Abstract
Expansion of a (CGG)n sequence in the 5′-UTR of the FMR1 gene to >200–2000 repeats abolishes its transcription and initiates fragile X syndrome (FXS). By contrast, levels of FMR1 mRNA are 5–10-fold higher in FXS premutation carriers of >55–200 repeats than in normal subjects. Lack of a corresponding increase in the amount of the product FMRP protein in carrier cells suggest that (CGG)>55–200 tracts thwart translation. Here we report that a (CGG)99 sequence positioned upstream to reporter firefly (FL) gene selectively diminished mRNA translation in coupled and separate T7 promoter-driven in vitro transcription and translation systems. The (CGG)99 tract similarly depressed mRNA utilization in HEK293 human cells transfected with plasmids bearing FMR1 promoter-driven FL gene. A (CGG)33 RNA tract formed a largely RNase T1-resistant intramolecular secondary structure in the presence of K+ ions. Expression of the quadruplex (CGG)n disrupting proteins hnRNP A2 or CBF-A in HEK293 cells significantly elevated the efficacy of (CGG)99 FL mRNA translation whereas hnRNP A2 or CBF-A mutants lacking quadruplex (CGG)n disrupting activity did not. Taken together, our results suggest that secondary structures of (CGG)n in mRNA obstruct its translation and that quadruplex-disrupting proteins alleviate the translational block.
INTRODUCTION
Fragile X syndrome (FXS), the most common cause of inherited mental retardation with a prevalence of 1:4000 in males and 1:6000 in females, is caused by an expansion of a (CGG)n sequence in the 5′ UTR of the FMR1 gene to >200–2000 repeats (1–5). Expansion of the (CGG)n tract beyond ∼200 repeats entails hypermethylation of the repeat sequence and of a neighboring CpG island that results in the transcriptional silencing of FMR1 and absence of the product FMRP protein (3). Although carriers of FMR1 premutation alleles that have >55–200 (CGG) repeats do not develop FXS, some do present various forms of clinical involvement such as minor alterations of physical features and emotional problems (6). Notably, ∼20% of the female premutation carriers develop premature ovarian failure (POF) (7) and one-third of the carrier males are affected by fragile X-associated tremor-ataxia syndrome (FXTAS) (8,9). In contrast to the absence of FMR1 transcripts in FXS cells, the levels of FMR1 mRNA in peripheral blood leukocytes at the upper range of FXS (CGG)n premutation were found to increase by 5- to 10-fold relative to normal subjects (10,11). However, the amounts of FMRP in these cells remain within the normal range or are somewhat below normal at higher premutation repeat sizes (10,12,13). A progressively diminishing association of FMR1 mRNA with polysomes that have increasing (CGG) repeat sizes was observed in lymphoblastoid cell lines of premutation carriers that over-express FMR1 mRNA (14). It appears, therefore, that expanded premutation (CGG)n tracts in FMR1 mRNA lower the efficiency of its translation in vivo. A clue to the underlying mechanism of the declining efficacy of translation was obtained in a study of the impeded translation of FMR1 mRNA that contained longer (CGG)n tracts. Although cells of a mildly affected FXS patient with FMR1 alleles of 57–285 CGG repeats exhibited normal steady-state levels of FMR1 mRNA, synthesis of FMRP from transcript with more than 200 repeats was markedly decreased and these transcripts were associated with stalled 40S ribosomal subunits (15). A plausible interpretation is that the longer (CGG)n sequences form secondary structures that block the migration of the 40S subunit. Thus, (CGG)55–200 premutation repeats may conceivably also fold into secondary structure(s) that slow down or obstruct ribosome progression, therefore lowering the efficiency of translation.
DNA and RNA (CGG) repeat sequences readily fold into hairpin structures (16–19) that may pair to generate stable intermolecular tetraplexes (20–23). Quadruplex (CGG)n structures in DNA templates were reported to hinder the progression in vitro of DNA polymerases (22,24). In a likely analogy, (CGG)n hairpin and quadruplex structures in mRNA may block polysome formation and impede ribosome progression and productive protein synthesis. If mRNA translation is obstructed by secondary structures of the moderately expanded premutation (CGG)n tracts, it should be enhanced by their destabilization. Indeed, several proteins and a low molecular size porphyrin were shown to efficiently disrupt in vitro tetraplex (CGG)n structures. The human Werner syndrome helicase was reported to catalyze the unwinding of intermolecular quadruplex structures of (CGG)n tracts in DNA (24,25). Member proteins of the hnRNP family; CBF-A (23,26,27), hnRNP A2 and modified hnRNP A1 (23) and UP-1 (28,29) were reported to mediate non-enzymatic destabilization of quadruplex forms of (CGG)n in both DNA and RNA. Additionally, the quadruplex DNA interacting cationic porphyrin TMPyP4 was shown to disrupt tetrahelical (CGG)n in DNA (30).
In this work, we report that upstream positioned mid-range premutation (CGG)n tracts preferentially suppress the in vitro and in vivo translation of a reporter firefly luciferase gene. Demonstrating that (CGG)n tract in RNA forms in vitro an intramolecular secondary structure which is likely to impede translation, we show that the tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A enhance in vivo the translation of mRNA molecules that contain 62 or 99 (CGG) repeats.
MATERIALS AND METHODS
Plasmids
pT7-FMR1-5′-UTR(CGG)n-FL—pSP6-5′-FMR1-UTR (CGG)n-FL (n = 0, 30, 62, 99) plasmids were prepared by modifications of previously described constructs (31). The no-repeat plasmid (high-copy origin) was digested with NaeI and BlpI, while the 30, 62 and 99 CGG-repeat plasmids (low-copy origin) were digested with BglII. The BglII overhang was blunted with mung bean nuclease and digested by BlpI. Thus regions containing the CMV promoter and the 5′-end of the FMR1 5′-UTR, up to the BlpI site were removed while creating blunt-end to BlpI-overhang linear vectors. Both high- and low-copy plasmids were ligated to a double-stranded oligomer, 5′-(GGCATTTAGGTGACACTATAGATCAGGCGC)-3′·3′-(CCGTAAATCCACTGTGATATCTAGTCCGCGAGT)-5′ that contained the SP6 promoter and restored the 5′-end of FMR1. The formed FMR1 5′-UTR sequence starting from transcription site 1 was as previously described (32) with the following modifications. The first two 5′ AG bases of the 5′-UTR were changed to GA for optimal SP6 transcription, the 3′-end base was C instead of G, and the no CGG-repeat 5′UTR was devoid of both the CGG repeat and the following GC dinucleotide. The (CGG)30 repeat sequence had two intervening (AGG) triplets at positions 11 and 21, the (CGG)62 repeat contained an (AGG) interruption at the 11th position and the (CGG)99 repeat had at least one AGG intervening trinucleotide of undetermined location. Next, bacteriophage T7 promoter was incorporated into the pSP6-5′-FMR1-UTR(CGG)n-FL plasmids. Double-stranded DNA adaptor including the T7 promoter sequence (underlined) 5′-d(TCACGCGTACTTAATACGACTCACTATAGGCTAGCCGC)-3′•3′-d(CGCATGAATTATGCTGAGTGATATCCGATCGGCGAGT)-5′ was ligated into the BlpI site upstream to the plasmid UTR-(CGG)n sequence. Plasmids without a (CGG) tract or that included a (CGG)30 insert were propagated in 500 ml of LB medium at 37°C for 12 h in Escherihcia coli DH10B (gift of Dr D. Gidoni, Volcani Center, Israel) and purified (Maxi-prep kit, Qiagen). Due to the instability of the (CGG)62 and (CGG)99 repeats, plasmids harboring these inserts were grown in multiple 3-ml cultures. Following plasmid purification (Mini-prep kit, Qiagen), lengths of the BlpI and NheI excised inserts were determined by agarose gel electrophoresis and only plasmids that contained validated (CGG)62 or (CGG)99 tracts were used.
pCMV-FMR1-5′-UTR (CGG)n-FL—pCMV-FMR1-5′-UTR(CGG)99-FL plasmid (31) was contributed by Dr P. Hagerman, UC Davies. To construct plasmids with 0, 30 or 62 repeats, the FMR1-5′-UTR(CGG)99 sequence was excised by BlpI and NheI digestion and the linearized plasmid was ligated with (CGG)0, (CGG)30 or (CGG)62-containg FMR1-5′-UTR sequences that were excised from the respective pT7-FMR1-5′-UTR(CGG)n-FL plasmids by BlpI and NheI digestion. Plasmid propagation and verification of correct repeat sizes were performed as described for the pT7-FMR1-5′-UTR(CGG)n-FL plasmids.
pCS107-FMR1-5′-UTR(CGG)33—The FMR1 5′ untranslated region was amplified from genomic DNA of EBV transformed B lymphoblastoid cell line of a normal subject (gift of Dr P. Chiurazzi, Catholic University, Italy). The UTR tract which included 33 (CGG) repeats interrupted by two (AGG) triplet units at trinucleotides 11 and 21 was PCR amplified using a 5′-FMR1 exon forward primer 5′-(CGCGGATCCCGCTCAGCTCCGTTTCGGTTTC)-3′ and FMR1 3′-end exon primer 5′-(CCGGAATTCTAGAAAGCGCCATTGGAGCCCC)-3′. The amplified fragment was restricted by BamHI and EcoRI digestion and cloned into SP6 promoter containing pCS107 plasmids that were propagated in E. coli DH10B and purified (Maxi-prep, Qiagen).
