Abstract
Oxanine having an O-acylisourea structure was explored to see if its reactivity with amino group is useful in DNA microarray fabrication. By the chemical synthesis, a nucleotide unit of oxanine (Oxa-N) was incorporated into the 5′-end of probe DNA with or without the -(CH2)n- spacers (n = 3 and 12) and found to immobilize the probe DNA covalently onto the NH2-functionalized glass slide by one-pot reaction, producing the high efficiency of the target hybridization. The methylene spacer, particularly the longer one, generated higher efficiency of the target recognition although there was little effect on the amount of the immobilized DNA oligomers. The post-spotting treatment was also carried out under the mild conditions (at 25 or 42°C) and the efficiencies of the immobilization and the target recognition were evaluated similarly, and analogous trends were obtained. It has also been determined under the mild conditions that the humidity and time of the post-spotting treatment, pH of the spotting solution and the synergistic effects with UV-irradiation largely contribute to the desired immobilization and resulting target recognition. Immobilization of DNA oligomer by use of Oxa-N on the NH2-functionalized surface without any activation step would be employed as one of the advanced methods for generating DNA-conjugated solid surface.
INTRODUCTION
The DNA microarray is one of the most powerful biotechnological tools used to conduct high-throughput analysis of DNA sequences, genetic variations and gene expressions (1–3). To develop efficient DNA microarray systems, one of the most essential and important subjects is how to immobilize probe DNA oligomers on the solid surface so that the resulting hybridization between the targets and the probe is detected clearly and the array can be stored for long periods without cleavage of the probes (4–6). For this purpose, fabrication systems generating covalent bondings between the probe and the solid surface is preferred since DNA probes, which are tightly immobilized on the surface by the covalent bond except in a few cases such as thiol-gold conjugating (7), provide high stability of the arrays and reproducibility of the data obtained (4–6,8,9).
For the fabrication of DNA microarray systems, both probe DNA oligomers and solid surfaces are usually modified with reactive organic functional groups (6,8,9), and then by utilizing the chemical activation employing appropriate reagents, covalent bonding is formed between the probe and surface (4–6,8,9). Several functional groups such as carboxyl, phosphate, aldehyde and amino groups are commonly introduced, and therefore, the relevant chemical activation steps have also been developed according to the combination of the introduced functional groups (6,8–15).
Amino groups, for instance, have been chiefly employed for both the probe and the surface because of its easy preparation, stable functionality and wide applicability. To attach the probe DNA oligomers covalently on the NH2-functionalized surface, the solid surface modified with amino groups are subsequently subjected to chemical activation by use of homobifunctional linkers such as disuccinimidyl glutarate (DSG), phenylene diisothiocyanate (PDC) (13,16), etc. Although such surface-activation strategies are frequently adopted for covalent bonding formation in DNA microarray fabrication, they have some drawbacks that the activated surface has no long life and the surface should therefore be activated just prior to use, and during the cross-linking reaction, undesirable by-products remain on the surface (6,16). Also, there is a high possibility for the activated groups to react with free amino groups on the same surface or with the amino groups of DNA nucleobases, which inactivate the immobilized probes (6,16).
As another approach, probe DNA oligomer is subjected to chemical activation step and then the activated probe DNA employed for the covalent bonding formation on the surface (6,8,10–12). For instance, when the probe DNA oligomers with carboxyl or phosphate groups at the ends are immobilized on the NH2-functionalized surface, dehydration reagents such as dicyclohexylcarbodiimide (DCC), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), etc. are employed usefully for their activation (17–19). Such probe-activation strategies, however, have several fundamental problems in DNA microarray fabrication. Since most of the DNA microarray fabrications are based on robotic-spotting process, the probe should be prepared as a reproducible or controllable form for obtaining stable and reliable immobilization performance. However, activated carboxyl or phosphate groups do not have a sufficiently long half-life in aqueous conditions, because the attached activator group is easily hydrolyzed (17) so that the activated DNA probes are inactivated. In addition, much excess amount of the dehydration reagent is required compared to the probe DNA oligomer, and therefore, by-products are formed. Also, the sample solution containing only the activated probe DNA oligomers, which would be ideal, cannot be obtained easily by general purification methods (10–12).
As mentioned above, in the processes of DNA microarray fabrication, activation strategies, which have been used widely for covalent bonding formation between the probe and surface, still possess technical problems. Consequently, simple and direct covalent bonding fabrication method, which does not require additional activation steps and leaves no by-products, is preferred.
In the present study, we employed oxanine (Oxa) as a new linker for mediating direct covalent bonding reaction for the immobilization of probe DNA oligomer on the NH2-functionalized surface in one-pot mode, as shown in Figure 1. Oxa was identified in 1996 as a unique lesion generated as one of the main deamination products of guanine (Gua) by NO- or HNO2-induced nitrosative oxidation (20–28) and the formation mechanism has been identified in detail (29–31). Since Oxa has an O-acylisourea structure, an activated-carboxyl group, as illustrated in Figure 2A, Oxa is expected to react with amino or thiol group of biomolecules (32,33). However, the practical use of such a unique function has not yet been focused on until date. Recently, we have developed a solid-phase chemical synthesis procedure for incorporating Oxa into DNA oligomers (34), so that it is possible to employ Oxa-containing DNA oligomers in the field of biotechnological applications such as DNA microarray fabrication. Probe DNA molecules were prepared by incorporation of phosphoramidite monomer of deoxyoxanosine (dOxo, deoxynucleoside of Oxa) into the 5′-end of DNA oligomers with or without fluorescence labeled at 3′-end by the chemical synthesis. They were then spotted on the NH2-functionalized glass slide, and subsequently subjected to the post-spotting treatments such as baking process (conventional method) or mild treatment (substitution method newly adopted in this study). Then, the performance of Oxa as a linker was evaluated by investigating the immobilization efficiency and hybridization efficiency. This study shows that use of Oxa as a linker is efficient for covalent attachment of probe DNA in one-pot mode even under the mild conditions and therefore practically appropriate for DNA microarray fabrication.
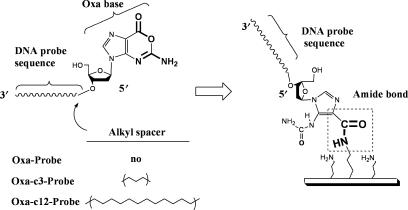
Figure 1.
Schematic diagrams for the use of Oxa as a linker to directly immobilize probe DNA oligomers on the NH2-functionalized surface. Through the activation-free reactivity of Oxa-nucleotide unit (Oxa-N) with amino groups, the probe DNA oligomers with Oxa-N (Oxa-Probe, Oxa-c3-Probe and Oxa-c12-Probe) are covalently attached on the surface in one-pot reaction.
