Abstract
When an essential amino acid is limited, a signaling cascade is triggered that leads to increased translation of the ‘master regulator’, activating transcription factor 4 (ATF4), and resulting in the induction of specific target genes. Binding of ATF4 to the amino acid response element (AARE) is an essential step in the transcriptional activation of CHOP (a CCAAT/enhancer-binding protein-related gene) by amino acid deprivation. We set out to identify proteins that interact with ATF4 and that play a role in the transcriptional activation of CHOP. Using a tandem affinity purification (TAP) tag approach, we identified p300/CBP-associated factor (PCAF) as a novel interaction partner of ATF4 in leucine-starved cells. We show that the N-terminal region of ATF4 is required for a direct interaction with PCAF and demonstrate that PCAF is involved in the full transcriptional response of CHOP by amino acid starvation. Chromatin immunoprecipitation analysis revealed that PCAF is engaged on the CHOP AARE in response to amino acid starvation and that ATF4 is essential for its recruitment. We also show that PCAF stimulates ATF4-driven transcription via its histone acetyltransferase domain. Thus PCAF acts as a coactivator of ATF4 and is involved in the enhancement of CHOP transcription following amino acid starvation.
INTRODUCTION
Mammals have evolved complex adaptative mechanisms that enable cells to survive many stressful environmental conditions including amino acid limitation. The signal transduction pathway that is triggered in response to amino acid starvation is referred to as the amino acid response (AAR) (1). The initial step in the AAR is the activation by uncharged tRNAs of GCN2 kinase which phosphorylates the α subunit of translation initiation factor eIF2 (eIF2α) on serine 51 (2,3). This phosphorylation decreases the translation of most mRNAs by inhibiting the delivery of the initiator Met-tRNAi to the initiation complex. However, eIF2α phosphorylation also triggers the translation of specific mRNAs including the activating transcription factor 4 (ATF4). Once induced, ATF4 directly or indirectly induces transcription of specific target genes (4).
Among the genes induced via the GCN2/ATF4 pathway, the CCAAT/enhancer-binding protein homologous protein (CHOP) encodes a ubiquitous transcription factor that heterodimerizes avidly with the other members of the C/EBP and jun/fos families (5–7). The amino acid regulation of CHOP transcription involves a cis-acting element in the promoter that has been named amino acid response element (AARE) (8). This element is essential for the induction of CHOP transcription by amino acid starvation and functions as an enhancer element. In the past few years, several functional AAREs have been described in other genes including asparagine synthetase (ASNS) (9,10) and activating transcription factor 3 (ATF3) (11). The AARE sites of CHOP, ASNS and ATF3 have a 9 bp core element (5′-A/GTTG/TCATCA-3′) but the sequences differ by one or two nucleotides between genes.
It is now established that in amino acid-starved cells, a multiproteic complex is bound to the AARE sequences including a number of regulatory proteins such as ATF4 (9,11,12), C/EBPβ (13), ATF3 (10,12), activating transcription factor 2 (ATF2) (11,14) or tribbles-related protein 3 (TRB3) (15). These factors are involved in either inducing or repressing transcription of target genes in response to amino acid starvation. Importantly, all of the known AARE sites bind ATF4 whereas the binding activity and the role of other AARE-binding factors appear to vary according to the AARE sequence and chromatin structure. For example, CHOP and ATF3 sequences also bind ATF2 whereas the ASNS site does not (8,11,12,16).
The key role of ATF4 in amino acid-regulated transcription has been clearly established in the past few years (1,10,11,14). It has been shown that (i) the expression of ATF4 and its binding to AARE sequences are increased following amino acid starvation, (ii) kinetic of ATF4 binding to AARE are similar between tested genes with a dramatic increase in the first hour after a single amino acid removal, sustained over the next two hours, (iii) in cells devoid of ATF4 expression, the induction of mammalian genes upon amino acid starvation is completely lost and (iv) when over-expressed, ATF4 by itself is able to activate the AARE-dependent transcription. One major role of ATF4 is to mediate part of cell response to stress signals such as ER stress or amino acid deprivation (17). Both transcription and translation of ATF4 are selectively increased in stress conditions (18,19), resulting in the induction of many genes involved in amino acid metabolism or transport and in resistance to oxidative stress (4).
ATF4 belongs to the basic region/leucine zipper (bZIP) family of transcription factors, which also includes members of the Jun/Fos (AP-1) family (20,21). This factor is known to form heterodimers with members of AP1 and CCAAT/enhancer-binding protein (C/EBP) families (22–24) rather than proteins of the ATF/CREB family (25–27). Heterodimerization of ATF4 represents a powerful means to regulate its transcriptional activity and consequently the expression of target genes. ATF4 also interacts with coactivators such as p300 and CBP (28,29) and with several general transcription factors such as TBP, TFIIB, RAP30 (28) and RPB3 (30). In the context of the AAR, the heterodimeric partner of ATF4 on AARE remains to be identified. It has been suggested that ATF4 may also interact with one or more cofactors to make the promoter more accessible to the general transcription machinery (10) but these cofactors also remain to be identified.
The present study was designed to identify proteins interacting with ATF4 and playing a role in the transcriptional activation of CHOP in response to amino acid starvation. Recent progress in mass spectrometric protein sequencing technology together with the rapid growth of protein and genome databases have made direct approaches to map protein–protein interactions feasible. Using a tandem affinity purification (TAP) tag approach, we identify p300/CBP-associated factor (PCAF) as a novel interaction partner of ATF4 in amino acid-starved cells. Our results provide evidence that PCAF acts as a coactivator of ATF4 and is involved in the enhancement of CHOP transcription following amino acid starvation.
MATERIALS AND METHODS
Plasmid constructions
2X-CHOP-AARE-TK-LUC was generated as previously described (8). To express PCAF in mammalian cells, plasmids for wild-type (pCX-Flag-PCAF) and HAT-defective PCAF (pCX-Flag-PCAF ΔHAT) were provided by Chao-Zhong Song (University of Washington, Seattle, WA). To express PCAF in vitro, pCI-Flag-PCAF was kindly given by Rosemary Kiernan (Montpellier, France). The expression plasmid for the ATF4 cDNA was a gift of Irina Lassot (Institut Cochin, Paris, France). To generate the GST–ATF4 (amino acids 1–351) and GST–ATF4 (amino acids 1–100) fusion proteins, the corresponding regions of human ATF4 cDNA were amplified by PCR and inserted into the BamHI/EcoRI sites of pGEX-4T-1 (Amersham). Constructs including other ATF4 deletion mutants fused to GST (glutathione-S-transferase) were kindly provided by Florence Margottin-Goguet (Institut Cochin, Paris, France). The mammalian ATF4 expression plasmid used in the TAP technique (pZome-1-N-TAP-ATF4) was generated by inserting the full-length coding region of human ATF4 cDNA into the EcoRI site of pZome-1-N (Euroscarf, Germany).
Cell culture and treatment conditions
HeLa cells, mouse embryonic fibroblasts (MEF) and retroviral packaging cell line BOSC23 were cultured at 37°C in Dulbecco's modified Eagle's medium F12 (DMEM F12) (Sigma) containing 10% fetal bovine serum. When indicated, DMEM F12 lacking leucine was used. In all experiments involving amino acid starvation, 10% dialyzed calf serum was used. MEF deficient in ATF4 were kindly given by David Ron (Skirball Institute of Biomolecular Medicine, New York) (4).