pCMV2-Flag-hnRNP A2 and pCMV2-Flag-CBF-A—Plasmid pCMV2-Flag was constructed to harbor and express cDNA encoding the quadruplex (CGG)n-disrupting protein hnRNP A2 or its mutant hnRNP A2 F[54]S that lost the destabilizing activity (23). The plasmids pGEX-2T-hnRNP A2 (gift of Dr Ralph C. Nichols, Dartmouth School of Medicine, NH) or pGEX-2T-hnRNP A2 F[54]S (23) were cut by EcoRI and SalI and the electrophoretically isolated cDNA were ligated into EcoRI and SalI-linearized pCMV2-Flag plasmid (Stratagene). To construct pCMV2-Flag plasmids that express the quadruplex (CGG)n destabilizing CBF-A protein or its mutant CBF-A ΔRNP11 that lost the tetraplex disrupting activity (26), pCMV2-Flag was cut by EcoRI and dephosphorylated with calf intestinal phosphatase (New England Biolabs). CBF-A or CBF-A ΔRNP11 encoding sequences that were cleaved by the same enzyme out of pGEX-2T plasmids (26) were ligated to the linearized pCMV2-Flag plasmids. The restriction digestion at the border of the Flag and the CBF-A or CBF-A ΔRNP11 encoding sequences caused loss of the reading frame. A proper reading frame was restored by PFU (Promega) catalyzed PCR amplification of the altered DNA region using a 5′ primer 5′-d(CTTGCGGCCGCGAATCCATGGCCGACC)-3′ and a 3′ primer 5′-d(GGTCGGCCATGGATTCGCGGCCGCAAG)-3′. The modified plasmids were cloned and restoration of the reading frame was verified by DNA sequencing.
Coupled in vitro transcription–translation
Coupled transcription and translation reactions were carried out using the TNT® transcription–translation system (Promega). Reaction conditions were essentially according to the manufacturer's specifications. Briefly, reaction mixtures contained in a final volume of 25 μl: 1 μl of 25× TNT reaction buffer; 0.5 μg XbaI-linearized pT7-FMR1-5′-UTR(CGG)n-FL plasmid; 12.5 μl 2× TNT rabbit reticulocyte lysate; 0.5 μl TNT T7 RNA polymerase; 5.0 μCi [α-32P] UTP (3000 Ci/mmol, Amersham); 20 μM of each amino acid; 40 U RNasin ribonuclease inhibitor (Promega). Following incubation at 30°C for 30 min, the samples were rapidly cooled to 4°C and 5 μl aliquots of each reaction mixture were mixed with 100 μl of luciferase assay Reagent II (Promega). Following a 3 s delay, firefly luciferase activity was determined for 30 s (20/20 luminometer, Turner Biosystems). To measure levels of the radiolabeled RNA, gel-loading dye containing SDS to a final concentration of 0.5% was added to 10 μl aliquots of the reaction mixtures and the samples were electrophoresed through 0.8% agarose gel. The identity of the radioactive RNA bands as full-length luciferase gene transcripts was verified by northern analysis using the luciferase 3′ gene probe 5′-(CTTTCCGCCCTTGGCCTTTATGAGGATC)-3′. The relative levels of the labeled luciferase RNA molecules that contained (CGG)n tracts of different lengths were determined by phosphor imager analysis.
Transcription in vitro
To investigate the effect of different lengths of (CGG)n tracts on transcription, pT7-FMR1-5′-UTR(CGG)n-FL plasmids (n = 0, 30, 62 or 99) were linearized by XbaI restriction digestion and transcribed in vitro by T7 RNA polymerase. Reaction mixtures contained in a final volume of 20 μl: 0.5 μg linear plasmid DNA; 1 mM each of the four rNTPs; 3.0 μCi [α-32P] UTP (3000 Ci/mmol, Amersham); 40 U of ribonuclease inhibitor (Takara); 80 mM potassium acetate; 40 U of T7 RNA polymerase and T7 polymerase reaction buffer (Promega). The reaction mixtures were incubated at 30°C for 30 min and the RNA polymerization reaction was terminated by the addition of 5 μl of 2.5% SDS in gel loading dye. Following resolution of the radiolabeled RNA by electrophoresis in 0.8% agarose, the gels were dried and radioactivity of the product RNA molecules was quantified by phosphor imaging analysis. That the radioactive RNA bands represented full-length transcripts was confirmed by northern analysis as described for the coupled transcription–translation system.
Translation in vitro
Equal amounts of radiolabeled RNA transcripts of the pT7-FMR1-5′-UTR(CGG)n-FL plasmids were translated in vitro. To prepare the RNA, 1 μg of each XbaI-linearized plasmid was transcribed at 37°C for 1.5 h in a final volume of 20 μl of Ampliscribe T7 transcription system (Epicenter Technologies) that contained 3.0 μCi [α-32P] UTP (3000 Ci/mmol, Amersham). Plasmid DNA was removed by digestion at 37°C for 15 min with 1 U of RNase-free DNase (Promega) and the RNA transcripts were extracted with ultra pure phenol:chloroform:isoamyl alcohol (25:24:1, Invitrogen) followed by chloroform extraction. The RNA was precipitated and washed with 70% ethanol and dissolved in RNase-free water. Approximately equal amounts of RNA as estimated by absorption at 260 mμ were resolved by agarose gel electrophoresis and the precise amounts of RNA were normalized by the radioactivity of the full-length RNA transcript bands. Equal amounts (0.5 μg each) of the FMR1-5′-UTR(CGG)n-FL transcripts were translated in rabbit reticulocyte lysate (Flexi® translation system, Promega) as follows. The RNA samples were incubated at 30°C for 30 min in a final volume of 25 μl of reticulocyte lysate translation system that contained 2.0 mM DTT; 1 mM magnesium acetate; 70 mM KCl and 20 U ribonuclease inhibitor (Takara). The translation reaction was terminated by rapid cooling of the samples to 4°C and the luciferase activity was determined in 10 μl aliquots of each reaction mixture as described for the coupled transcription–translation system.
Ribonuclease T1 protection analysis
One microgram of XhoI-linearized pCS107-FMR1-5′-UTR(CGG)33 plasmid DNA was transcribed in AmpliScribe SP6 system (Epicenter Biotechnologies) according to the manufacturer's instructions. The RNA transcript was precipitated and washed with 70% ethanol and resolved by denaturing gel electrophoresis through 10% polyacrylamide, 8 M urea. The gel band that contained the RNA transcript was visualized by UV adsorption, excised and agitated overnight at 4°C in a solution of 0.3 M sodium acetate pH 5.2; 0.5 mM EDTA; 0.1% SDS. Following ethanol precipitation and wash, the eluted RNA transcripts were dephosphorylated at 37°C for 1 h by shrimp alkaline phosphatase (USB). The phosphatase was heat-inactivated at 65°C for 15 min and the RNA was 5′-end labeled using T4 polynucleotide kinase and [γ-32P] ATP (33). The labeled RNA was resolved by denaturing gel electrophoresis through 10% polyacrylamide, 8 M urea and recovered and isolated by ethanol precipitation as described above. Duplex regions of RNA were distinguished from unpaired tracts by the resistance of the former to digestion by the single-stranded RNA-specific nuclease RNase T1. Labeled RNA transcripts were dissolved in RNase-free water and supplemented with unlabeled yeast RNA carrier to a final concentration of 8 μM. Digestion of native RNA with a specified amount of RNase T1 (Ambion) was conducted at 20°C for 20 min in a reaction mixture that contained in 10 μl final volume 10 mM MgCl2 and 80 mM potassium acetate in 10 mM Tris-HCl buffer, pH 7.4. Digestion of heat-denatured RNA by RNase T1 was conducted at 55°C for 15 min in the same reaction mixture that also contained 3.5 M urea. The reactions were terminated by adding to each mixture 20 μl of inactivation-precipitation buffer as specified by the manufacturer. Products of the ribonuclease digestion reactions were resolved by denaturing gel electrophoresis in 12% polyacrylamide, 8 M urea.
Formation and resolution of intramolecular secondary structure of (CGG)33 RNA
FMR1-5′-UTR(CGG)33 RNA was transcribed and internally labeled in reaction mixtures that contained in a final volume of 20 μl: 1.0 μg XhoI-linearized pCS107-FMR1-5′-UTR(CGG)33 plasmid DNA, 10 mM DTT, 40 U RNasin (Promega), 0.5 mM ATP, 3.0 mM CTP, 6.0 mM GTP or 0.5 mM 7-deazaGTP (TriLink Biotechnologies), 8.3 μM UTP, 50 μCi [α-32P] UTP (800 mCi/mmol, Amersham), 20 U SP6 polymerase and 4 μl 5× SP6 transcription buffer (Promega). Transcription was conducted at 37°C for 60 min, the DNA was digested by 1 U DNase I at the same temperature for 15 min and the product RNA was phenol extracted and precipitated and washed with ethanol.
An intramolecular secondary structure of the RNA was generated by incubating at 4°C for 15 min, mixtures that contained in a final volume of 10 μl: <2.5 nM 5′-32P FMR1-5′-UTR(CGG)33 RNA, 120 mM KCl, 1 mM EDTA, 0.5 mM DTT, 20% glycerol in 20 mM Tris-HCl buffer, pH 7.8. Control reactions were performed under identical conditions except that KCl was omitted from the reaction mixture. The RNA was resolved by electrophoresis in non-denaturing 4% polyacrylamide gel in 0.5× TBE buffer, pH 8.2 that contained 10 mM KCl.