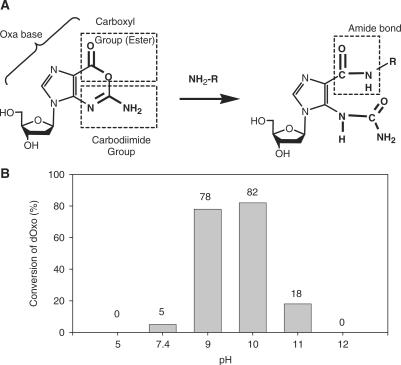
Figure 2.
(A) Covalent bond-forming reaction of Oxa with amine-containing molecules. Oxa has an O-acylisourea structure, in which carboxyl group is activated by carbodiimide in its base-ring (left), so that Oxa reacts with amino group to make amide bond without any additional activation (right). (B) The pH-dependent change of the reactivity of Oxa with hexylamine. 2′-deoxyoxanosine (dOxao) (2.5 mM) was incubated with hexylamine (25 mM) for 4 h in several universal buffer systems with different pH (pH 5.0, 7.4, 9.0, 10.0, 11.0 and 12.0) composed of borate, ascorbate and phosphate. Uracil was added to the reaction mixture as an internal standard and the components were separated by RP–HPLC and the conversions were estimated on the basis of the peak areas of the product and uracil. RP–HPLC condition: elution; a gradient of 0% (0 min)–20% (10 min)–60% (20 min) of CH3CN in 100 mM TEAA buffer (pH 7.4) at a flow rate of 1 ml/min, column; Ultorn VX-ODS column (150 × 4.6 mm, particle size being 5 μm), temperature; ambient.
MATERIALS AND METHODS
Materials
Reagents for DNA oligomer synthesis including CPG column and appropriately protected normal nucleosides were obtained from Glen Research (Sterling, VA, USA) and solvents for the synthesis from Applied Biosystems (Foster, CA, USA). Other organic solvents were purchased from Nacalai Tesques (Osaka, Japan) and all other chemicals from Wako Pure Chemicals (Osaka, Japan). The glass slides and several NH2-functionalized glass slides [25.4 × 76.2 mm (±0.05 mm)] were obtained from Matsunami (Osaka, Japan), Corning (Corning, NY) and Nunc (Wiesbaden, Germany).
Instrument systems
For purification of synthesized probes and target DNA oligomers, an RP-HPLC system consisting of a Tosoh PX-8020 (controller), DP-8020 (pump), CO-8020 (temperature controller) and PD-8020 (diode detector) and Ultron VX-ODS column [150 × 4.6 mm (for analysis) or 150 × 6.0 mm (for purification), 5 μm; Shinwa Co. (Kyoto, Japan)] were used. Chemical synthesis of DNA probes and target DNA oligomers was carried out on an Applied Biosystems 3400 DNA synthesizer [Applied Biosystems (Foster, CA)]. UV spectra of DNA oligomers were measured on a Shimadzu UV-260 UV-Vis spectrophotometer equipped with an SPR-5 temperature controller. The DNA oligomers were spotted on the glass slides with an inkjet spotter [custom built by NGK Insulators (Nagoya, Japan)]. The spotted glass slides were scanned with DNA MicroArray Scanner Model G2505A of Agilent Technologies (Palo Alto, CA, USA) or Array WoRx of Applied Precision (Issaquah, WA, USA).
Probe and target DNA oligomers preparation
Probe DNA oligomers with Oxa-nucleotide unit (Oxa-N) at the 5′-end were prepared by the solid-phase chemical synthesis procedure, as reported previously (34). The probe sequence, 5′-d(TGTTGTCGAAAATGTCAACG)-3′ (XDHp), was designed from the antisense part of the xylitol dehydrogenase gene (XDH) from Pichia stipitis. After the synthesis of XDHp, Oxa-N was incorporated at the 5′-end in the final coupling step. Three kinds of probe DNA oligomers with Oxa-N were synthesized; one is the wild type (Oxa-Probe) containing Oxa-N at the 5′-end and the others are those in which carbon 3 (–(CH2)3–) and carbon 12 (–(CH2)12–) spacers are inserted between XDHp and Oxa-N (namely, Oxa-c3- and Oxa-c12-Probes, respectively) as shown in Figure 1. For the analysis of the immobilization efficiency, probe DNA oligomers labeled with fluorescein [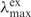 = 495 nm,
= 495 nm, 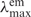 = 519 nm, εmax = 75 000 (M−1 cm−1)] at the 3′-end were prepared by using fluorescein-bound CPG column (Glen Research).
= 519 nm, εmax = 75 000 (M−1 cm−1)] at the 3′-end were prepared by using fluorescein-bound CPG column (Glen Research).
For the analysis of the hybridization efficiency of the probe DNA oligomers immobilized on the glass slide, two types of XDH target sequences, 5′-d(CGTTGACATTTTCGACAACA)-3′ (short XDH target; XDHt-S) and 5′d(CTGCTGCTGTCGCCAAGACCTTCGGTGCTAAGGGTGTCATCGTCGTTGACATTTTCGACAACA AGTTGAAGATGGCCAAGGACATTGGTGCTGCTACTCACACCTT)-3′ (long XDH target; XDHt-L, underlined bases are the same sequence as XDHt-S) were prepared and their 5′-ends were labeled with Cy3 [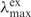 = 547 nm,
= 547 nm, 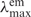 = 563 nm, εmax = 136 000 (M−1 cm−1)]. The probe and target DNA oligomers used in this study are listed in Table 1.
= 563 nm, εmax = 136 000 (M−1 cm−1)]. The probe and target DNA oligomers used in this study are listed in Table 1.
Table 1.