Generation of stable cell lines
Retroviral infection was performed as described (31). BOSC23 cells were transfected with either pZome-1-N (mock) or pZome-1-N-TAP-ATF4. After 48 h of transfection, the medium containing retroviruses was collected, filtered, treated by polybrene (1 mg/ml) and transferred on ATF4 −/− MEF. Infected cells were selected with puromycin (2 mg/ml) for 3 weeks. The expression of TAP-ATF4 was analyzed by immunoblotting analysis with an anti-ATF4 antibody.
Antibodies
The antibodies against ATF4 (sc-200), PCAF (sc-8999) and β-actin (sc-7210) were purchased from Santa Cruz Biotechnology, Inc (Santa Cruz, CA, USA).
Nuclear extract preparation
Nuclear extracts were prepared from HeLa cells and MEF as described previously (8).
TAP purification
TAP–ATF4 complexes were purified using a published procedure (32) with minor modifications. Nuclear extracts were prepared from thirty 150-mm plates of mock or ATF4–TAP-transfected cells and subsequently adjusted to IgG-binding conditions: 180 mM NaCl, 10 mM Tris–HCl pH 8.0, 0.2% NP-40, 0.5 mM dithiothreitol (DTT), complete protease inhibitors (Sigma), 10 mM β-glycerophosphate and 20 mM NaF. Diluted extracts were rotated overnight at 4°C with 100 μl of IgG matrix (Amersham Biotech), after which the beads were washed extensively in binding buffer. Washed beads were re-suspended in TEV cleavage buffer (10 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.3% NP-40, 0.5 mM EDTA, 0.5 mM DTT), and 5–15 μl of recombinant TEV enzyme (Invitrogen) was added to the mixture. After 2 h of rotation at 16°C, the TEV eluate from the IgG column was recovered and adjusted to calmodulin-binding conditions: 150 mM NaCl, 45 mM Tris–HCl pH 8.0, 0.7 mM Mg-acetate, 0.7 mM imidazole, 2.5 mM CaCl2, 0.2% NP-40, 10 mM β-mercaptoethanol and rotated for 2 h at 4°C with 50 μl of calmodulin affinity resin (Stratagene). After binding, sedimented beads were washed extensively with calmodulin-binding buffer. Bound proteins were recovered by boiling the calmodulin beads for 5 min in protein sample buffer.
Sample preparation and mass spectrometric analysis
The protein complexes recovered from TAP purification were fractionated on a 7% SDS–polyacrylamide gel. Proteins were detected by silver staining. The protein bands on 1D gel were excised from gels using blade of scapel. The bands were washed with 100 µl of 25 mM NH4HCO3 for 30 min, destained with 100 µl of 25 mM NH4HCO3/acetonitrile (v/v) twice 30 min and dehydrated in acetonitrile. Bands were completely dried using a speed vac before trypsin digestion. The dried gel volume was evaluated and three volumes of trypsin (V5111; Promega, Madison, WI, USA), 10 ng/µl in 25 mM NH4HCO3 were added. Digestion was performed at 37°C during 5 h. The gels pieces were centrifuged and 8–12 µl of acetonitrile (depending of gel volume) were added to extract peptides. The mixture was sonicated for 5 min and centrifuged. For MALDI-TOF MS analysis, 1 µl of supernatant was loaded directly onto the MALDI target. The matrix solution (5 mg/ml -cyano-4-hydroxycinnamic acid in 50% acetonitrile/0.1% trifluoroacetic acid) was added immediately and allowed to dry at room temperature. A Voyager DE-Pro model of MALDI-TOF mass spectrometer (Perseptive BioSystems, Farmingham, MA, USA) was used in positive-ion reflector mode for peptide mass fingerprinting. External calibration was performed with a standard peptide solution (Proteomix, LaserBio Labs, Sophia-Antipolis, France). Internal calibration was performed using peptides resulting from auto-digestion of porcine trypsin. Monoisotopic peptide masses were assigned and used from NCBI database searches with the ‘Mascot’ and ‘Profound’ softwares (http://www.matrixscience.com and http://prowl.rockefeller.edu).
GST pull-down experiments
Fresh overnight cultures of BL21 pLysS Escherichia coli strain transformed with GST-fused constructs were diluted 1:10 in LB medium containing ampicillin (100 μg/ml). Isopropyl-1-thio-β-d-galactopyranoside was added in growing exponential bacterial culture to a final concentration of 1 mM and incubated for 4 h at 30°C. Cells were re-suspended in STE buffer (10 mM Tris pH 8, 150 mM NaCl, 1 mM EDTA). After 10 000g centrifugation during 10 min at 4°C, pellets were frozen during 5 min, re-suspended in STE buffer containing 1 mM dithiothreitol and 10% sarcosyl. Lysates were sonicated for 1 min and clarified at 10 000g for 10 min at 4°C. The bacterial supernatant was rocked overnight at 4°C with glutathione-Agarose resin (Sigma) and beads were washed three times in PBS containing Triton X-100 (0.1%) and PMSF (1 mM). 35S-labeled PCAF protein was generated in vitro using the TNT T7-coupled reticulocyte lysate system (Promega) according to the manufacturer's instructions. 35S-labeled PCAF protein or 1 mg of nuclear extracts were incubated with the beads in binding buffer (20 mM HEPES pH 7.4, 125 mM NaCl, 0.1% Triton X-100, 2 mM DTT), 2 mM ethylenediamine tetra-acetic acid (EDTA), 10 μM ZnCl2, complete protease inhibitors (Sigma) by rocking 2 h at 4°C. The glutathione-Agarose beads were then washed four times in PBS buffer containing 0.1% Triton X-100 and 160 mM NaCl and re-suspended in Laemmli buffer. Proteins were released from beads by boiling 5 min, and subjected to SDS–PAGE analysis. Fractionated proteins were visualized by western blot using anti-PCAF or anti-ATF4 antibodies or by autoradiography using a PhosphorImager and IMAGEQUANT software (Molecular Dynamics).
Transient transfection and luciferase (LUC) assay
Cells were plated in 12-well dishes and transfected by the calcium phosphate coprecipitation method as described previously (12). One microgram of luciferase plasmid was transfected into the cells along with 0.05 µg of pCMV-βGal, a plasmid carrying the bacterial β-galactosidase gene fused to the human cytomegalovirus immediate-early enhancer/promoter region, as an internal control. Cells were then exposed to the precipitate for 16 h, washed twice in phosphate-buffered saline (PBS), and then incubated with DMEM F12 containing 10% fetal bovine serum. Two days after transfection, cells were harvested in 100 µl of lysis buffer (Promega) and centrifuged at 13 000g for 2 min. Twenty micro liters of the supernatant were assayed for luciferase activity (YELEN, Ensue La Redonne, France). For all the transfection experiments presented, a plasmid pCMV-βGal was used as an internal control. β-Galactosidase activity was measured as described previously (12). Relative luciferase activity was given as the ratio of relative luciferase unit/relative β-Gal unit. All values are the means calculated from the results of at least three independent experiments performed in triplicate.
