Abstract
RNA interference is mediated by small interfering RNAs (siRNAs) that upon incorporation into the RNA-induced silencing complex (RISC) can target complementary mRNA for degradation. Standard siRNA design usually feature a 19–27 base pair contiguous double-stranded region that is believed to be important for RISC incorporation. Here, we describe a novel siRNA design composed of an intact antisense strand complemented with two shorter 10–12 nt sense strands. This three-stranded construct, termed small internally segmented interfering RNA (sisiRNA), is highly functional demonstrating that an intact sense strand is not a prerequisite for RNA interference. Moreover, when using the sisiRNA design only the antisense strand is functional in activated RISC thereby completely eliminating unintended mRNA targeting by the sense strand. Interestingly, the sisiRNA design supports the function of chemically modified antisense strands, which are non-functional within the context of standard siRNA designs. This suggests that the sisiRNA design has a clear potential of improving the pharmacokinetic properties of siRNA in vivo.
INTRODUCTION
RNA interference (RNAi) was initially discovered in Caenorhabditis elegans by Fire and colleagues, who showed that introduction of long double-stranded RNA (dsRNA) caused a nearly complete inhibition of genes harboring the same sequence (1). It was subsequently demonstrated that short ∼21 bp dsRNAs, termed small interfering RNA (siRNA), were functional triggers of RNAi without inducing the innate immune responses associated with longer dsRNA in mammalian cells (2). Natural siRNAs are processed from longer dsRNA species derived from, e.g. virus, mobile elements or transgenic RNA by the cytoplasmic RNAse III enzyme Dicer (3). Similarly, exogenous 19–27 bp siRNAs are functional if introduced into the cytoplasm (2). Here, the siRNA will be incorporated into the RNA-induced silencing complex (RISC) by a RISC loading complex (RLC), which is best described in Drosophila (4), but likely also exists in humans (5). By sensing the thermodynamic asymmetry of siRNA duplex ends, RLC distinguishes the siRNA guiding antisense strand from the sense strand, thereby dictating the so-called pre-RISC to assemble asymmetrically on the siRNA duplex (4,6). Although both strands of the siRNA duplex are initially incorporated into pre-RISC, the RLC-tagged sense strand is subsequently cleaved and released thereby establishing activated RISC which contains only the single stranded antisense strand. Recent data suggest that the catalytic core protein of RISC, the Ago2 endonuclease, initiates sense strand elimination by cleaving it 9 nt from its 5′ end during RISC activation (7–9). Although the helicase activity for unwinding the duplex remains unidentified, these events expose the antisense strand in RISC to the mRNA target, which is subsequently cleaved probably by a similar mechanism.
The use of synthetic siRNAs in vivo is currently hampered by lack of efficient means of siRNA delivery, low biostability in biological fluids and low specificity of action due to inherent gene off-target effects caused by the microRNA-like behavior of all investigated siRNAs (10–12). Several attempts to reduce off-target effects through chemical modification of synthetic siRNA have been made (13,14). Since both strands of a siRNA duplex can contribute to off-target effects (10), minimizing sense strand incorporation into activated RISC should significantly increase targeting specificity. It is well established that the siRNA strand with the thermodynamically least stable 5′ end is preferentially utilized as antisense strand in activated RISC (6,15). Accordingly, selective thermodynamic stabilization of sense strand 5′ ends by incorporation of locked nucleic acids (LNA) has been shown to reduce unwarranted gene silencing by the sense strand (13,16). Here, we apply a radically different design characterized by an intact antisense strand complemented with two shorter 9–13 nt sense strands, together named small internally segmented interfering RNA (sisiRNA, Table 1). We show that only the antisense strand of this construct is capable of gene silencing thereby significantly increasing targeting specificity. Moreover, incorporation of LNA nucleotides into the disrupted sense strand significantly increases serum stability which may be important for in vivo applications. Interestingly, the sisiRNA design can functionally accommodate heavily modified antisense strands that are non-functional as standard siRNAs. This potentially allows the application of more highly functionalized siRNA designs.
Table 1.
Oligonucleotide sequences and chemical modifications
Oligonucleotide sequences and chemical modifications. Top panel. List of RNA oligoes used in this study. SS and AS denote sense strand and antisense strand, respectively. SS1 corresponds to a continuous version of 5′SS1 and 3′SS1. 5′SS3/ 3′SS3 and 5′SS4/ 3′SS4 are variants of the 5′SS1 and 3′SS1 pair where the nick has been shifted one position towards the 3′ end and 5′ end, respectively. 5′SS2 and 3′SS2 are the RNA versions of 5′SS1 and 3′SS1. 3′SS4 is equivalent to 3′SS1 but without a 3′ terminal U-residue. Bold underlined letters indicate the position of the LNA nucleotides. C: LNA-5-Me cytosine, G: LNA-Guanine, T: LNA-Thymine, aT: N2′-adamantylmethylcarbonyl 2′-amino-LNA-thymine, pT: N2′-pyren-1-ylmethyl 2′-amino-LNA-thymine. Bottom panel: Selected examples of duplexes used in this study. Sense strand is at the top.