Probe and target DNA oligomers used in this study
| Probe DNA oligomers | |
|---|---|
| Oxa-Probe | 5′-d(OTGTTGTCGAAAATGTCAACG)-3′ |
| Oxa-c3-Probe | 5′-d(Oc3TGTTGTCGAAAATGTCAACG)-3′ |
| Oxa-c12-Probe | 5′-d(Oc12TGTTGTCGAAAATGTCAACG)-3′ |
| Oxa-Probe-F | 5′-d(OTGTTGTCGAAAATGTCAACGF)-3′ |
| Oxa-c3-Probe-F | 5′-d(Oc3TGTTGTCGAAAATGTCAACGF)-3′ |
| Oxa-c12-Probe-F | 5′-d(Oc12TGTTGTCGAAAATGTCAACG F)-3′ |
| Target DNA oligomers | |
| Cy3-XDHt-S | 5′-d(Cy3 CGTTGACATTTTCGACAACA)-3′ |
| Cy3-XDHt-L | 5′-d(Cy3 CTGCTGCTGTCGCCAAGACCT |
| TCGGTGCTAAGGGTGTCATCGT | |
| CGTTGACATTTTCGACAACAAG | |
| TTGAAGATGGCCAAGGACATT | |
| GGTGCTGCTACTCACACCTT)-3′ | |
| Non-complementary | 5′-d(Cy3 GCAACTGTAAAAGCTGTTGT)-3′ |
O: Oxanine, F: Fluorescein, c3: Carbon 3 spacer (–(CH2)3–), c12: Carbon 12 spacer (–(CH2)12–), Cy3: Cy3 fluorescence label; In Cy3-XDHt-L, underlined bases are the same sequence as Cy3-XDHt-S.
Preparation of the amine-functionalized surface
The NH2-functionalized surface was prepared on glass slides as follows. After 10 min ultrasonication in acetone and 10 min vacuum drying, to prepare a hydroxyl group-enriched surface, treatment in Piranha solution (70% H2SO4, 30% H2O2) was carried out at 55°C for 30 min. After cleaning by sonication in methanol, methanol/toluene [1:1 (v/v)] and toluene (for 10 min each), silanization was performed by immersing the glass slides in 3-aminopropyl-triethoxysilane (APTS) solution (2% in toluene). During the silanization, containers were placed in an orbital shaker with gentle shaking (70 r.p.m.) at room temperature. The glass slides were then cleaned in an ultrasound bath in toluene, toluene/methanol [1:1(v/v)] and methanol (for 10 min each). The glass slides were baked at 110°C for 1 h. The prepared NH2-functionalized glass slides were stored in vacuo at room temperature immediately prior to use.
Spotting, post-spotting treatment and analyses of immobilization and hybridization efficiencies: procedures for evaluation of the modifier as a linker
To see the effect of the modifiers attached to the 5′-end on how efficiently probe DNA oligomers were formed on the NH2-functionalized surface (the performance of the modifier as a linker), two different evaluations were employed. One is the immobilization efficiency evaluated by immobilizing probes labeled with fluorescein at the 3′-end (Oxa-Probe-F, Oxa-c3-Probe-F and Oxa-c12-Probe-F) and by measuring the fluorescence intensity of each spot. The other is the hybridization efficiency of the target DNA oligomers, labeled with Cy3 at the 5′-end, with the probes without the fluorescein-label (Oxa-Probe, Oxa-c3-Probe and Oxa-c12-Probe). The resulting Cy3-fluorescence intensity of each spot was measured, assuming that the Cy3-fluorescence intensity is correlated with the quantity of the active probe DNA oligomers immobilized on the glass slides.
First, on the NH2-functionalized glass slides, probe DNA oligomers were spotted as follows; aqueous solutions of probe DNA oligomers (100 pmol/µl) were mixed in the ratio of 1:1 (v/v) with an inkjet-spotting solution consisting of glycerin, glycerol and disaccharides (35), the pH of which was controlled by using sodium-phosphate buffers (10 mM). About 100 pl of these spotting probe solutions (50 pmol/µl) was spotted at 3 mm spacing on the NH2-functionalized glass slides with an inkjet spotter. As shown in Figure 3A, the spotted DNA probes were round in shape (∼80 µm in diameter) and clearly separated from each other. No satellite spot was observed.
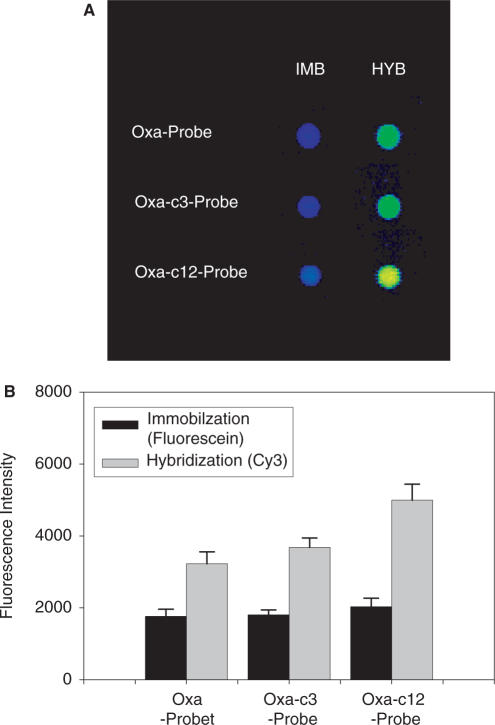
Figure 3.
(A) Fluorescence intensity data obtained for the spots. After the baking process (incubation for 1 h at 80°C) of the spotted glass slide was employed as conventional post-spotting treatment, the immobilization efficiency and hybridization efficiency of each spot were analyzed for the evaluation of the performance of Oxa as a linker, as explained in Materials and Methods section. The spots in the line of IMB show the fluorescence from fluorescein attached to the probe DNA oligomers, which indicates the efficiency of the immobilization, and those in the line of HYB the fluorescence from Cy3 attached to the target DNA oligomers (Cy3-XDHt-L, a long XDH target of 106-mer) hybridized with the probe, which is referred to the hybridization efficiency and also indicates the efficiency of the probe immobilization. It should be noted that the spots are named according to the name of the probe DNA oligomers as listed in Table 1 and that in the case of IMB and HYB, probes with and without fluorescein label were used, respectively. (B) The bar graphs representation of the average fluorescence intensities (arbitrary unit) of the spots in (A) [εmax (fluorescein) = 75 000 (M−1 cm−1) and εmax (Cy3) = 136 000 (M−1 cm−1)]. The error bar represents the SD of fluorescence intensity. The meaning of immobilization and hybridization in the box is the same as explained in (A).
After the inkjet spotting of the probes, the post-spotting treatment, which is required for the efficient attachment of probe DNA oligomer, was employed on the spotted glass slides. Baking process carried out at 80°C for 1h was employed as the conventional method. As substitution method for baking process, the mild conditions (e.g. at 25 and 42°C) were also employed as described in the the next section (Exploration of new conditions of post-spotting treatments).
After the post-spotting treatments, in order to remove the unreacted DNA probes from the slides, the incubated slides were rinsed with 2× SSC (saline sodium citrate) containing 0.1% SDS (sodium dodecyl sulfate, 15 min), soaked in 2× SSC containing 0.2% SDS (5 min), washed with water, soaked in ethanol (1 min) and air-dried at room temperature (30 min). To inactivate unspotted areas, the rinsed slides were incubated in a blocking solution containing 1% bovine serum albumin (BSA), 4× SSC and 0.5% SDS at 42°C for 45 min. The prepared slides with the immobilized probe DNA oligomers were stored in a desiccator, and used to see the effects of the modifiers on the efficiency of the immobilization (see IMB of Figure 3A).
