Abstract
The double-strand DNA break repair pathway, non-homologous DNA end joining (NHEJ), is distinctive for the flexibility of its nuclease, polymerase and ligase activities. Here we find that the joining of ends by XRCC4-ligase IV is markedly influenced by the terminal sequence, and a steric hindrance model can account for this. XLF (Cernunnos) stimulates the joining of both incompatible DNA ends and compatible DNA ends at physiologic concentrations of Mg2+, but only of incompatible DNA ends at higher concentrations of Mg2+, suggesting charge neutralization between the two DNA ends within the ligase complex. XRCC4-DNA ligase IV has the distinctive ability to ligate poly-dT single-stranded DNA and long dT overhangs in a Ku- and XLF-independent manner, but not other homopolymeric DNA. The dT preference of the ligase is interesting given the sequence bias of the NHEJ polymerase. These distinctive properties of the XRCC4-DNA ligase IV complex explain important aspects of its in vivo roles.
INTRODUCTION
Pathologic double-strand DNA breaks (DSBs) arise when ionizing radiation passes near DNA or when a replication fork encounters a nick. In mitotic cells, physiologic double-strand breaks are generated in lymphocytes during V(D)J recombination and class switch recombination (1,2). Many pathologic DSBs and nearly all physiologic DSBs in mitotic cells are repaired by the non-homologous DNA end joining (NHEJ) pathway. Given this, it is not surprising that individuals born with defects in non-homologous DNA end joining (NHEJ) are sensitive to ionizing radiation and have severe combined immunodeficiency (SCID) (3). Many factors involved in NHEJ have been identified based on analysis of human and mouse SCID (4–8), and ∼15–20% of human SCID is due to NHEJ defects.
The proteins involved in NHEJ include Ku70, Ku80, DNA-PKcs, Artemis, XRCC4, DNA ligase IV, pol mu, pol lambda and XLF (or Cernunnos) (1). NHEJ is thought to begin with Ku binding to the two newly created DNA ends at the double-strand break. Ku improves the affinity of a nuclease complex (Artemis:DNA-PKcs), a ligase complex (XRCC4:DNA ligase IV) and POL X polymerases (pol μ and pol λ). XLF (or Cernunnos) (5–7) stimulates the XRCC4-DNA ligase IV complex in ligation assays (9–12).
We and others have recently described the ability of XRCC4-DNA ligase IV to ligate incompatible DNA ends (12,13) and the ability of the XLF (Cernunnos) protein to stimulate the ligation of DNA ends (9,10). However, two studies observed that XLF (Cernunnos) stimulated ligation by XRCC4-DNA ligase IV at compatible DNA ends (9,10), whereas one study only observed stimulation at incompatible DNA ends (12).
Here, we find a marked influence of terminal DNA end sequence on end joining by XRCC4-DNA ligase IV. Nearly all aspects of the sequence effect conform to a model in which the two DNA ends initially anneal at any shared sites of complementarity. However, some aspect of the joining process places the base at position N on the top strand in a sterically constrained position relative to the N+1 base on the anti-parallel strand. In addition, we find that at physiologic Mg2+ concentrations, XLF (Cernunnos) stimulates all DNA end joining, regardless of the extent of terminal microhomology between the two DNA ends, though inefficiently joined ends benefit most by the presence of XLF (Cernunnos, but hereafter called XLF, for simplicity). Finally, we find that XRCC4-DNA ligase IV is capable of ligating single-stranded DNA and long single-stranded overhangs. We discuss how these properties of the NHEJ ligase complex explain aspects of the flexibility of mammalian NHEJ.
MATERIALS AND METHODS
Oligonucleotides
Oligonucleotides used in this study were synthesized by Operon Biotechnologies, Inc. (Huntsville, AL, USA) and are listed in the Supplementary Data. We purified the oligonucleotides using 12% or 15% denaturing polyacrylamide gel electrophoresis (PAGE) and determined the concentration spectrophotometrically.
Oligonucleotides were labeled at the 5′-end with [γ-32P]ATP (3000 Ci/mmol) (PerkinElmer Life Sciences, Boston, MA, USA) and T4 polynucleotide kinase (New England Biolabs, Beverly, MA, USA) according to the manufacturer's instructions. Unincorporated radioisotope was removed by using G-25 Sephadex (Amersham Biosciences, Inc., Piscataway, NJ, USA) spin-column chromatography. To make the double-stranded DNA substrate, labeled oligonucleotides were mixed with an equal amount of unlabeled complementary oligonucleotide in a buffer containing 10 mM Tris–hydrochloride, pH 8.0, 1 mM EDTA, pH 8.0 and 100 mM NaCl. The mixture was heated at 100°C for 5 min, allowed to cool to room temperature for 3 h, and then incubated at 4°C overnight.
