cells expressing the embryonic ε and fetal γ-globin genes and from non-erythroid HeLa cells that do not express these genes, were digested with different concentrations of MNase, the digests combined, and mono and di-nucleosomes purified by sucrose gradient fractionation (Figure 1B) (see Methods section). Sensitivity to MNase was determined by comparing nucleosome-associated DNA with genomic DNA at and between LCR HSs using quantitative real-time PCR with TaqMan probes (Figure 1A).
Figure 1.
The human β-globin locus. (A) Locations of DNase I hypersensitive sites of the LCR and individual globin genes are drawn to scale. Amplicons used in real-time PCR are represented by vertical bars under the corresponding LCR sequences. PCR amplicons are directed to known nuclease hypersensitive regions of the LCR HSs that have been fine mapped [e.g (3) and see Figure 3A for sequence details]. (B) Nuclei of K562 and HeLa cells were digested with MNase and mono and di-nucleosomes were purified on a 5–30% sucrose gradient. DNA extracted from the nucleosomes was run on a 2% agarose gel. Lanes 1 and 2, K562 DNA; lanes 3 and 4, HeLa cell DNA; M, marker DNA (bp). (C) MNase sensitivity was determined across the LCR by real-time PCR, quantitatively comparing 1 ng of nucleosomal DNA with the same amount of genomic DNA prepared by brief restriction enzyme digestion. The results are averages of three independent preparations ± SEM.
HSs in the LCR cores showed much higher MNase sensitivity than intervening regions between the HSs sites in K562 cells indicating nucleosome loss at these sites as expected (Figure 1C). However, the magnitude of nucleosome loss was variable at different HSs. Notably, the core regions of HS1, HS2 and HS4 were highly depleted of nucleosomes, whereas DNA corresponding to HS5 was less depleted although all these sites were equally DNase I sensitive (25). MNase sensitivity was not notable at HS3. The lack of nucleosome remodeling at HS3 in K562 cells is consistent with a very weak DNase I sensitive site there (25,33). We note that the relative intensity of nucleosome loss indicated by MNase sensitivity at the LCR core HSs is in good agreement with results obtained from ChIP assays of formaldehyde cross-linked chromatin using an antibody against total histone H3 (25) (see Supplementary Figure 1) as would be predicted if a fully representative pool of nucleosomes had been obtained by graded MNase digestion. None of the HSs were depleted of nucleosomes in HeLa cells (Figure 1C).
Histone modifications in the LCR
To investigate directly the histone modifications present on nucleosomes that remained associated with the LCR, we analyzed by ChIP H3 and H4 hyperacetylation and di-methylation of H3 K4 using non-cross-linked mono- and di-nucleosomes. Purified nucleosomes were reacted with antibodies to acetylated H3 and H4 and di-methyl H3 K4 and immunoprecipitated DNA was analyzed by quantitative real-time PCR across the β-globin LCR using the TaqMan probes shown in Figure 1A.