Then, the hybridization efficiency was analyzed in another method for evaluating the performance of the modifier as a linker. The prepared slides were treated with hybridization buffer (5× SSC containing 0.5% SDS) containing the target DNA oligomers (10 pM). After covering with a cover slip, the glass slides were kept at 42°C for 16 h. Then, to remove non-hybridized target DNA oligomers, the incubated slides were immersed in 2× SSC containing 0.1% SDS (5 min), and finally agitated in a gently shaking bath in 1× SSC (5 min) and 0.1× SSC (5 min). After spin-drying, the slides were stored in a desiccator, and used to see the hybridization efficiency of the immobilized probe DNA oligomers (see HYB of Figure 3A).
The glass slides were scanned with a microarray scanner such as DNA MicroArray Scanner Model G2505A or Array WoRx, and the fluorescent image intensities and the location of each analyte spot on the slides were measured using the mapping software, GenePixPro Ver 5.0 or 5.2 of Molecular Devices (Sunnyvale, CA, USA). The fluorescence intensity data obtained for 90 spots for each type of probe DNA oligomer were used for the calculation of the statistical data, which are shown in Figure 3B.
Exploration of new conditions of post-spotting treatments
As described in the previous section, baking process of the spotted slide carried out at 80°C for 1 h, was used as conventional post-spotting treatment. In this study, some other substitution methods, in particular, incubated under the mild conditions (e.g. 25 and 42°C), were explored as new post-spotting treatments and their effects on the performance of the modifier as a linker were compared with those by conventional baking process.
The same procedures of spotting, washing and hybridization steps were carried out on the same probe and target DNA oligomers as adopted in the previous section, except for the employment of the new conditions or combinations of post-spotting treatments. As shown in Figure 4, the temperature, humidity and time of the post-spotting treatments, pH of the spotting solution and the combination of conventional or new post-spotting treatments were investigated. For the evaluation of the performance of the modifier as a linker, the efficiencies of immobilization and hybridization were analyzed, and their analyses procedures were conducted in the same way as described in the previous section, unless otherwise specified.
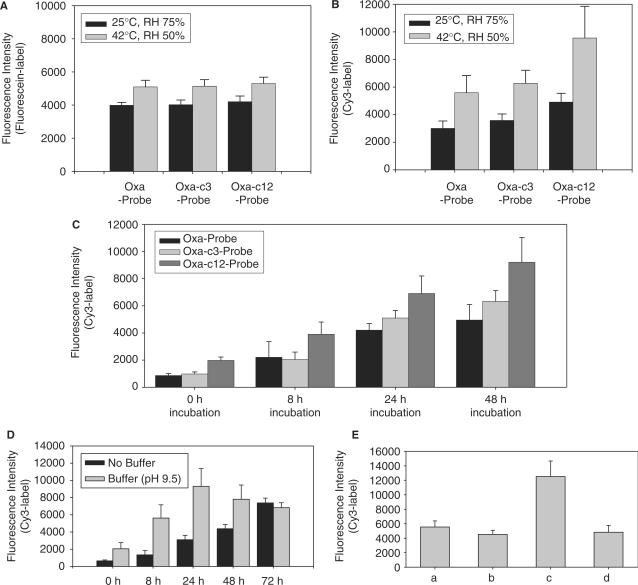
Figure 4.
Effects of the experimental parameters, such as temperature, relative humidity, reaction time, pH and additional UV irradiation in the employment of post-spotting treatment, on the performance of the modifier as a linker on the NH2-functionalized glass slide. (A) Immobilization efficiency obtained for the probes with Oxa-N at the 5′-end by measuring fluorescein-label at the 3′-end (Oxa-Probe-F, Oxa-c3-Probe-F and Oxa-c12-Probe-F). The mild conditions of post-spotting treatment were employed on the spotted glass slide and then, the immobilization efficiency of each spot was analyzed for the evaluation of Oxa as a linker. Gray bars were obtained by one mild condition of post-spotting treatment (at 25°C and RH 75% for 48 h), and the black bars by another mild condition of post-spotting treatment (at 42°C and RH 50% for 24 h). [εmax (fluorescein) = 75 000 (M−1 cm−1)]. (B) Hybridization efficiency obtained for the probes with Oxa-N at the 5′-end (Oxa-Probe, Oxa-c3-Probe and Oxa-c12-Probe). The hybridization efficiency of each spot was analyzed for the evaluation of the performance of the modifier as a linker. The representation of gray and black bars in the figure and the mild conditions of post-spotting treatment are the same as explained in (A). [εmax (Cy3) = 136 000 (M−1 cm−1)]. (C) Time-dependent performance of Oxa as a linker under the mild conditions of post-spotting treatment (at 42°C and RH 50%). The resultant hybridization efficiency was analyzed for the evaluation. Probes with Oxa-N at the 5′-end (Oxa-Probe, Oxa-c3-Probe and Oxa-c12-Probe) were used and the target was Cy3-XDHt-L. The vertical axis shows the Cy3-fluorescence intensities of the spots including the duplexes between the probes and the target. (D) Effect of the spotting solution pH on the covalent bonding formation between Oxa and amino group on the surface. The resultant hybridization efficiency and its time-dependent change were analyzed for the evaluation of the performance of Oxa as a linker. The probe was Oxa-Probe and the target Cy3-XDHt-L. The black bars were the results obtained without the pH control, and the gray bars were the results obtained by adding the same amount of phosphate buffer solution to the spotting solution (final pH 9.5). (E) Effect of the combinations of the post-spotting treatments on the performance of Oxa as a linker. First, the mild conditions of the post-spotting treatment (72 h incubation at 42°C and RH 50%) were employed on the glass slide, in which Oxa-Probe in original spotting solution was spotted and then, the glass slide were subsequently subjected to other conventional post-spotting treatments such as (a) none, (b) baking (at 80°C for 2 h) and (c) UV irradiation at 600 mJ. Bar (d) was obtained without the employment of the mild treatment but only by UV irradiation at 600 mJ. All the spots were then hybridized with the target Cy3-XDHt-L and measured for Cy3-fluorescence intensities. The resultant hybridization efficiency was used for the evaluation of the performance of Oxa as a linker.
