Abstract
DNA damage checkpoints are signal transduction pathways that are activated after genotoxic insults to protect genomic integrity. At the site of DNA damage, ‘mediator’ proteins are in charge of recruiting ‘signal transducers’ to molecules ‘sensing’ the damage. Budding yeast Rad9, fission yeast Crb2 and metazoan 53BP1 are presented as mediators involved in the activation of checkpoint kinases. Here we show that, despite low sequence conservation, Rad9 exhibits a tandem tudor domain structurally close to those found in human/mouse 53BP1 and fission yeast Crb2. Moreover, this region is important for the resistance of Saccharomyces cerevisiae to different genotoxic stresses. It does not mediate direct binding to a histone H3 peptide dimethylated on K79, nor to a histone H4 peptide dimethylated on lysine 20, as was demonstrated for 53BP1. However, the tandem tudor region of Rad9 directly interacts with single-stranded DNA and double-stranded DNAs of various lengths and sequences through a positively charged region absent from 53BP1 and Crb2 but present in several yeast Rad9 homologs. Our results argue that the tandem tudor domains of Rad9, Crb2 and 53BP1 mediate chromatin binding next to double-strand breaks. However, their modes of chromatin recognition are different, suggesting that the corresponding interactions are differently regulated.
INTRODUCTION
To protect the integrity of their DNA against the attacks from various endogenous and environmental sources, cells have evolved a genome surveillance network that carefully coordinates DNA repair with cell cycle progression. DNA double-strand breaks (DSBs) are considered the most toxic type of DNA damage. If left unrepaired or repaired improperly, they cause chromosomal aberrations, which may be lethal or result in oncogenic transformation. A prominent cellular response to DSBs is the focal assembly of a large number of DNA repair proteins and checkpoint proteins at the site of damage. In particular, in budding yeast, induction of HO endonuclease in G1 results in a Tel1-dependent histone H2A S129 phosphorylation, the subsequent retention of the checkpoint adaptor protein Rad9 to DSBs, and the phosphorylation of its N-terminal region by the Ddc2/Mec1 complex (1). More generally, Rad9 is phosphorylated in a Mec1- and Tel1-dependent manner after cell treatment with various DNA-damaging agents [UV, γ-rays, MMS; (2,3)]. Subsequently, Rad9 binds to the checkpoint effector kinase Rad53, which transautophosphorylates and becomes active (4,5). Similarly, in fission yeast, after creation of DNA damage by either ionizing radiation (IR) or the site-specific HO endonuclease, the checkpoint adaptor Crb2 forms distinct nuclear foci at DSBs (6). It is also hyperphosphorylated by the Rad3 protein kinase and is required for the activation of the effector kinase Chk1 (7). In metazoa, upon exposure to DNA-damaging agents, 53BP1 undergoes a rapid relocalization to sites of DSBs and is phosphorylated by the ATM kinase in its N-terminal region (8–11). All three proteins Rad9, Crb2 and 53BP1 possess two C-terminal BRCA1 C Terminus (BRCT) motifs. This motif is found in a number of proteins implicated in various aspects of cell cycle control, recombination and DNA repair (12,13). Crb2 and 53BP1 also exhibit a tandem tudor domain between their N-terminal region, rich in phosphorylation motifs, and the C-terminal BRCT domains (14–16). Such a domain is predicted in the case of Rad9, despite the lack of sequence identity in this region between the three proteins (16). Furthermore, in vitro, the tandem tudor domain of 53BP1 specifically binds with a micromolar affinity to H4K20me2, a histone H4 peptide dimethylated on K20 (14,17). In vivo, the interaction between 53BP1 and the correspondingly modified H4 is necessary for the accumulation of 53BP1 to DSBs (14). A millimolar interaction is found when looking at the binding of Crb2 tudor region with the same H4K20me2 peptide by NMR (14). Histone H4 K20 methylation by Set9 methyltransferase is required for formation of Crb2 foci in Schizosaccharomyces pombe (18). However, Set9 is not required to arrest division in response to DNA damage.
Based on these properties, Rad9, Crb2 and 53BP1 are proposed to play similar roles in DNA damage signaling and repair. In particular, the current thinking is that Rad9 recognizes modified histones close to DSBs through its predicted tudor domains (16,19). Indeed, cells lacking the H3 methylase Dot1 or carrying a mutant allele of Rad9 (Y798Q) supposed to be defective in K79 dimethylated H3 binding are G1 checkpoint-defective and fail to phosphorylate Rad9 or activate Rad53.
However, there are clearly some differences between metazoan 53BP1 and yeast Crb2/Rad9. For example, the BRCT motifs of Crb2 are required for both homo-oligomerization and foci formation at sites of damage (20), whereas the regions sufficient for these functions lie upstream of the BRCT domains in 53BP1 (21). Moreover, Rad9 and Crb2 play a major role in cell cycle checkpoint control, whereas 53BP1 has limited checkpoint functions (21). It was recently proposed that 53BP1 might rather act as an adaptor in the repair of DSBs (21).
Here, we investigate the functional role of the predicted tudor region of Rad9. We confirm that this region is important for the resistance of Saccharomyces cerevisiae to different stresses. We also analyze the molecular mechanisms linking Rad9 to chromatin. We determined the 3D structure of the proposed tudor region and assessed its binding properties in vitro. Our results show that ScRad9[754–947] indeed folds into a tandem tudor domain. However, the five-residue histone-binding cage found in 53BP1 is only partially conserved in Rad9. Moreover, the tandem tudor region of Rad9 does not directly recognize the K79 dimethylated histone H3 peptide or the K20 dimethylated histone H4 peptide reported to bind 53BP1. Our results rather support a mechanism in which Rad9 directly recognizes DNA at the site of damage.
MATERIALS AND METHODS
Strain, plasmids and media
The yeast strain rad9Δ-L157 harbors a complete deletion of the RAD9 gene and is described in (22). To test the resistance of yeast cells to genotoxic stresses, overnight precultures grown in YPD medium (1% yeast extract, 2% Bacto peptone, 2% glucose) were diluted to an OD of 0.1 and grown for an additional 4–5 h. Tenfold dilutions were then spotted on YPD plates with or without drugs. Sets of YPD plates were irradiated by UV light at 120 J/m2 using a Stratalinker 1800 or X-irradiated (150 gray) using a X-irradiator 130 kV Faxitron at 2.5 Gy/min.