Transfection of cultured human cells
Human Embryonic Kidney 293 cells (HEK293) were seeded in 0.1% gelatin-coated 10 cm plates and grown to 80–90% confluence at 37°C in 5% CO2 atmosphere in Dulbecco Modified Eagle's Medium (DMEM) supplemented with 4.5 g/l d-glucose, 5 mM l-glutamine, 10% fetal calf serum, 83.3 U/ml each Penicillin and Streptomycin and 0.2 μg/ml Amphotericin B (Biological Industries, Israel). Cells that were reseeded at 3.3 × 105 cells per 6 cm gelatin-coated plate, were grown overnight and transiently co-transfected with three plasmids: a vector harboring reporter firefly luciferase (FL) without or with an upstream (CGG) repeat tract; plasmid that encoded normalizing Renilla luciferase (RL) and plasmids expressing quadruplex destabilizing proteins or their inactive mutants. The growth medium was replaced with 4 ml of fresh DMEM to which 100 μl of DMEM were added that contained 6 μl Fugene 6.0 (Roche), 0.2 μg pCMV-FMR1-5′-UTR(CGG)n-FL DNA and 0.02 μg pCMV-RL plasmid (Promega) and 1.8 μg of DNA of one of the following expression plasmids: pCMV2-Flag; pCMV2-Flag-hnRNP A2 pCMV2-Flag-hnRNP A2 F[54]S; pCMV2-Flag CBF-A or pCMV2-Flag-CBF-A ΔRNP11. The cells were harvested 24 h after transfection, washed once with 1.5 ml cold phosphate buffered saline (PBS) and resuspended in 3 ml PBS. Aliquots of each sample were used to determine FL and RL activities and to conduct semi-quantitative RT-PCR measurements of the levels of their mRNA transcripts.
Correction for variations in cell viability and transfection efficiency was performed for each experiment as described (31) except that the RL-corrected FL activity was normalized to FL activity of cells transfected with reporter pCMV-FMR1-5′-UTR-FL plasmid with no upstream (CGG) repeat tract.
Dual luciferase assay
FL and RL activities were determined in lysates of transfected HEK293 cells according to the manufacturer's instructions using the dual luciferase reporter assay system, (Promega). Briefly, the cells were lysed in passive lysis buffer (Promega) and 20 μl of the cell lysate were added to 100 μl luciferase reagent II. Following a 3 s delay, FL activity was measured for 30 s using the 20/20 luminometer. The reaction was terminated by adding 100 μl Stop and Glo reagent to quench the FL activity and after a 3 s delay, the activity of RL was determined for 30 s. In each sample, triply measured values of FL activity were normalized to RL activity.
Semi-quantitative RT-PCR determination of relative mRNA levels
RNA was isolated from HEK293 cells using the total RNA isolation kit (Cartagen, USA) and genomic and plasmid DNA were removed by use of the Turbo DNA-free kit (Ambion). Reverse transcription and amplification reactions were conducted with the Reverse-it one-step RT-PCR kit (ABgene, UK). Each reaction mixture contained in a final volume of 25 μl: 0.05 μg total RNA; 0.3 or 0.9 μCi [α-32P] dCTP (3000 Ci/mmol) for the reverse transcription of FL or RL mRNA, respectively, and 5.0 pmol each of FL forward primer 5′-(CTATGAAGAGATACGCCCTGGTTCCTGG)-3′ and FL reverse primer 5′-(GGCAGTTCTATGAGGCAGAGCGAC)-3′ or RL forward primer 5′-(GGGATGAATGGCCTGATATTGAAGAAG)-3′ and RL reverse primer 5′-(CAATTTGTACAACGTCAGGTTTACCACC)-3′. FL or RL RNA were amplified by RT-PCR for 26 or 29 cycles, respectively, of 94°C for 20 s, 53°C for 25 s and 72°C for 1 min. Based on measurements of the levels of radiolabeled FL and RL reverse transcripts for each cycle between the 20th and 35th cycles, it was determined that 26th or 29th cycles were at the middle of the respective linear range of amplification of FL and RL RNA. Every set of reactions included a negative control of a mixture without RT to verify that all the amplification products were copies of RNA template and not of contaminating DNA. Equal aliquot volumes of the FL or RL RT-PCR 32P-labeled DNA products were electrophoresed at 12 V/cm through 4 or 6% non-denaturing polyacrylamide gels in 0.5× TBE buffer (2.4 mM EDTA in 1.08 mM Tris-borate buffer, pH 8.3) until a bromophenol blue marker reached the end of the gel. DNA size markers (T4 PNK end-labeled 5′-32P peqGOLD 100 bp DNA ladder, PeqLab Biotechnologies) were used to identify full-length FL and RL RT-PCR DNA product bands. The identity of these DNA products was verified by parallel electrophoresis of radiolabeled PCR products of the original plasmids DNA with the FL or RL primers. Amounts of the electrophoretically resolved DNA samples were quantified in the dried gels by phosphor imaging analysis.
Western analysis
Equal amounts of HEK293 cell lysates protein were resolved by 10% SDS-PAGE. Following transfer to nitrocellulose membrane, the expression of functional or mutant Flag-hnRNP A2 or FLAG CBF-A proteins was detected by use of murine anti-Flag monoclonal antibody (1:1000, Sigma) as the primary antibody and horseradish peroxidase-conjugated goat anti mouse IgG (H + L, 1:10 000, Pierce) as secondary antibody. Horseradish peroxidase activity was detected by use of the Super Signal Wes Pico chemiluminescence substrate (Pierce).
RESULTS
Premutation size (CGG)n repeat tract in DNA and RNA selectively diminish translation in vitro
We first evaluated in model T7 promoter driven coupled and separate in vitro transcription and translation systems, the effects of increasing sizes of normal and premutation-range (CGG)n tracts on FL gene expression.
Premutation-range (CGG)n repeat sequences selectively depress translation in vitro
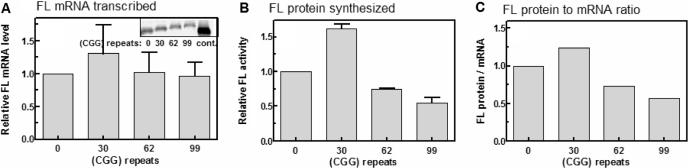
The effect of increasing lengths of upstream (CGG)n tracts was first examined in a coupled in vitro transcription–translation system using as template linearized pT7-FMR1-5′-UTR(CGG)n-FL plasmids that harbor in the 5′ to 3′ direction bacteriophage T7 promoter, the GC-rich 5′-UTR of FMR1, tracts of 0, 30, 62 or 99 (CGG) repeats and an FL reporter gene. Following transcription of the plasmids and coupled translation, FL mRNA that was identified by northern analysis, was resolved by agarose gel electrophoresis and quantified and FL protein activity was determined as described in Materials and Methods section and in the legend to Figure 1. Average results of three independent measurements are summarized in Figure 1. As is evident, transcription driven by the T7 promoter was only minimally affected by the expanding length of the (CGG) repeat sequence (Figure 1, first panel). Relative to FL transcription with no (CGG)n upstream tract, a cluster of 30 (CGG) trinucleotides that represented the most frequent normal size of the repeat sequence (34), enhanced on average the accumulation of FL transcripts by ∼1.3-fold whereas 62 and 99 repeats did not change the initial level of FL mRNA. Unlike transcription, the extent of translation was significantly affected by the (CGG)n stretches. Relative to FL transcripts with no upstream (CGG) repeat tract, a stretch of 30 (CGG) trinucleotides enhanced on average the translation of FL mRNA by 1.6-fold (Figure 1, second panel). By contrast, 62 or 99 (CGG) repeats depressed the relative translation of FL by 1.3- and 1.8-fold, respectively. Moreover, when compared with RNA that contained 30 (CGG) triplets, tracts of 62 or 99 (CGG) triplets lowered the level of FL protein by 2.2- or 3-fold, respectively. The decreased efficiency of translation of FL mRNA molecules as a function of the increasing length of the (CGG)n tract is best reflected by the ratios of FL protein to mRNA. As shown in the third panel of Figure 1, relative to transcripts devoid of the (CGG) repeat sequence, the efficiency of translation of mRNA that contained 62 or 99 repeats was diminished by 1.4- or 1.75-fold, respectively. Further, when compared to the maximally translated RNA molecules that included 30 (CGG) repeats, the utilization of mRNA molecules with 62 or 99 (CGG) repeats was diminished by 1.7- or 2.2-fold, respectively.
Figure 1.
Effect of (CGG) repeat size on the synthesis of FL mRNA and protein in a coupled transcription–translation in vitro system. Coupled transcription and translation of linearized pT7-FMR1-5′-UTR(CGG)n-FL plasmid DNA (n = 0, 30, 62 or 99) were conducted in a TNT® (Promega) reaction mixture containing [α-32P] UTP as described under Materials and Methods section. Presented are histograms of the measured levels of FL mRNA and protein activity and of their calculated ratios. The inset in sub-panel A is a typical phosphor image of agarose gel resolved radiolabeled FL transcripts containing increasing lengths of the (CGG)n repeat sequence. The control lane has PCR product of the pT7-FMR1-5′-UTR(CGG)0-FL DNA. RNA and protein were quantified as detailed in the Materials and Methods section. Each data point is an average of the results of three independent experiments. FL protein to mRNA ratios (panel C) were calculated by dividing for each (CGG)n repeat size the average value of FL protein activity by the corresponding average level of FL mRNA.