Protein purification
The purification of Ku has been described (14). XRCC4:DNA ligase IV complex was purified from baculovirus-insect cell system as described (15). Native DNA polymerase mu was expressed and purified from Escherichia coli, as described previously (13). Soluble human XLF-myc-his protein was expressed in 293T cells and purified by Ni-NTA agarose beads and Mono Q column as described (9). T4 DNA ligase was purchased from New England Biolabs (Beverly, MA, USA).
DNA ligation assay
The DNA ligation assay was performed in a 10 μl reaction. DNA substrates were first incubated with or without Ku and/or XLF in 1× ligation reaction buffer (25 mM Tris–hydrochloride, pH 7.5, 75 mM NaCl, 72.5 mM KCl, 2 mM DTT, 0.025% Triton X-100 and 100 μM EDTA) supplemented with 10% PEG (MW > 8000 kD), 50 μg/ml BSA and 5% glycerol at room temperature for 15 min. The EDTA was used to eliminate effects of any possible trace divalent cations, but is not necessary. Mg2+ is added where specified above this low level of EDTA. PEG improves the ligation efficiency substantially. The low level of Triton X-100 is irrelevant to the ligation efficiency. Ligation was initiated by adding 10 mM MgCl2 with the combinations of proteins designated on the gels (XRCC4-DNA ligase IV, and pol mu, where indicated). Reactions were then incubated at 37°C for 30 min or 2.5 min as indicated. After incubation, reactions were stopped, deproteinized with organic extraction and analyzed by 8% or 10% denaturing PAGE gel. Gels were dried, exposed in a PhosphorImager cassette and scanned.
RESULTS
Influence of terminal DNA sequence on ligation by XRCC4-DNA ligase IV and stimulation by XLF
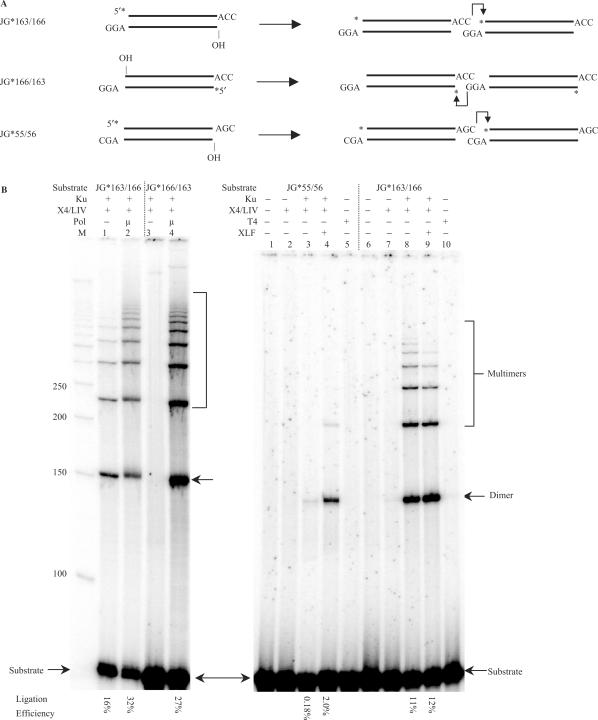
We have found that there is a wide variation in the efficiency of DNA end ligation by XRCC4-DNA ligase IV due to minor variations in the DNA end sequences. For example, the end sequences illustrated in Figure 1B, left panel (–ACC3′ joined to 3′ GGA–) are joined well (on the top) strand by Ku plus XRCC4-ligase IV (lane 1), and this is only slightly increased by the addition of pol mu (lane 2). However, the bottom strand of those same DNA ends is not ligated well (lane 3), unless pol mu is present to fill in the 1 nt gap (lane 4). This illustrates that the two strands of the same pair of DNA ends are not ligated with equal efficiency by XRCC4:DNA ligase IV and some aspect of the sequence at the two ligation points determines this difference. (Note that when we test the top strand for ligation, the bottom strand has an unligatable 5′OH at the junction. Likewise, when we test the bottom strand for ligation, the top strand has an unligatable 5′OH at the junction.)
Figure 1.