RESULTS AND DISCUSSION
Performance of Oxa as a linker
Covalent bonding formation with amines
As a preliminary trial, Oxa was explored for its property as a linker, for instance, its reactivity with primary amines and the stability of the resulting product (Supplementary Data, Table S1 and Figures S1 and S2). Since Oxa contains an O-acylisourea structure in the 6-membered ring in which the carboxyl group moiety is bound to the carbodiimide group, as shown in Figure 2A, Oxa is expected to react with amino groups readily to result in the amide-bond formation without any further activation step of carboxyl group. When 2.5 mM dOxo was incubated in the presence of 25 mM hexylamine, a peak due to the product was observed in RP–HPLC chromatogram (Supplementary Data, Figure S1). Spectroscopic analysis including NMR (Supplementary Data, Figure S2) and mass spectroscopy (data not shown) indicated that O-acylisourea structure in Oxa of dOxo reacted with the amino group of hexylamine, resulting in the ring-opened product (dOxo-hexylamine, 1-(2-deoxy-β-d-ribofuranosyl)-5-ureido-1H-imidazole-4-carboxylic acid hexylamide), as shown in Figure 2A. This amide-bond formation was observed in the confined pH range of ∼6.5 − 11 as shown in Figure 2B. The rate constant for this reaction was measured at pH 9.5 and 25°C as 2.30 × 10−4 mM−1 min−1 (second-order reaction). Further, the stability of the N-glycosidic bond was analyzed and for instance, the sufficiently long half-life at pH 9.5 and 42°C was obtained at 841 h (Supplementary Data, Table S1).
It should be noted that the O-acylisourea structure of Oxa can be maintained as ring-closed structure up to relatively high pH conditions [pKa = 9.4 (36)]. Considering both that the optimum condition for the active amino groups is above pH 9 and that Oxa is stable in alkali conditions, the use of Oxa as a linker is suitable for the covalent attachment of the probe DNA oligomers on the NH2-functionalized surface. Based on these data and the functional merits, Oxa was employed as a new linker, in the present study.
Direct immobilization of Oxa-containing DNA oligomers onto NH2-functionalized glass slides
As shown in the previous section and Figure 2, Oxa reacted with amino groups without any activation step and the amide bond was formed in relatively high pH conditions (pH 9–10). It was expected, therefore, that Oxa could provide efficient covalent bonding formation with amino groups, that is, it could be utilized as a new linker for simple and direct covalent attachment of the probe DNA oligomers on the NH2-functionalized surface. To see if this is the case, fluorescein-labeled probe DNA oligomers (Oxa-Probe-F, Oxa-c3-Probe-F and Oxa-c12-Probe-F) were spotted on the NH2-functionalized glass slides, and the glass slides were subjected to the baking process (1 h incubation at 80°C), conventional post-spotting treatment. The slides were washed to remove the unreacted probe DNA oligomers and then the fluorescence intensity of each spot was measured for estimating the amount of DNA probes immobilized. The typical fluorescein fluorescence of the spot is shown in the line of IMB in Figure 3A and the intensities of the spots were measured by the scanner. It was found, as represented in Figure 3B, that the fluorescence intensities of the spots of probe DNA oligomers with Oxa-N were 1761 (Oxa-Probe), 1805 (Oxa-c3-Probe) and 2032 (Oxa-c12-Probe) (black bars). Although the longer spacer such as –(CH2)12– was found to increase the immobilization efficiency, it was not remarkable. The spotted probe DNA oligomers with Oxa-N were further washed by the use of high-salt 5× SSC for over 16 h. In the cases of the probe DNA oligomers with Oxa-N, the changes in fluorescence intensities were negligible while in the case of those without Oxa-N, significant decrease in the intensities observed (data not shown), indicating that Oxa-N covalently immobilized the probes.
The efficiencies for the hybridization of the target DNA oligomers with the probes immobilized on the glass slide were also investigated. Oxa-Probe, Oxa-c3-Probe and Oxa-c12-Probe, which have no fluorescein-label, were spotted analogously on the NH2-functionalized glass slide and the spotted slides were also subjected to baking process (1 h incubation at 80°C). After such a conventional post-spotting treatment and washing steps were performed, the target DNA oligomer, a 106-mer (Cy3-XDHt-L) that has a 20-base sequence in the middle complementary to the probe and whose 5'-end was labeled with Cy3 was hybridized. Cy3-fluorescence intensities of each spot were measured as shown in the line of HYB in Figure 3A. As depicted by the bar graphs in Figure 3B, the three types of probe DNA oligomers with Oxa-N showed high Cy3-fluorescence intensities. The intensities were 3227 (Oxa-Probe), 3678 (Oxa-c3-Probe) and 4992 (Oxa-c12-Probe) (gray bars). It was suggested that the longer alkyl spacer would produce the larger efficiency for the target recognition. Analogous results were obtained when Cy3-XDHt-S was used as a target, and when non-complementary was utilized as a target, non-specific adsorption of the target was not detected. Several kinds of commercially available aminosilane-modified glass slides were also tested, and analogous results in immobilization and hybridization efficiencies were obtained (data not shown).
Exploration of new post-spotting treatments
Effects of temperature and humidity on the performance of Oxa as a linker
If the probe DNA oligomer and the functionalized surface are prepared in the same ways, the post-spotting treatment becomes the determinant step, which influences the performance of the modifier as linker on the solid surface. As described in the previous section, baking process of the spotted slide carried out at 80°C for 1 h was employed as conventional post-spotting treatment. However, such a harsh condition does not seem to be proper for the natural property of probe DNA oligomers, so that mild conditions would be ideal for post-spotting treatment, if covalent attachment is possible. In this study, some other substitution methods, which are more compatible to probe DNA oligomer, were investigated as new post-spotting treatments because Oxa is expected to show sufficient reactivity with amino group in the mild conditions even without any activation step.
Two kinds of mild treatments, in which the spotted glass slide was incubated at 25 and 42°C, were employed as the post-spotting treatment and the performance of the modifier as a linker was evaluated by investigating its immobilization and hybridization efficiencies. The three probe DNA oligomers with Oxa-N at the 5′-end and fluorescein-label at the 3′-end were spotted on the NH2-functionalized glass slides and then, each mild treatment was employed on the spotted slides as the post-spotting treatment by controlling relative humidity (RH). First, in the case of mild treatment at 25°C, it was found that at this temperature condition, the fluorescence intensities increased with increase in RH and reached a plateau when RH was 75% (data not shown). The data obtained at 25°C and RH 75% for 48 h are depicted by the black bars in Figure 4A. The average fluorescein intensities of the immobilized probe DNA oligomers with Oxa-N were 3991 (Oxa-Probe-F), 4025 (Oxa-c3-Probe-F) and 4203 (Oxa-c12-Probe-F). In the experiments at 42°C, the immobilization efficiencies of the probes were also investigated by controlling RH. In the experiments at 42°C for 24 h, the highest immobilization data was obtained at RH 50%, as represented by the gray bars in Figure 4A. The average fluorescein intensities obtained were 5096 (Oxa-Probe-F), 5139 (Oxa-c3-Probe-F) and 5298 (Oxa-c12-Probe-F).