MATERIALS AND METHODS
Constructs and cells
The human lung cancer cell line H1299 produced to stably express EGFP (EGFP half-life 2 h) was a gift from Dr Anne Chauchereau (CNRS, Villejuif, France). H1299 and T98G cells were grown in RPMI-1640 containing 10% FBS, 1% penicillin/streptomycin.
The two reporter constructs pISOantisense-target and pISOsense-target were constructed by annealing equimolar amounts of the following DNA oligoes 5′-GCGACGTAAACGGCCACAAGTTC-3′ and 3′-TCGACGCTGCATTTGCCGGTGTTCAAGGATC-5 (antisense target) or 5′-CTAGGCGACGTAAACGGCCACAAGTTCAGCT-3′ and 3′-CGCTGCATTTGCCGGTGTTCAAG-5′ (sense target) into SacI/NheI digested pISO (kindly provided by David Bartel) (17) downstream of the firefly luciferase coding sequence.
siRNA synthesis
Non-modified and LNA-modified RNA oligoes were prepared on an automated DNA synthesizer as described earlier (16). The synthesis of adamantyl and pyrenyl containing RNA oligoes is described elsewhere (J.W., S.W.L., B.R.B., manuscript in preparation).
Sense and antisense strands where mixed in annealing buffer (10 mM Tris–HCl, pH 7.3, 50 mM NaCl) at 20 mM equimolar concentration and incubated at 95°C for 1 min and at 1 h at 37°C.
Quantification of EGFP and GAPDH
Cells used for EGFP northern, western and flow-cytometry analysis were seeded at ∼20% confluency and transfected using Bio-Rad Silentfect transfection reagent (50 nM final RNA concentration) according to manufacturer's instructions. Twenty-four hours later cells were replenished with fresh medium and incubated for another 24 h before either re-transfection using Lipofectamine2000 (50 nM final RNA concentration) according to manufactures directions or harvested for western blot analysis, northern blot analysis or flow-cytometry analysis (counting approximately 5 × 104 cells and averaged). Western blotting was performed as follows: cells were washed twice in PBS and an equal amount of cells were lysed in 2× SDS sample buffer [4% Sodium Dodecyl-Sulphate (SDS), 20% glycerol, 125 mM Tris/HCl pH 6.8, 0.01 mg/ml Bromphenol Blue, 10% b-mercaptoethanol] at 90°C for 2 × 10 min separated by gentle pippeting. Proteins were separated in 12% SDS acrylamide gels and electroblotted overnight onto a PVDF membrane (Immobilon). The filter was blocked for 1 h with PBS containing 10% w/v milk. EGFP protein was detected using a 1:1000 dilution of a rabbit polyclonal EGFP antibody. The mouse hnRNP C1 antibody was a gift from Seraphin Pinol-Roma. A horseradish peroxidase (hrp) conjugated secondary antibody (DAKO) was used with ECL reagent (Amersham Biosciences) for visualization. EGFP mRNA was analyzed by northern blotting according to standard procedures.
GAPDH mRNA expression was quantified by northern blotting; wild-type H1299 cells were transfected at 50% confluency using TransIT-TKO® transfection reagent (Mirus) according to the manufactures protocol (10 nM final RNA concentration). RNA was harvested 48 h post-transfection using TRIzol® reagent (Invitrogen) and northern blotting was performed according to standard procedures.
Interferon response assay
siRNA-variants (80 nM) or poly(I:C) (0.8 μg/ml) were transfected into T98G cells using the TransIT-TKO® transfection reagent (Mirus) according to the manufactures protocol. Total RNA was purified using Trizol® reagent (Invitrogen), DNase treated and subjected to oligo-dT-primed reverse transcription. qPCR was performed using the platinum SYBR®Green qPCR supermix (Invitrogen) on a Stratagene Mx3005p qPCR system. Primers used for amplification of ISG56: 5′-AAGGCAGGCTGTCCGCTTA-3′ and 5′-TCCTGTCCTTCATCCTGAAGCT-3′. Primers for amplifying GAPDH: 5′-GAAGGTGAAGGTCGGAGT-3′ and 5′-GAAGATGGTGATGGGATTTC-3′. The PCR conditions are: 1 cycle: 95°C 10 min, 40 cycles: 95°C 30 s, 55°C 1min, 72°C 30 s, 1 cycle: 95°C 30 s, 55°C 1 min, 95°C 1 min. Relative quantification of mRNAs levels were done by using the ▵▵CT-method. The experiments were done in triplicates and ISG56 levels for siRNA-treated cells were normalized to TransIT-TKO treated controls.
siRNA stability assay
Annealed LNA-modified sisiRNAs, LNA-modified siRNAs or siRNAs were incubated at 37°C in either 10% or 80% fetal calf serum in DMEM (Gibco). Aliquots of 5 μl (each containing 20 pmol of siRNA) were diluted in 25 μl 1.2× TBE loading buffer (1.2× TBE, 10% glycerol, bromphenol blue) and snap-frozen on dry ice immediately upon sample taking. Samples were run on a 15% native polyacrylamide gel and stained using SYBR Gold® (Invitrogen).