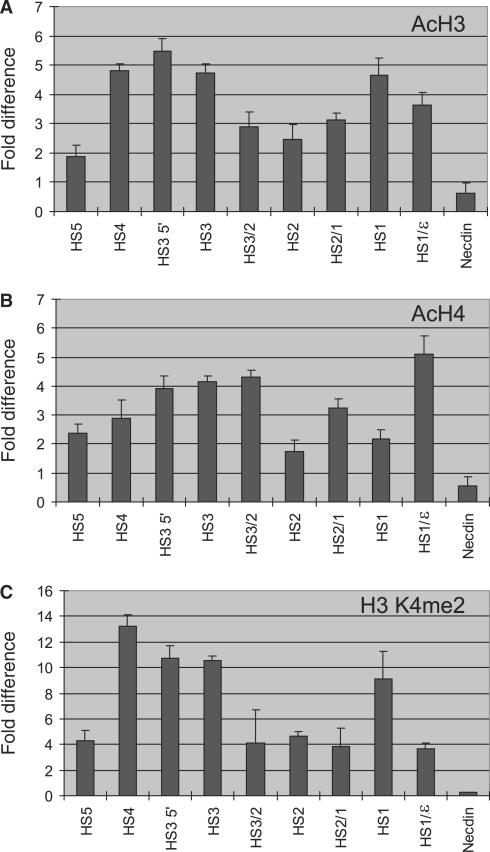
Across the LCR sites tested, histones H3 and H4 were hyperacetylated and di-methylated at K4 compared to the repressed necdin gene (Figure 2A–C). The patterns of the modifications were quite similar and variable across the locus, and we found that peak modification sometimes occurred outside the HSs cores (i.e. HS3 5′, Figure 2A) consistent with data on histone modification in the murine LCR (34). Compared to other sites, HS1, HS3 and HS4 were highly modified and HS2 and HS5 were less so. Thus, there was an imperfect correlation between histone modifications and histone depletion. For example, HS1, HS2 and HS4 were the most highly depleted sites but only HS1 and HS4 appeared as peaks of histone modifications. In addition, HS3 was as highly modified as HS1 and HS4 but was not depleted of nucleosomes. HS5 significantly retained nucleosomes and was only weakly marked by histone acetylation and H3 K4 di-methylation. Weak acetylation of HS5 nucleosomes compared to the other LCR HSs contrasts with findings that the homologous chicken 5′HS4 insulator site and an ectopic chicken 5′HS4 in human cells are very strongly marked by acetylated H3 (35,36). However, recent studies showed that murine HS5 was not hyperacetylated (37).
Figure 2.
Histone acetylation and methylation. Modified histones were immunoprecipitated from mono- and dinucleosomes using antibodies specific to (A) acetylated H3, (B) acetylated H4 or (C) di-methylated H3 K4. Fold difference was calculated by comparing the amount of target sequence in 1 ng of immunoprecipitated DNA to the amount of target sequence in the same amount of input DNA. The results of at least two chromatin preparations are shown ± SEM.
Distinct association of transcriptional activators in LCR DNase I HSs
The nucleosome preparation used to obtain the data in Figures 1 and 2 without cross-linking of nuclei results in MNase digestion and loss of sequences not bound to histones. Nucleosome loss in LCR HSs might be a prerequisite for or occur concomitantly with the binding there of transcriptional activators or activator binding might preclude re-deposition of nucleosomes following cell division. To investigate whether the nucleosome-depleted sequences were occupied instead by transcription activators, ChIP was carried out with nuclei cross-linked by formaldehyde to stabilize protein–DNA interactions. The HSs of the human β-globin LCR contain multiple cis-elements for binding of erythroid and ubiquitous transcriptional activators (Figure 3A). We focused on in vivo binding of the erythroid factors NF-E2 and GATA-1 in K562 cells using ChIP assays and antibodies against these proteins. Cells were cross-linked by formaldehyde treatment and chromatin was fragmented by sonication and MNase digestion to 100–200 bp size, comparable to the nucleosomal chromatin used for the data in Figures 1 and 2 (Supplementary Figure 2). DNA obtained after ChIP was analyzed across the LCR by quantitative real-time PCR using the TaqMan probes shown in Figure 1A.