DNA substrates with a gap are ligated by XRCC4-Ligase IV in a sequence-dependent manner. (A) Three 73 bp substrates with different 3′ overhangs were designed and tested for the efficiency of ligation over a gap by XRCC4-Ligase IV. A star indicates the position of the radioisotope label. JG*163/166 and JG*166/163 are the same duplex DNA substrate but with one strand labeled or the other. In the ligations, a dimer, e.g. involves two molecules of the same species joined head to tail. (B) In each reaction, 20 nM substrate was incubated with the protein(s) indicated above the lane in a 10 μl reaction for 30 min at 37°C. After incubation, reactions were deproteinized and analyzed by 8% denaturing PAGE. Protein concentrations are as follows: Ku, 25 nM; XRCC4-Ligase IV, 50 nM; pol mu, 25 nM; T4 DNA ligase, 120 nM; XLF, 100 nM. Twenty-five micromolar of each dNTP was added to the reactions where pol mu was present. Hundred micromolar of ATP was also added in each reaction shown in the left panel, and 1 mM of ATP was added to the reactions where T4 DNA ligase was present. ‘M’ indicates 50 bp DNA ladder. Ligation products were quantified, and ligation efficiencies were provided under each lane. DNA sequence analysis confirmed that the junctional sequences conformed to those shown in A. Note that when the top strand is assayed for ligation, the bottom strand has an unligatable 5′OH at the junction, and likewise when the bottom strand is assayed for ligation, the top strand has an unligatable 5′OH at the junction. If we do permit ligation of both strands, then the more readily ligated strand now markedly facilitates the ligation of the anti-parallel strand (unpublished data). The same results for the right panel are obtained when a lower amount of ligase is used (data not shown).
There are hundreds of variations of the two partially complementary DNA ends of Figure 1A, and we have not analyzed all of them. However, a 1 nt change in the complementary portion, such that the ends are –AGC3′ and 3′ CGA– now results in a substantial reduction (60-fold) in the ability of Ku plus XRCC4-ligase IV to join the top strand of these ends (Figure 1B, right panel, lanes 3 versus 8). DNA sequence analysis confirmed the junctional sequence. These results illustrate that 1 nt changes in otherwise identical overhangs can markedly affect the joining.
Importantly, XLF is able to permit joining to a much greater level for a pair of inefficiently joined ends (Figure 1B, right panel, lanes 3 versus 4). In contrast, ends joined efficiently are not stimulated further by XLF (lanes 8 versus 9). [Assays done using shorter incubation times and less ligase rule out the possibility that the lack of stimulation is due to the reaction reaching a plateau (unpublished data).] This raises the possibility that base pairings (2 bp here) that are accommodated well by the ligase complex are sufficient to stabilize those ends in a manner that XLF cannot improve upon, whereas more weakly accommodated end sequences benefit from XLF. The following sections describe studies to further explore both the sequence effects on ligation and the XLF effects on ligation efficiency.
The relative efficiency of DNA end joining conforms to a model invoking steric constraints on the ends within the ligase complex
For ligation with XRCC4-DNA ligase IV and Ku (with or without XLF), we observed a certain pattern of ligation efficiencies for pairs of ends with partial complementarity (2 bp of terminal microhomology with a 1 nt gap on each strand). After annealing at any chance terminal microhomologies, the order of ligation efficiencies for the various pairs of DNA ends fits a pattern most consistent with some steric limitations due to purines at positions 2 and 3 nt from one 3′ end being in conflict with purines on the anti-parallel strand, but at positions that are 1 nt shifted from the initial base pairing. The ligation efficiency is optimal when the purine:purine conflict (R:R) is minimal within a slanted 2 nt region covering each of the two overhangs (Supplementary Figure 1A; see Supplementary Figure 2 for a diagram of the model).