Then, the hybridization efficiency was also analyzed for the probe DNA oligomers obtained by the employment of the same mild treatments (post-spotting treatment) as performed in the experiment for the immobilization efficiency. The probes without fluorescein-label were used and Cy3-XDHt-L was the target. At 25°C and RH 75% for 48 h, the Cy3-fluorescence intensities of the targets were measured to be 3005, 3582 and 4915 for Oxa-Probe, Oxa-c3-Probe and Oxa-c12-Probe, respectively, as shown by the black bars in Figure 4B. The average values obtained at 42°C and RH 50% for 24 h were 5590, 6268 and 9551 for Oxa-Probe, Oxa-c3-Probe and Oxa-c12-Probe, respectively, as represented by the gray bars in Figure 4B. The results obtained at 42°C and RH 50% for 24 h were found to be higher than the results obtained by the baking process as post-spotting treatment (Figure 3A and B). These results indicate that the performance of Oxa to make a covalent bonding formation with amino groups on the surface is effective even in such mild conditions, resulting in the high-hybridization efficiency. Analogous results were obtained when Cy3-XDHt-S was used as a target, and when non-complementary was utilized as a target, non-specific adsorption of the target was not detected.
Effects of treatment time and pH on the performance of Oxa as a linker
Another parameter responsible for the performance of Oxa as a linker may be the time of post-spotting treatments. Several spotted glass slides were identically prepared by spotting the probes (Oxa-Probe, Oxa-c3-Probe and Oxa-c12-Probe) on the NH2-functionalized surface. Then, under the employment of post-spotting treatments at 42°C and RH 50% (mild treatment), the treated slides were taken according the treatment time. For each slide sample, the resultant hybridization efficiency measured by Cy3-fluorescence intensities (the target: Cy3-XDHt-L) was investigated for the evaluation of the time-dependent performance of Oxa as a linker. As shown in the bar graphs in Figure 4C, the hybridization efficiency increased depending on the time of the post-spotting treatment. Analogous results were obtained when Cy3-XDHt-S was used as a target, and when non-complementary was utilized as a target, non-specific adsorption of the target was not obtained.
Next, the influences of the pH values on the covalent bonding formation between Oxa and amino group on the surface were investigated by controlling the pH of the spotting solutions. The resultant hybridization efficiency was used for the evaluation of the performance of Oxa as a linker. The probe DNA oligomers employed were Oxa-Probe and the target Cy3-XDHt-L. The pH value of the original spotting solutions was ∼7.5. The pH of the spotting solution was adjusted by adding the same portion of 10 mM sodium phosphate buffer with different pHs to the original spotting solutions. After the spotting of the probes, the glass slides were subjected to the post-spotting treatment at 42°C and RH 50%. As the pH was raised from ∼6 to 10, the efficiency was increased at the higher pH, that is, the reaction of Oxa and amino group on the surface was increased as the pH of the spotting solution increased (data not shown). However, as shown in Figure 4D, as the incubation time increased, the efficiency decreased after 24 h at high pH, such as pH 9.5, while in the original spotting solution, which does not contain additional salts of sodium phosphate, the efficiency increased with time. These results indicated that although high pH and salts enhance the formation of amide bonding between Oxa and amino group on the surface, they may also cause undesirable influence when the spots are formed and the concentrations of the components raised accordingly as the time of post-spotting treatments elapsed.
Effects of the combination of post-spotting treatments on the performance of Oxa as a linker
As mentioned above, when Oxa is employed as a linker of probe DNA oligomers, the mild conditions (25 or 42°C) can be employed efficiently as new post-spotting treatment. Such a mild condition of new post-spotting treatment (mild treatment), which is currently adopted here, could be more adequate for the quality or the nature of probe DNA oligomer than the harsh condition of conventional method (baking process). In the present section, the new mild treatment or its combination with the conventional post-spotting treatments such as baking process or UV-irradiation were employed on the spotted glass slides. Then, the synergistic effects were investigated by analyzing the hybridization efficiency, which is closely correlated with the linkage performance of the spotted probe DNA. Oxa-Probe and Cy3-XDHt-L were employed as a probe and a target, respectively. The original spotting solution was used for the spotting, and the new mild treatment of post-spotting treatment was carried out at 42°C and RH 50% for 72 h. Compared to the result obtained by the new mild treatment only (Figure 4E-a), as shown in Figure 4E-b, the combination of the new mild treatment and the following baking process (at 80°C for 2 h) did not create any synergistic effects but lowered the efficiency. On the other hand, as presented in Figure 4E-c, the combination of new mild treatment with UV irradiation with a total energy of 600 mJ, which mediates reactive radicals to induce the cross-linkage among the probes, was observed to show synergistic effect, more than double efficiency than the result obtained by the new mild treatment only. This result is probably due to the increased amount of immobilized probe DNA oligomers on the surface, in which first, the covalently attached probes are formed though the reactivity of Oxa and then, the non-covalently attached probes are cross-linked non-specifically by the UV irradiation. UV irradiation alone (Figure 4E-d) did not show such desirable results. Consequently, such sequential employments of post-spotting treatments, in particular, a combination of the new mild treatment and UV irradiation, will be usefully used for the fabrication of highly efficient DNA microarray systems.
CONCLUSION
To fabricate DNA microarray systems that recognize targets with high reproducibility and without error and are stable during storage, the development of efficient methods by which probe DNA oligomers are immobilized on the solid surface through the covalent bonding formation between the probe and the solid surface is required. Also, it is recommended that such a covalent bonding reaction is performed under mild conditions so that undesirable by-products are not produced on the surface.
In the present study, Oxa, which is one of the unique lesions generated from Gua by NO- or HNO2- induced nitrosative oxidation and has an O-acylisourea structure reactive to amines, was investigated as a functional linker to see if it is useful in DNA microarray fabrication carried out under mild conditions. By solid-state chemical synthesis, a nucleotide unit of Oxa (Oxa-N) was incorporated into the 5′-end of probe DNA oligomers with or without the spacers such as –(CH2)3– and –(CH2)12– between Oxa-N and the probe sequence, and utilized to immobilize the probes onto the NH2-functionalized glass slide surface.