To construct the plasmid YCp50-Rad9ΔTudor expressing the mutant Rad9 protein deleted for the tudor domain (aa 754–947), the fragments A (bp 2110–2262) and B (bp 2842–2887) of S. cerevisiae RAD9 gene encoding the Rad9 sequences surrounding the tudor domain were amplified by PCR using the primers 5′-gatacaatag agatcggtga-3′ (A5′) and 5′-acttcagtat gcgtatttat gcccgaacct gtctcccctg-3′ (A3′) and the primers 5′-caggggagac aggttcgggc ataaatacgc atactgaagt-3′ (B5′) and 5′-cctgttctga tttcaccaga-3′ (B3′), respectively. The fragment AB was constructed by PCR sewing the fragments A and B, and amplified using the primers A5′ and B3′. The URA3-marked plasmid YCp50-Rad9 (23) harboring the wild-type RAD9 gene under the control of its own promoter was digested at a unique SnaBI site located within the tudor-encoding sequence and cotransformed into the yeast strain rad9Δ-L157 along with the fragment AB so as to promote its repair by homologous recombination with the fragment AB and consequently the deletion of the tudor-encoding sequence. Ura+ transformants containing the repaired YCp50-Rad9 plasmids were recovered and tested by PCR for the presence of the rad9ΔTudor allele. Plasmids encoding Rad9ΔTudor were then isolated and verified by sequencing.
Analysis of Rad53 phosphorylation
Rad9Δ-L157 transformants containing either an empty vector, the YCp50-Rad9 or the YCp50-Rad9ΔTudor plasmids were grown to exponential phase in YPD, arrested in G1 with α-factor (0.5 μM final concentration), UV irradiated with 120 J/m2 and harvested at different time points after irradiation. Analysis of Rad53 phosphorylation was performed as described previously (24).
Preparation of proteins and peptides
Preparation and purification of wild-type and mutant 53BP1 and Rad9 domains, as well as isotope enrichment with 15N and 15N/13C follow previously published procedures (15,23). The peptides H3-K79, H3K79me2, metazoan H4-K20me3 and yeast H4-K20me2 were purchased from Peptide Speciality Laboratories GmbH (Heidelberg). The metazoan peptides H4-K20me2 and H4-K20me2-long were synthesized in the laboratory, purified by HPLC (cationic and reverse column) and checked by mass spectroscopy. The following peptides were obtained:
| H3-K79: EIAQDFKTDLR | |
| H3-K79me2: EIAQDFK*TDLR | K* dimethylated |
| Yeast H4-K20me2: KRHRK*ILRD | K* dimethylated |
| Metazoan H4-K20me3: KRHRK**VLRD | K** trimethylated |
| Metazoan H4-K20me2: KRHRK*VLRD | K* dimethylated |
| Metazoan H4-K20me2-long: GKGGAKRHRK*VLRDNIQGK | K* dimethylated |
NMR experiments for the structure determination
Rad9 NMR samples were prepared in 50 mM MES buffer (pH 6) containing 50 mM NaCl in either 90%H2O/10% D2O or in 100% D2O. 1 mM EDTA, a protease inhibitor cocktail (SIGMA), 1 mM NaN3 and 1 mM 3-(trimethylsilyl)[2,2,3,3,-2H4]propionate (TSP) were added to the samples. All assignment experiments were performed at 30°C on a Bruker DRX-600 equipped with a triple resonance TXI cryoprobe. The 1H-15N HSQC NOESY and 1H-13C HSQC NOESY were recorded on a 800 MHz Varian spectrometer at the IBS in Grenoble, France. All spectra were processed with the programs Xwinnmr (Bruker) or NMRPipe (25) and analyzed using Sparky.
NMR titrations with peptides and DNA
NMR titrations were carried out by recording 1H-15N HSQC experiments at 600 MHz, using 15N-labeled protein sample at concentrations of 0.2–0.5 mM. The peptides H3-K79me, H4-K20me3, H4-K20me2 and H4-K20me2-long were added up to 9-, 7-, 5- and 2 molar excess to the protein samples, respectively. Two different oligonucleotides were tested: a 10 bp oligonucleotide 5′-AACTCGAGTT-3′ (PROLIGO, Paris) and a 12 bp oligonucleotide 5′-CGATCAATTACT-3′ (EUROBIO, Courtaboeuf). Both oligonucleotides were annealed with their complementary strand prior to NMR experiments and added up to a 6- and 2-fold molar excess to the protein sample, respectively. Dissociation constants were estimated by fitting the titration curves with the Kaleidagraph software, using the following equation:  , where y is the weighted chemical shift displacement |Δ δ (1H)| + 0.1 × |Δ δ(15N)|, x is the ligand concentration, c is the initial concentration of the protein, Δδmax is the maximum variation of the weighted chemical shift displacements and Kd is the estimated dissociation constant.
, where y is the weighted chemical shift displacement |Δ δ (1H)| + 0.1 × |Δ δ(15N)|, x is the ligand concentration, c is the initial concentration of the protein, Δδmax is the maximum variation of the weighted chemical shift displacements and Kd is the estimated dissociation constant.
Isothermal titration calorimetry
All ITC measurements were recorded at 30°C with a MicroCal MCS instrument (MicroCal, Inc., Northampton, MA, USA). The mouse 53BP1 tudor fragment (1463–1617), H4-K20me2 and H4K20-me2-long were equilibrated in the same buffer containing 50 mM Tris/HCl at pH 7.2, and 50 mM NaCl. The protein (18–33 µM) in the 1.337 ml calorimeter cell was titrated by the peptide (generally 10–15 times more concentrated) by automatic injections of 5–10 µl each. The first injection of 2 µl was ignored in the final data analysis. Integration of the peaks corresponding to each injection and correction for the baseline were done using Origin-based software provided by the manufacturer. Curve fitting was done with a standard one-site model and gives the stoichiometry (n), equilibrium binding constant (Ka) and enthalpy of the complex formation (ΔH). Control experiments, consisting of injecting peptide solutions into the buffer, were performed to evaluate the heat of dilution.
Electrophoresis mobility shift assays
DNA preparation
The 357, 211 and the 146 bp DNA fragments were generated by polymerase chain reaction with a thermostable DNA polymerase (Promega) using a PTC-100 PCR System (MJ Research, Inc.). The 357, 211 and the 146 bp DNA fragments, obtained from the BamH1 digest, the Dra I and BamH1 double digest, and the BamH1 and Dra I double digest of the plasmid pUC(357.4) (26) respectively, were used as templates. The 5′-GATCCTCTAGAGTCCGGCTAC-3′ oligonucleotide was used as sense primer for the 146 and the 357 bp fragments whereas the 5′-AAAGGGTCAGGGATGTTATGACG-3′ and the 5′- CCCGGGCGAGCTCGAATTCC-3′ oligonucleotides were used as antisense primers for the 146 and the 357 bp fragments, respectively. The 5′-AAATAGCTTAACTTTCATCAAGCAAG-3′ and 5′-CCCGGGCGAGCTCGAATTCC-3′ oligonucleotides were used as sense and antisense primers for the 211 bp fragment. The five DNA fragments of 35 bp or nucleotides long were obtained by annealing of oligonucleotides (see Table below) purchased from MWG or Eurobio. The 35 bp fragment was obtained by annealing oligonucleotide 1 and 2, the 35 bp fragment containing a GT mismatch was obtained by annealing oligonucleotide 1 and 3, the 35 bp fragment containing a nick was obtained by annealing oligonucleotide 1, 4 and 5, the 35 bp fragment containing a gap was obtained by annealing oligonucleotide 1 and 5, the 35 bp fragment containing one biotin was obtained by annealing oligonucleotide 2 and 6, the 35 bp fragment containing two biotins was obtained by annealing oligonucleotide 6 and 7, the single-stranded DNA of 35 nt was formed by oligonucleotide 1. Annealing was performed in 10 mM Tris–HCl pH 8, 1 mM EDTA, 200 mM NaCl, by heating at 90°C for 10 min followed by slow cooling at room temperature.