Analysis of gene expression using real-time RT–PCR
Total RNA was prepared using a RNeasy mini kit (Qiagen) and treated with DNase I, Amp Grade (InVitrogen) prior to cDNA synthesis. RNA integrity was electrophoretically verified by ethidium bromide staining. RNA (0.5 μg) was reverse transcribed with 100 U of Superscript II plus RNase H- Reverse Transcriptase (InVitrogen) using 100 μM random hexamer primers (Amersham Biosciences), according to the manufacturer's instructions. To measure the relative amount of human CHOP, ATF4 and PCAF mRNA, primers were the following: hCHOP (forward primer, 5′-cagaaccagcagaggtcaca-3′ reverse primer, 5′-agctgtgccactttcctttc-3′), hATF4 (forward primer, 5′-aaccgacaaagacaccttcg-3′; reverse primer, 5′-acccatgaggtttgaagtgc-3′) and hPCAF (Qiagen, QT00092267 #). All the primers yielded PCR products 200 bp in size. To control for RNA quality and cDNA synthesis, β-actin mRNA was also amplified with forward (5′-ctcgcaggtcaagagcaag-3′) and reverse primers (5′-gacagctgctccaccttctt-3′). To measure the transcriptional activity from the CHOP gene, oligonucleotides derived from CHOP intron 1 and exon 1 were used to measure the short-lived unspliced transcript (hnRNA, heterogenous nuclear RNA). This procedure for measuring transcriptional activity is based on that described by Lipson and Baserga (33). The primers for amplification were: forward primer, 5′-aaggcactgagcgtatcatgt-3′; reverse primer, 5′-ctctcggacggtccctaact-3′. Quantification involved the use of standard curves that had been prepared with plasmids containing specific sequences of each gene. We cloned all the PCR products into the pGEM-T easy vector (Promega) according to the manufacturer's instructions. For the construction of standard curves, pGEM-T easy plasmids were prepared as 10-fold serial dilution in water, from 4 ng to 0.4 pg. PCR was carried out using a LightCycler™ System (Roche) as described previously (12). LightCycler quantification software (version 3.5) was used to compare amplification in experimental samples during the log–linear phase to the standard curve from the dilution series of control plasmids. Relative results were displayed in nanograms of target gene per 100 ng of β-actin. Each experiment was repeated three times to confirm the reproducibility of the results.
Chromatin immunoprecipitation analysis (ChIP)
ChIP analysis was performed according to the protocol of Upstate Biotechnology, Inc. (Charlottesville, VA, USA) with minor modifications. Cells were seeded at 1 × 106/100 mm dish with DMEM F12 and grown for 24 h. Cells were transferred to fresh DMEM F12 12 h before transfer to either complete DMEM F12 or DMEM F12 lacking leucine for the time period indicated in each figure. Protein–DNA was cross-linked by adding formaldehyde directly to the culture medium to a final concentration of 1% and then stopped 8 min later by the addition of glycine to a final concentration of 0.125 M. Cross-linked chromatin was sonicated using a Vibra cell sonicator (Biobloc Scientific Technology) for 10 bursts of 30 s at power 2 with 1-min cooling on ice between each burst to obtain DNA fragments of an average of 400 bp. Extracts from 1 × 106 cells were incubated with 5 µg of antibody. A rabbit anti-chicken IgG was used as the nonspecific antibody control. The antibody-bound complex was precipitated by protein A-Agarose beads (Upstate Biotechnology). The DNA fragments in the immunoprecipitated complex were released by reversing the cross-linking overnight at 65°C and purified using a phenol/chloroform extraction and ethanol precipitation. Real-time quantitative PCR was performed by using a LightCycler (Roche) and a SYBR-Green-I-containing PCR mix (Qiagen), following the recommendations of the manufacturer. The immunoprecipitated material was quantified relative to a standard curve of genomic DNA. Primers used for human sequences: hCHOP amplicon A, 5′-gcagcctaaccaaagacctg-3′ and 5′-ggaggcaacttgaccaaaag-3′; hCHOP amplicon B (AARE), 5′-aagaggctcacgaccgacta-3′ and 5′-atgatgcaatgtttggcaac-3′; hCHOP amplicon C, 5′-agtgccacggagaaagctaa-3′ and 5′-ccatacagcagcctgagtga-3′. Primers used for mouse sequences: mCHOP AARE, 5′-gggcagacaagttcaggaag-3′ and 5′-atgatgcaatgtttggcaac-3′. The reactions were incubated at 95°C for 15 min to activate the polymerase, followed by amplification at 95°C for 15 s, 55°C for 20 s and 72°C for 20 s for 45 cycles. After PCR, melting curves were acquired by stepwise increases in the temperature from 65 to 95°C to ensure that a single product was amplified in the reaction. The results are expressed as the percentage of antibody binding versus the amount of PCR product obtained using a standardized aliquot of input chromatin. Samples are the means from at least three independent immunoprecipitations.
SiRNA preparation and transfection
SiRNA corresponding to PCAF mRNA (5′-ucgccgugaagaaagcgcadTdT-3′ and 5′-ugcgcuuucuucacggcgadTdT-3′) (1024864 #) and to control (1027280 #) were from Qiagen. Annealing was performed as described by the manufacturer: the complementary two strands (each 5 nmol) in 250 μl of siRNA suspension buffer (Qiagen) were heated 1 min at 90°C and then incubated for 1 h at 37°C. One day before transfection with siRNA, HeLa cells were plated in 6-well plates at 25% confluency. Then 30 pmol of siRNA were introduced into the cells using the calcium phosphate precipitation as described above. Forty-eight hours after transfection, the expression level of PCAF was analyzed by western blotting.
Immunoblot analysis
Cells were lyzed in radioimmune precipitation assay buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, 50 mM NaF, 2 mM Na3VO4, 100 nM acid okadaic, 25 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, protease inhibitor cocktail from Sigma), then proteins were resolved by SDS–PAGE and transferred onto a Hybond-P PVDF membrane (Amersham Biosciences). Membranes were blocked for 1 h at room temperature with a solution of 5% nonfat milk powder in TN (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 0.1% Tween-20). The blots were then incubated with primary antibody in blocking solution overnight at 4°C. Antibodies were diluted according to the manufacturer's instructions. The blots were washed three times in TN and incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (1:5000) (Santa Cruz, CA, USA) in blocking buffer for 1 h at room temperature. After three washes, the blots were developed using the enhanced chemiluminescence (ECL) detection system (Amersham Biosciences).
RESULTS
Identification of PCAF as an interaction partner of ATF4 by TAP
To identify ATF4 interacting proteins in mammalian cells, we performed TAP coupled with mass spectrometry (34). We developed a mammalian expression vector coding for a fusion protein consisting of amino acids 1–351 of ATF4 linked to the TAP tag (Supplementary Figure 1A). In this construct, the TAP tag consists of the protein A (Prot.A) and the calmodulin-binding peptide affinity sequences that are separated by the recognition sequence for tobacco etch virus (TEV) protease, permitting proteolytic elution of the fusion protein from the IgG affinity resin (32,34). The constructs expressing the ATF4 fusion protein (TAP-ATF41–351) or the tag alone (TAP) were stably transfected into ATF4 −/− mouse MEF. We chose ATF4-deficient cells because absence of endogenous ATF4 expression was expected to increase purification efficiency. Supplementary Figure 1B shows that in ATF4 −/− MEF expressing TAP proteins, the endogenous form of ATF4 was not detected in the absence of leucine (compare lanes 3–5 with lane 2). To check the functionality of TAP-ATF41–351 protein, a LUC reporter driven by two copies of the CHOP AARE was transiently transfected into ATF4 −/− MEF. Supplementary Figure 1C shows that this fusion protein activated the AARE-dependent transcription and produced about the same transcriptional response as obtained with the wild-type form of ATF4. Cells expressing TAP proteins were incubated for 2 h in control medium or in medium lacking leucine, and nuclear extracts were prepared and applied to dual affinity chromatography according to the TAP protocol (32). Bands representing putative ATF4-binding proteins were purified by SDS–PAGE and analyzed by MALDI-TOF (data not shown). Comparison of the obtained peptide sequences with protein databases identified several proteins that had been previously linked to RNA transcription. Here we report the identification of p300/CBP-associating factor (PCAF) as a novel interaction partner of ATF4 in leucine-starved cells.