Dual luciferase assay
H1299 cells were plated in 6-well plates in RPMI supplemented with 10% fetal bovine serum and grown ON to 40–60% confluency. pISOantisense-target and pISOsense-target (1 μg) were co-transfected with 0.002 μg pRluc-N2 (Perkin–Elmer) and the siRNA duplexes (10 nM final concentration) by simultaneous use of 6 μl TransIT-LT1 (Mirus) and 6 μl TransIT-TKO (Mirus) according to the manufactures protocol. The Dual-luciferase assay was done 48 h posttransfection using the ‘Dual-luciferase reporter assay system’ (Promega) according to the manufactures protocol. The luciferase activities were measured on a Lumat LB 950 luminometer (Berthold) and normalized to the renilla luciferase signal.
RESULTS
A siRNA containing a nicked sense strand is fully functional
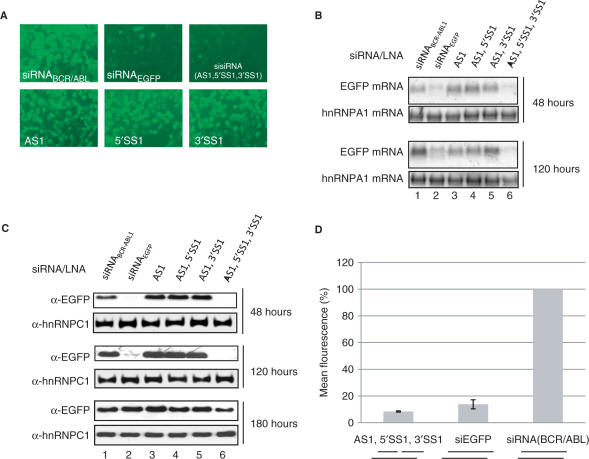
To eliminate sense strand incorporation into activated RISC, we applied a novel siRNAs design characterized by an intact antisense strand complemented with two shorter sense strands. We anticipated that by incorporating LNA nucleotides into such tri-molecule construct, sufficient stability and dsRNA structural mimicry would be achieved to allow RNA interference activity. We initially designed a sisiRNA composed of a 10 and 12 nt sense strand directed towards a previously established functional target in the mRNA encoding enhanced green fluorescent protein (EGFP)(18). To stabilize the sisiRNA construct we incorporated LNA at two and four positions in the sense 5′ and 3′ half-strands, respectively, and near the 3′ end of the antisense strand and assembled the construct from these three strands [AS1 + 5′SS1 + 3′SS1 (sisiRNA), Table 1]. Together with a standard siRNA and unrelated control siRNA, the constructs were tested by transfection into an H1299 lung carcinoma cell line that stably expresses destabilized EGFP. Subsequently, the level of EGFP mRNA and protein expression was monitored on the basis of fluorescence microscopy (Figure 1A), northern blotting (Figure 1B), western blotting (Figure 1C) and flow cytometry (Figure 1D). Treating cells with 50 nM LNA-modified sisiRNA or siRNA yielded a comparable 10-fold knock down after 48 h (Figure 1B–D). The duration of the knock down effect by the sisiRNA was similar or slightly superior to unmodified siRNA at 120 (5 days) or 180 h (7.5 days) (Figure 1B and C). Hence, sisiRNA exhibits similar silencing activity in cell culture as compared to standard siRNAs. The activity of the LNA-modified sisiRNA was strictly depending on the presence of all three strands as omitting one or both of the short sense strands (5′SS1 or 3′SS1) eliminated the activity of the sisiRNA (Figure 1A–C).
Figure 1.
Testing the knock down of EGFP by sisiRNA and related constructs. The EGFP expression was assessed both at the RNA and protein level in H1299 cells. (A) Fluorescence microscopy analysis of EGFP expression in H1299 cells. Cells were treated with 50 nM of the indicated combinations of RNA/LNA oligoes and analyzed 48 hours after for EGFP expression. (B) Northern blot showing EGFP mRNA expression 48 and 120 hours after initial transfection with 50 nM of the indicated combinations of oligonucleotides. (C) Western blot showing the expression of EGFP protein in cells at 48, 120 and 180 hours after transfection with 50 nM of the indicated RNA constructs. The filter was reprobed with an antibody specific to hnRNPC1 protein as a loading control. (D) Flow cytometry analysis showing the mean green fluorescence of 50.000 cells 48 hours posttransfection (based on three experiments).
To investigate if the sisiRNA design is applicable to other target sequences, we additionally targeted the endogenous gene GAPDH in H1299 cells using two sisiRNA designs differing only in the position of the LNA residues in passenger fragments [AS8 + 5′SS6 + 3′SS6 (siGAPDH 1) and AS8 + 5′SS6*+3′SS6* (siGAPDH 2), Table 1] and compared them to unmodified siRNA targeting GAPDH [AS7+SS7 (siGAPDH), Table 1] and LNA-modified siRNA [AS8+SS6 (siGAPDH, LNA), Table 1]. All four constructs resulted in a similar ∼60% reduction in GAPDH mRNA levels (Figure 2A and B, columns 1–4) as compared to cells transfected with EGFP-specific siRNA or non-transfected controls (Figure 2A and B, columns 5 and 6). Competitive knock down levels have also been observed with sisiRNA directed towards H-Ras in HeLa cells (M.B.L. unpublished data). Hence, the sisiRNA design has proven highly functional for all tested targets with efficiencies similar to unmodified or LNA-modified siRNA.