The effect of (CGG) repeat length on the efficiency of mRNA and protein synthesis in vitro was next investigated in separate transcription or translation systems. Linearized pT7-FMR1-5′-UTR(CGG)n-FL DNA (n = 0, 30, 62 or 99) was transcribed using T7 RNA polymerase and the amounts of full-length FL transcripts as identified by northern analysis were quantified. Results presented in Figure 2A show that the presence of increasing lengths of an upstream (CGG)n tract upstream to the FL gene lowered the synthesis of mRNA to a limited extent. Relative to DNA with no (CGG)n tract, 30, 62 or 99 (CGG) repeats decreased the average levels of FL gene transcripts by 9, 27 or 33%, respectively. The effect of an increasing number of mRNA (CGG) repeats on protein synthesis in vitro, was measured by translating in a reticulocyte lysate system equal amounts of full-length FL mRNA molecules that contained 0, 30, 62 or 99 (CGG) repeats and determining the activity of the product FL protein (see Material and Methods section). As shown in Figure 2B, relative to the translation of mRNA devoid of a (CGG) repeat sequence, the translation of FL mRNA molecules that included 30, 62 or 99 (CGG) repeats declined on average by 5, 75 or 72%, respectively. Since the translation system was provided with equal amounts of each mRNA type, these results indicated a diminishing efficacy of the translation of mRNA molecules that contained progressively increasing length of the (CGG) repeat sequence. Hence, results obtained in vitro with coupled or separate transcription and translation systems both demonstrated preferential decline in the efficacy of mRNA translation as a function of (CGG)n length.
Figure 2.
Effect of (CGG) repeat size on T7 polymerase catalyzed FL transcription and on the translation in vitro of FL mRNA. (A) Levels of FMR1-5′-UTR(CGG)n-FL mRNA (n = 0, 30, 62 or 99) transcribed in vitro by T7 polymerase. Reaction conditions and resolution and quantification of the radiolabeled transcripts were as described under Materials and Methods section. The height of each column represents an average of the results of four independent experiments with the SD indicated by error bars. (B) Levels of the in vitro translated FL protein. Equal amounts of pT7-FMR1-5′-UTR(CGG)n-FL RNA transcripts containing the indicated (CGG) repeat sizes were translated in vitro in a rabbit reticulocyte lysate Flexi® system (Promega) section and relative FL protein activities were determined as described in the Materials and Methods section. Shown are average results and SD of four independent experiments.
(CGG) repeat tract in RNA forms secondary structure
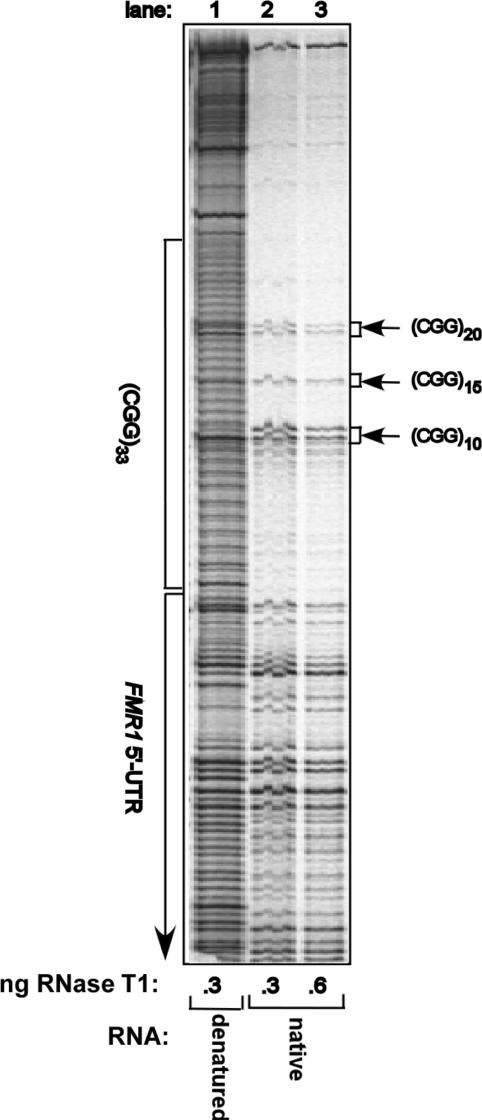
DNA and RNA (CGG) repeat sequences were shown to readily form hairpin (16,17,19,20,22,35), tetraplex (20–22) and slipped-strand (36) secondary structures. A likely cause for the observed declining efficiency of translation as a function of the size of the upstream (CGG)n mRNA sequence could be the formation of secondary structures by the RNA repeat sequence. To probe the structure of a (CGG)n tract in RNA, denatured or native 5′-32P labeled RNA transcripts of pCS107-FMR1-5′-UTR(CGG)33 DNA were digested by RNase T1 and products of the nucleolytic digestion were resolved by denaturing polyacrylamide gel electrophoresis (see Materials and Methods section). RNase T1 cleaves phosphodiester bonds in single-stranded RNA 3′ to unpaired guanine residues whereas paired guanines resist digestion. As seen in Figure 3, the FMR1-5′-UTR(CGG)33 denatured RNA transcript in lane 1 was cleaved by RNase T1 at each and every guanine residue. By contrast, as indicated in lanes 2 and 3, the enzyme did not digest portions of the native RNA GC-rich FMR1 5′-UTR sequence and particularly of the (CGG)33 repeat. In fact, nearly all the (CGG)33 stretch in native RNA-resisted digestion by RNase T1 except for cleavage at the tenth and twentieth (CGG) trinucleotides that, respectively, precede the (AGG)11 and (AGG)21 intervening triplets and a faint digestion band at the fifteenth (CGG) trinucleotide. Thus, except for selected unpaired (CGG) trinucleotides, the majority of the guanine residues were engaged in base pairing. These data indicated, therefore, that most of the repeat sequence folded into a secondary structure. Notably, a large portion of the FMR1 5′ UTR sequence also resisted digestion by RNase T1 indicating that it too was largely in secondary structure.
Figure 3.
Ribonuclease T1 protection analysis of FMR1-5′-UTR(CGG)33 RNA. Linearized pCS107- FMR1-5′-UTR(CGG)33 was transcribed in AmpliScribe SP6 system (Epicenter Biotechnologies). The RNA transcripts were isolated, purified and [5′-32P] end labeled as described under Materials and Methods section. Native or denatured radiolabeled RNA samples were digested by the indicated amounts of RNase T1 under conditions specified in the ‘Materials and Methods’ section. Shown is a representative phosphor image of ribonuclease digests of RNA samples resolved by 12% polyacrylamide, 8 M urea denaturing gel electrophoresis.
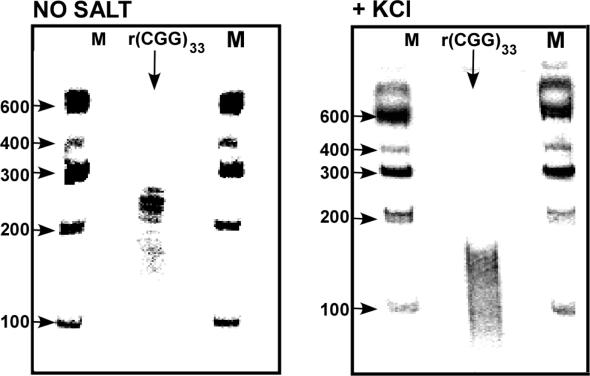
We next sought to directly demonstrate the ability of the RNA (CGG)33 tract to form an intramolecular secondary structure. Aliquots of <2.5 nM 5′-32P labeled FMR1-5′-UTR(CGG)33 RNA were incubated at 4°C for 15 min without or in the presence of 120 mM KCl and resolved by electrophoresis in non-denaturing polyacrylamide gels that were, respectively, devoid of salt or contained 10 mM KCl. Representative electrophoregrams shown in Figure 4 indicated that the 365-bases-long single-stranded (CGG)33 RNA migrated in the absence of salt between DNA size markers of 200 and 250 bp. By contrast, in the presence of KCl, the RNA migrated between DNA markers of 100 and 200 bp. Thus, the (CGG)33 RNA sequence formed in the presence of K+ ions a rapidly migrating compact structure. The zero-order kinetics of the formation of this species was consonant with it being an intramolecular secondary structure that may represent either a hairpin or quadruplex formation (see Discussion section). Being positioned at the 5′ end of FMR1 mRNA, such structure could well retard its translation.
Figure 4.
FMR1-5′-UTR(CGG)33 RNA forms K+-ion-dependent compact intramolecular secondary structure. 5′-32P FMR1-5′-UTR(CGG)33 RNA was incubated at 4°C for 15 min in intramolecular quadruplex formation mixtures that were devoid of salt or contained 120 mM KCl as described under Materials and Methods section. The RNA mixtures were resolved by electrophoresis in non-denaturing 6% polyacrylamide gels in 0.5× TBE buffer, pH 8.2 that were, respectively, devoid of or contained 10 mM KCl. Shown are autoradiograms of electrophoretically resolved RNA without or with KCl. The salt-dependent different electrophoretic mobility of the RNA can be assessed by their different migration relative to 5′-[32P] labeled DNA size markers (M).
Repression of translation by (CGG)n tracts in mRNA is alleviated in vivo by tetraplex (CGG)n destabilizing proteins
We next examined in HEK293 cells the in vivo effects of premutation-range (CGG) repeat tracts on FMR-1 promoter-driven transcription and translation of an FL reporter gene. In subsequent experiments, we inquired whether tetraplex (CGG)n destabilizing proteins affect the expression of an FL gene preceded by (CGG) repeats of different lengths.