The N-terminal region of ATF4 is required for a direct interaction with PCAF
To confirm that PCAF can be a partner of ATF4, in vitro pull-down assays were performed. Bacterially expressed GST-ATF4 (amino acids 1–351) fusion protein was immobilized on glutathione beads and later incubated with nuclear extracts from HeLa cells starved for 2 h with leucine. Immunoblot analysis revealed that PCAF present in nuclear extracts from leucine-starved HeLa cells was retained specifically with the full-length ATF4 (Figure 1A). No interaction of PCAF with GST alone could be detected. These results show that the interaction observed by the TAP method can also be reconstituted in vitro.
Figure 1.
PCAF interacts directly with the N-terminal region of ATF4. (A) Bacterially expressed GST-ATF4 (amino acids 1–351) fusion protein was immobilized on glutathione beads and incubated with nuclear proteins from leucine-starved HeLa cells (2 h). After extensive washing, proteins bound to the beads were eluted in protein sample buffer and analyzed by western blotting with anti-PCAF (top) or anti-ATF4 (bottom) antibodies. The expression of the endogenous (50 kDa) and the recombinant ATF4 proteins (75 kDa) are visualized. Ten percent of the nuclear extracts used in the pull-down experiments were loaded in the Input lane 3. (B) Schematic representation of the ATF4 deletions employed in the experiments reported in (C) and (D). The basic region/leucine zipper (bZip) and the N-terminal (N-ter region) domains are indicated. (C) Nuclear extracts from leucine-starved HeLa cells (2 h) were used in GST pull-down experiments with bacterially expressed full length ATF4 wt (lane 1) or various deletion mutants fused to GST (lanes 2–5). PCAF bound to the GST–ATF4 constructs was detected by western blot with anti-PCAF antibody. Ten percent of the nuclear extracts used in the pull-down experiments were loaded in the Input lane 7. The amount of GST-fused recombinant proteins was monitored by Coomassie blue staining. (D) In vitro pull-down assay of 35S-labeled PCAF against full-length (lane 2), or two ATF4-deletion mutants (lanes 3 and 4) fused to GST. The input (lane 1) was loaded with the amount of 35S-labeled proteins used in the binding reactions. The amount of GST-fused recombinant proteins was monitored by Coomassie blue staining, and radioactive signals of radiolabeled proteins were analyzed using a phosphorimaging device.
To find which domain of ATF4 is required for the interaction with PCAF, truncated ATF4 derivatives were used in GST pull-down assays (Figure 1B). As shown in Figure 1C, ATF4 deleted of residues 282–351 including the bZIP domain (lane 2) retained PCAF-binding capacity. By contrast, ATF4 deleted of residues 1–85 (lane 3) or other larger deletion mutants (lanes 4 and 5) did not interact with PCAF like GST alone (lane 6) used as a control, suggesting that the N-terminal region of ATF4 is required for the interaction with PCAF. From these results, we cannot exclude the possibility that the ATF4–PCAF interaction is not direct and requires specific accessory factors present in eukaryotic cell nuclear extracts. To demonstrate the direct interaction between PCAF and the N-terminus of ATF4, we monitored the binding of 35S-PCAF produced in vitro to full-length, N-terminal (amino acids 1–100) or ATF4 deleted of residues 1–85 fused to GST (Figure 1D). Only the full-length and the 1–100 N terminal derivatives (lanes 2 and 3) showed consistent interaction with PCAF. These data demonstrate that ATF4–PCAF interaction occurs through a direct interaction involving the N-terminal region of ATF4.
Effect of amino acid starvation on PCAF mRNA and protein
To further identify the role of PCAF in amino acid-regulation of CHOP transcription in human cells, we first examined the effect of leucine starvation on the PCAF mRNA and protein levels in HeLa cells. Kinetic analysis of mRNA level indicated that PCAF mRNA was not affected by amino acid starvation while ATF4 and CHOP mRNA were increased (Figure 2A). Protein analysis showed that the expression of PCAF was not significantly affected by 1–2 h of leucine starvation. However, PCAF level was greatly reduced following 4–8 h of amino acid starvation while ATF4 level was markedly increased (Figure 2B).
Figure 2.
Measurement of PCAF mRNA and protein accumulation in amino acid-starved cells. HeLa cells were incubated either in control (+) or leucine-free medium (−) and harvested for protein extraction and RNA isolation after the indicated incubation times. (A) Total RNA was extracted and real-time RT–PCR was performed as described in Materials and Methods section. The mRNA induction level is defined as the ratio of the relative mRNA level of leucine-starved cells to that of non-starved cells. (B) PCAF, ATF4 and β-Actin protein contents were analyzed by western blots as described in Materials and Methods section.
Binding of ATF4 to the CHOP AARE in vivo is associated with binding of PCAF in response to amino acid starvation
Using a ChIP analysis, we had previously demonstrated that following amino acid starvation, ATF4 binds to the AARE sequence within the CHOP promoter in vivo (14). To determine whether PCAF also targets the CHOP AARE, HeLa cells were incubated in control or leucine-free medium for 2 h and ChIP assays were performed with primer sets covering either the 5′ region (amplicon A), the AARE (amplicon B) or the first intron (amplicon C) of the CHOP gene (Figure 3A). The results show recruitment of both PCAF and ATF4 to the AARE following 2 h of leucine deprivation (Figure 3B). In addition, bindings of PCAF and ATF4 were not detected in the 5′ region or in the first intron of CHOP, confirming that both factors are specifically engaged on the AARE.
Figure 3.
PCAF recruitment to CHOP AARE in response to leucine starvation. (A) Scheme of the human CHOP gene indicating the different amplicons produced for the ChIP analysis: A (−1678 to −1478), B (−472 to −301) and C (+1163 to +1372). The AARE is boxed in gray. (B) HeLa cells were incubated 2 h either in control (+leu) or leucine-free medium (−leu) and harvested. ChIP analysis was performed as described under Materials and Methods section using antibodies specific for PCAF and ATF4 and different sets of primers to produce amplicon A, B or C. Data were plotted as the percentage of antibody binding versus the amount of PCR product obtained using a standardized aliquot of input chromatin. Each point represents the mean value of three independent experiments, and the error bars represent the SEM. (C) Time course of PCAF and ATF4 recruitments during leucine starvation. HeLa cells were incubated either in control (+leu) or leucine-free medium (−leu) and harvested for 0–8 h. ChIP analysis was performed using antibodies specific for PCAF and ATF4 and a set of primers to amplify amplicon B (see above). Total RNA was extracted and the CHOP transcriptional activity was determined by real-time RT–PCR analysis of CHOP pre-mRNA as described under Materials and Methods section. The dotted line represents the increase in CHOP pre-mRNA induction level.
We then investigated the kinetics of PCAF engagement on the CHOP AARE in response to amino acid starvation. PCAF recruitment increased slightly after 1 h of leucine deprivation, peaked at 2 h and fell within 2–8 h of amino acid deprivation (Figure 3C). Comparison of these kinetics with those obtained for ATF4 binding reveals a similarity in the time courses of recruitment of these two factors. Also, by plotting the pre-mRNA content on the same graph, it is apparent that the engagement of PCAF and ATF4 closely paralleled the increase in CHOP transcription in the first 2 h.