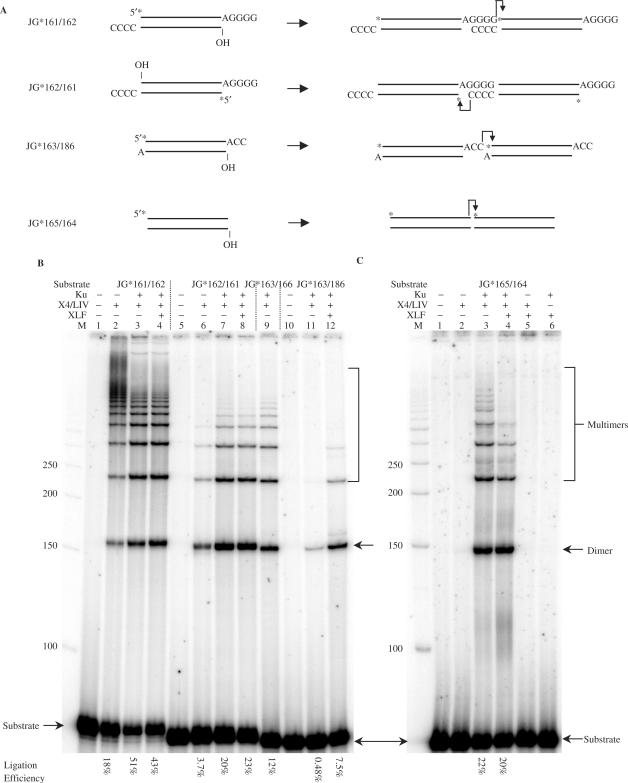
Figure 2.
XLF stimulates incompatible end ligation by XRCC4-Ligase IV but not compatible and blunt end ligations. (A) Substrates with compatible, incompatible 3′ overhangs and blunt ends were designed and tested for their ligation efficiency with or without XLF. A star indicates the position of the radioisotope label. In the ligations, a dimer, e.g. involves two molecules of the same species joined head to tail. (B and C), In each reaction, 20 nM substrate was incubated with the protein(s) indicated above the lane in a 10 μl reaction for 30 min at 37°C. After incubation, reactions were deproteinized and analyzed by 8% denaturing PAGE. Protein concentrations: Ku, 25 nM; X4-LIV, 50 nM; XLF, 100 nM. ‘M’ indicates 50 bp DNA ladder. DNA sequence analysis confirmed that the junctional sequences conformed to those shown in A.
In order to test this model, we created many of the end pair configurations for 3 bp 3′ overhangs that share 2 bp of terminal microhomology. Among the 11 substrate pairs created, nearly the entire set conformed to the model (Supplementary Figure 1B and Figure 1; see model in Supplementary Figure 2). Specifically, the substrates with the least R:R conflict within the 2 nt region are most efficiently ligated (Figure 1B, right panel, lane 8 and Supplementary Figure 1B, lane 13). The substrates with the most R:R conflict within the 2 nt region were least efficiently joined (Figure 1B, left panel, lane 3; right panel, lane 3 and Supplementary Figure 1, lane 3). Hence, the two DNA ends behave according to a model where they initially anneal and then are subject to steric constraints on what bases can occupy the 3′ overhangs, and these limitations affect ligation across any gaps in the top or bottom strands (Supplementary Figure 2).
XLF effectively stimulates incompatible or inefficient end ligation by XRCC-DNA ligase IV, in addition to compatible end joining done at physiologic Mg2+ concentrations
As mentioned above, we find that ligation of pairs of DNA ends by XRCC4-DNA ligase IV are highly stimulated for ligation by XLF. However, compatible DNA ends (4 bp 3′ overhangs) and pairs of blunt DNA ends are not stimulated for ligation by XLF (Figure 2). This suggests that substantial terminal homology achieves the same effect as XLF for incompatible DNA ends, namely, stabilization.
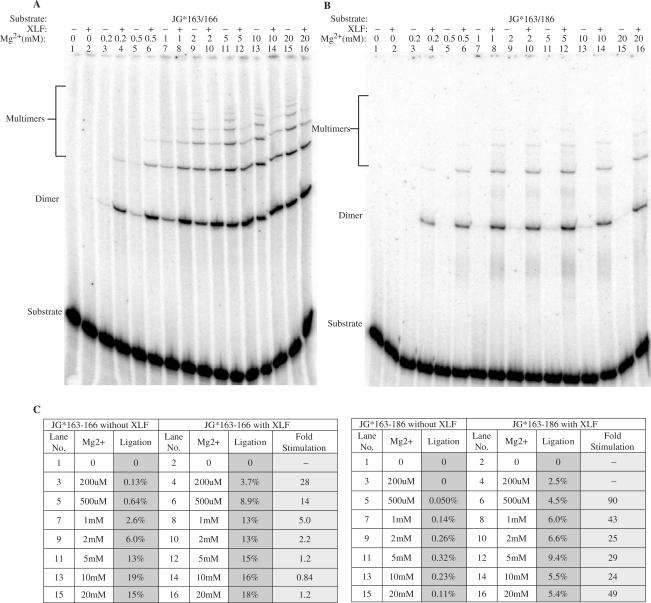
In previous work, we and others had shown that XLF can stimulate ligation of compatible DNA ends (9,10). However, our studies were done at concentrations of Mg2+ (2.5 mM) that are closer to physiologic [∼0.3 mM; (16)]. We reasoned that our failure to detect XLF stimulation for compatible DNA ends here (Figure 2B) might be due to use of a higher Mg2+ concentration (10 mM) in the ligation buffer in the current study. To test this, we studied the ligation as a function of the Mg2+ concentration. At concentrations of Mg2+ of 0.2, 0.5, 1 or 2 mM, XLF stimulated ligation of the compatible DNA ends. XLF failed to stimulate at concentrations ranging from 5 to 20 mM Mg2+ (Figure 3A).