Figure 2.
Knockdown of GAPDH mRNA by sisiRNA. (A) Northen blot analysis of GAPDH mRNA expression in H1299 cells 48 h after transfection of either siGAPDH, siGAPDH (LNA), sisiGAPDH 1, siGAPDH 2 or siEGFP as indicated. Upper panel: GAPDH mRNA as evaluated by northern blotting using a GAPDH probe. Lower panel: ethidium bromide staining of 18S rRNA (loading control). Experiments were performed in triplicates. (B) Quantification of GAPDH mRNA levels in (A). All values are normalized to siEGFP transfected cells.
LNA modifications are essential for the sisiRNA design, increases sisiRNA serum stability and are non-immunogenic in cell culture
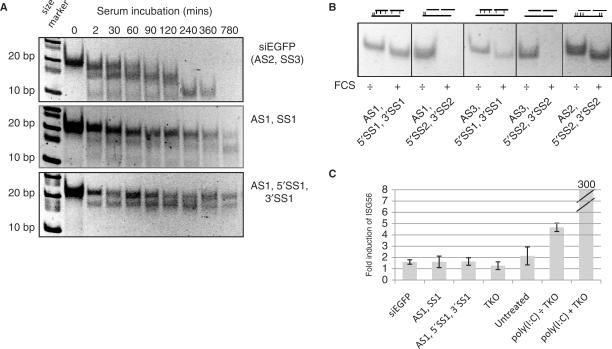
We speculated that the LNA-modified siLNA and sisiRNA designs may have greater stability compared to unmodified siRNAs, both in terms of premature strand separation and resistance to RNases. In accordance, we found that upon incubation in 80% FCS, unmodified siRNAs were rapidly degraded within 1½ h whereas a large proportion of both LNA-modified sisiRNA and an identical construct, but with continuous sense strand (AS1+SS1, Table 1), remained intact for 780 min (13 h) (Figure 3A). Notably, we have observed no significant knock down when using a sisiRNA containing only unmodified residues (data not shown) suggesting that LNA modifications are not only beneficial but also essential for the integrity of the sisiRNA design in biological fluids. This is compatible with the observation that LNA-modifications in the complementary part of either the antisense or sense strand are essential for sisiRNA serum stability (Figure 3B). Hence, we conclude that the sisiRNA design is highly stable in serum despite of the introduction of an internal nick in the sense strand.
Figure 3.
Serum stability of the sisiRNA design. (A) LNA-modified sisiRNA and LNA-modified siRNA have increased serum stability compared to unmodified siRNA. The siRNA variants were incubated in 80% FCS and aliquots taken at indicated time points. Serum-stability was evaluated by PAGE followed by SYBR Gold® staining. Whereas siRNA is degraded within 1–1½ h of serum incubation, significant amount of LNA-modified sisiRNA and LNA-modified siRNA are still present after 13 h of incubation. A size marker is indicated to the left. (B) LNA-base pairing is essential for the integrity of sisiRNA molecules upon incubation in 10% FCS. The indicated sisiRNA molecules carrying different or no LNA modifications were incubated in the presence (+) or absence (−) of 10% FCS for 24 h and duplex stability was evaluated by PAGE followed by SYBR Gold® staining. sisiRNA constructs with LNA in both strands exhibited full stability whereas sisiRNA containing only RNA were completely degraded upon serum incubation. The position of the LNA modifications are indicated schematically (vertical lines). (C) LNA-modified sisiRNA and LNA-modified siRNA do not activate the interferon system as evaluated by induction of ISG56 in T98G cells. The glioblastoma cell line T98G was transfected with 80 nM of the siRNA variants or 0.8 μg poly(I:C) as indicated and ISG56 mRNA levels evaluated by qPCR analysis 48 h posttransfection. Only transfection of poly(I:C) (pos. control) lead to high levels of ISG56 induction, whereas ISG56 levels for all siRNA-variants were indistinguishable from untreated cells or cells treated with transfection reagent alone (Mirus Trans-IT TKO®).
Chemical modifications of nucleic acids can have a dramatic influence on the cellular immune response in cultured cells and in animals (19,20). We did, however, not observe any cytotoxic side-effects or growth inhibition in sisiRNA-treated cells as compared to standard siRNAs (data not shown). To analyze that the sisiRNA design does not trigger cellular interferon responses, we transfected the human glioblastoma T98G cell line using 80 nM of the different siRNA constructs and measured the induction of ISG56, which is strongly induced by both types of IFNs (21) and dsRNA (22). ISG56 induction has previously been reported in T98G cells upon siRNA transfection (23), yet no significant differences in ISG56 induction were observed between LNA-modified sisiRNA, LNA-modified siRNA and unmodified siRNA (Figure 3C). In contrast, poly(I:C) induced the ISG56 several hundred fold. Hence, at least in cell culture, sisiRNA appears to be immunologically similar to standard siRNAs.