ATF4 is essential for PCAF recruitment to the CHOP AARE following amino acid starvation
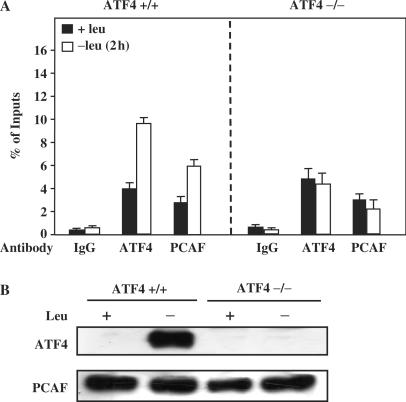
In previous studies, ATF4 was shown to be essential for CHOP induction in response to leucine starvation (12). The results described above suggest that ATF4 may be involved in PCAF recruitment to CHOP AARE following amino acid starvation. To investigate the link between binding of ATF4 to the CHOP AARE and the recruitment of PCAF, ChIP experiments were performed in MEFs deficient in ATF4 and in the corresponding wild-type cells. The ChIP results obtained with wild-type MEFs are consistent with those described above with HeLa cells (Figure 4A). By contrast, in cells lacking ATF4, the increase in PCAF binding to the CHOP AARE was lost. Protein analysis shows that the lack of ATF4 did not affect the level of PCAF expression (Figure 4B). Taken together, these results demonstrate that ATF4 is essential for the recruitment of PCAF on the CHOP AARE following amino acid starvation.
Figure 4.
Role of ATF4 in PCAF recruitment to CHOP AARE in response to leucine starvation. ATF4 +/+ and ATF4 −/− MEF were incubated 2 h either in control (+leu) or leucine-free medium (−leu) and harvested. (A) ChIP analysis was performed as described under Materials and Methods section using antibodies specific for PCAF and ATF4 and a set of primers to produce amplicon B (Figure 3A). Data were plotted as the percentage of antibody binding versus the amount of PCR product obtained using a standardized aliquot of input chromatin. Each point represents the mean value of three independent experiments and the error bars represent the SEM. We note that there remains 5% of ATF4 antibody binding in ATF4 KO cells. ChIP experiments were also performed with primer sets reaching much farther upstream or downstream from the CHOP gene (data not shown). The results indicate that the amount of ATF4 binding in ATF4-deficient cells was due to the background observed for the ATF4 antibody. (B) Western blot analysis of ATF4 and PCAF was performed from nuclear extracts.
PCAF stimulates ATF4-driven transcription via its histone acetyltransferase (HAT) domain
Several studies have shown that PCAF is a transcription coactivator with intrinsic acetylase activity (35). Having established that ATF4 recruits PCAF on the CHOP AARE, we sought to determine whether PCAF functioned as a coactivator of ATF4 in AARE-dependent transcription. Cotransfection experiments were carried out in HeLa cells using a LUC reporter driven by two copies of the CHOP AARE and the expression plasmids for ATF4 and PCAF or their respective empty vectors. This assay revealed that PCAF stimulated ATF4-driven transcription but had no effect by itself on luciferase expression (Figure 5). By contrast, an HAT-defective PCAF containing a deletion of amino acids 497–526 (36) failed to stimulate ATF4-driven transcription. These results demonstrate that PCAF functions as a coactivator of ATF4 and show that PCAF HAT activity is required for ATF4/PCAF synergistic activation of the AARE-dependent transcription.
Figure 5.
Role of PCAF in the stimulation of ATF4 transcriptional activity. HeLa cells were transiently transfected with a luciferase construct containing two copies of the CHOP AARE inserted 5′ to the TK promoter (2X-CHOP-AARE-TK-LUC), the expression vector for ATF4, PCAF or HAT-defective mutant PCAF (PCAFΔHAT) or the empty vector, as indicated. Two days after transfection, cells were harvested for preparation of cell extracts and determination of LUC activity. Results are given as the ‘fold induction’ relative to the cells transfected with empty vector. For all the transfection experiments presented, a plasmid pCMV-βGal was used as an internal control. Relative luciferase activities were determined as described in Materials and Methods section. Each data represents the mean of at least three independent experiments performed in triplicate.
PCAF is required for the full transcriptional response of CHOP to leucine starvation
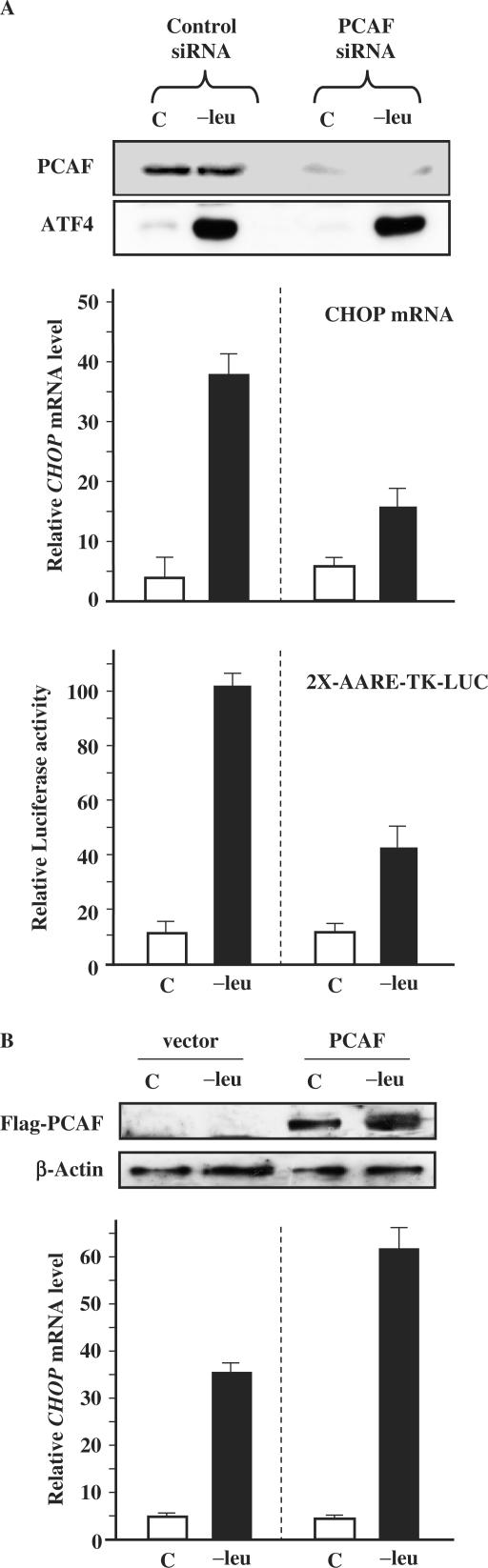
To assess the role of PCAF in the amino acid regulation of CHOP expression, we first measured the effect of leucine starvation on both CHOP mRNA content and AARE-dependent transcription in PCAF-deficient cells. We employed small interfering double-stranded RNA (siRNA) transfection to specifically inhibit the endogenous expression of PCAF. HeLa cells were transfected with either siPCAF or control siRNA, and then incubated with either control or leucine-free medium for 2 h. Figure 6A shows that PCAF-siRNA transfection dramatically decreased the PCAF protein content but did not affect the increase in ATF4 expression. Lack of PCAF affected the response of CHOP transcription to leucine depletion: the induction of CHOP mRNA and the AARE-dependent transcription were significantly reduced. In control siRNA-transfected cells, the response to leucine starvation was not affected. We then examined the effect of over-expressing PCAF on the increase in CHOP mRNA in response to leucine starvation (Figure 6B). HeLa cells were transiently transfected with PCAF or the empty vector and then incubated either with control or leucine-free medium for 2 h. Over-expression of PCAF protein leads to a significant increase in the amino acid inducibility of CHOP. Taken together, these results demonstrate that PCAF is required to obtain maximal induction of CHOP by leucine starvation.