Figure 3.
XLF stimulation of ligation as a function of Mg2+ concentration. (A and B) Compatible substrate JG*163/166 and incompatible substrate JG*163/186 were tested for the XLF stimulation at various Mg2+ concentrations. In each reaction, 20 nM substrate was incubated with 25 nM Ku, 50 nM XRCC4-Ligase IV and with or without 100 nM XLF in a 10 μl reaction with Mg2+ concentrations varying from 0 to 20 mM. For substrate JG*163/166, reactions were carried out at 37°C for 2.5 min; for substrate JG*163/186, reactions were carried out at 37°C for 30 min. After incubation, reactions were deproteinized and analyzed by 8% denaturing PAGE. (C), Ligation products were quantified and ligation efficiencies were provided along with each substrate under different Mg2+ concentrations. The fold of XLF stimulation was calculated.
For incompatible DNA end joining, XLF stimulated ligation by XRCC4-DNA ligase IV at all Mg2+ concentrations between 0.2 and 20 mM (Figure 3B and Supplementary Figure 3). These observations help to unify the disparate findings reported by various laboratories in which some found XLF stimulation of compatible DNA end ligation (9,10) and others did not (12). Like terminal microhomology, XLF and Mg2+ may help to stabilize the pair of DNA ends within the ligase complex. For ends that are already stabilized by terminal microhomology and high Mg2+ (5 mM or above), XLF may have no additional stabilizing effect.
XRCC4-DNA ligase IV can ligate single-stranded DNA
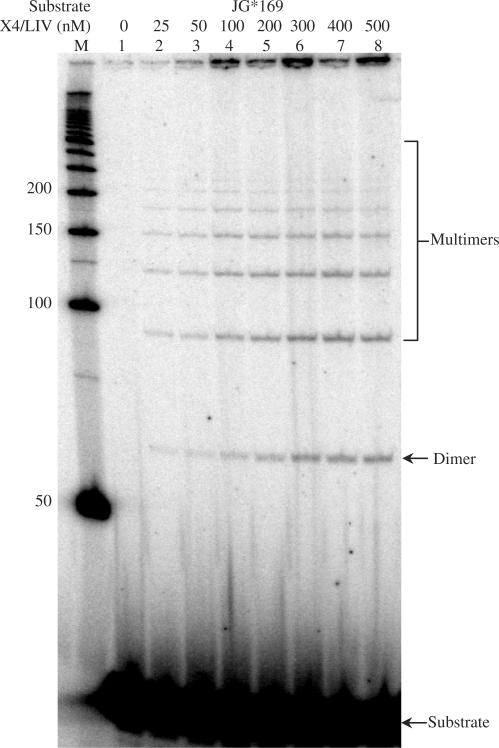
In our analysis of incompatible DNA end joining, we examined progressively longer overhangs (up to 15 nt poly-dT overhangs). For comparison, we tested for the ligation of poly-dT as single-stranded DNA. We were surprised to detect a ladder of ligation products exceeding 6 unit molecules in length for the poly-dT substrate (Figure 4). Notably, poly-dA,-dG,-dC and -rU were not ligatable (data not shown). Moreover, a single terminal dT at the 5′ or 3′ end of the single-strand was not sufficient to permit ligation. Five dT nucleotides at both the 5′ and 3′ end were readily ligated, regardless of the internal sequence (data not shown). Neither Ku nor XLF could stimulate this single-strand ligation, probably because Ku does not bind single-stranded DNA and XLF binds it very inefficiently (9). In related studies, we have also observed ligation of a DNA duplex with a long poly-dT overhang to a single-stranded poly dT molecule (Supplementary Figure 4, lanes 2 and 3) and ligation of two duplex molecules, one with a long 3′ dT overhang and the other with a long 5′ dT overhang (lanes 4–8). Hence, the XRCC4-ligase IV complex has remarkable flexibility to ligate single-stranded DNA and long 5′ or 3′ overhangs. Yet the interactions of the single-stranded substrate near the active site of the ligase complex may require more than one dT nucleotide near the 5′ and the 3′ end.