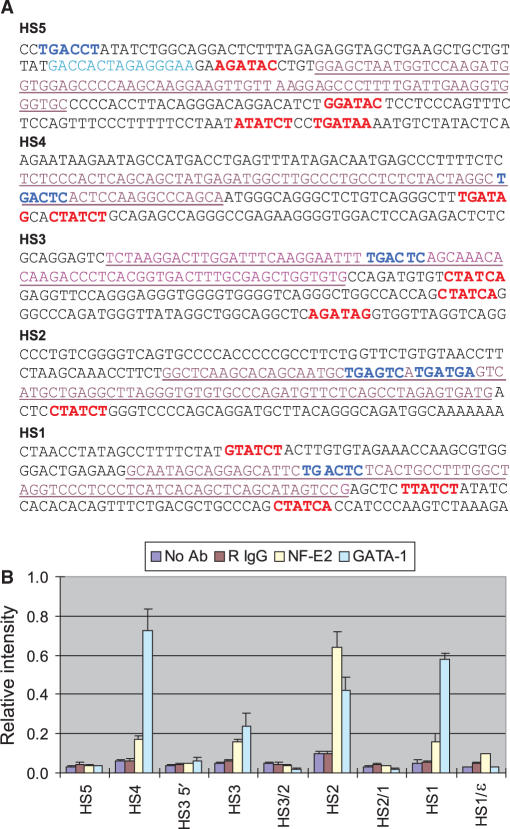
Figure 3.
Association of transcriptional activators in the LCR. (A) Sequences of LCR HSs cores are presented. Potential sites with which transcription activators could interact are indicated. Red, GATA-1 motifs; blue, NF-E2/AP1 motifs; turquoise, CTCF site in HS5; purple, underlined sequences represents the TaqMan amplicons for each HS. (B) ChIP was performed with antibodies specific to NF-E2 and GATA-1 and chromatin fixed with 1% formaldehyde. Relative intensity was determined by quantitatively comparing the amount of target sequence in immunoprecipitated DNA to the amount of target sequence in input DNA. Values for GATA-1 were re-scaled by dividing by 4 to present alongside the results for NF-E2. Signals obtained without antibody and with normal rabbit IgG were included as controls. The results of three independent experiments ± SEM are depicted.
The analysis showed that NF-E2 and/or GATA-1 were specifically bound in vivo at HS1-HS4 (Figure 3B). NF-E2 association was essentially limited to HS2. Association of GATA-1 was observed at HS1, HS2 and HS4: the GATA-1 signal at HS3 was very low, which might be related to the retention of nucleosomes at that site. At HS5, there was no significant signal for these activators even though GATA-1 and NF-E2 motifs are present in HS5 (Figure 3A). In earlier work, we showed that the insulator factors CTCF and USF occupy this site (12,25). The results show that the HSs in the human β-globin LCR have distinct associations with transcriptional activators as they do in the murine β-globin locus and that not all motifs for an activator are occupied in chromatin (11,27).
Recruitment of HATs and HMT to HS2
Histone acetylation and methylation are carried out by acetyltransferase (HAT) and methyltransferase (HMT) complexes, respectively. CBP and p300 HATs are important for histone acetylation in the globin locus (9,36,38). Human H3K4 methyltransferase complexes, of which Ash2L is a shared component (39–42), have not been detected in human globin locus sequences although the mono-, di- and tri-methylated forms of H3 K4 are variably detected across the LCR and human β-globin locus (25). We examined the distribution of co-activators CBP, p300 and Ash2L protein across the human β-globin LCR using chromatin prepared by the same procedure used for ChIP assays of transcriptional activators and immunoprecipitation using antibodies against these proteins.
The distributions of the three co-activators in the LCR were highly consistent with one another (Figure 4A and B). A strong signal with all antibodies was detected in HS2, where NF-E2 interaction was strong, while the other LCR HSs showed weak signals. Interestingly, the association of CBP and p300 at HS1 and HS4 was very weak in comparison to HS2 even though strong association of GATA-1 was observed at those sites and GATA-1 is implicated in CBP and p300 recruitment to the LCR (9,12,14). To investigate whether GATA-1 and/or NF-E2 were important for recruitment of Ash2L to HS2, we performed a ChIP assay using K562 cell clones with stable, chromatinized episomes that contain wild-type or GATA-1 or NF-E2-mutated HS2 linked to a complete ε-globin gene. Figure 4C shows that mutation of the HS2 GATA-1 site did not affect Ash2L recruitment to HS2. However, loss of NF-E2 binding resulted in failure to recruit Ash2L to HS2. Thus, NF-E2 is implicated in recruitment of H3 K4 histone methyltransferase activity to the β-globin LCR.