Position and size of the sisiRNA sense strand nick
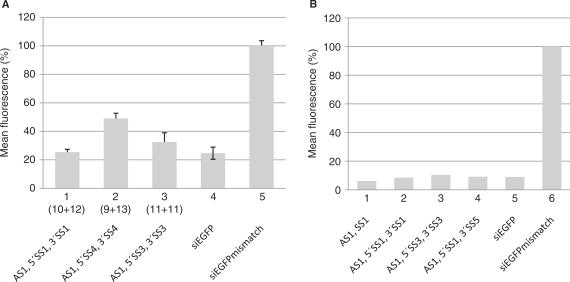
To further optimize the sisiRNA design, we tested a series of different sense and antisense strands. In one experiment, the position of the gap in the sense strand was either shifted one position towards the 3′ end [AS1 + 5′SS3 + 3′SS3 (sisiRNA11+11), Table 1] or one position towards the 5′ end [AS1 + 5′SS4 + 3′SS4 (sisiRNA9+13), Table 1] which since the initiation of this work has been reported to be the natural cleavage site for Ago2 in RISC (7–9). For the sisiRNA11+11 design, only a minor decline in silencing was observed whereas the sisiRNA9+13 design was slightly less efficient in gene silencing (Figure 4A, compare columns 1–3). A similar minor decline in knock down efficiency was seen for a sisiGAPDH9+13 design as compared to siGAPDH 1 and siGAPDH 2 (data not shown). These data underscore the notion that the position of LNA modifications need not be fixed within the sisiRNA (Figure 2A and B) and the position of the nick in the passenger strand need not to mimic the natural cleavage site (Figure 4A). Increasing the gap size of the sense strand of the sisiRNA duplex to 1–2 nt resulted in a dramatic decline in sisiRNA activity irrespectively of gap position (data not shown). In conclusion we find flexibility in positioning of both the LNA modifications and the sense strand central nick, yet the sisRNA10+12 design has proven most efficient among the sisiRNAs combinations tested (Figure 4A, data not shown).
Figure 4.
Optimization the sisiRNA design. The knock down efficiencies between different sisiRNA and siRNA designs were compared by targeting EGFP mRNA. (A) Analyzing the effect of different gap positions in the sense strand. The numbers correspond to the size of the 5′ and 3′ fragment of the sense strand, respectively. (B) Analyzing the effect of modifications at the 3′ terminal nucleotide on the sense strand. The mean fluorescence of approximately 50 000 cells was measured by flow cytometry. The siRNA mismatch represents a siRNA that contains four mismatches to the EGFP target (Table 1).
The 3′SS1 strand was initially designed to contain an additional U-residue at the 3′ end in order to ease the chemical synthesis. To test whether this residue affects sisiRNA activity, 3′SS1 was synthesized without this terminal U-residue (3′SS5; Table 1). This alteration did not alter the activity of the construct significantly (compare columns 2 and 4, Figure 4B).
The discontinuity of the sense strand completely eliminates its guide activity
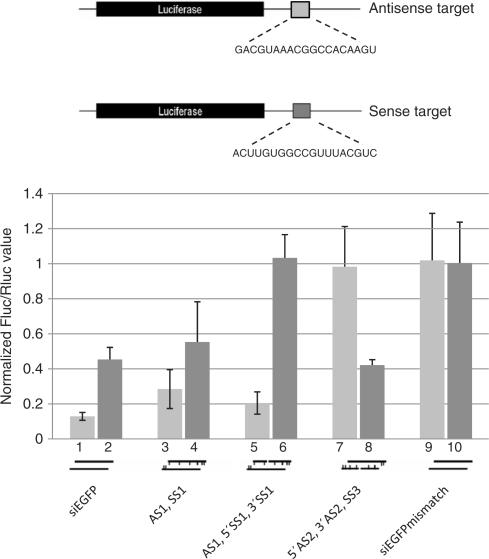
To test whether the discontinuity of the intended sense strand eliminates its contribution to gene silencing we inserted the EGFP target sequence for either the siRNAEGFP antisense or sense strand within the 3′ UTR of a firefly luciferase reporter construct (Figure 5). This strategy allowed us to differentially assess the knock down effect derived from the antisense and the sense strand incorporation into activated RISC. As predicted, the LNA-modified sisiRNA construct was significantly more specific than the equivalent siRNA and LNA-modified siRNA duplexes since ∼50% knockdown was constantly seen from the sense strand with standard siRNA design (columns 2 and 4, Figure 5). In contrast, the sisiRNA design completely abrogated the silencing of the ‘sense target’ as compared to mismatch controls without compromising the potent knockdown mediated by the antisense strand (columns 5 and 6, Figure 5). To test whether it is possible to abrogate the silencing function of an otherwise optimal antisense strand with a nick, another LNA-modified sisiRNA with an intact sense strand and a discontinuous antisense strand (5′AS2, 3′AS2, SS3) was tested for the ability to knock down the antisense and sense targets (Table 1). This design completely eliminated silencing of the antisense target yet retained silencing of the sense target to a level comparable to the standard siRNA (columns 7 and 8, Figure 5). Collectively these data clearly demonstrate that the sisiRNA construct exhibits a much higher level of specificity for the intended target compared to the usual siRNA design.
Figure 5.