Correct annealing was controlled on 8 or 10% native polyacrylamide gels. 5′ End labeling with 32P-ATP and T4 polynucleotide kinase was performed at 25°C for the 35 bp DNA fragments and at 37°C for the 146, 211 and 357 bp fragments according to standard protocols (27).
| Oligonucleotide 1 | 5′-GGGGCATGCCTGCAGGTCGACT CTAGAGGATCCCC-3′ |
| Oligonucleotide 2 | 3′-CCCCGTACGGACGTCCAGCTGA GATCTCCTAGGGG-5′ |
| Oligonucleotide 3 | 3′-CCCCGTACGGACGTCCAGtTGA GATCTCCTAGGGG-5′ |
| Oligonucleotide 4 | 3′-CCCCGTACGGACGTCCAGCT-5′ |
| Oligonucleotide 5 | 3′-GAGATCTCCTAGGGG-5′ |
| Oligonucleotide 6 | 5′ GGGGCATGCCTGCAGGTCGACT CTAGAGGATCCCC 3′ biotin |
| Oligonucleotide 7 | biotin 3′CCGTACGGACGTCCAGCTG AGATCTCCTAGGGG 5′ |
Protein–DNA interactions
In reactions in which streptavidin was present, the double-stranded 35 bp DNA with one or two biotins was preincubated with the streptavidin (0.5 mg/ml) for 10 min prior to addition of the protein, in order to allow conjugation of biotin. Proteins diluted at the indicated concentrations, in buffer containing 50 mM Tris–HCl, pH 8.0, 1 mM EDTA, 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride, 1 mM DTT, 50 mM NaCl and 0.1% triton X-100 were incubated with radioactive DNA fragments for 2 h at 22°C. Protein–DNA complexes were analyzed on 6% polyacrylamide gels at an acrylamide to bis-acrylamide ratio of 29/1 (w/w), in 0.5× TEG [12.5 mM Tris–HCl (pH 8.4), 95 mM glycine and 0.5 mM EDTA] as indicated in the figure legends. After one hour pre-electrophoresis, samples were loaded onto the gels and resolved at 70 V by a 1–4 h electrophoresis depending upon the size of the DNA. DNA retardation was detected by autoradiography of the dried polyacrylamide gels at –80°C using Biomax MR films (Kodak) and an intensifying screen. For affinity measurements, dried polyacrylamide gels were exposed to a phosphor screen, and measurements of the radioactive signals were performed with a STORM 860 scanner (Amersham Bioscience) using ImageQuant software (GE Healthcare).
RESULTS
Towards the definition of a new globular domain in S. cerevisiae Rad9
From the analysis of Rad9 and 53BP1 sequences, it was predicted that the fragment 754–947 of Rad9 corresponds to a tudor region similar to that of 53BP1 (23). We showed that this fragment, which we will hereafter call ScRad9[754–947], is folded, contains 73% of β-sheet and 27% of random coil and has a heat transition midpoint of 52°C (23). These structural characteristics are similar to those of the tandem tudor domain of mouse 53BP1. However, the Rad9 fragment is prone to aggregation. Various attempts to optimize its solubility by modifying its limits or by mutating its cysteines failed. In particular, ScRad9[754–931] showed the same aggregation propensity than ScRad9[754–947] and ScRad9[754–931,C789A,C812S,C853S] aggregated 1.5 times faster. Cys863 could not be mutated without strongly affecting the solubility of the Rad9 fragment in Escherichia coli. Screening of buffer conditions was carried out to limit the aggregation of ScRad9[754–947] and led to the selection of MES 50 mM, NaCl 50 mM, 2 mM TCEP at pH 6, in which 50% of the fragment is aggregated after three days at 0.2 mM and 30°C.
Under these conditions, the NMR 15N-HSQC spectra of ScRad9[754–931] and ScRad9[754–947] are superimposable, while the spectrum of ScRad9[754–931,C789A,C812S,C853S] shows frequency shifts at peaks further assigned to residues spatially close to the three mutated cysteines (Supplementary Figure S1A and B). All these fragments share a common fold characterized by a large dispersion of the 1H and 15N NMR signals.
ScRad9[754–947] exhibits highly flexible regions on various timescales
The 3D solution structure of ScRad9[754–947] was characterized by heteronuclear double and triple resonance NMR spectroscopy. Between residues 756 and 895, 81% of the backbone NMR signals were assigned; moreover, the NMR signals of 67 and 8% of the side chains were completely and partially assigned, respectively. Unassigned signals were clustered in residues K755, C789, I814, C863 and segments 776–777, 794–798, 824–830, 832–833, 896–897. In the C-terminal part of ScRad9[754–947], between residues 896 and 947, the NMR signals corresponding to regions 896–910 and 918–929 were also unassigned. Most of the unassigned fragments are in conformational exchange on a millisecond timescale, as only few peaks could correspond to these fragments in the 3D spectra. A heteronuclear 2D 15N→1H nOe experiment was carried out in order to identify faster motions, i.e. picosecond to nanosecond timescale dynamics, in exposed loops or unstructured segments. This experiment shows that the backbone of residues 756–762 and 930–947, corresponding to the N- and C-terminal parts, is essentially unstructured (15N→1H nOe ≤0). Within and after the globular core, residues 806–811, 879–882 and 911–916 belong to highly flexible loops (0 < 15N→1H nOe < 0.5).