Figure 4.
Association of HATs and HMT in the LCR. Chromatin fixed with 1% formaldehyde was reacted with antibodies specific to CBP and p300 (A) or Ash2L (B). Immunoprecipitated DNA was quantitatively compared with input DNA as described in the legend to Figure 3. Signals obtained without antibody and with normal rabbit IgG served as experimental controls. The results are the averages of three independent experiments ± SEM. (C) ChIP was performed with Ash2L antibody using K562 cell clones containing episomes with either an intact HS2 or HS2 mutated at the NF-E2 motif (NF-E2m) or GATA-1 motif (GATAm).
DISCUSSION
This study reveals the chromatin structure of individual LCR DNase I HSs of the human β-globin locus to be unique. We found no perfect correlations among the characteristics of hypersensitive sites that we studied. For example, although strength of DNase I hypersensitivity correlated well with nucleosome loss at HS1, HS2 and HS4, there was a poor correlation at HS5. Nucleosome depletion at HS1 and HS4 correlated with histone hyperacetylation at remaining nucleosomes, consistent with acetylation preceding histone eviction (43). However, HS2 and HS5 did not fit this model. Transcription activators GATA-1 and NF-E2 occupied the HSs that were depleted of nucleosomes but to different extents, and did not occupy HS5 at all, even though motifs for these activators are present at all the sites. Co-activators CBP and p300 HATs and the Ash2L HMT component were recruited essentially only to HS2 despite the presence of at least GATA-1 at HS1 and HS4. Thus, we observed a surprising diversity among the β-globin LCR HSs. We suggest that HS2 rather than other LCR HSs may play a pivotal role in LCR organization and/or activity, at least at stages when the embryonic and fetal globin genes are expressed as reflected in K562 cells.
DNase I sensitivity and nucleosome loss
LCRs contain clusters of DNase I HSs as revealed by Southern blotting but it is highly problematic to quantify and compare these sensitivities. The application of quantitative, real-time PCR to this problem revealed that HS1-4 in the mouse globin LCR in fetal liver cells had very similar kinetics and end points of digestion by DNase I (44). In human K562 cells, qPCR analysis showed that HS1, HS2, HS4 and HS5 were equally highly sensitive to DNase I but that HS3 was significantly less sensitive and retained histone H3 (25); it remains to be determined whether or not weak DNase I sensitivity is a feature of HS3 in normal human erythroid tissue at the embryonic stage.
We used quantitative methods to measure nucleosome loss at the HSs of the human β-globin LCR and found that HS1, HS2 and HS4 were significantly and similarly depleted of nucleosomes in agreement with their similar DNase I sensitivities. HS3 was not depleted of nucleosomes, also in accord with the DNase results and the retention of nucleosomes at HS3 is consistent with H3 retention there (25). Interestingly, HS5 was only modestly depleted of nucleosomes, even though its DNase hypersensitivity was similar to HS1, HS2 and HS4. We suggest that nucleosomes remain at HS5 but may exist in an ‘altered’ state such that they are quite sensitive to DNase I digestion (15).
Nucleosome loss and histone acetylation and K4 di-methylation
Studies in complex loci, such as the human growth hormone locus (5), the human MHC class II HLA-DRA locus (6), the mouse Th2 cytokine locus (45) and the human and mouse β-globin loci (25,46–48), show that histone hyperacetylation and H3 K4 di-methylation are continuous through the LCRs but that the DNase I HSs have varying histone acetylation levels. Our studies provide some insight into the relationship between histone modifications and histone loss at the HSs of an LCR but reveal a complex picture.