Figure 4.
Single-stranded DNA can be ligated by XRCC4-Ligase IV. Single-stranded 30 dT substrate (JG*169) was tested for direct ligation by XRCC4-Ligase IV. In each reaction, 50 nM substrate was incubated with different amounts of XRCC4-Ligase IV as indicated above each lane in a 10 μl reaction for 30 min at 37°C. After incubation, reactions were deproteinized and analyzed by 10% denaturing PAGE. ‘M’ indicates 50 bp DNA ladder. Quantification shows that 0.1% of the substrate is converted to the ligated products in lanes 6–8. No ATP is present in the incubations, consistent with the pre-charged status of DNA ligase IV. [The amount of material in the well is small relative to the amount of substrate and does not vary linearly with the amount of ligase (compare lanes 3 versus 7).]
DISCUSSION
Several new facets of the XRCC4-DNA ligase IV and XLF-XRCC4-DNA ligase IV complexes have been described here. First, XRCC4-DNA ligase IV can ligate single-stranded DNA molecules and long 3′ or 5′ overhangs. Second, the XLF stimulation of XRCC4-DNA ligase IV is greater for pairs of ends that have a low efficiency due to sequence preferences of the ligase, due to gaps or due to lack of terminal microhomology, suggesting that XLF contributes to the end-to-end stability in the absence of other stabilizing factors. Mg2+ may also stabilize the approximation of the two DNA ends. Third, ligation of partially compatible ends by XRCC4-DNA ligase IV is influenced by the overhang sequence in ways that suggest steric constraints.
Single-stranded DNA ligation
The single-stranded DNA ligation is noteworthy among DNA ligases. T4 RNA ligase has the ability to ligate single-stranded DNA (17,18). Among DNA ligases, T4 DNA ligase has been reported to ligate single-stranded DNA at a level that can only be detected by PCR (∼1 part in 100 000); however, the possibility of fold-back at the DNA termini (double-strandedness) was not ruled out in that study, making even this trace level of activity uncertain (19). Therefore, the single-stranded DNA ligation by XRCC4-DNA ligase IV is quite remarkable, especially given that this ligase is also capable of acting on a variety of duplex DNA end configurations. This suggests that the mode of binding by this ligase to the substrate DNA is quite flexible, accommodating either two single-strands or two duplex strands.
Sequence effects on the ligation by XRCC4-DNA ligase IV
Regardless of any flexibility in the association of the DNA ends with the ligase complex, it is clear that sequence effects are surprisingly large for both duplex DNA ends and for single-stranded DNA. We do not know the bases for all aspects of the unusual sequence preferences. Some of these can be accounted for by the R:R conflict considerations suggested above. However, the poly-dT preference relative to other sequences for the single-strand DNA end ligation is unusual. Pol μ is known to add nucleotides in a template-independent manner [as well as in a template-dependent manner (20–24)], and pol μ prefers to add pyrimidines, particularly dT (13). It is possible that ligase IV evolved to have preferred non-covalent bonding sites for poly-dT overhangs, and this may also apply for single-stranded poly-dT DNA. In other words, the pol μ dT addition preference may have lead to a selective advantage for a ligase with the dT sequence preference. That is, the sequence preference of the ligase may have evolved to match that of the polymerase or vice versa. Though there is no direct evidence that the properties described in this purified system correspond to in vivo events, there are pathologic and physiologic junctional sequences that have remained without explanation, and which might now be explained by the features reported here. The propensity of XRCC4-ligase IV to ligate dT overhangs (and the propensity of pol mu to add dT and dC) may explain the pyrimidine-rich, particularly dT, additions at many NHEJ junctions at chromosomal translocations sites [the translocations from cells that do not express TdT (25)]. In addition these propensities may explain the pyrimidine-rich, particularly dT-rich, signal joint additions in pre-B cells that lack TdT expression (26). These pyrimidine-rich additions have heretofore been perplexing. The evolution of a polymerase and a ligase with preferences for such sequences may provide an explanation for them.
The unique ability of DNA ligase IV to join incompatible or gapped DNA ends may depend on its ability to contort the ends into a configuration that can be joined, or to accept a contorted substrate. Based on our observations, we propose that the purine/pyrimidine structure of the DNA ends is a key factor. Specifically, the active site of the ligase may not be sized or structured to accept DNA ends that place purines near one another on opposing strands, or to contort these ends sufficiently to ligate them. However, the contortion capability of the ligase and the flexibility of the DNA ends are two aspects of the same enzyme:substrate interaction.