For evaluating the performance of Oxa as a linker, two analyses such as immobilization efficiency and hybridization efficiency were performed. The former was estimated by measuring the fluorescence intensity of the spots for the probe DNA oligomers whose 3′-end was labeled with fluorescein and the latter by immobilizing the probe DNA oligomers without fluorescein-labeling and measuring the fluorescence intensity after the hybridization with the Cy3-labeled target DNA oligomers.
Mild conditions were also explored as a new post-spotting treatment for the efficient attachment of probe DNA oligomer on the surface. The temperature and humidity of the post-spotting treatment were controlled and their effects on the performance of Oxa as a linker were investigated. Under the mild conditions of post-spotting treatment such as a temperature of 25°C and a RH of 75%, and 42°C and RH 50%, it was found that probe DNA oligomers with Oxa-N produce high efficiencies of immobilization and hybridization, and that longer alkyl chain spacers lead to better recognition of target DNA oligomers. In addition, the effects of some other parameters such as the time of post-spotting treatment and pH of spotting solutions were investigated to enhance the covalent bonding formation of Oxa with amino group on the surface. It was found that under the mild conditions of post-spotting treatment, the performance of Oxa as a linker increased linearly with the time of post-spotting treatment, but that pH of spotting solution did not give rise to the desirable improvement of the results although at higher pH of the spotting solution, the covalent bonding formation of Oxa with amino group on the surface was accelerated. The newly adopted mild treatment and other conventional post-spotting treatments were sequentially employed on the spots of probe DNA oligomer with Oxa-N, and the synergistic effect was found in the combination of the mild treatment (42°C and RH 50%) and UV irradiation with a total energy of 600 mJ.
Consequently, Oxa has been found to be useful to covalently immobilize probe DNA oligomers onto the NH2-functionalized glass slide by one-pot reaction under the mild conditions. For covalently immobilizing probe DNA oligomers by the conventional methods on the solid surface, additional activation steps for probe and surface are commonly employed, which require some additional steps such as stringent washing, etc. and produce undesirable by-products which reduce the immobilization efficiency of the spotted DNA probe and results in the low-hybridization efficiency. On the other hand, the currently developed system in which Oxa-N is employed as a linker and immobilization of probe DNA oligomers can be performed under mild conditions that requires no activation step and produces no by-products. Also the resulting high target recognition efficiency is notable. The incorporation of Oxa-N in the probe DNA oligomers as a linker would be employed as one of the advanced methods for the immobilization of the widely used NH2-functionalized glass slides. Preparation of various types of DNA molecules with Oxa-N would also be a useful tool for the investigation of physiologically active DNA conjugates with other biological high molecules such as proteins, which is one of the hottest but the most difficult subjects in the current biochemistry and molecular biology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work is partially supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan to K.M. (No. 18350083), by Grants-in-Aid for Regional Science and Technology Promotion ‘Kyoto Nanotechnology Cluster’ project from MEXT, Japan, and by Grants-in-Aid for post-doctoral fellowship for foreign researcher (P04404) and his host from the Japan Society for the Promotion of Science. This work was also supported by CREST of the Japan Science and Technology Agency and by the Korean Research Foundation (No. M01-2005-000-10436-0). Funding to pay the Open Access publication charges for this article was provided by MEXT, Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Light-directed, spatially addressable parallel chemical synthesis. Science. 1991;251:767–773. doi: 10.1126/science.1990438. [DOI] [PubMed] [Google Scholar]
- 2.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 3.Brown PO, Bostein D. Exploring the new world of the genome with DNA microarrays. Nat, Genet. Suppl. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 4.Pirrung MC. How to make a DNA chip. Angew. Chem. Intl Ed. 2002;41:1276–1289. doi: 10.1002/1521-3773(20020415)41:8<1276::aid-anie1276>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Heller MJ. DNA microarray technology: devices, systems, and applications. Annu. Rev. Biomed. Eng. 2002;4:129–153. doi: 10.1146/annurev.bioeng.4.020702.153438. [DOI] [PubMed] [Google Scholar]
- 6.Dufva M. Fabrication of high quality microarrays. Biomol. Eng. 2005;22:173–184. doi: 10.1016/j.bioeng.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 7.Ulman A. Formation and structure of self-assembled monolayers. Chem. Rev. 1996;96:1533–1554. doi: 10.1021/cr9502357. [DOI] [PubMed] [Google Scholar]
- 8.Beaucage SL. Strategies in the preparation of DNA oligonucleotide arrays for diagnostic applications. Curr. Med. Chem. 2001;8:1213–1244. doi: 10.2174/0929867013372463. [DOI] [PubMed] [Google Scholar]
- 9.Schena M. Microarray Analysis. Hoboken, NJ, USA: John Wiley & Sons; 2002. [Google Scholar]
- 10.Bischoff R, Coull JM, Regnier FE. Introduction of 5-terminal functional groups into synthetic oligonucleotides for selective immobilization. Anal. Biochem. 1987;164:336–344. doi: 10.1016/0003-2697(87)90502-1. [DOI] [PubMed] [Google Scholar]
- 11.Zammatteo N, Jeanmart L, Hamels S, Courtois S, Louette P, Hevesi L, Remacle J. Comparison between different strategies of covalent attachment of DNA to glass surfaces to build DNA microarrays. Anal. Biochem. 2000;280:143–150. doi: 10.1006/abio.2000.4515. [DOI] [PubMed] [Google Scholar]
- 12.Joos B, Kuster H, Cone R. Covalent attachment of hybridizable oligonucleotides to glass supports. Anal. Biochem. 1997;247:96–101. doi: 10.1006/abio.1997.2017. [DOI] [PubMed] [Google Scholar]