HS1 and HS4 were depleted of nucleosomes but those remaining were highly acetylated consistent with kinetic analysis in yeast that show histones are first acetylated and then lost from a DNase I HS at the PHO5 promoter (43). However, other sites including HS3 were also highly acetylated but not depleted of nucleosomes, indicating that acetylation alone is insufficient for nucleosome loss. Furthermore, HS2, which is the most nucleosome depleted site, was not hyperacetylated. Thus, the order of nucleosome remodeling and histone modification events at these HSs is unclear and might be variable (49). In addition, HS5, which was moderately depleted of nucleosomes, was not a peak of histone acetylation. This contrasts with data for the homologous 5′HS4 insulator site that flanks the chicken β-globin locus (35). It was proposed that chicken 5′HS4 hyperacetylation and K4 di-methylation function to block encroachment of repressive histone modifications from a heterochromatic region 5′ of the chicken globin locus. This heterochromatic region is not present 5′ of the human or mouse globin loci and, possibly, this function may not have been maintained by HS5 (50).
Nucleosome loss and activator and co-activator interaction
We found that HS2 is occupied by NF-E2 and GATA-1 as well as the co-activators CPB/p300 and Ash2L. At HS1 and HS4, nucleosomes are similarly depleted as at HS2 but only GATA-1 was detected at these sites; we cannot rule out the possibility that other factors that we did not assay co-occupy these sites with GATA-1 to the exclusion of nucleosomes. In any case, co-activators were not substantially recruited to HS1 or HS4 despite the presence of GATA-1. At HS2, NF-E2 interaction is required for recruitment of Ash2L, consistent with a physical interaction between these two proteins (51). The relatively higher level of histone acetylation and H3 K4 di-methylation both up- and downstream of HS2 suggests these modifications spread from HS2 where co-activator complexes are recruited.
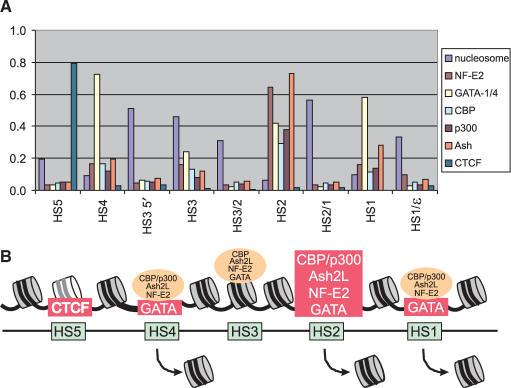
Figure 5 depicts the nucleosome landscape through the β-globin LCR and the sites of activator and co-activator detection. The data suggest that HS2 is particularly important to LCR functions at least when the embryonic and fetal globin genes are active. HS2 is the only LCR HS that fulfills the classical definition of an enhancer. Despite this apparent importance of HS2, its activities within the murine LCR can at least be partially compensated by the other HSs when it is deleted (52). It would be informative to investigate histone and factor/co-factor interactions in what remains of the LCR under these circumstances.
Figure 5.
Nucleosome and factor binding landscape in the β-globin LCR. (A) The ChIP data from Figures 3 and 4 are depicted graphically together with the values for nucleosome retention as shown in Figure 1C. Data for CTCF binding was previously reported (25). (B) A model illustrating nucleosome and factor occupancy at the β-globin LCR HSs. The picture is not drawn to scale. Major protein interactions in chromatin are boxed in red. Weak associations are depicted in orange ovals.
In considering our nucleosome and factor occupancy data, two considerations merit discussion. The first is that these profiles represent an average over all the templates analyzed. Thus, retention of nucleosomes and weak detection of activators such as seen at HS3 could indicate that stable nucleosomes occupy the majority of templates while a minority is occupied by factors. Alternatively, there may be a dynamic interchange of nucleosomes and factors (53) but that most often nucleosomes prevail. We favor the latter explanation. According to this interpretation, at other sites such as HS1, HS2 and HS4, the dynamic interchange results in relatively greater occupancy (possibly cooperatively) by factors and exclusion of nucleosomes. Chromatin disruption resulting in nucleosome loss provides the opportunity for non-replication coupled replacement of H3 by the variant H3.3 (54,55). It would be interesting to determine to what extent H3.3 replacement of H3 is observed across the LCR HSs. These proteins are so closely related that such experiments require stable expression of a tagged version of H3.3 to which an antibody can be directed. Quite recently, it has been shown that GAGA factor induces DNase I hypersensitivity, H3 K4 di-methylation and a high H3.3 to H3 ratio at a site that is important for maintenance of white gene expression in Drosophila (56).