Degree of XLF stimulation is largest for pairs of DNA ends that are ligated with the lowest efficiency
At physiologic concentrations of Mg2+, XLF stimulates the ligation of pairs of DNA ends by XRCC4-DNA ligase IV, regardless of any stability due to terminal base pairing. However, at higher Mg2+ concentrations (>5 mM), terminal base pairing makes any stabilization by XLF negligible [Figure 2B and (12)]. It is important to note that both physiologic breaks [due to V(D)J recombination or class switch recombination] or pathologic breaks typically do not have very much terminal base pairing beyond the chance 1 or 2 nt. Given the cellular concentration of free Mg2+ [∼0.3 mM; (16)], it is clear that the stimulation provided by XLF is a major contribution to incompatible DNA end joining. Given that supra-physiologic concentrations of Mg2+ plus terminal base pairing make XLF dispensible, there is the possibility that XLF, high Mg2+ and terminal base pairing all serve to stabilize the binding of the two DNA ends within the XRCC4-DNA ligase IV complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by grants from NIH to M.R.L. Funding to pay the Open Access publication charges for this article was provided by M.R.L.
REFERENCES
- 1.Lieber MR, Yu K, Raghavan SC. Roles of nonhomologous DNA end joining, V(D)J recombination, and class switch recombination in chromosomal translocations. DNA Repair. 2006;5:1234–1245. doi: 10.1016/j.dnarep.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 2.Dudley DD, Chaudhuri J, Bassing CH, Alt FW. Mechanism and control of V(D)J recombination versus class switch recombination: similarities and differences. Adv. Immunol. 2005;86:43–112. doi: 10.1016/S0065-2776(04)86002-4. [DOI] [PubMed] [Google Scholar]
- 3.Schwarz K, Ma Y, Pannicke U, Lieber MR. Human severe combined immune deficiency. Bioessays. 2003;25:1061–1070. doi: 10.1002/bies.10344. [DOI] [PubMed] [Google Scholar]
- 4.Moshous D, Callebaut I, Chasseval RD, Corneo B, Cavazzana-Calvo M, Diest FL, Tezcan I, Sanal O, Bertrand Y, et al. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell. 2001;105:177–186. doi: 10.1016/s0092-8674(01)00309-9. [DOI] [PubMed] [Google Scholar]
- 5.Callebaut I, Malivert L, Fischer A, Mornon JP, Revy P, Villartay JPD. Cernunnos interacts with the XRCC4/DNA-ligase IV complex and is homologous to the yeast nonhomologous end-joining factor NEJ1. J. Biol. Chem. 2006;281:13857–13860. doi: 10.1074/jbc.C500473200. [DOI] [PubMed] [Google Scholar]
- 6.Buck D, Malivert L, deChasseval R, Barraud A, Fondaneche M.-C, Xanal O, Plebani A, Stephan J.-L, Hufnagel M, et al. Cernunnos, a novel nonhomologous end-joining factor, is mutated in human immunodeficiency with microcephaly. Cell. 2006;124:287–299. doi: 10.1016/j.cell.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 7.Ahnesorg P, Smith P, Jackson SP. XLF interacts with the XRCC4-DNA ligase IV complex to promote nonhomologous end-joining. Cell. 2006;124:301–313. doi: 10.1016/j.cell.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 8.Hartley KO, Gell D, Smith GCM, Zhang H, Divecha N, Lees-Miller SP, Anderson CW, Jackson SP. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell. 1995;82:849–856. doi: 10.1016/0092-8674(95)90482-4. [DOI] [PubMed] [Google Scholar]
- 9.Lu H, Pannicke U, Schwarz K, Lieber MR. Length-dependent binding of human XLF to DNA and stimulation of XRCC4.DNA ligase IV activity. J. Biol. Chem. 2007;282:11155–11162. doi: 10.1074/jbc.M609904200. [DOI] [PubMed] [Google Scholar]
- 10.Hentges P, Ahnesorg P, Pritcher RS, Bruce CK, Kysela B, Green AJ, Bianchi J, Wilson TE, Jackson SP, et al. Evolutionary and functional conservation of the DNA nonhomologous end joining protein, XLF/Cernunnos. J. Biol. Chem. 2006;281:36952–36959. doi: 10.1074/jbc.M608727200. [DOI] [PubMed] [Google Scholar]