Figure 6.
Effect of PCAF knockdown or PCAF over-expression on the amino acid regulation of CHOP expression. (A) Effect of PCAF knockdown. HeLa cells were transfected with PCAF siRNA or control siRNA. Two days after siRNA transfection, cells were incubated for 2 h in control (C) or in medium lacking leucine (−leu) and then harvested to extract RNA and proteins. Total RNA was extracted and real-time RT–PCR was performed as described in Materials and Methods section. PCAF and ATF4 protein contents were analyzed by western blot. One day after siRNA transfection, cells were transfected with 2X-CHOP-AARE-TK-LUC reporter construct to measure the AARE-dependent transcription. After two days, cells were incubated for 16 h in control (C) or in medium lacking leucine (−leu) and cells were then harvested. LUC activity was measured as described in Materials and Methods section. (B) Effect of PCAF over-expression. HeLa cells were transiently transfected with Flag-PCAF expression construct (PCAF) or with empty vector (vector). Two days after transfection, cells were incubated for 2 h in control (C) or in medium lacking leucine (−leu) and then harvested to extract RNA and proteins. Total RNA was extracted and real-time RT–PCR was performed as described in Materials and Methods section. Each data represents the mean of at least three independent experiments performed in triplicate. Flag-PCAF and β-Actin protein contents were analyzed by western blot.
DISCUSSION
Mammalian cells have evolved complex cellular responses to stress conditions. Both transcription and translation of ATF4 are selectively increased in response to amino acid deprivation (17), even when global protein synthesis is repressed, resulting in the induction of a wide variety of ATF4 target genes (1). The data reported in the present study yield several novel findings regarding the mechanisms by which ATF4 activates gene transcription upon amino acid starvation: (i) we have found evidence that the N-terminal region of ATF4 interacts directly with PCAF in amino acid-starved cells, (ii) we demonstrate that PCAF is involved in enhancing the transcriptional response of CHOP by amino acid starvation, (iii) we establish that PCAF is recruited on the CHOP AARE in response to amino acid starvation and that ATF4 is essential for its recruitment and (iv) we show that PCAF enhances ATF4-driven transcription via its HAT domain.
PCAF has been described as a coactivator that mediates the transcription of many genes (37). Like a number of transcriptional coactivators, this factor possesses an intrinsic histone acetylase activity (38,39). The role of PCAF in transcription has been investigated in multiple studies, and its requirement as a HAT and coactivator has been described for nuclear receptor- (40,41) and growth factor-mediated (42) activations and for myogenesis (43) among other processes. Here we report evidence that PCAF functions as a coactivator of ATF4 in the transcriptional response of CHOP following amino acid starvation. Like several nuclear proteins such as CBP and p300, PCAF interacts directly with the ATF4 N-terminal domain, shown to be a transcriptional activation domain (28). We also show that the HAT activity of PCAF is required for enhancing the activation of the AARE-dependent transcription by ATF4. PCAF preferentially acetylates lysine 14 of histone H3 but also less efficiently acetylates lysine 8 of histone H4 (44). At present, the exact role of the HAT activity of PCAF in promoting the AARE-dependent transcription by ATF4 remains to be established. However, there are several lines of evidence suggesting that PCAF is not involved in histone acetylation at the CHOP promoter. First, we recently reported that the acetylation status of histone H3 remained unchanged at the CHOP AARE region within 1 h of removal of leucine from the medium while the acetylation of H4 is increased (14). Second, in ATF4-deficient cells, where the recruitment of PCAF was completely lost, we had also previously shown that the level of histone H4 acetylation remained elevated (14). Last, we now show that the HAT activity of PCAF is required to stimulate the transcriptional activity of ATF4 on a non-integrated CHOP AARE-reporter bacterial plasmid. In addition to histones, a number of transcription factors are also substrates for acetylation by nuclear HAT (45–47). The consequences of acetylation on protein function range from one protein to another depending on where in the protein the acetylation takes place. Acetylation has been reported to modulate protein–protein interactions, inhibit nuclear export (48) and alter protein stability (29). CBP and p300 were described as acetylating ATF4 in its bZIP domain and enhancing its transcriptional activity (28,29). In addition, Gachon et al. (49) have found evidence that acetylation of ATF4 in vitro is mediated by p300 but not by PCAF. In our model, the target of the PCAF HAT domain in the transcriptional activation of CHOP upon amino acid starvation remains to be identified.
The present experiments demonstrate that ATF4 binding is essential for the transitory recruitment of PCAF on the CHOP AARE following amino acid starvation. PCAF recruitment on the CHOP AARE falls within 2–8 h of leucine deprivation, while CHOP transcription is still increased. Therefore, ATF4-mediated PCAF recruitment is essential in enhancing the transcription of CHOP in response to a short period of amino acid starvation. It is possible that another cofactor may be involved in the ATF4-dependent response of CHOP during long-term amino acid deprivation. The drop in PCAF recruitment might be explained by the decrease in PCAF expression level observed following 4–8 h of amino acid starvation, while ATF4 is still bound to the AARE. The ubiquitin-proteasome degradation pathway plays an important role in transcription regulation to assure the controlled and timely termination of signaling by irreversible destruction of the activated transcription regulators. PCAF has been shown to be a target for the E3 ubiquitin ligase MDM2 (50). However, the enhancement of the PCAF degradation by leucine starvation remains to be demonstrated.
Our present findings provide evidence that PCAF is required to obtain maximal induction of CHOP transcription in response to leucine starvation. We have recently reported that following amino acid starvation, phosphorylation of ATF2 at the CHOP AARE occurs prior to ATF4 binding, histone acetylation, and increase in CHOP mRNA (14). We have further shown that ATF2 is involved in promoting the modification of the chromatin structure to enhance CHOP transcription (14). ATF2 was not identified in our ATF4-TAP screen. We have recently examined the formation of the ATF2/ATF4 heterodimer by in vitro translated proteins in gel shift assays. ATF2 and ATF4 do not form a heterodimer that binds the CHOP AARE sequence (data not shown). Therefore, it is unlikely that ATF2 and ATF4 interact in vivo on the CHOP promoter. Although it is well established that ATF2 can interact directly with p300 and CBP coactivators (51,52), the interaction of PCAF with ATF2 on the AARE remains to be shown. ATF4 may also participate in the trigger mechanism of CHOP transcriptional activation, promoting further recruitment of activator proteins such as PCAF to the promoter. Among the potential ATF4-binding proteins we have identified, PCAF is the only one with HAT activity. Using a ChIP approach, we have recently observed that p300 and CBP are present constitutively in the CHOP AARE-binding complex (data not shown). Whether these coactivators interact directly with ATF4 is unanswered. Further experiments will be required to study their role in the amino acid regulation of CHOP transcription. Taken together, the results demonstrate that following amino acid starvation there is a highly coordinated time-dependent program of interaction between a precise set of ATF subfamily members and coactivators leading to transcriptional activation of CHOP.