Translation in vivo of FL mRNA is repressed by an upstream (CGG)99 RNA tract
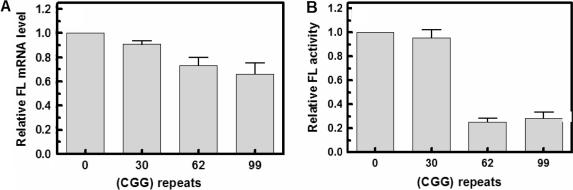
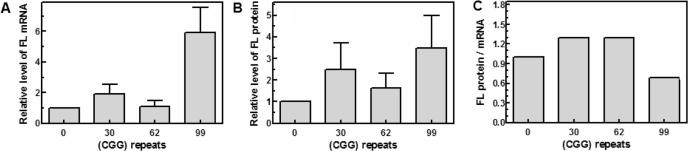
To investigate the effect of different lengths of upstream (CGG)n tracts on the in vivo expression of the FL reporter gene, HEK293 cells were transfected with pCMV-FMR1-5′-UTR(CGG)n-FL (n = 0, 30, 62 or 99). Following 24 h growth, the cells were harvested and lysed and the RL-adjusted relative amounts of FL mRNA and protein were determined and normalized to the transfection efficiency as described under Materials and Methods section. Results of these experiments indicated that relative to a pCMV-FMR1-5′-UTR(CGG)n-FL plasmid with no (CGG)n tract, the amounts of transcribed FL mRNA for plasmids with 30, 62 or 99 (CGG) repeats increased on average by 1.9-, 1.1- or 5.9-fold, respectively (Figure 5A, see also Table 1). Parallel comparison indicated that the relative activity of FL protein encoded by plasmids that had 30, 62 or 99 repeats increased by 2.5-, 1.4- or 3.5-fold, respectively (Figure 5B and Table 1). Thus, transcription of the FL gene was enhanced in vivo in the presence of 30 or 99 (CGG) repeats but was essentially unaffected by 62 repeats. By contrast (CGG)n tracts of all sizes raised the levels of FL protein to different extents. Most notable, however, was the different effect of different (CGG)n sizes on the efficacy of FL mRNA utilization. Tracts of 30 or 62 (CGG) repeats increased the relative efficiency of FL translation by 1.3-fold. However, the longer (CGG)99 sequence reduced the relative efficacy of FL translation to 0.6-fold (Figure 5C and Table 1). Thus, as revealed by comparing Figures 1 and 2 to Figure 5 and Table 1, the (CGG)99 tract lowered the relative efficacy of FL mRNA translation in both in vitro and in vivo systems.
Figure 5.
Effect of (CGG)n repeat size on the synthesis of FL mRNA and protein in HEK293 cells. Cultured HEK293 cells that were co-transfected with FL reporting pCMV-FMR1-5′-UTR(CGG)n-FL (n = 0, 30, 62 or 92) and RL normalizing pCMV-RL plasmids were harvested after 24 h, lysed and aliquots of the cell lysates were used to conduct semi-quantitative RT-PCR measurements of FL and RL mRNA levels and to determine the FL protein activity (Materials and Methods section). (A) Histogram of the measured relative levels of FL mRNA as a function of the (CGG)n tract size. Each column represents an average of the results of nine independent experiments. (B) Histogram of the measured relative levels of FL protein activity as a function of the (CGG)n tract size. Each column represents an average of the results of ten independent experiments. (C) Histogram of the calculated ratios of FL protein to mRNA.
Table 1.
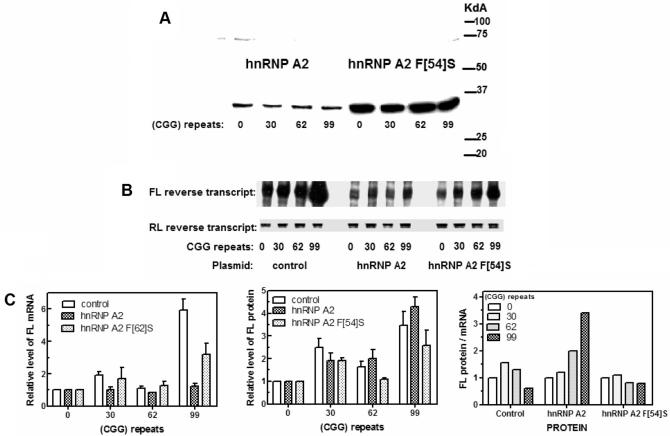
Expression of hnRNP A2 in HEK293 cells increases the efficiency of translation of mRNA that contains 62 or 99 (CGG) repeats
| Expressed protein | None | hnRNPA2 | hnRNP A2 F[54]S | ||||||
|---|---|---|---|---|---|---|---|---|---|
| (CGG) repeats | FL mRNA | FL protein | FL protein/ mRNA | FL mRNA | FL protein | FL protein/ mRNA | FL mRNA | FL protein | FL protein/ mRNA |
| 0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 30 | 1.9 ± 0.6 (9) | 2.5 ± 1.2 (10) | 1.3 | 1.3 ± 0.7 (4) | 1.6 ± 1.2 (3) | 1.2 | 1.7 ± 1.1 (3) | 1.9 ± 0.2 (2) | 1.1 |
| 62 | 1.1 ± 0.3 (8) | 1.4 ± 0.7 (7) | 1.3 | 1.0 ± 0.2 (4) | 2.0 ± 0.8 (4) | 2.0 | 1.3 ± 0.4 (3) | 1.1 ± 0.1 (3) | 0.8 |
| 99 | 5.9 ± 1.7 (6) | 3.5 ± 1.5 (6) | 0.6 | 1.4 ± 0.3 (4) | 4.3 ± 0.7 (4) | 3.1 | 3.2 ± 1.1 (3) | 2.6 ± 1.2 (3) | 0.8 |
The calculated average ratios of FL protein to mRNA are in bold.
The tetraplex (CGG)n destabilizing protein hnRNP A2 alleviates the repression of FL mRNA by (CGG)n RNA tracts
It is reasonable that the source for the diminished efficiency of translation in vivo by a (CGG)99 tract in mRNA (Figure 5) was the formation of an intramolecular secondary structure that impeded the progression of the translation machinery. Member proteins of the hnRNP family; CBF-A (23,26,27), hnRNP A2 (23) and UP-1 (28,29) were shown to destabilize in vitro tetraplex structures of the (CGG)n sequence. We thus inquired whether the tetraplex (CGG)n-disrupting protein hnRNP A2 affected the (CGG)n-modulated transcription or translation of FL in vivo. The plasmid pCMV2-Flag-hnRNP A2 which expressed the tetraplex RNA and DNA (CGG)n-unfolding protein hnRNP A2 under the control of a CMV promoter, was co-transfected into HEK293 cells with the reporter plasmid pCMV-FMR1-5′-UTR(CGG)n-FL (n = 0, 30, 62 or 99) and the RL encoding vector pCMV-RL. Transcription and translation of FL and RL in hnRNP A2 expressing cells were compared to their levels in cells that were transfected with the control vectors pCMV2-Flag lacking an hnRNP A2 coding sequence or pCMV2-Flag-hnRNP A2 F[54]S that harbored F to S mutation which obliterated the tetraplex (CGG)n destabilizing activity of hnRNP A2 (23). Following 24 h growth, the cells were harvested and lysed and the expression of hnRNP A2 or hnRNP A2 F[54]S was monitored in the cell lysates by western analysis and normalized levels of FL mRNA and protein were measured (see Materials and Methods section). Results of a set of these experiments are summarized in Figure 6 and Table 1. Representative western blots of hnRNPA2 and hnRNP A2 F[54]S and typical agarose gel resolution of RT-PCR amplified FL and RL reverse transcripts are shown in Figure 6A and B, respectively. It is evident that although the amounts of expressed hnRNP A2 F[54]S exceeded those of hnRNPA2, the level of each protein was unaffected by the number of (CGG) repeats in the co-transfected FL reporter gene (Figure 6A). Quantified results of multiple similar determinations of RL normalized levels of FL mRNA and protein activity are summarized in Table 1 and Figure 6C.
Figure 6.
Effect of hnRNP A2 and its inactive mutant hnRNP A2 F[54]S on the transcription and translation of FL reporter gene preceded by different sizes of (CGG)n repeat tract. Cultured HEK293 cells were co-transfected with FL reporting plasmid pCMV-FMR1-5′-UTR(CGG)n-FL (n = 0, 30, 62 or 99) and RL normalizing pCMV-RL vector and with one of the following plasmids: empty control pCMV2-Flag vector, hnRNP A2 expressing plasmid pCMV2-Flag-hnRNPA2 or pCMV2-Flag-hnRNPA2 F[54]S plasmid that expresses mutant hnRNP A2 F[54]S. The transfected cells were grown and lysed as detailed in the legend to Figure 5. Aliquots of the cell lysates were used to detect by western analysis the expressed hnRNP A2 proteins, to determine the levels of FL mRNA by semi-quantitative RT-PCR analysis and to measure the activity of FL protein as described under Materials and Methods section. (A) Representative western analysis of hnRNP A2 or hnRNP A2 F[54]S expressed in cells co-transfected with pCMV-FMR1-5′-UTR(CGG)n-FL plasmids bearing different lengths of the repeat tract. (B) Typical electrophoretic separation of FL and RL reverse transcript products of semi-quantitative RT-PCR assay. The FL mRNA RNA molecules, bearing (CGG)n tracts of the indicated sizes, were produced in cells transfected by empty control pCMV2-Flag vector or in cells that expressed hnRNP A2 or hnRNP A2 F[54]S. (C) Graphic presentation of average levels of FL mRNA (first panel) and FL protein (second panel) that were produced in control cells and in cells that expressed hnRNP A2 or hnRNP A2 F[54]S. Calculated average ratios of FL protein to mRNA in control and hnRNP A2 or hnRNP A2 F[54]S expressing cells are shown in the third panel.