ScRad9[754–947] contains a tandem tudor domain, similar to that of S. pombe Crb2 and human/mouse 53BP1
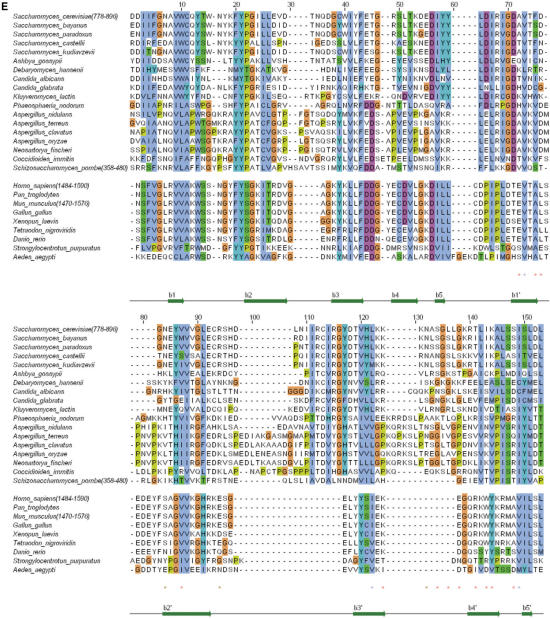
Analysis of the NMR frequencies by TALOS (28) gave access to 65 Φ and Ψ values, and observation of long-range nOes on 1H-15N HSQC NOESY and 1H-13C HSQC NOESY experiments provided 985 inter-residual 1H-1H proximities within the globular core. Calculation of 3D structures consistent with these experimental data using CNS (29) enabled us to determine the global fold of region 762–896. Additional refinement took into account 42 hydrogen bond restraints corresponding to slowly exchanging amide protons. Nine hundred structures of the fragment 762–896 were calculated, and the ten structures of lowest energy were analyzed (Supplementary Table S1). These structures have a root-mean-square deviation around the average backbone structure of 1.7 Å. They consist of two β-barrels formed by residues 786–790 (β1), 797–806 (β2), 811–816 (β3), 819–824 (β4), 828–830 (β5) and residues 768–771 (β0′), 837–841 (β1′), 844–853 (β2′), 869–874 (β3′), 884–889 (β4′), 893–895 (β5′) (Figure 1A). Such a tandem β-barrel structure is reminiscent of the 53BP1 and Crb2 tandem tudor structure (14–16). Consistently, we obtained a significant structural alignment between Rad9 sequence and the sequences of the tandem tudor domains of Crb2 and 53BP1 (Figure 1E). This alignment reveals that Rad9/Crb2 and Rad9/53BP1 sequences share only 18 and 15% of identical residues in the tudor region, respectively. However, the structural fit on the 60 Cα atoms of the 10 common β-strands (i.e. β1 to β5 and β1′ to β5′) yields 3.2 and 3.1 Å between Rad9/Crb2 and Rad9/53BP1, respectively (Figure 1B and C). The individual β-barrels of Rad9 are particularly similar to those of the tudor folds of Crb2 and 53BP1, with Cα root-mean-square deviations within each barrel comprised between 1 and 1.5 Å. Moreover, the relative positioning of the β-barrels in Rad9 is close to that found for Crb2 and 53BP1.
Figure 1.
(A) Ribbon representation of the ScRad9[754–947] 3D structure. Only fragment 762–896 is displayed. Assigned regions are in green, unassigned regions are in purple. The β-sheets are colored in magenta, except the strand β0′ which is in cyan. (B) Superimposition of the 3D structures of ScRad9[754–947] (magenta) and Crb2[358–507] (cyan). (C) Superimposition of the 3D structures of ScRad9[754–947] (magenta) and Mm53BP1[1463–1617] (yellow). (D) Ribbon representation of the 3D structure of the Rad9 fragment 762–896, calculated with three additional hydrogen bond restraints deduced from the structural comparison with Crb2 (see text). Colors are the same as in (A). (E) Alignment of the Rad9 sequence 778–896 with sequences of analogous proteins from 17 yeast species and 9 metazoans. This alignment was deduced from the structural alignment of ScRad9[754–947] with human 53BP1 tandem tudor domain [PDB reference 1XNI, (16); PDB reference 2G3R, (14)], mouse 53BP1 tandem tudor domain [PDB reference 1SSF, (15,31)] and fission yeast Crb2 tandem tudor domain [PDB reference 2FHD, (14)]. Red/blue stars indicate Rad9 solvent-exposed/buried residues whose backbone 15N or 1Hn NMR signals are affected by addition of a 10 mer oligonucleotide. Brown stars indicate Rad9 residues whose side chain 15N or 1Hn NMR signals are affected by the oligonucleotide addition.
The relative positioning of the tudor folds in Rad9 is poorly defined because several protein fragments are unassigned at the interface
The major structural differences in the tudor region between Rad9 and its potential homologs Crb2 and 53BP1 reside in the poorly defined structure of loop 786–801 (β1β2) and in the slightly different positioning of one barrel relatively to the other (Figure 1B and C). In fact, as region 824–829, which contains β5, is not assigned (as shown by the purple color of its backbone on Figure 1A), it is not possible to demonstrate the presence of a β-sheet between β1 and β5. Similarly, because the region 789–798 is essentially unassigned (Figure 1A), the 3D structure of loop β1β2 is unknown, and the presence of a β-sheet between β2 and β5′ cannot be proven. Finally, a C-terminal α-helix is found after β5′ in Crb2 and 53BP1, which interacts with β2, but the region corresponding to this helix is unassigned in Rad9. Thus, because several NMR frequency assignments in loop β1β2, in strand β5 and in the region following β5′ are lacking, it is not possible to describe the conformation of the interface between the two β-barrels in the β5/β1/β2/β5′ region. Conformational exchange on a microsecond to millisecond timescale at this interface is one possible explanation for the lack of NMR data.
We tested if the relative positioning of the tudor folds observed in 53BP1 and Crb2 was consistent with the NMR data obtained on Rad9. Therefore, we calculated a set of 3D structures of the Rad9 fragment 762–896, using three additional hydrogen bond restraints linking the oxygen of Q790 (β1) to the nitrogen of D827 (β2), and the oxygen/nitrogen pair of F797 (β2) to the corresponding pair in L895 (β5′) (Figure 1D). These structures are as consistent as the first set of structures with the experimental NMR data (Supplementary Table S2). The second β-barrel of these structures is structurally similar to that of the first set of structures. However, the conformation of the first barrel as well as the relative positioning of the two barrels is closer from Crb2: the structural fit on the 60 Cα atoms of the 10 common β-strands (i.e. β1 to β5 and β1′ to β5′) yields 2.7 and 2.4 Å between Rad9/Crb2 and Rad9/53BP1, respectively. Thus, the relative positioning of the tudor folds observed in 53BP1 and Crb2 is one of the possible relative positioning of Rad9 tudor folds.