The sisiRNA design increases the specificity of gene silencing. The knockdown activity of the two strands was assessed by measuring luciferase expression from reporter constructs containing either the target sequence for the sense or antisense strand of the EGFP specific duplex (light and dark gray bars, respectively). The reporter constructs are drawn above (not to scale) and the siRNA constructs are indicated schematically below. The values are averaged over three completely independent experiments. The luciferase values of each experiment are normalized to make the sums of the luciferase activities in each of the experiments equal. For each reporter construct, the firefly luciferase (Fluc)/renilla luciferase (Rluc) ratio was normalized to mismatch controls.
The sisiRNA design is compatible with higher levels of antisense modification
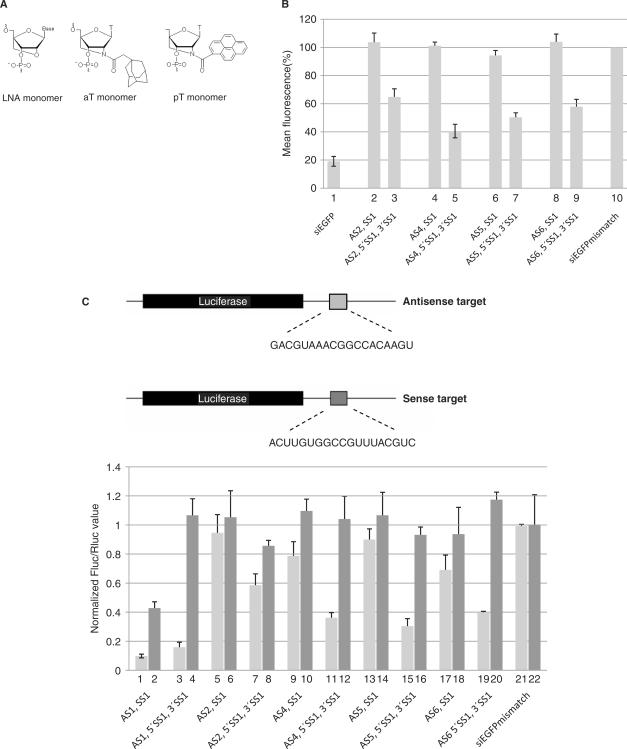
We and others have previously found that extensive LNA-modification of antisense strands strongly interfere with RNAi activity in standard siRNA designs (Figure 6B, column 2 and data not shown) (13,24). Therefore, we initially designed AS1 to contain only two LNA residues near the 3′ end. To investigate whether the discontinuity of the sense strand influences the requirement for unmodified residues in the body of the antisense strand, we tested an LNA-modified sisiRNA with a highly modified antisense strand containing six LNA residues (AS2). This antisense strand is essentially inactive when paired with an all-RNA sense strand (data not shown) or an LNA-modified sense strand (LNA-modified siRNA duplex AS2+SS1, Table 1; Figure 6B, column 2). Interestingly, the requirement for unmodified residues in the antisense strand was less stringent when using the sisiRNA design (compare columns 2 and 3, Figure 6B). A similar improvement in knock down efficiency was observed using the 3′ end shortened sense construct and the LNA-modified sisiRNA11+11 design (data not shown). To test if a similar effect applies to other types of chemical modifications that do not increase siRNA thermodynamic stability, we tested three designs containing either additional single N2′-adamantyl (AS4 and AS5, Table 1) (synthesis will be described elsewhere) or N2′-pyren-1-yl 2′-amino-LNA-T (26) (AS6, Table 1) modifications in the antisense strand (Table 1, aT and pT, respectively). These types of modifications render the siRNA almost non-functional when paired to SS1 in a standard (LNA-modified) siRNA design (Figure 6B, columns 4, 6 and 8). However, in the context of the sisiRNA design, both the adamentyl and pyrenyl antisense strands resulted in a 40–60% knock down of EGFP expression (Figure 6, columns 5, 7 and 9). Hence, the sisiRNA design can accommodate a wide variety of bulky chemical modifications that otherwise are incompatible with the activity of standard duplex siRNA.
Figure 6.
The sisiRNA design supports the silencing effects of chemically modified antisense strands. (A) Molecular structure of N2′-adamantylmethylcarbonyl 2′-amino-LNA-Thymine and N2′-pyren-1-ylmethyl 2′-amino-LNA-thymine. (B) Analyzing the effect of the sisiRNA design on the silencing efficiency of heavily modified antisense strands. The mean fluorescence of approximately 50 000 cells was measured by flow cytometry. The siRNA mismatch represents a siRNA that contains four mismatches to the EGFP target (Table 1). (C) The knockdown activity of the two siRNA duplex strands was assessed by measuring luciferase expression from reporter constructs containing either the target sequence for the sense or antisense strand of the EGFP specific duplex (light and dark gray bars, respectively). The reporter constructs are drawn above (not to scale). The experiment was performed in triplicate and for each reporter construct the firefly luciferase (Fluc)/renilla luciferase (Rluc) ratio was normalized to mismatch controls.