Second, the close interactions among the HSs in the chromatin hub need to be taken into account (21). While the weak association of non-DNA binding co-activators CBP/p300 and Ash2L with HS1, HS3 and HS4 may be indicative of low level independent association at those sites, it may instead be attributable to fortuitous cross-linking due to close proximity with HS2 where the most extensive activator and co-activator occupancy was observed. While dynamic and spatial considerations deserve further attention, our ‘snapshot’ of nucleosome and activator/co-activator occupancy at the β-globin LCR DNase I HSs nevertheless indicates a remarkable level of diversity among the sites and a prominent role for HS2 in LCR organization and function when embryonic and fetal genes are being expressed.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank members of the Dean lab for discussions and Dr Cecelia Trainor for critical comments on the manuscript. This research was supported by the Intramural program of the NIDDK, NIH (A.D.) and the National Cancer Institute of Canada (M.B.). Funding to pay the Open Access publication charges for this article was provided by the Intramural program of the NIDDK, NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dean A. On a chromosome far, far away: LCRs and gene regulation. Trends Genet. 2006;22:38–45. doi: 10.1016/j.tig.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Pomerantz O, Goodwin AJ, Joyce T, Lowrey CH. Conserved elements containing NF-E2 and tandem GATA binding sites are required for erythroid-specific chromatin structure reorganization within the human β-globin locus control region. Nucleic Acids Res. 1998;26:5684–5691. doi: 10.1093/nar/26.24.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim A, Dean A. A human globin enhancer causes both discrete and widespread alterations in chromatin structure. Mol. Cell. Biol. 2003;23:8099–8109. doi: 10.1128/MCB.23.22.8099-8109.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ho Y, Elefant F, Cooke N, Liebhaber S. A defined locus control region determinant links chromatin domain acetylation with long-range gene activation. Mol. Cell. 2002;9:291–302. doi: 10.1016/s1097-2765(02)00447-1. [DOI] [PubMed] [Google Scholar]
- 6.Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nat. Immunol. 2003;4:132–137. doi: 10.1038/ni883. [DOI] [PubMed] [Google Scholar]
- 7.Armstrong JA, Emerson BM. NF-E2 disrupts chromatin structure at human β-Globin locus control region hypersensitive site 2 in vitro. Mol. Cell. Biol. 1996;16:5634–5644. doi: 10.1128/mcb.16.10.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 9.Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol. Cell. Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gui CY, Dean A. A major role for the TATA box in recruitment of chromatin modifying complexes to a globin gene promoter. Proc. Natl Acad. Sci. USA. 2003;100:7009–7014. doi: 10.1073/pnas.1236499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Im H, Grass JA, Johnson KD, Kim SI, Boyer ME, Imbalzano AN, Bieker JJ, Bresnick EH. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl Acad. Sci. USA. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao H, Kim A, Song SH, Dean A. Enhancer blocking by chicken beta -globin 5′ HS4: role of enhancer strength and insulator nucleosome depletion. J. Biol. Chem. 2006;281:30573–30580. doi: 10.1074/jbc.M606803200. [DOI] [PubMed] [Google Scholar]
- 13.Kim A, Zhao H, Ifrim I, Dean A. β-globin intergenic transcription and histone acetylation dependent on an enhancer. Mol. Cell. Biol. 2007;27:2980–2986. doi: 10.1128/MCB.02337-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SI, Bultman SJ, Jing H, Blobel GA, Bresnick EH. Dissecting molecular steps in chromatin domain activation during hematopoietic differentiation. Mol. Cell. Biol. 2007;27:4551–4565. doi: 10.1128/MCB.00235-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorch Y, Cairns BR, Zhang M, Kornberg RD. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 16.Aalfs JD, Kingston RE. What does ‘chromatin remodeling' mean? Trends Biochem. Sci. 2000;25:548–555. doi: 10.1016/s0968-0004(00)01689-3. [DOI] [PubMed] [Google Scholar]