The tandem tudor region ScRad9[754–947] is required for cell resistance to genotoxic stresses and for Rad53 phosphorylation in G1 phase
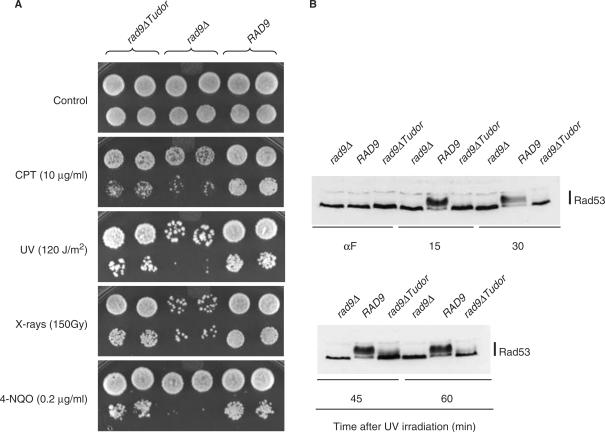
To analyze the functional importance of Rad9 tandem tudor region for resistance to DNA damage, rad9Δ cells complemented with either the wild-type RAD9 gene or the rad9ΔTudor allele deleted for the tudor-encoding sequence were tested for their resistance to various genotoxic stresses, including UV- and X-irradiation, camptothecin (CPT, an inhibitor of DNA topoisomerase I) and 4-nitroquinoline 1-oxide (4-NQO, a drug producing bulky base adducts of the type that is mainly repaired by the nucleotide excision repair system). As shown in Figure 2A, the deletion of Rad9 tandem tudor region resulted in a hypersensitivity to DNA damage which was particularly pronounced in cases of UV-irradiation and camptothecin treatment. However, the rad9ΔTudor allele retained much of the functionality of RAD9 since rad9ΔTudor cells were more resistant to DNA damage than rad9Δ mutants.
Figure 2.
ScRad9[754–947] is required for cell resistance to DNA damage and for Rad53 phosphorylation in G1 phase. (A) rad9Δ (rad9Δ -L157) transformants containing either an empty vector, the YCp50-Rad9 or the YCp50-Rad9Δ Tudor plasmids were grown to exponential phase in YPD medium and 10-fold serial dilutions were spotted on YPD plates supplemented or not with camptothecin (CPT) or 4-nitroquinoline 1-oxide (4-NQO). Sets of YPD plates were also UV- or X-irradiated. (B) rad9Δ (rad9Δ -L157) transformants containing either an empty vector, the YCp50-Rad9 or the YCp50-Rad9Δ Tudor plasmids were arrested in G1 with α-factor (αF), UV irradiated (120 J/m2) and maintained in G1 phase in the continuous presence of α-factor. Aliquots were taken at the indicated times after UV irradiation and analyzed by western blotting using anti-Rad53 antibodies.
To clarify further the biological role of the tandem tudor domain of Rad9, we analyzed Rad53 phosphorylation status after UV irradiation in wild-type, rad9Δ and rad9ΔTudor cells. When asynchronously growing cells were UV irradiated, a slight delay in Rad53 phosphorylation was observed in rad9Δ and rad9ΔTudor mutants 30 min after UV irradiation, but this defect disappeared 60 min after irradiation (data not shown). In contrast, when cells were synchronized in G1 with α-factor before UV irradiation and maintained in G1 thereafter, rad9Δ and rad9ΔTudor cells exhibited a strong defect in Rad53 phosphorylation (Figure 2B), which suggests that the presence of the tandem tudor domain in Rad9 is critical for the phosphorylation of Rad53 under these conditions.
ScRad9[754–947] tandem tudor domain is not sufficient for binding to H3 K79 and H4 K20 dimethylated peptides
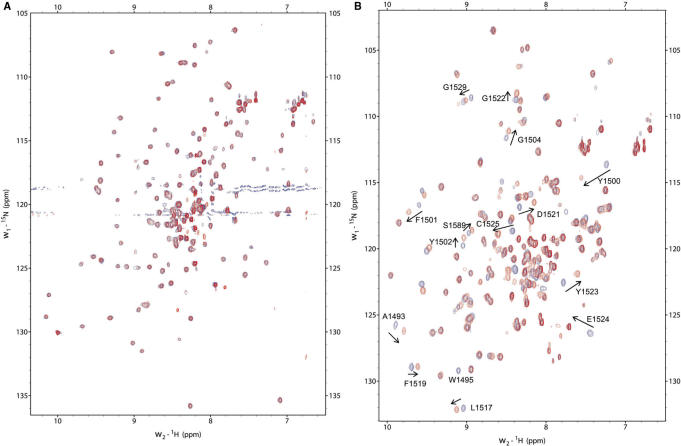
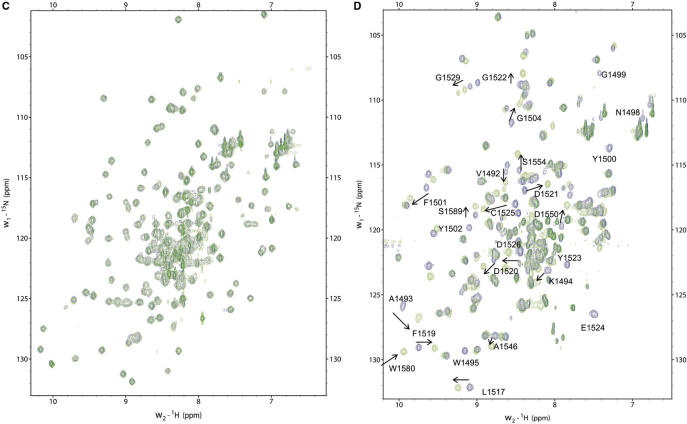
53BP1 and Crb2 tandem tudor domains interact with a histone H4 peptide specifically dimethylated at K20 (H4K20me2) with 2.10−5 M and 10−3 M affinity, respectively (14,17). 53BP1 tandem tudor domain also interacts with a histone H3 peptide dimethylated on K79 (H3K79me2) with a 10−3 M affinity (14). Interactions of 53BP1 and Crb2 with H4K20me2 seem to play a preponderant role in targeting these proteins to DNA damage foci (14,18,30). Thus, we looked for similar in vitro interactions in the case of ScRad9[754–947] by performing NMR titrations. All the experiments were carried out in parallel on mouse 53BP1 fragment 1463–1617 (hereafter called Mm53BP1[1463–1617]) as a control, by performing NMR titrations and ITC studies. The peptides tested in this study were H3K79, H3K79me2, H4K20me2, H4K20me2-long and H4K20me3. First, we added each histone peptide to 15N-labeled ScRad9[754–947] and Mm53BP1[1463–1617] samples and followed the chemical shift perturbations of 15N and 1Hn nuclei by recording 1H-15N HSQC experiments. Surprisingly, the 1Hn and 15N chemical shifts of ScRad9[754–947] were not modified by the addition of the following histone peptides: H3K79me2, yeast and metazoan H4K20me2, metazoan H4K20me3 (Figure 3). Thus, our Rad9 fragment alone cannot bind to yeast H3K79me2 and H4K20me2 peptides and metazoan H4K20me2 and H4K20me3 peptides.
Figure 3.