ATF4 has been shown to be a master regulator of a number of amino acid-regulated gene transcription such as CHOP and ASNS (1). Although CHOP and ASNS AARE sequences exhibit some structural and functional similarities, there are significant differences in the molecular mechanisms involved in the induction of CHOP following amino acid starvation and those described for ASNS. Using a ChIP approach, Chen et al. (10) did not observe any significant recruitment of PCAF to the ASNS promoter in response to amino acid limitation. We show here that PCAF is recruited specifically to the CHOP AARE to enhance the ATF4 transcriptional activity. By contrast, in the context of the ASNS AARE sequences, we also observe that ATF4 did not require PCAF to activate ASNS transcription in response to amino acid starvation (data not shown). As suggested by Chen et al. (10), ATF4 may act as a recruiting factor for an unknown HAT activity making the ASNS promoter more accessible to RNA Pol II and the general transcription machinery, but it is clear that PCAF is not involved. It is possible that another ATF4-interacting factor not present in the ASNS AARE-binding complex may also be essential for PCAF recruitment on the CHOP AARE. All these data suggest that although most of the amino acid-responsive genes have AARE sites that are similar in sequence, the key regulator ATF4 and other distinct transcription factors and coactivators may be involved in modulating transcriptional activation. These differences in mechanism would permit flexibility among amino acid-regulated genes in the rapidity and magnitude of the transcriptional response for the same initial signal. Further insight into how the transcriptional machinery assembles at the amino acid-responsive gene promoter and modulates transcription will improve our understanding of the molecular steps required for nutritional control by the amino acid response pathway.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Sylvère Baron and Laurent Leotoing for helpful discussions about GST pull-down assays. This work was supported by grants from the Institut National de la Recherche Agronomique and the Région Auvergne. Funding to pay the Open Access publication charges for this article was provided by the Institut National de la Recherche Agronomique.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kilberg MS, Pan YX, Chen H, Leung-Pineda V. Nutritional control of gene expression: how mammalian cells respond to amino acid limitation*. Annu. Rev. Nutr. 2005;25:59–85. doi: 10.1146/annurev.nutr.24.012003.132145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimball SR, Jefferson LS. Amino acids as regulators of gene expression. Nutr. Metab. (Lond.) 2004;1:3. doi: 10.1186/1743-7075-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wek RC, Jiang HY, Anthony TG. Coping with stress: eIF2 kinases and translational control. Biochem. Soc. Trans. 2006;34:7–11. doi: 10.1042/BST20060007. [DOI] [PubMed] [Google Scholar]
- 4.Harding HP, Zhang Y, Zeng H, Novoa I, Lu PD, Calfon M, Sadri N, Yun C, Popko B, et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell. 2003;11:619–633. doi: 10.1016/s1097-2765(03)00105-9. [DOI] [PubMed] [Google Scholar]
- 5.Ron D, Habener JF. CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant-negative inhibitor of gene transcription. Genes Dev. 1992;6:439–453. doi: 10.1101/gad.6.3.439. [DOI] [PubMed] [Google Scholar]
- 6.Luethy JD, Holbrook NJ. Activation of the gadd153 promoter by genotoxic agents: a rapid and specific response to DNA damage. Cancer Res. 1992;52:5–10. [PubMed] [Google Scholar]
- 7.Sylvester SL, Ap Rhys CM, Luethy-Martindale JD, Holbrook NJ. Induction of GADD153, a CCAAT/enhancer-binding protein (C/EBP)-related gene, during the acute phase response in rats. Evidence for the involvement of C/EBPs in regulating its expression [published erratum appears in J. Biol. Chem., 1995 Jun 16; 270, 14842] J. Biol. Chem. 1994;269:20119–20125. [PubMed] [Google Scholar]
- 8.Bruhat A, Jousse C, Carraro V, Reimold AM, Ferrara M, Fafournoux P. Amino acids control mammalian gene transcription: activating transcription factor 2 is essential for the amino acid responsiveness of the CHOP promoter. Mol. Cell. Biol. 2000;20:7192–7204. doi: 10.1128/mcb.20.19.7192-7204.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siu F, Bain PJ, LeBlanc-Chaffin R, Chen H, Kilberg MS. ATF4 is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 2002;277:24120–24127. doi: 10.1074/jbc.M201959200. [DOI] [PubMed] [Google Scholar]
- 10.Chen H, Pan YX, Dudenhausen EE, Kilberg MS. Amino acid deprivation induces the transcription rate of the human asparagine synthetase gene through a timed program of expression and promoter binding of nutrient-responsive basic region/leucine zipper transcription factors as well as localized histone acetylation. J. Biol. Chem. 2004;279:50829–50839. doi: 10.1074/jbc.M409173200. [DOI] [PubMed] [Google Scholar]
- 11.Pan YX, Chen H, Thiaville MM, Kilberg MS. Activation of the ATF3 gene through a co-ordinated amino acid-sensing response programme that controls transcriptional regulation of responsive genes following amino acid limitation. Biochem. J. 2007;401:299–307. doi: 10.1042/BJ20061261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Averous J, Bruhat A, Jousse C, Carraro V, Thiel G, Fafournoux P. Induction of CHOP expression by amino acid limitation requires both ATF4 expression and ATF2 phosphorylation. J. Biol. Chem. 2004;279:5288–5297. doi: 10.1074/jbc.M311862200. [DOI] [PubMed] [Google Scholar]
- 13.Siu F, Chen C, Zhong C, Kilberg MS. CCAAT/enhancer-binding protein-beta is a mediator of the nutrient-sensing response pathway that activates the human asparagine synthetase gene. J. Biol. Chem. 2001;276:48100–48107. doi: 10.1074/jbc.M109533200. [DOI] [PubMed] [Google Scholar]
- 14.Bruhat A, Cherasse Y, Maurin AC, Breitwieser W, Parry L, Deval C, Jones N, Jousse C, Fafournoux P. ATF2 is required for amino acid-regulated transcription by orchestrating specific histone acetylation. Nucleic Acids Res. 2007;35:1312–1321. doi: 10.1093/nar/gkm038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jousse C, Deval C, Maurin AC, Parry L, Cherasse Y, Chaveroux C, Lefloch R, Lenormand P, Bruhat A, et al. TRB3 inhibits the transcriptional activation of stress-regulated genes by a negative feedback on the ATF4 pathway. J. Biol. Chem. 2007;282:15851–15861. doi: 10.1074/jbc.M611723200. [DOI] [PubMed] [Google Scholar]
- 16.Bruhat A, Averous J, Carraro V, Zhong C, Reimold AM, Kilberg MS, Fafournoux P. Differences in the molecular mechanisms involved in the transcriptional activation of the CHOP and asparagine synthetase genes in response to amino acid deprivation or activation of the unfolded protein response. J. Biol. Chem. 2002;277:48107–48114. doi: 10.1074/jbc.M206149200. [DOI] [PubMed] [Google Scholar]
- 17.Harding HP, Novoa II, Zhang Y, Zeng H, Wek R, Schapira M, Ron D. Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell. 2000;6:1099–1108. doi: 10.1016/s1097-2765(00)00108-8. [DOI] [PubMed] [Google Scholar]