To further characterize the mechanism for the relaxed stringency of antisense strand modification, we investigated the incorporation of the sense strand (SS1) into RISC in lightly modified antisense siRNA (AS1+SS1) as compared to the siRNAs with heavily modified antisense (AS2+SS1, AS4+SS1, AS5+SS1, AS6+SS1). The LNA-modified siRNA (AS1+SS1) caused an ∼60% reduction of reporter levels from the sense target (Figure 6C, column 2), confirming that the sense strand is indeed incorporated into activated RISC and active. In contrast, the function of the sense strand was virtually lost when it was paired to highly modified antisense strands (AS2, SS1; AS4, SS1; AS5, SS1; AS6, SS1) (Figure 6C, columns 6, 10, 14 and 18). Hence, the poor silencing by the heavily modified antisense strands (AS2, AS4, AS5 and AS6) is not simply be due to a shifted strand selection (towards SS1) during RISC activation. Instead, heavy modification of siRNA duplexes in the antisense strand seems to abrogate its function at steps prior to RISC activation. The sisiRNA design, however, seems to partly rescue such defects thereby allowing heavily modified antisense strand to be efficiently loaded into activated RISC. To ensure that the knock down effects obtained using siRNA with lightly and heavy modified sisiRNA or siRNA are specific, we synthesized the equivalent set of LNA-modified siRNA and sisiRNA containing five mismatched positions (SS8+AS9, SS8+AS10 and 5′SS8 + 3′SS8+AS9, 5′SS8 + 3′SS8+AS10, respectively; Table 1) and tested them in the luciferase reporter assay (Supplementary Figure 1). The results confirmed the increased potency of heavily modified antisense strands when situated in a sisiRNA design and showed that the knock down was specific to the wild type EGFP target sequence.
DISCUSSION
In this study, we have developed a radical new siRNA design composed of an intact antisense strand complemented with two shorter 10–12 nt sense strands. We show that this three-stranded construct is fully functional and that it has several advantages over the standard 21 nt duplex siRNA designs. (i) The LNA-modified sisiRNAs have similar high potency as compared to standard siRNAs in cell culture, yet has greatly enhanced stability in serum which is expectably important for in vivo applications. (ii) The segmented nature of the passenger strand completely alleviates its contribution to unwarranted gene knock down thereby greatly increasing targeting specificity and expectably reducing off-target effects. (iii) The sisiRNA design has the ability to rescue the function of chemically modified antisense strands that are non-functional within the context of a standard siRNA duplex thereby allowing more chemical modification to be incorporated into the antisense strand. (iv) The sisiRNA design has six terminal ends compared to four in normal siRNA which can conveniently be used for tethering functional chemical groups to enhance, e.g. cellular delivery. For instance, it is possible to tether bulky groups like cholesterol to the 5′ end of the downstream sense strand without loosing activity (M.B.L., J.K., J.W., unpublished data). (v) As the yield of synthesis is usually higher for shorter RNA strands, the cost of large-scale synthesis in connection with therapeutic application may be reduced using a sisiRNA design.
An important feature of the sisiRNA design is the ability to completely eliminate the contribution of the segmented strand to gene silencing while leaving the RNAi activity of the opposing strand intact (Figure 5). The resultant increase in gene silencing specificity can be expected to reduce the genome-wide off-targets effects from the sense strand that has been observed for other investigated siRNAs (10). Furthermore, as strand selection is primarily determined by the thermodynamic asymmetry of siRNA duplex ends, highly efficient siRNA may be difficult to design if the target sequence is restricted to a thermodynamically unfavorable region, e.g. when the intension is to target single nucleotide mutation or junctions between fused genes. In these instances, the sisiRNA design will ensure that only the unsegmented strand can contribute to gene silencing irrespectively of the thermodynamic profile of the sisiRNA duplex and will thereby eliminate the significant unwarranted silencing conferred by the thermodynamically favored opposing strand.
Leuchner et al. (8) have previously demonstrated that pre-cleaved siRNA, similar to our unmodified sisiRNA, is capable of RISC loading and target cleavage in a cell extract. However, we find that sisiRNAs without LNA residues are non-functional in a cellular context, even if 2′ OMe modified residues are introduced in the short sense strands (data not shown). Based on our stability assays (Figure 3B), the most likely explanation is that the unmodified strands in sisiRNA are dissociating and degraded in vivo and that only the significant increase in Tm, provided by the LNA residues, renders the duplex sufficiently stable under these conditions.
An interesting observation is that sisiRNA function does not rely strictly on exact structural mimicry of an intermediate Ago2-cleavage product as the strand nick can be moved 1–2 nt without major loss of silencing efficiency (Figure 4A, data not shown). In particular, the sisiRNA design mimicking the ‘natural’ Ago2-cleavage product (sisiRNA9+13) seems less efficient than when moving the nick 1 and 2 nt towards the 3′ end of the sense strand (sisiRNA10+12 and sisiRNA11+11). Based on in vitro data from Leuchner et al. (8) these constructs are most likely cleaved by Ago2, liberating one or two nucleotides, respectively. It is therefore possible that allowing a ‘natural’ Ago2-cleavage event in the sisiRNA10+12 and sisiRNA11+11 designs may further help RISC activation by facilitating subsequent steps such as, e.g. sense strand elimination. Hence, we believe that the sisiRNA10+12 design introduces novel improvements in siRNAs function beyond those offered by the structural mimicry of natural intermediates in the RNAi pathway.