NMR titrations with dimethylated histone peptides. In (A) is displayed the superimposition of the 1H-15N HSQC spectra of H3K79me2 bound (red) versus free (blue) ScRad9[754–947]. In (B), the same superimposition is shown for Mm53BP1[1463–1617]. Similarly, in (C) is displayed the superimposition of the 1H-15N HSQC spectra of yeast H4K20me2 bound (green) versus free (blue) ScRad9[754–947]. In (D), the same superimposition is shown for metazoan H4K20me2 bound (green) versus free (blue) Mm53BP1[1463–1617]. Peptides were added to the protein fragments up to peptide:protein ratios of (A) 4:1 (B) 7:1 (C) 8:1 (D) 4:1. Peaks shifting or disappearing after histone peptide binding are labeled.
On the opposite, when adding the same H3K79me2 or H4K20me3 peptides to Mm53BP1[1463–1617], several 1H and 15N chemical shifts moved from their native values to values corresponding to a bound state (Figure 3). The exchange rate between the free and bound states of the 53BP1 fragment was fast on the NMR timescale, suggesting that the affinity between Mm53BP1[1463–1617] and the histone peptides is in the millimolar range. Fitting the variation of the weighted chemical shift displacements against the peptide concentration yielded a Kd value of 1.5 ± 0.5 mM and 1.7 ± 0.8 mM for H3-K79me2 and H4-K20me3, respectively. After addition of the H4K20me2 or H4K20me2-long peptides to the same 53BP1 fragment, the 1H-15N peaks that shifted in the previous interactions either disappeared or shifted (Figure 3). In these two latter cases, the exchange between the free and bound states of the 53BP1 fragment was intermediate to fast on the NMR timescale, suggesting that the affinity of Mm53BP1[1463–1617] for these histone peptides is in the micromolar range. NMR estimation of the affinity was confirmed by ITC measurements. A Ka of 1.3 × 105 ± 1.6 × 104 M−1 (Kd of 7.7 µM) with a stoichiometry of 1, a ΔH of –2.2 ± 0.1 kcal/mol, a ΔG of –7.1 kcal/mol and a TΔS of 4.9 kcal/mol was obtained for H4-K20me2 (Supplementary Figure S2A). A Ka of 1.1 × 105 ± 1.6 × 104 M−1 (Kd of 8.8 µM) with a stoichiometry of 1, a ΔH of –2.3 ± 0.1 kcal/mol, a ΔG of –6.9 kcal/mol and a TΔS of 4.6 kcal/mol was obtained for H4-K20me2-long (Supplementary Figure S2B). These values are similar to that previously measured by Botuyan et al. (14) for the interaction between human 53BP1 fragment 1484–1603 (similar to mouse 1470–1589) and the K20 dimethylated H4 peptide fragment 12–25 (14). Interestingly, the main driving term in the free energy of interaction is the positive entropic change, likely due to the dehydration of the binding interface upon complex formation. NMR titration using unmethylated H3K79 showed that this peptide does not interact with Mm53BP1[1463–1617], thus underlying the critical role of lysine methylation in the binding.
Residues whose 15N and 1Hn chemical shifts are sensitive to H4K20me2 peptide binding are colored on the 3D representation of Mm53BP1[1463–1617] (Supplementary Figure S3). The peptide-binding region is arranged around an aromatic cage formed by W1495, Y1500, F1519 and Y1523, as found by Mer and co-workers (21). In order to verify that these residues, as well as two close polar residues D1521 and C1525, are also involved in binding to the whole modified H3, several of them were mutated in Mm53BP1[1463–1617], and the binding to H3 from calf thymus was checked by GST-pulldown experiments. Mutations W1495A and D1521A completely abolished the binding, Y1500A and F1519A reduced the binding, and Y1523A and C1525A did not change significantly the GST-pull-down results (Supplementary Figure S4).
ScRad9[754–947] tandem tudor domain recognizes different DNA fragments with a micromolar affinity
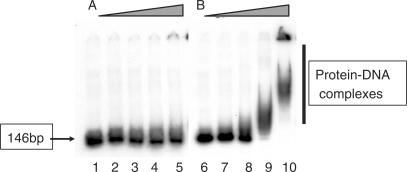
53BP1 and Crb2 tandem tudor domains were suggested not only to recognize modified histones, but also to bind DNA, in particular in the context of nucleosomes (14,15). However, if Mm53BP1[1463–1617] binds with a millimolar affinity to a short 10 bp oligonucleotide, as seen by NMR (15), we could not detect any interaction with a double-stranded DNA of 146 bp by gel retardation assays (Figure 4A). On the opposite, ScRad9[754–947] forms complexes with this 146 bp double-stranded DNA as demonstrated by delayed migration compared to naked DNA (Figure 4B).
Figure 4.
EMSA experiments between a 146 bp double-stranded oligonucleotide and (A) Mm53BP1[1463–1617], (B) ScRad9[754–947]. Increasing concentrations of proteins were incubated with a 146 bp DNA fragment at a concentration of 26 nM: 2-fold (lanes 2 and 7), 8-fold (lanes 3 and 8), 32-fold (lanes 4 and 9) and 128-fold (lanes 5 and 10) molar excess of proteins were used. Lanes 1 and 6 indicate the mobility of naked DNA.
In order to check if ScRad9[754–947] binds to a particular DNA structure, we measured its affinity for different 35 bp oligonucleotides: single, double-stranded containing a mismatch, a nick or a gap (Table 1; Figure 5A and B). Clearly, ScRad9[754–947] recognizes with a similar affinity the double-stranded oligonucleotides with or without mismatch, nick or gap. Its affinity for one of the single-stranded oligonucleotide is 2.5 times lower. The shorter ScRad9[754–931] binds to the 35 bp double-stranded DNA with a twice lower affinity (Figure 5C; Table 1). The ScRad9[754–931,C789A,C812S,C853S] mutant does not recognize DNA anymore when tested by gel retardation assay (Figure 5D). In parallel, surface plasmon resonance experiments confirmed that the ScRad9[754–931,C789A,C812S,C853S] mutant looses the DNA-binding capacity of wild-type ScRad9[754–931] (data not shown).
Table 1.
Affinity constants (µM) of ScRad9[754–947] and ScRad9[754–931] for different oligonucleotides. Experiments with ScRad9[754–947] were carried out with and without 1 mM DTT to measure the influence of cysteine oxidation on DNA binding. However, similar results were found in both conditions. Interactions for which the affinity was not determined are displayed as ‘nd’.
| Kd (µM) | 357 bp ds | 211 bp ds | 146 bp ds | 35 bp ds | 35 bp Nick | 35 bp mismatch GT | 35 bp Gap | 35 bp ss | 35 bp ds with a 3′ terminal biotin | 35 bp with a 3′ terminal biotin + 0.5 µg/µl streptavidin |
|---|---|---|---|---|---|---|---|---|---|---|
| ScRad9[754–947] with DTT | 0.42 ± 0.06 | 0.50 ± 0.07 | 0.34 ± 0.07 | 0.60 ± 0.14 | nd | nd | nd | 2.74 ± 0.27 | 0.83 ± 0.05 | 1.39 ± 0.08 |
| ScRad9[754–947] without DTT | nd | nd | nd | 0.96 ± 0.08 | 0.62 ± 0.06 | 0.89 ± 0.07 | 0.96 ± 0.13 | 2.50 ± 0.18 | nd | nd |
| ScRad9[754–931] with DTT | nd | nd | nd | 2.07 ± 0.27 | nd | nd | nd | 5.02 ± 0.67 | nd | nd |
Figure 5.