- 17.Reinke H, Horz W. Anatomy of a hypersensitive site. Biochim. Biophys. Acta. 2004;1677:24–29. doi: 10.1016/j.bbaexp.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 18.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 19.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 20.Levings PP, Bungert J. The human beta-globin locus control region. Eur. J. Biochem. 2002;269:1589–1599. doi: 10.1046/j.1432-1327.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- 21.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 22.Li Q, Zhang M, Han H, Rohde A, Stamatoyannopoulos G. Evidence that DNase I hypersensitive site 5 of the human beta-globin locus control region functions as a chromosomal insulator in transgenic mice. Nucleic Acids Res. 2002;30:2484–2491. doi: 10.1093/nar/30.11.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanimoto K, Sugiura A, Omori A, Felsenfeld G, Engel JD, Fukamizu A. Human beta-globin locus control region HS5 contains CTCF- and developmental stage-dependent enhancer-blocking activity in erythroid cells. Mol. Cell. Biol. 2003;23:8946–8952. doi: 10.1128/MCB.23.24.8946-8952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wai AW, Gillemans N, Raguz-Bolognesi S, Pruzina S, Zafarana G, Meijer D, Philipsen S, Grosveld F. HS5 of the human beta-globin locus control region: a developmental stage-specific border in erythroid cells. EMBO J. 2003;22:4489–4500. doi: 10.1093/emboj/cdg437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim A, Kiefer CM, Dean A. Distinctive signatures of histone methylation in transcribed coding and noncoding human β-globin sequences. Mol. Cell. Biol. 2007;27:1271–1279. doi: 10.1128/MCB.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson KD, Grass JA, Boyer ME, Kiekhaefer CM, Blobel GA, Weiss MJ, Bresnick EH. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl Acad. Sci. USA. 2002;99:11760–11765. doi: 10.1073/pnas.192285999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the β-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 29.McDowell JC, Dean A. Structural and functional cross-talk between a distant enhancer and the ε-globin gene promoter shows interdependence of the two elements in chromatin. Mol. Cell. Biol. 1999;19:7600–7609. doi: 10.1128/mcb.19.11.7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Litt MD, Simpson M, Recillas-Targa F, Prioleau MN, Felsenfeld G. Transitions in histone acetylation reveal boundaries of three separately regulated neighboring loci. EMBO J. 2001;20:2224–2235. doi: 10.1093/emboj/20.9.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Litt MD, Simpson M, Gaszner M, Allis CD, Felsenfeld G. Correlation between histone lysine methylation and developmental changes at the chicken beta-globin locus. Science. 2001;293:2453–2455. doi: 10.1126/science.1064413. [DOI] [PubMed] [Google Scholar]
- 32.Johnson KD, Bresnick EH. Dissecting long-range transcriptional mechanisms by chromatin immunoprecipitation. Methods. 2002;26:27–36. doi: 10.1016/S1046-2023(02)00005-1. [DOI] [PubMed] [Google Scholar]
- 33.Dhar V, Nandi A, Schildkraut CL, Skoultchi AI. Erythroid-specific nuclease-hypersensitive sites flanking the human β-globin domain. Mol. Cell. Biol. 1990;10:4324–4333. doi: 10.1128/mcb.10.8.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson KD, Grass JA, Park C, Im H, Choi K, Bresnick EH. Highly restricted localization of RNA polymerase II within a locus control region of a tissue-specific chromatin domain. Mol. Cell. Biol. 2003;23:6484–6493. doi: 10.1128/MCB.23.18.6484-6493.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell. 2004;16:453–463. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 36.Zhao H, Dean A. An insulator blocks spreading of histone acetylation and interferes with RNA polymerase II transfer between an enhancer and gene. Nucleic Acids Res. 2004;32:4903–4919. doi: 10.1093/nar/gkh832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kooren J, Palstra RJ, Klous P, Splinter E, von Lindern M, Grosveld F, de Laat W. beta-globin Active Chromatin Hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J. Biol. Chem. 2007;282:16544–16552. doi: 10.1074/jbc.M701159200. [DOI] [PubMed] [Google Scholar]