Data presented in Table 1 and in the first panel of Figure 6C show that the respective levels of FL (CGG)99 mRNA relative to FL (CGG)0 mRNA were 5.9- or 3.2-fold higher in cells that did not express hnRNP A2 or that produced mutant hnRNP A2 F[54]S protein. By contrast, expression of functional hnRNP A2 brought down FL (CGG)99 mRNA to its level in the control cells while augmenting the efficiency of its translation. Relative to RNA with no (CGG) repeat tract, FL (CGG)99 translation was increased by 3.5- and 2.6-fold without hnRNP A2 or in the presence of mutant hnRNP A2 F[54]S (Table 1, Figure 6C, second panel). As a result, in cells that did not have hnRNP A2 or that produced inactive hnRNP A2 F[54]S, the ratio of FL protein activity to FL (CGG)62 or FL (CGG)99 mRNA remained essentially unchanged or was lower by 20–40% relative to control. By clear contrast, in cells that expressed the active tetraplex (CGG)n resolving protein hnRNP A2, translation of FL (CGG)62 or (CGG)99 mRNA was elevated 1.4- and 1.2-fold and the ratio of FL protein to mRNA increased by 2.0- or 3.1-fold, respectively (Table 1, Figure 6C, third panel). In fact, the stimulation of translation by functional hnRNP A2 may be of even higher relative magnitude since the amounts of expressed hnRNP A2 F[54]S significantly exceeded those of hnRNP A2 (Figure 6A).
The tetraplex (CGG)n destabilizing protein CBF-A also alleviates the repression of translation by (CGG)n tracts
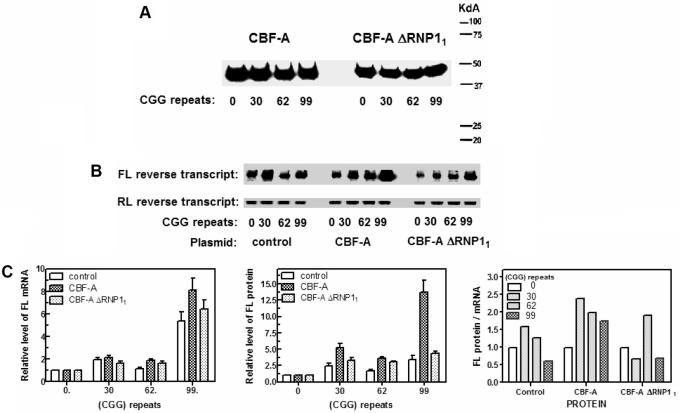
Similar to hnRNP A2, CBF-A another member of the hnRNP family, was shown to disrupt in vitro tetraplex structures of (CGG)n (23,26,27). We inquired, therefore, whether this protein was also capable of alleviating in vivo the repression of translation by premutation-range (CGG)n mRNA tracts. HEK293 cells were co-transfected with reporter plasmids pCMV-FMR1-5′-UTR(CGG)n-FL (n = 0, 30, 62 or 99) and a normalizing pCMV-RL vector. A third co-transfected plasmid was pCMV2-Flag-CBF-A that expressed functional CBF-A protein or one of two control plasmids, pCMV2-Flag devoid of a CBF-A encoding sequence or pCMV2-Flag-CBF-A ΔRNP11 that contained ΔRNP11 mutation in the CBF-A gene that abolished its tetraplex (CGG)n destabilization activity (26). Western analyses of active CBF-A and mutant CBF-A ΔRNP11 indicated that both proteins were expressed at similar levels which were unaffected by the size of the (CGG)n tract in the reporter FL gene (Figure 7A). Typical patterns of agarose gel resolved RT-PCR amplified reverse transcripts of FL and RL mRNA are shown in Figure 7B. Results of multiple measurements of RL normalized levels of FL mRNA and protein as a function of the (CGG)n size and of the expression or lack thereof of functional CBF-A are summarized in Table 2 and in Figure 7C. Data indicated that relative to FL gene with no (CGG)n tract, 30 or 62 (CGG) repeats modestly increased the levels of FL mRNA by up to ∼2-fold whether or not CBF-A or its mutant CBF-A ΔRNP11 were expressed. Moreover, the longer (CGG)99 tract affected a steeper increase of FL mRNA of 5.9-, 7.2- or 6.4-fold in cells that were, respectively, transfected with empty vector, expressed CBF-A or harbored CBF-A ΔRNP11 (Table 1, Figure 7C, first panel). Thus, by promoting FL (CGG)99 mRNA transcription, active CBF-A differed from functional hnRNP A2 which brought down its production to the (CGG)0 mRNA level (Table 1, see Discussion section). Relative to FL gene with no repeat tract, the presence of 30 or 62 (CGG) repeats moderately increased the levels of FL protein whether or not the cells expressed CBF-A or CBF-A ΔRNP11. By contrast, whereas the relative translation of FL (CGG)99 mRNA was elevated ∼4-fold in cells without or with CBF-A ΔRNP11, the level of FL protein increased by almost 14-fold in cells that expressed CBF-A (Table 2 and Figure 7C, second panel). As a result, whereas the relative efficacy of FL mRNA translation as manifested by the FL protein to mRNA ratio was depressed to 0.6- and 0.7-fold in control or CBF-A ΔRNP11 producing cells, respectively, it increased by 1.9-fold in functional CBF-A expressing cells (Table 2 and Figure 7C, third panel). Hence, despite their different effects on the transcription of FL (CGG)99 mRNA, both the tetraplex (CGG)n destabilizing proteins hnRNP A2 and CBF-A improved the efficiency of its translation.
Figure 7.
Effect of CBF-A and its inactive mutant CBF-A ΔRNP11 on the transcription and translation of FL reporter gene preceded by different sizes of (CGG)n repeat tract. Cultured HEK293 cells were co-transfected with FL reporting plasmid pCMV-FMR1-5′-UTR(CGG)n-FL (n = 0, 30, 62 or 99) and RL normalizing pCMV-RL vector and with one of the following plasmids: empty pCMV2-Flag vector, CBF-A expressing plasmid pCMV2-Flag-CBF-A or pCMV2-Flag-CBF-A ΔRNP11 plasmid that expressed mutant CBF-A ΔRNP11. The cells were grown and lysed as detailed in the legend to Figure 5. Aliquots of the cell lysates were used to detect by western analysis the expressed CBF-A proteins, to determine the levels of FL mRNA by semi-quantitative RT-PCR analysis and to measure the activity of FL protein as described under Materials and Methods section. (A) Representative western analysis of CBF-A or CBF-A ΔRNP11 expressed in cells co-transfected with pCMV-FMR1-5′-UTR(CGG)n-FL bearing different lengths of the repeat tract. (B) Typical electrophoretic separation of FL and RL reverse transcript products of semi-quantitative RT-PCR assay. The FL mRNA RNA molecules, bearing (CGG)n tracts of the indicated sizes, were produced in cells transfected by empty control pCMV2-Flag vector or in cells that expressed CBF-A or CBF-A ΔRNP11. (C) Graphic presentation of average levels of FL mRNA (first panel) and FL protein (second panel) that were produced in control cells and in cells that expressed hnRNP A2 or hnRNP A2 F[54]S. The third panel is a fit spline plot of the ratios of FL protein to mRNA. Calculated average ratios of FL protein to mRNA in control and CBF-A or CBF-A ΔRNP11 expressing cells are shown in the third panel.
Table 2.
Expression of CBF-A increases the efficiency of translation in HEK293 cells of mRNA that contains 62 or 99 (CGG) repeats
| Expressed protein | None | CBF-A | CBF-A ΔRNP11 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| (CGG) repeats | FL mRNA | FL protein | FL protein/ mRNA | FL mRNA | FL protein | FL protein/ mRNA | FL mRNA | FL protein | FL protein/ mRNA |
| 0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 |
| 30 | 1.9 ± 0.6 (9) | 2.5 ± 1.2 (10) | 1.3 | 2.1 ± 0.3 (5) | 5.0 ± 1.2 (5) | 2.3 | 1.6 ± 0.4 (5) | 3.3 ± 0.8 (3) | 2.1 |
| 62 | 1.1 ± 0.3 (8) | 1.4 ± 0.7 (7) | 1.3 | 1.8 ± 0.3 (4) | 3.6 ± 0.4 (3) | 2.0 | 1.6 ± 0.3 (4) | 3.1 ± 0.1 (2) | 1.9 |
| 99 | 5.9 ± 1.7 (6) | 3.6 ± 1.5 (6) | 0.6 | 7.2 ± 1.7 (4) | 13.7 ± 3.7 (4) | 1.9 | 6.4 ± 1.9 (5) | 4.4 ± 0.5 (3) | 0.7 |
The calculated average ratios of FL protein to mRNA are in bold.
DISCUSSION
Whereas FMR1 transcription is completely silenced in fragile X full mutation cells (3), premutation carriers produce 5- to 10-fold higher amounts of FMR1 mRNA than normal cells (10,11). Yet, despite the elevated levels of FMR1 mRNA in carrier cells, they produce FMRP at amounts that are within or below the norm (10,12,13). It thus appears that FMR1 translation is repressed by the moderately expanded premutation (CGG) repeat sequence. Indeed, evidence was presented to show that premutation (CGG) repeat sizes are linked to lowered association of FMR1 mRNA with lymphoblastoid cell polysomes (14) and that the 40S ribosomal subunits stall along FMR1 mRNA molecules that contain more than 200 repeats (15). Data indicated that (CGG)n tracts in RNA fold into hairpin (16,17,19,35) and tetraplex (23) structures. A reasonable conjecture, therefore, is that the diminished efficacy of the translation of premutation FMR1 mRNA is due to retarded ribosome association and stalled progression at secondary structures of the (CGG)n tract in premutation FMR1 mRNA. In this work, we showed that premutation-range (CGG)n tracts selectively reduced the efficiency of mRNA translation both in vitro and in vivo. Second, following a demonstration that (CGG)n sequence in RNA was capable of forming quadruplex structures, we showed that the expression in vivo of proteins that untangle tetraplex structures of (CGG)n increased the efficacy of mRNA utilization.