- 18.Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J. Cell. Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl Acad. Sci. USA. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karpinski BA, Morle GD, Huggenvik J, Uhler MD, Leiden JM. Molecular cloning of human CREB-2: an ATF/CREB transcription factor that can negatively regulate transcription from the cAMP response element. Proc. Natl Acad. Sci. USA. 1992;89:4820–4824. doi: 10.1073/pnas.89.11.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ameri K, Harris AL. Activating transcription factor 4. Int. J. Biochem. CellBiol. 2007 doi: 10.1016/j.biocel.2007.01.020. Jan 28; [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 22.Kato Y, Koike Y, Tomizawa K, Ogawa S, Hosaka K, Tanaka S, Kato T. Presence of activating transcription factor 4 (ATF4) in the porcine anterior pituitary. Mol. Cell. Endocrinol. 1999;154:151–159. doi: 10.1016/s0303-7207(99)00078-7. [DOI] [PubMed] [Google Scholar]
- 23.Gachon F, Gaudray G, Thebault S, Basbous J, Koffi JA, Devaux C, Mesnard J. The cAMP response element binding protein-2 (CREB-2) can interact with the C/EBP-homologous protein (CHOP) FEBS Lett. 2001;502:57–62. doi: 10.1016/s0014-5793(01)02646-1. [DOI] [PubMed] [Google Scholar]
- 24.Podust LM, Poulos TL, Waterman MR. Crystal structure of cytochrome P450 14alpha -sterol demethylase (CYP51) from Mycobacterium tuberculosis in complex with azole inhibitors. Proc. Natl Acad Sci. USA. 2001;98:3068–3073. doi: 10.1073/pnas.061562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl Acad. Sci. USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vinson CR, Hai T, Boyd SM. Dimerization specificity of the leucine zipper-containing bZIP motif on DNA binding: prediction and rational design. Genes Dev. 1993;7:1047–1058. doi: 10.1101/gad.7.6.1047. [DOI] [PubMed] [Google Scholar]
- 27.Gachon F, Thebault S, Peleraux A, Devaux C, Mesnard JM. Molecular interactions involved in the transactivation of the human T-cell leukemia virus type 1 promoter mediated by Tax and CREB-2 (ATF-4) Mol. Cell. Biol. 2000;20:3470–3481. doi: 10.1128/mcb.20.10.3470-3481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang G, Hai T. Characterization of human activating transcription factor 4, a transcriptional activator that interacts with multiple domains of cAMP-responsive element-binding protein (CREB)-binding protein. J. Biol. Chem. 1997;272:24088–24095. doi: 10.1074/jbc.272.38.24088. [DOI] [PubMed] [Google Scholar]
- 29.Lassot I, Estrabaud E, Emiliani S, Benkirane M, Benarous R, Margottin-Goguet F. p300 modulates ATF4 stability and transcriptional activity independently of its acetyltransferase domain. J. Biol. Chem. 2005;280:41537–41545. doi: 10.1074/jbc.M505294200. [DOI] [PubMed] [Google Scholar]
- 30.De Angelis R, Iezzi S, Bruno T, Corbi N, Di Padova M, Floridi A, Fanciulli M, Passananti C. Functional interaction of the subunit 3 of RNA polymerase II (RPB3) with transcription factor-4 (ATF4) FEBS Lett. 2003;547:15–19. doi: 10.1016/s0014-5793(03)00659-8. [DOI] [PubMed] [Google Scholar]
- 31.Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–1156. doi: 10.1016/0092-8674(94)90006-x. [DOI] [PubMed] [Google Scholar]
- 32.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 33.Lipson KE, Baserga R. Transcriptional activity of the human thymidine kinase gene determined by a method using the polymerase chain reaction and an intron-specific probe. Proc. Natl Acad. Sci. USA. 1989;86:9774–9777. doi: 10.1073/pnas.86.24.9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, Seraphin B. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 35.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Song CZ, Keller K, Murata K, Asano H, Stamatoyannopoulos G. Functional interaction between coactivators CBP/p300, PCAF, and transcription factor FKLF2. J. Biol. Chem. 2002;277:7029–7036. doi: 10.1074/jbc.M108826200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marmorstein R. Structure of histone acetyltransferases. J. Mol. Biol. 2001;311:433–444. doi: 10.1006/jmbi.2001.4859. [DOI] [PubMed] [Google Scholar]
- 38.Yang XJ, Ogryzko VV, Nishikawa J, Howard BH, Nakatani Y. A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature. 1996;382:319–324. doi: 10.1038/382319a0. [DOI] [PubMed] [Google Scholar]
- 39.Berger SL. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 40.Blanco JC, Minucci S, Lu J, Yang XJ, Walker KK, Chen H, Evans RM, Nakatani Y, Ozato K. The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev. 1998;12:1638–1651. doi: 10.1101/gad.12.11.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korzus E, Torchia J, Rose DW, Xu L, Kurokawa R, McInerney EM, Mullen TM, Glass CK, Rosenfeld MG. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- 42.Xu L, Lavinsky RM, Dasen JS, Flynn SE, McInerney EM, Mullen TM, Heinzel T, Szeto D, Korzus E, et al. Signal-specific co-activator domain requirements for Pit-1 activation. Nature. 1998;395:301–306. doi: 10.1038/26270. [DOI] [PubMed] [Google Scholar]
- 43.Puri PL, Sartorelli V, Yang XJ, Hamamori Y, Ogryzko VV, Howard BH, Kedes L, Wang JY, Graessmann A, et al. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell. 1997;1:35–45. doi: 10.1016/s1097-2765(00)80005-2. [DOI] [PubMed] [Google Scholar]
- 44.Schiltz RL, Mizzen CA, Vassilev A, Cook RG, Allis CD, Nakatani Y. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 1999;274:1189–1192. doi: 10.1074/jbc.274.3.1189. [DOI] [PubMed] [Google Scholar]
- 45.Sartorelli V, Puri PL, Hamamori Y, Ogryzko V, Chung G, Nakatani Y, Wang JY, Kedes L. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell. 1999;4:725–734. doi: 10.1016/s1097-2765(00)80383-4. [DOI] [PubMed] [Google Scholar]
- 46.Martinez-Balbas MA, Bauer UM, Nielsen SJ, Brehm A, Kouzarides T. Regulation of E2F1 activity by acetylation. EMBO J. 2000;19:662–671. doi: 10.1093/emboj/19.4.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Giandomenico V, Simonsson M, Gronroos E, Ericsson J. Coactivator-dependent acetylation stabilizes members of the SREBP family of transcription factors. Mol. Cell. Biol. 2003;23:2587–2599. doi: 10.1128/MCB.23.7.2587-2599.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madison DL, Yaciuk P, Kwok RP, Lundblad JR. Acetylation of the adenovirus-transforming protein E1A determines nuclear localization by disrupting association with importin-alpha. J. Biol. Chem. 2002;277:38755–38763. doi: 10.1074/jbc.M207512200. [DOI] [PubMed] [Google Scholar]
- 49.Gachon F, Devaux C, Mesnard JM. Activation of HTLV-I transcription in the presence of Tax is independent of the acetylation of CREB-2 (ATF-4) Virology. 2002;299:271–278. doi: 10.1006/viro.2002.1501. [DOI] [PubMed] [Google Scholar]
- 50.Jin Y, Zeng SX, Lee H, Lu H. MDM2 mediates p300/CREB-binding protein-associated factor ubiquitination and degradation. J. Biol. Chem. 2004;279:20035–20043. doi: 10.1074/jbc.M309916200. [DOI] [PubMed] [Google Scholar]
- 51.Sano Y, Tokitou F, Dai P, Maekawa T, Yamamoto T, Ishii S. CBP alleviates the intramolecular inhibition of ATF-2 function. J. Biol. Chem. 1998;273:29098–29105. doi: 10.1074/jbc.273.44.29098. [DOI] [PubMed] [Google Scholar]
- 52.Kawasaki H, Song J, Eckner R, Ugai H, Chiu R, Taira K, Shi Y, Jones N, Yokoyama KK. p300 and ATF-2 are components of the DRF complex, which regulates retinoic acid- and E1A-mediated transcription of the c-jun gene in F9 cells. Genes Dev. 1998;12:233–245. doi: 10.1101/gad.12.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.