- 38.Forsberg EC, Johnson K, Zaboikina TN, Mosser EA, Bresnick EH. Requirement of an E1A-sensitive coactivator for long-range transactivation by the β-globin locus control region. J. Biol. Chem. 1999;274:26850–26859. doi: 10.1074/jbc.274.38.26850. [DOI] [PubMed] [Google Scholar]
- 39.Wysocka J, Myers MP, Laherty CD, Eisenman RN, Herr W. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 2003;17:896–911. doi: 10.1101/gad.252103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, et al. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol. Cell. Biol. 2003;23:140–149. doi: 10.1128/MCB.23.1.140-149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, et al. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol. Cell. 2004;13:587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 42.Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 43.Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 44.McArthur M, Gerum S, Stamatoyannopoulos G. Quantification of DNaseI-sensitivity by real-time PCR: quantitative analysis of DNaseI-hypersensitivity of the mouse beta-globin LCR. J. Mol. Biol. 2001;313:27–34. doi: 10.1006/jmbi.2001.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 46.Forsberg EC, Downs KM, Christensen HM, Im H, Nuzzi PA, Bresnick EH. Developmentally dynamic histone acetylation pattern of a tissue- specific chromatin domain. Proc. Natl Acad. Sci. USA. 2000;97:14494–14499. doi: 10.1073/pnas.97.26.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bulger M, Schubeler D, Bender MA, Hamilton J, Farrell CM, Hardison RC, Groudine M. A complex chromatin landscape revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim A, Dean A. Developmental stage differences in chromatin sub-domains of the β-globin locus. Proc. Natl Acad. Sci. USA. 2004;101:7028–7033. doi: 10.1073/pnas.0307985101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 50.Dillon N, Sabbattini P. Functional gene expression domains: defining the functional unit of eukaryotic gene regulation. Bioessays. 2000;22:657–665. doi: 10.1002/1521-1878(200007)22:7<657::AID-BIES8>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 51.Brand M, Ranish JA, Kummer NT, Hamilton J, Igarashi K, Francastel C, Chi TH, Crabtree GR, Aebersold R, Groudine M. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat. Struct. Mol. Biol. 2004;11:73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- 52.Fiering S, Epner E, Robinson K, Zhuang Y, Telling A, Hu M, Martin DIK, Enver T, Ley TJ, Groudine M. Targeted deletion of 5′HS2 of the murine β-globin LCR reveals that it is not essential for proper regulation of the β-globin locus. Genes Dev. 1995;9:2203–2213. doi: 10.1101/gad.9.18.2203. [DOI] [PubMed] [Google Scholar]
- 53.Nagaich AK, Walker DA, Wolford R, Hager GL. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol. Cell. 2004;14:163–174. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- 54.Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell. 2002;9:1191–1200. doi: 10.1016/s1097-2765(02)00542-7. [DOI] [PubMed] [Google Scholar]
- 55.Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 56.Nakayama T, Nishioka K, Dong YX, Shimojima T, Hirose S. Drosophila GAGA factor directs histone H3.3 replacement that prevents the heterochromatin spreading. Genes Dev. 2007;21:552–561. doi: 10.1101/gad.1503407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.