Relative to FL reporter gene with no (CGG)n upstream tract, the introduction of 30 (CGG) repeats upstream to the gene stimulated its in vitro transcription and translation in coupled T7 promoter-driven transcription–translation system (Figure 1) and in FMR1 promoter-driven transcription and translation in vivo (Figure 5). The most frequent normal number of (CGG) repeats in the FMR1 gene and its transcript in normal subjects is 29–33 (34). In vivo expression of the FL gene (Figure 5) suggested that secondary structures of the (CGG)29–33 tracts may promote maximum efficacy of translation FMR1 mRNA in cells. Such requirement may exert positive selective pressure that results in the preservation of the (CGG)29–33 repeat sequence in the human population. As our results showed, 62 or 99 (CGG) repeats that represent the lower range of premutation size slightly repressed (Figure 2) or did not affect (Figure 1) the in vitro transcription of the FL gene. Most significantly, both 62 and 99 (CGG) repeats did sharply suppress FL translation both in coupled or in separate in vitro systems (Figures 1 and 2). The observed declining ratio of FL protein to mRNA indicated that the premutation repeat tracts reduced the efficiency of utilization of the FL mRNA. Results presented in Figures 3 and 4 demonstrated that the (CGG)33 RNA repeat maintained in vitro a secondary structure and that a large portion of the guanine-rich FMR1 5′ UTR sequence itself also folded into secondary configuration (Figure 3). It is plausible that the nature and geometry of secondary structures that (CGG)n RNA tracts formed independently or together with the 5′ UTR tract differed for different sizes of the repeat sequence. Thus, the spatial arrangement of secondary structures of the normal 30 repeats may have promoted the association of mRNA with polysomes and enhanced ribosome progression whereas the different geometries of secondary structures of premutation sized 62 or 99 repeats impeded the translation machinery and limited the efficacy of mRNA utilization. Formation and stabilization of quadruplex nucleic acids require the presence of alkali ion. Thus, the potassium ion dependence of the formation of intramolecular complex by the (CGG)33 RNA sequence (Figure 4) was consistent with tetrahelical nature of this secondary structure. In contrast to tetraplex formation, hairpins can be generated in the absence of ions (16–19). However, the possibility remained that under our experimental conditions K+ ions were critical for the stability of hairpins. Hence, although the data presented in Figure 4 were consonant with a tetraplex structure of the (CGG)33 RNA sequence, the possibility that the observed secondary structure was a hairpin could not be ruled out.
Transcription in vivo of FMR1 promoter-driven FL reporter gene and translation of the transcripts in HEK293 cells respond differently to the presence of upstream premutation (CGG)n tracts than do T7 promoter-driven in vitro transcription and translation. Yet in both the in vitro and in vivo systems, premutation size repeat tracts ultimately lowered the efficacy of mRNA utilization. Unlike the essentially unaltered extent of transcription in vitro (Figures 2 and 3), the amount of FL (CGG)30 mRNA relative to FL (CGG)0 mRNA increased 2-fold, FL (CGG)62 mRNA remained unchanged and FL (CGG)99 mRNA was elevated by nearly 6-fold (Figure 5). It is plausible that transcription efficiency was differentially affected by distinct (CGG)n configurations in DNA such that different lengths of the repeat sequence formed secondary structures geometries that were better targets for the transcription factors than others. The modulation of transcription by different lengths of the (CGG) repeat in DNA could also reflect different degree of the in vivo chromatinization of the different lengths of the repeat tracts and dissimilar nucleosome positioning along these sequences. The observed higher amounts of transcripts that were produced in vivo in the presence of 99 (CGG) triplets was in line with the reported increased accumulation of FMR1 mRNA in leukocytes of premutation carriers (10,11). The contrasting unchanged or reduced transcript accumulation in vitro was likely to be due to the use of T7 in place of the FMR1 promoter and to the dissimilar reaction conditions of the in vitro and in vivo transcription systems. The (CGG)n tracts had also opposing effect on translation in vitro and in vivo. Whereas the relative extent of translation of FL mRNA molecules that contained 62 or 99 (CGG) repeats was sharply diminished in vitro relative to 30 repeats (Figures 1 and 2), it decreased in HEK293 cells for 62 repeats but was somewhat increased at 99 repeats (Figure 5 and Table 1). The observed elevated level of protein in the transfected cultured cells was inconsistent with the reported unchanged or slightly reduced amounts of FMRP in cells of premutation carriers (10,12,13). Although the reason for this discrepancy is not clear, cells of premutation carriers (14), transfected HEK293 cells (Figure 5 and Table 1) and in vitro translation systems (Figures 1 and 2) all displayed reduced translation efficiency of premutation mRNA. A likely common denominator for all the systems may, therefore, be the impediment to the translation machinery by tetraplex structures of the intermediately expanded (CGG)n tract.
Results summarized in Figures 6 and 7 and Tables 1 and 2 indicated that the RNA binding and tetraplex DNA and RNA-disrupting proteins hnRNP A2 and CBF-A increased the efficacy of (CGG)n mRNA translation in vivo. Based on our observation that (CGG)33 RNA formed intramolecular secondary structure (Figures 3 and 4), we assumed that a stable (CGG)99 unimolecular folded domain in FL mRNA was responsible for its retarded translation in vitro (Figures 1B, C and 2B) and in vivo (Figure 6). In such a case, untangling of the secondary structure should increase the efficiency of translation. We demonstrated in the past that intermolecular tetraplex structures of (CGG)n in DNA were disrupted in vitro by members of the hnRNP family CBF-A and hnRNP A2 (23,26,27). We also showed that mutations in each of the two conserved domains of both proteins, the RNP11 box and the ATP/GTP-binding fold, specifically abolished their in vitro quadruplex disruption activity (23,26). Results described here showed that expression of functional hnRNP A2 in HEK293 cells, but not of its inactive hnRNP A2 F[54]S mutant, alleviated the repression of FL mRNA translation by a (CGG)99 tract (Figure 6C and Table 1). Also, active CBF-A, but not its ΔRNP11 mutant, increased the efficiency of the in vivo translation of (CGG)99 FL mRNA (Figure 7C and Table 2). Although the hnRNP A2 F[54]S and CBF-A ΔRNP11 mutants lost their capacity to disrupt tetraplex structures of the (CGG)n sequence, they maintained their nucleic acids binding activity (Cohen, E. and Weisman-Shomer, P., unpublished data). It is likely, therefore, that the enhancement of (CGG)n mRNA translation by hnRNP A2 and CBF-A was due to their quadruplex destabilizing activity rather than to their RNA-binding capacity. Whereas both hnRNP A2 and CBF-A were shown to disrupt quadruplex structures of (CGG)n in DNA, only hnRNP A2 but not CBF-A destabilized in vitro a bimolecular quadruplex structure of synthetic (CGG)7 RNA (23). Because the in vitro and in vivo conditions for CBF-A action were different, since the likely RNA structure that formed in vivo was intra- rather than intermolecular (Figure 4), and as the CBF-A ΔRNP11 mutant failed to stimulate translation, we assume that similar to hnRNP A2, CBF-A also enhanced the translation of FL (CGG)99 mRNA by untangling its monomolecular secondary structure. The observed correlation between the ability of hnRNP A2 or CBF-A to unfold quadruplex structures of RNA or DNA and their capacity to alleviate the blocking of translation by the (CGG)99 tract, was consistent with the folded repeat sequence being a tetrahelix. However, as discussed above, the observed intramolecular secondary structure of the (CGG)n repeat may represent a hairpin rather than quadruplex. Such scenario raises the untested prospect that hnRNP A2 and CBF-A can also destabilize hairpins and that similar to their tetraplex unfolding activity, the unwinding of hairpins also requires that the proteins possess intact RNP1 and ATP/GTP motifs.
Taken together, our results lent credence to the notion that secondary structures of the expanded (CGG) repeat tract in fragile X premutation mRNA hindered translation and that their disruption alleviated this impediment. This conclusion gained support recently by the report that a quadruplex motif in the 5′ UTR of the human NRAS oncogene mRNA modulated its translation in vitro. Furthermore, bioinformatics analysis predicted that 5′ UTR elements in 2922 other human mRNA species can assume tetraplex structure that may potentially regulate their translation (37). These results, as well as the present report on the capacity of quadruplex destabilizing proteins to alleviate the impediment to translation that a folded (CGG)n tract posed, are in line with the notion that tetrahelical motifs in mRNA may modulate its translation.
Although functional hnRNP A2 and CBF-A augment the translation of FL (CGG)99 mRNA, their mechanisms of action appeared to be dissimilar. Whereas the level of FL (CGG)99 mRNA was 5.9-fold higher than that of FL mRNA with no repeat tract, hnRNP A2 lowered the amount of FL (CGG)99 mRNA to that of the FL (CGG)0 mRNA. At the same time, hnRNP A2 did not affect the amounts of accumulated FL protein that FL (CGG)99 mRNA encoded (Table 1 and Figure 6C). It is possible that hnRNP A2 suppressed transcription by affecting the conformation and chromatinization of the FMR1 promoter through its direct interaction with the adjacent (CGG)99 repeat tract. Yet, although hnRNP A2 lowered the amount of FL (CGG)99 mRNA, it increased the relative efficacy of its utilization by 5.2-fold, as reflected by the rising ratio of FL protein to mRNA from 0.6 to 3.1 (Table 1 and Figure 6C). By contrast, functional CBF-A increased the relative amount of FL (CGG)99 transcripts and amount of FL protein by 7.2- and 13.7-fold, respectively (Table 2 and Figure 7C). As a result of the accumulation of FL protein in excess over FL mRNA, CBF-A augmented the efficacy of mRNA usage and increased the ratio of FL protein to mRNA by 3.2-fold from 0.6 to 1.9 (Table 2 and Figure 7C). The different patterns of translation enhancement by hnRNP A2 and CBF-A is not surprising. In addition to its quadruplex (CGG)n untangling activity, CBF-A also binds single-stranded DNA and tetraplex forms of some sequences whereas hnRNP A2 also binds RNA and interacts with some DNA structures. It is plausible, therefore, that because of their binding to different targets in DNA or RNA in the cell in addition to their direct interaction with tetraplex (CGG)n in RNA, each protein differently affects transcription and translation.