DNA-binding properties of Rad9 tudor region. (A) EMSA experiment between a 35 bp single-stranded oligonucleotide and ScRad9[754–947]. (B–D) EMSA experiments between a 35 bp double-stranded oligonucleotide and (B) ScRad9[754–947], (C) ScRad9[754–931], (D) ScRad9[754–931,C788A,C811S,C852S]. (E) EMSA experiment between a 357 bp double-stranded oligonucleotide and ScRad9[754–947]. (F) EMSA experiment between a 35 bp double-stranded oligonucleotide with a 3′ terminal biotin and ScRad9[754–947]. For this experiment, DNA is preincubated with 0.5 mg/ml streptavidin 10 min prior to addition of ScRad9[754–947] in order to allow conjugation of biotin. Increasing concentrations of proteins were incubated with a 357 or 35 bp DNA fragment at a concentration of 26 nM: 9-fold (lane 22), 16-fold (lanes 2, 7, 12, 17 and 27), 18-fold (lane 23), 32-fold (lanes 3, 8, 13, 18 and 28), 36-fold (lane 24), 64-fold (lanes 4, 9, 14, 19 and 29), 72-fold (lane 25) and 128-fold (lanes 5, 10, 15, 20 and 30) molar excess of proteins were used. Lanes 1, 6, 11, 16, 21 and 26 indicate the mobility of naked DNA.
Because various studies suggest that Rad9 intervenes in the case of DSBs, we tested DNA molecules of various lengths, in order to determine if ScRad9[754–947] binds preferentially at the ends of DNA. In the absence of any specificity, for the same concentration of DNA, the percentage of complexes should be inversely proportional to the length of the tested DNA fragment. Measurement of the affinity of ScRad9[754–947] for the 35 bp double-stranded DNA, as well as for three other double-stranded DNAs of different lengths, shows that the affinity does not depend on the tested DNA: calculation of the ratios between bound DNA and free DNA measured on the gels enabled us to estimate an apparent affinity constant of 0.4 (± 0.1) µM, 0.5 (± 0.1) µM, 0.3 (± 0.1) µM and 0.6 (± 0.1) µM for DNAs of respectively 357, 211, 146 and 35 bp (Table 1; Figure 5B and E).
Furthermore, we tested the binding of ScRad9[754–947] to a 35 bp double-stranded oligonucleotide biotinylated at one or two 3′-OH ends. Clearly, the presence of biotine does not modify the affinity of ScRad9[754–947] for the oligonucleotide (Table 1). However, after binding of streptavidin to one of the ends of the 35 bp DNA, the affinity drops by a factor of two [i.e. 1.4 (± 0.1) µM; Figure 5F and Table 1]. After binding of streptavidin to both biotinylated extremities, the gel shift pattern is largely modified (data not shown). These results are consistent with the hypothesis of Rad9 tudor domain binding to DSBs directly, but it must be stressed that obstruction by streptavidin might also have prevented the formation of the previously observed Rad9–DNA interactions.
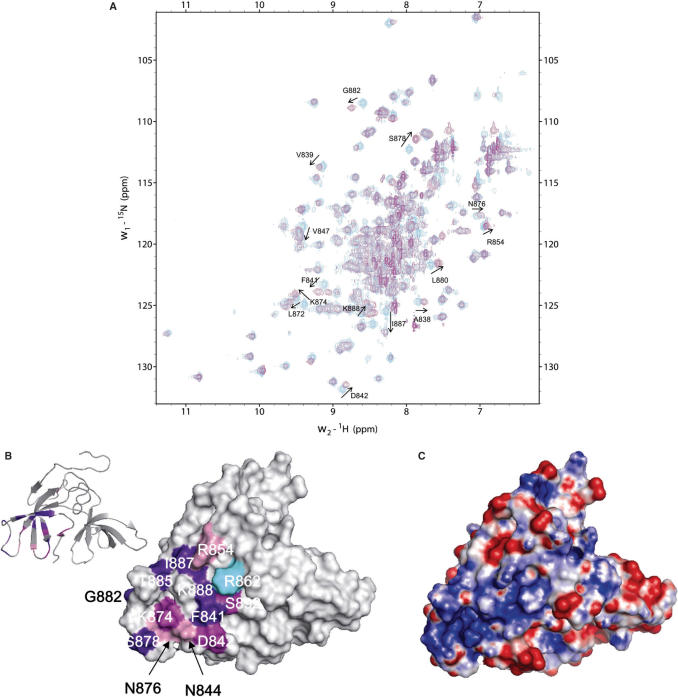
Finally, we observed by NMR the binding of a 10 bp oligonucleotide 5′-AACTCGAGTT-3′ or a 12 bp oligonucleotide 5′-CGATCAATTACT-3′ to an NMR sample of 15N labeled ScRad9[754–947]. Therefore, we added progressively the DNA to an NMR sample of ScRad9[754–947], and we followed the chemical shifts of labeled 15N nuclei and 15N attached 1H nuclei by recording 1H-15N HSQC spectra. Fifteen HSQC peaks corresponding to the backbone nuclei (15N, 1Hn) of A838, V839, F841, D842, V847, L872, K874, S878, L880, G882, T885, I887, K888, S892, I893 and three peaks corresponding to the side-chain nuclei (15N, Hn) of N844, R854, N876 shifted after addition of DNA (Figure 6A). All these changes correspond to residues located around the highly flexible loop β3′β 4′, in the second tudor fold and in the main positively charged region of ScRad9[754–947] (Figure 6B and C). Fitting the variation of weighted chemical shift displacements against the oligonucleotide concentration for these peaks yielded a Kd value comprised between 3 and 20 µM.
Figure 6.
Interaction of ScRad9[754–947] with a 10 bp oligonucleotide. (A)Superimposition of the 1H-15N HSQC spectra of DNA bound (magenta) versus free (cyan) ScRad9[754–947]. DNA/peptide ratio yielded 2.2 in the DNA bound ScRad9[754–947] spectrum. Peak shifting after DNA addition are labeled. (B) Surface representation of ScRad9[754–947] showing the DNA-binding region. When the backbone NH-group chemical shift perturbation (|Δ δ (1H)| + 0.1 × |Δ δ (15N)|) of a residue is higher than the average of all combined shifts plus one SD (0.050 p.p.m.), the residue is colored in purple. When the backbone chemical shift perturbation is comprised between the average value (0.023 p.p.m.) and the average plus 1 SD, the residue is colored in magenta. When the side-chain chemical shift perturbation is higher than 0.023 p.p.m., the residue is colored in pink. The side chain of R862 whose amide is not assigned is shown in cyan. (C) Surface representation of the electrostatic potential of ScRad9[754–947]. The binding and electrostatic surfaces are shown in the same orientation.