We and others have observed that extensive chemical modifications in the antisense strand of siRNAs generally are incompatible with their function in gene silencing (Figure 6, data not shown)(24). Yet, the specific steps in the RNAi pathway, which are incompatible with extensive siRNA modification, are only poorly defined. Interestingly, the sisiRNA design can significantly enhance the efficiency of heavily modified antisense strands. We demonstrate here that the inability of extensively LNA-, LNA/adamantyl- and LNA/pyrenyl-modified antisense strands to support RISC activity can be partially rescued by the sisiRNA-design, whereas both strands of similarly modified ordinary siRNAs are non-functional (Figure 6). This shows that heavy modification of an antisense strand abrogates the function of both sense and antisense strands that are individually functional within the context of activated RISC (Figure 6B and C). This suggests that extensive modification of antisense strands may lead to impairments prior to RISC activation, e.g. siRNA recruitment by the RLC or structural rearrangements within pre-RISC. It may be speculated that the segmented sense strand in the sisiRNA design facilitates the preferential loading of the intact opposing antisense strand into activated RISC and thereby enhance their silencing potential. However, no increase in silencing by the unsegmented strand in neither AS1 + 5′SS1 + 3′SS1 nor 5′AS2 + 3′AS2+SS3 designs was observed as compared to AS1+SS1 (Figure 5). In agreement, no enhancement in silencing by the sisiRNA design as compared to siRNA was seen in titration assays (0.01–100 nM) suggesting that strand selection is not affected (data not shown). Furthermore, the rescue of silencing by the sisiRNA design seems not to rely on alteration of the siRNA thermodynamic profile as the adamantyl and pyrenyl modifications, if anything, slightly destabilize siRNA duplexes in contrast to the stabilizing effect of the LNA-residues (J.W., unpublished data). Instead, heavily modified siRNAs may be too inflexible for structural rearrangements within pre-RISC during RISC loading or activation. The central strand nick in the sisiRNA design may provide more structural flexibility to the sisiRNA duplex allowing it to better position itself during RISC activation.
The possibility to incorporate more extensive chemical modifications into the sisiRNA design as compared to standard siRNAs may have beneficial properties for steps both upstream and downstream of RISC activation in the RNAi pathway. Introducing lipophilic groups like adamantyl and pyrenyl may increase cellular uptake of siRNA duplexes and unnatural modifications in general will increase siRNA stability in intra- and extracellular compartments. Furthermore, modifications in the seed region (nucleotides 2–8 of the antisense strand) may prove essential to minimize inherent gene off-target effects by siRNAs as it has been previously been demonstrated for position 2 in the antisense strand (14). Finally, it is possible that increased numbers of LNA residues in the antisense strand may improve the target specificity and affinity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by the Danish National Research Foundation, the Danish Technical Research Council, the Danish Strategic Research Council, the Danish Cancer Society and the EU-FP6 RIGHT project (no. LSHB-CT-2004-005276). The authors would like to thank Claus Bus for excellent technical assistance, David Bartel for plasmid constructs, Anne Chauchereau for EGFP expressing cell line and Serafin Pinol-Roma for antibodies. M.B.L., J.B.B., C.K.D., B.R.B. and S.W.L. performed research; J.B.B., C.K.D., M.B.L., J.K. and J.W. designed research and J.K., J.W. and J.B.B. wrote the article. Funding to pay the Open Access publication charges for this article was provided by the EU-FP6 RIGHT project (no. LSHB-CT-2004-005276).
Conflict of interest statement. None declared.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 3.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 4.Pham JW, Sontheimer EJ. Molecular requirements for RNA-induced silencing complex assembly in the Drosophila RNA interference pathway. J. Biol. Chem. 2005;280:39278–39283. doi: 10.1074/jbc.M509202200. [DOI] [PubMed] [Google Scholar]
- 5.Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 7.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 8.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 10.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G, Linsley PS. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- 11.Birmingham A, Anderson EM, Reynolds A, Ilsley-Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- 12.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Orum H, et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jackson AL, Burchard J, Leake D, Reynolds A, Schelter J, Guo J, Johnson JM, Lim L, Karpilow J, et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006 doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- 16.Petersen M, Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 17.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 18.Lewis DL, Hagstrom JE, Loomis AG, Wolff JA, Herweijer H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat. Genet. 2002;32:107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- 19.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- 20.Morrissey DV, Lockridge JA, Shaw L, Blanchard K, Jensen K, Breen W, Hartsough K, Machemer L, Radka S, et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 21.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl Acad. Sci. USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geiss G, Jin G, Guo J, Bumgarner R, Katze MG, Sen GC. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 2001;276:30178–30182. doi: 10.1074/jbc.c100137200. [DOI] [PubMed] [Google Scholar]
- 23.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat. Cell Biol. 2003;5:834–839. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 24.Braasch DA, Jensen S, Liu Y, Kaur K, Arar K, White MA, Corey DR. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- 25.Hrdlicka PJ, Babu BR, Sorensen MD, Wengel J. Interstrand communication between 2′-N-(pyren-1-yl)methyl-2′-amino-LNA monomers in nucleic acid duplexes: directional control and signalling of full complementarity. Chem. Commun. (Camb) 2004:1478–1479. doi: 10.1039/b404446k. [DOI] [PubMed] [Google Scholar]