ACKNOWLEDGEMENTS
The authors are grateful to Dr Paul Hagerman, UC Davies for fruitful discussions. We wish to thank the three expert referees and the Executive Editor for their excellent critique of this article. This study was supported by grants to M.F. from Conquer Fragile X Foundation, USA, the Israel Science Foundation and the United States-Israel Binational Science Foundation. S.K. was supported by an Israel Science Ministry doctoral scholarship. Funding to pay the Open Access publication charges for the article were provided by the Israel Science Foundation and a VATAT doctoral fellowship to S.K.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fu YH, Kuhl DP, Pizzuti A, Pieretti M, Sutcliffe JS, Richards S, Verkerk AJ, Holden JJ, Fenwick R.G., Jr, et al. Variation of the CGG repeat at the fragile X site results in genetic instability: resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- 2.Oberlè I, Rousseau F, Heitz D, Kretz C, Devys D, Hanauer A, Boue J, Bertheas MF, Mandel JL. Instability of a 550-base pair DNA segment and abnormal methylation in fragile X syndrome. Science. 1991;252:1097–1102. doi: 10.1126/science.252.5009.1097. [DOI] [PubMed] [Google Scholar]
- 3.Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 4.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 5.Yu S, Pritchard M, Kremer E, Lynch M, Nancarrow J, Baker E, Holman K, Mulley JC, Warren ST, et al. Fragile X genotype characterized by an unstable region of DNA. Science. 1991;252:1179–1181. doi: 10.1126/science.252.5009.1179. [DOI] [PubMed] [Google Scholar]
- 6.Hagerman RJ, Hagerman PJ. The fragile X premutation: into the phenotypic fold. Curr. Opin. Genet. Dev. 2002;12:278–283. doi: 10.1016/s0959-437x(02)00299-x. [DOI] [PubMed] [Google Scholar]
- 7.Allingham-Hawkins DJ, Babul-Hirji R, Chitayat D, Holden JJ, Yang KT, Lee C, Hudson R, Gorwill H, Nolin SL, et al. Fragile X premutation is a significant risk factor for premature ovarian failure: the International Collaborative POF in Fragile X study – preliminary data. Am. J. Med. Genet. 1999;83:322–325. [PMC free article] [PubMed] [Google Scholar]
- 8.Hagerman RJ, Leehey M, Heinrichs W, Tassone F, Wilson R, Hills J, Grigsby J, Gage B, Hagerman PJ. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- 9.Berry-Kravis E, Lewin F, Wuu J, Leehey M, Hagerman R, Hagerman P, Goetz CG. Tremor and ataxia in fragile X premutation carriers: blinded videotape study. Ann. Neurol. 2003;53:616–623. doi: 10.1002/ana.10522. [DOI] [PubMed] [Google Scholar]
- 10.Tassone F, Hagerman RJ, Chamberlain WD, Hagerman PJ. Transcription of the FMR1 gene in individuals with fragile X syndrome. Am. J. Med. Genet. 2000;97:195–203. doi: 10.1002/1096-8628(200023)97:3<195::AID-AJMG1037>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 11.Tassone F, Hagerman RJ, Loesch DZ, Lachiewicz A, Taylor AK, Hagerman PJ. Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. Am. J. Med. Genet. 2000;94:232–236. doi: 10.1002/1096-8628(20000918)94:3<232::aid-ajmg9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 12.Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kenneson A, Zhang F, Hagedorn CH, Warren ST. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- 14.Primerano B, Tassone F, Hagerman RJ, Hagerman P, Amaldi F, Bagni C. Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA. 2002;8:1482–1488. [PMC free article] [PubMed] [Google Scholar]
- 15.Feng Y, Zhang F, Lokey LK, Chastain JL, Lakkis L, Eberhart D, Warren ST. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995;268:731–734. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- 16.Chen X, Mariappan SV, Catasti P, Ratliff R, Moyzis RK, Laayoun A, Smith SS, Bradbury EM, Gupta G. Hairpins are formed by the single DNA strands of the fragile X triplet repeats: structure and biological implications. Proc. Natl Acad. Sci. USA. 1995;92:5199–5203. doi: 10.1073/pnas.92.11.5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadel Y, Weisman-Shomer P, Fry M. The fragile X syndrome single strand d(CGG)n nucleotide repeats readily fold back to form unimolecular hairpin structures. J. Biol. Chem. 1995;270:28970–28977. doi: 10.1074/jbc.270.48.28970. [DOI] [PubMed] [Google Scholar]
- 18.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Handa V, Saha T, Usdin K. The fragile X syndrome repeats form RNA hairpins that do not activate the interferon-inducible protein kinase, PKR, but are cut by Dicer. Nucleic Acids Res. 2003;31:6243–6248. doi: 10.1093/nar/gkg818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fry M, Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kettani A, Kumar RA, Patel DJ. Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol. 1995;254:638–656. doi: 10.1006/jmbi.1995.0644. [DOI] [PubMed] [Google Scholar]
- 22.Usdin K, Woodford KJ. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23:4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khateb S, Weisman-Shomer P, Hershco I, Loeb LA, Fry M. Destabilization of tetraplex structures of the fragile X repeat sequence (CGG)n is mediated by homolog-conserved domains in three members of the hnRNP family. Nucleic Acids Res. 2004;32:4145–4154. doi: 10.1093/nar/gkh745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamath-Loeb AS, Loeb LA, Johansson E, Burgers PM, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase delta specifically facilitate copying of tetraplex and hairpin structures of the d(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- 25.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 26.Weisman-Shomer P, Cohen E, Fry M. Distinct domains in the CArG-box binding factor A destabilize tetraplex forms of the fragile X expanded sequence d(CGG)n. Nucleic Acids Res. 2002;30:3672–3681. doi: 10.1093/nar/gkf506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weisman-Shomer P, Cohen E, Fry M. Interruption of the fragile X syndrome expanded sequence d(CGG)n by interspersed d(AGG) trinucleotides diminishes the formation and stability of d(CGG)n tetrahelical structures. Nucleic Acids Res. 2000;28:1535–1541. doi: 10.1093/nar/28.7.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuda H, Katahira M, Tsuchiya N, Enokizono Y, Sugimura T, Nagao M, Nakagama H. Unfolding of quadruplex structure in the G-rich strand of the minisatellite repeat by the binding protein UP1. Proc. Natl Acad. Sci. USA. 2002;99:12685–12690. doi: 10.1073/pnas.152456899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fukuda H, Katahira M, Tanaka E, Enokizono Y, Tsuchiya N, Higuchi K, Nagao M, Nakagama H. Unfolding of higher DNA structures formed by the d(CGG) triplet repeat by UP1 protein. Genes Cells. 2005;10:953–962. doi: 10.1111/j.1365-2443.2005.00896.x. [DOI] [PubMed] [Google Scholar]
- 30.Weisman-Shomer P, Cohen E, Hershco I, Khateb S, Wolfovitz-Barchad O, Hurley LH, Fry M. The cationic porphyrin TMPyP4 destabilizes the tetraplex form of the fragile X syndrome expanded sequence d(CGG)n. Nucleic Acids Res. 2003;31:3963–3970. doi: 10.1093/nar/gkg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen LS, Tassone F, Sahota P, Hagerman PJ. The (CGG)n repeat element within the 5′ untranslated region of the FMR1 message provides both positive and negative cis effects on in vivo translation of a downstream reporter. Hum. Mol. Genet. 2003;12:3067–3074. doi: 10.1093/hmg/ddg331. [DOI] [PubMed] [Google Scholar]
- 32.Beilina A, Tassone F, Schwartz PH, Sahota P, Hagerman PJ. Redistribution of transcription start sites within the FMR1 promoter region with expansion of the downstream CGG-repeat element. Hum. Mol. Genet. 2004;13:543–549. doi: 10.1093/hmg/ddh053. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Russel DW. Molecular Cloning, A Laboratory Manual. 3rd. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2001. edn. [Google Scholar]
- 34.Brown WT, Houck G.E., Jr, Jeziorowska A, Levinson FN, Ding X, Dobkin C, Zhong N, Henderson J, Brooks SS, et al. Rapid fragile X carrier screening and prenatal diagnosis using a nonradioactive PCR test. J. Am. Med. Assoc. 1993;270:1569–1575. [PubMed] [Google Scholar]
- 35.Jasinska A, Michlewski G, de Mezer M, Sobczak K, Kozlowski P, Napierala M, Krzyzosiak WJ. Structures of trinucleotide repeats in human transcripts and their functional implications. Nucleic Acids Res. 2003;31:5463–5468. doi: 10.1093/nar/gkg767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinden RR, Pytlos MJ, Potaman VN. Mechanisms of DNA repeat expansion. In: Fry M, Usdin K, editors. Human Nucleotide Expansion disorders. 2006. Vol. 19. Springer, Berlin, Heidelberg, New York, pp. 3–53. [Google Scholar]
- 37.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]