- 11.Budman J, Kim SA, Chu G. Processing of DNA for nonhomologous end-joining is controlled by kinase activity and XRCC4/ligase IV. J. Biol. Chem. 2007;282:11950–11959. doi: 10.1074/jbc.M610058200. [DOI] [PubMed] [Google Scholar]
- 12.Tsai CJ, Kim SA, Chu G. Cernunnos/XLF promotes the ligation of mismatched and noncohesive DNA ends. Proc. Natl Acad. Sci. USA. 2007;104:7851–7856. doi: 10.1073/pnas.0702620104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu J, Lu H, Tippin B, Shimazaki N, Goodman MF, Lieber MR. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. EMBO J. 2007;26:1010–1023. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma Y, Pannicke U, Schwarz K, Lieber MR. Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in nonhomologous end joining and V(D)J recombination. Cell. 2002;108:781–794. doi: 10.1016/s0092-8674(02)00671-2. [DOI] [PubMed] [Google Scholar]
- 15.Nick McElhinny SA, Snowden CM, McCarville J, Ramsden DA. Ku recruits the XRCC4-ligase IV complex to DNA ends. Mol Cell Biol. 2000;20:2996–3003. doi: 10.1128/mcb.20.9.2996-3003.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang DL, Yen CF, Nadler JL. Insulin increases intracellular magnesium transport in human platelets. J. Clin. Endocrinol. Metab. 1993;76:549–553. doi: 10.1210/jcem.76.3.8445010. [DOI] [PubMed] [Google Scholar]
- 17.Snopek TJ, Sugino A, Agarwal KL, Cozzarelli NR. Catalysis of DNA joining by bacteriophage T4 RNA ligase. Biochem. Biophys. Res. Commun. 1976;68:417–424. doi: 10.1016/0006-291x(76)91161-x. [DOI] [PubMed] [Google Scholar]
- 18.Brennan CA, Manthey AE, Gumport RI. Using T4 RNA ligase with DNA substrates. Methods Enzymol. 1983;100:38–52. doi: 10.1016/0076-6879(83)00044-0. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn H, Frank-Kamenetskii MD. Template-independent ligation of single-stranded DNA by T4 DNA ligase. FEBS J. 2005;272:5991–6000. doi: 10.1111/j.1742-4658.2005.04954.x. [DOI] [PubMed] [Google Scholar]
- 20.NickMcElhinny SA, Havener JM, Garcia-Diaz M, Juarez R, Bebenek K, Kee BL, Blanco L, Kunkel TA, Ramsden DA. A gradient of template dependence defines distinct biological roles for family x polymerases in nonhomologous end joining. Mol. Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 21.NickMcElhinny SA, Ramsden DA. Polymerase mu is a DNA-directed DNA/RNA polymerase. Mol. Cell. Biol. 2003;23:2309–2315. doi: 10.1128/MCB.23.7.2309-2315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominguez O, Ruiz JF, Lera TLD, Garcia-Diaz M, Gonzalez MA, Kirchhoff T, Martinez C, Bernad A, Blanco L. DNA polymerase mu (Pol mu), homologous to TdT, could act as a DNA mutator in eukaryotic cells. EMBO J. 2000;19:1731–1742. doi: 10.1093/emboj/19.7.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moon AF, Garcia-Diaz M, Bebenek K, Davis BJ, Zhong X, Ramsden DA, Kunkel TA, Pedersen LC. Structural insight into the substrate specificity of DNA polymerase mu. Nat. Struct. Mol. Biol. 2007;14:45–53. doi: 10.1038/nsmb1180. [DOI] [PubMed] [Google Scholar]
- 24.Ramadan K, Shevelev IV, Maga G, Hubscher U. De novo DNA synthesis by human DNA polymerase lambda, DNA polymerase mu, and terminal deoxynucleotidyl transferase. J. Mol. Biol. 2004;339:395–404. doi: 10.1016/j.jmb.2004.03.056. [DOI] [PubMed] [Google Scholar]
- 25.Roth DB, Chang XB, Wilson JH. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol. Cell. Biol. 1989;9:3049–3057. doi: 10.1128/mcb.9.7.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieber MR, Hesse JE, Mizuuchi K, Gellert M. Lymphoid V(D)J recombination: nucleotide insertion at signal joints as well as coding joints. Proc. Natl Acad. Sci. USA. 1988;85:8588–8592. doi: 10.1073/pnas.85.22.8588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.