- 13.Benters R, Niemeyer NM, Whorle D. Dendrimer-activated solid supports for nucleic acid and protein microarrays. ChemBiochem. 2001;2:686–694. doi: 10.1002/1439-7633(20010903)2:9<686::AID-CBIC686>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 14.Tao S, Gao H, CaoF., Ma X, Cheng J. Blocking oligo – a novel approach for improving chip-based DNA hybridization. Mol. Cell. Probes. 2003;17:197–202. doi: 10.1016/s0890-8508(03)00053-7. [DOI] [PubMed] [Google Scholar]
- 15.Kamisetty NK, Pack SP, Nonogawa M, Devarayapalli KC, Kodaki T, Makino K. Development of an efficient amine-functionalized glass platform by additional silanization treatment with alkylsilane. Anal. Bioanal. Chem. 2006;386:1649–1656. doi: 10.1007/s00216-006-0741-6. [DOI] [PubMed] [Google Scholar]
- 16.Ynag M, Kong RYC, Kazmi N, Leung AKC. Covalent immobilization of oligonucleotides on modified glass/silicon surfaces for solid-phase DNA hybridization and amplification. Chem. Lett. 1994;27:257–258. [Google Scholar]
- 17.Hermanson GT. Bioconjugate Techniques. 1996. Academic Press, San Diego, CA, pp. 169–186. [Google Scholar]
- 18.Garbarek Z, Gergely J. Zero-length crosslinking procedure with the use of active esters. Anal. Biochem. 1990;185:131–135. doi: 10.1016/0003-2697(90)90267-d. [DOI] [PubMed] [Google Scholar]
- 19.Staros JV, Wright RW, Swingle DM. Enhancement by N-hydroxysulfosuccinimide of water-soluble carbodiimide-mediated coupling reactions. Anal. Biochem. 1986;156:220–222. doi: 10.1016/0003-2697(86)90176-4. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki T, Yamaoka R, Nishi M, Ide H, Makino K. Isolation and characterization of a novel product, 2′-deoxyoxanosine, from 2′-deoxyguanosine, oligodeoxyribonucleotide, and calf thymus DNA treated by nitrous acid and nitric oxide. J. Am. Chem. Soc. 1996;118:2515–2516. [Google Scholar]
- 21.Suzuki T, Matsumura Y, Ide H, Kanaori K, Tajima K, Makino K. Deglycosylation susceptibility and base-pairing stability of 2′-deoxyoxanosine in oligodeoxynucleotide. Biochemistry. 1997;36:8013–8019. doi: 10.1021/bi970166l. [DOI] [PubMed] [Google Scholar]
- 22.Suzuki T, Yoshida M, Yamada M, Ide H, Kobayashi M, Kanaori K, Tajima K, Makino K. Misincorporation of 2′-deoxyoxanosine 5′-triphosphate by DNA polymerases and its implication for mutagenesis. Biochemistry. 1998;37:11592–11598. doi: 10.1021/bi980971f. [DOI] [PubMed] [Google Scholar]
- 23.Lucas LT, Gatehouse D, Shuker DG. Efficient nitroso group transfer from N-nitrosoindoles to nucleotides and 2′-deoxyguanosine at physiological pH. A new pathway for N-nitrosocompounds to exert genotoxicity. J. Biol. Chem. 1999;274:18319–18326. doi: 10.1074/jbc.274.26.18319. [DOI] [PubMed] [Google Scholar]
- 24.Hernandez B, Soliva R, Luque FJ, Orozco M. Misincorporation of 2′-deoxyoxanosine into DNA: a molecular basis for NO-induced mutagenesis derived from theoretical calculations. Nucleic Acids Res. 2000;28:4873–4883. doi: 10.1093/nar/28.24.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terato H, Masaoka A, Asagoshi K, Honsho A, Ohyama Y, Suzuki T, Yamada M, Makino K, Yamamoto K, et al. Novel repair activities of AlkA (3-methyladenine DNA glycosylase II) and endonuclease VIII for xanthine and oxanine, guanine lesions induced by nitric oxide and nitrous acid. Nucleic Acids Res. 2002;30:4975–4984. doi: 10.1093/nar/gkf630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hitchcock TM, Dong L, Connor EE, Meira LB, Samson LD, Wyatt MD, Cao W. Oxanine DNA glycosylase activity from mammalian alkyladenine glycosylase. J. Biol. Chem. 2004;279:38177–38183. doi: 10.1074/jbc.M405882200. [DOI] [PubMed] [Google Scholar]
- 27.Nakano T, Katafuchi A, Shimizu R, Terato H, Suzuki T, Tauchi H, Makino K, Skorvaga M, Van Houten B, et al. Repair activity of base and nucleotide excision repair enzymes for guanine lesions induced by nitrosative stress. Nucleic Acids Res. 2005;33:2181–2191. doi: 10.1093/nar/gki513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakano T, Asagoshi K, Terato H, Suzuki T, Ide H. Assessment of genotoxic potential of nitric oxide-induced guanine lesions by in vitro reactions with Escherichia coli DNA polymerase I. Mutagenesis. 2005;20:209–216. doi: 10.1093/mutage/gei027. [DOI] [PubMed] [Google Scholar]
- 29.Glaser R, Son MS. Pyrimidine ring opening in the unimolecular dediazoniation of guanine diazonium ion. An ab initio theoretical study of the mechanism of nitrosative guanosine deamination. J. Am. Chem. Soc. 1996;118:10942–10943. [Google Scholar]
- 30.Rayat S, Glaser R. 5-Cyanoimino-4-oxomethylene-dihydroimidazole and nitrosative guanine deamination. A theoretical study of geometries, electronic structures and N-Protonation. J. Org. Chem. 2003;68:9882–9892. doi: 10.1021/jo0351522. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki T, Ide H, Yamada M, Endo N, Kanaori K, Tajima K, Morii T, Makino K. Formation of 2′-deoxyoxanosine from 2′-deoxyguanosine and nitrous acid: mechanism and intermediates. Nucleic Acids Res. 2000;28:544–551. doi: 10.1093/nar/28.2.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakano T, Terato H, Asagoshi K, Masaoka A, Mukuta M, Ohyama Y, Suzuki T, Makino K, Ide H. DNA-protein cross-link formation mediated by oxanine. A novel genotoxic mechanism of nitric oxide-induced DNA damage. J. Biol. Chem. 2003;278:25264–25272. doi: 10.1074/jbc.M212847200. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki T, Yamada M, Ide H, Kanaori K, Tajima K, Morii T, Makino K. Identification and characterization of a reaction product of 2′-deoxyoxanosine with glycine. Chem. Res. Toxicol. 2000;13:227–230. doi: 10.1021/tx990164x. [DOI] [PubMed] [Google Scholar]
- 34.Pack SP, Nonogawa M, Kodaki T, Makino K. Chemical synthesis and thermodynamic characterization of oxanine-containing oligodeoxynucleotides. Nucleic Acids Res. 2005;33:5771–5780. doi: 10.1093/nar/gki865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamoto T, Suzuki T, Yamamoto N. Microarray fabrication with covalent attachment of DNA using bubble jet technology. Nat. Biotechnol. 2000;18:438–441. doi: 10.1038/74507. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki T, Yamada M, Ide H, Kanaori K, Tajima K, Morii T, Makino K. Influence of ring opening-closure equilibrium of oxanine, a novel damaged nucleobase, on migration behavior in capillary electrophoresis. J. Chromatogr. A. 2000;877:225–232. doi: 10.1016/s0021-9673(00)00178-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.