DISCUSSION
Our results show that the tandem tudor domain of Rad9 is important for yeast resistance to DNA damage, and is critical for Rad53 phosphorylation in G1 after UV irradiation. Similarly, Wysocki et al. (19) observed the in vivo consequences of the mutation of the strictly conserved Y798 into Q in the tandem tudor domain of Rad9, which probably destabilizes the domain. Expression of the rad9Y798Q allele in a rad9Δ background could not restore the checkpoint function nor Rad53 phosphorylation when cells were treated in G1 with ionizing radiation. All these results indicate that the tandem tudor domain of Rad9 is important for yeast survival after various genotoxic stresses and is involved in the G1 checkpoint response to IR and UV. We characterized its 3D structure and tested its interaction with different components of chromatin.
Structural differences between Rad9 and Crb2/53BP1 in the tudor region
ScRad9[754–947] exhibits a tandem tudor domain structurally similar to that of 53BP1 and Crb2. However, the N- and C-terminal regions surrounding the tudor domain show different structural characteristics. First, the C-terminal α-helix observed in Crb2 and to a larger extent in 53BP1 corresponds to an unassigned fragment in Rad9, suggesting that either the fragment exhibits local conformational exchange, or that its position relatively to the tudor core fluctuates. Circular dichroïsm studies favor the first hypothesis: they give a 6% helix content for Mm53BP1[1463–1617] whereas ScRad9[754–947] has no detectable helix signal (23). Second, ScRad9[754–947] exhibits supplementary loops and β-strands at its N-terminus, between residues 754 and 782. This region is not present in the 53BP1/Crb2 tandem tudor domains analyzed by NMR or X-ray crystallography. In the case of Rad9, it contains several hydrophobic residues that are related by nOe interactions to the second tudor fold (Y761, I767, T769, A770, I781, F782). Interestingly, only one regular β-sheet is found between the additional N-terminal region and the tudor folds. This β-sheet involves residues 768–771 of β0′ and the four last residues of β2′ (Figure 1). In the X-ray structure of human 53BP1 tandem tudor domain, the corresponding four residues form a β-sheet with the same residues of another 53BP1 molecule (16). Thus, the propensity of the four residues to be involved in a β-sheet is conserved from 53BP1 to Rad9, but the functional significance of this structural property is not known.
The loops observed in ScRad9[754–947] are particularly flexible in solution, either on a microsecond to millisecond timescale, as loops β1β2 and β4β5, or on a picosecond to nanosecond timescale, as loop β3′β4′. Alternative conformations have been described in the 1.2 Å resolution X-ray structure of human 53BP1 tudor region, in particular in loop β1β2 (14). NMR analysis of Mm53BP1[1463–1617] showed that, in solution, loop β1β2 of Mm53BP1[1463–1617] exhibits picosecond to nanosecond timescale motions of particularly large amplitude (15). Furthermore, 15 residues were found in two different chemical environments, and these residues are mainly located at the interface between the first tudor fold and the C-terminal α-helix (31). Thus, in Rad9 as in 53BP1, the large loops of the tudor folds and the interface between the first tudor and the region following the second tudor seem to be highly dynamical, both on a rapid (ps–ns) and slower (μs–ms) timescale.
Interestingly, calculation of the electrostatic potential surfaces of the tudor region of 53BP1, Crb2 and Rad9 shows that, if 53BP1 and Crb2 exhibit many well-dispersed positive and negative patches, Rad9 has a largely negatively charged surface and a unique positively charged patch centered on loop β3β4′ (Figure 6C). This loop is more than 8 residue-long and rich in positively charged residues in most Rad9 yeast analogs, while it is only 2 or 3 residue-long in fission yeast and in metazoans (Figure 1E). Moreover, several residues of loop β2′β3′ participate to the positively charged patch in Rad9, and this loop is <9 residue-long in metazoans, while it is between 14 and 26 residue-long in Rad9 yeast analogs (Figure 1E). These observations suggest that DNA recognition by the tudor region is a property shared by most Rad9 yeast analogs and involves a large positively charged patch around loops β2′β3′ and β3′β4′.
Why the tudor region of Rad9 does not bind to the tested methylated histone peptides?
According to our study, it appears that Mm53BP1[1463–1617] interacts with methylated histone peptides and in particular with H4K20me2, whereas ScRad9[754–947] does not interact with any of these peptides. Consistently, GST-pull-down experiments using calf thymus H3 showed that, after elution from the beads, H3 is revealed by immunoblotting together with GST-Mm53BP1[1463–1617], but not together with GST-ScRad9[754–947] (data not shown). Two different groups reported the capture of histone H3 by GST-ScRad9[750–917] (16,32). Our effort in using such Rad9 construction was stopped by the poor stability of this fragment: 40% of the corresponding NMR signal was lost after 2.5 h at 30°C. From our results, the tandem tudor domain of Rad9 does not bind to H3 as modified in calf thymus cells, does not recognize H3K79me2, and is not sufficient to bind to H4K20me2 or H4K20me3.
It has been recently shown that the structure of 53BP1/H4-K20me2 complex uncovers a unique five residue-binding cage, remarkably conserved in Crb2, that best accommodates a dimethyllysine but excludes a trimethyllysine, thus explaining the methylation state-specific recognition of H4-K20 (14). This five residue-binding cage includes four aromatic residues (W1495, Y1502, F1519, Y1523) and an aspartic acid (D1521), all from the first tudor domain of 53BP1. On the basis of the structural alignment between Rad9 and 53BP1 in the tudor region (Figure 1E), a dimethyllysine-binding pocket partially similar to that of 53BP1 is predicted in Rad9. Within the five residues in contact with K20me2, W1495, Y1502 and F1519 correspond to aromatic amino acids in Rad9, D1521 correspond to a threonine, and Y1523 is absent (it corresponds to a residue deleted in Rad9). We and others (16) have shown that replacement of D1521 by an alanine abolished binding of 53BP1 tudor region to H3. In the absence of D1521, the favorable coulombic interaction in the form of a salt bridge between the carboxylate of the aspartic acid and the dimethylammonium ion of H4-K20me2 can no more contribute to the affinity and high specificity of the protein for H4-K20me2 (14). Interestingly, the aspartic acid is present in Crb2. Similarly, 53BP1 Y1500 which interacts with R19 of the peptide H4-K20me2 in the complex is replaced by a lysine in Rad9. So the guanidinium group of R19 cannot make a cation-π interaction with the phenyl ring of the tyrosine. Such differences may account for the loss of histone binding in the case of Rad9. Another argument comes from the conformational exchange exhibited by loop β1β2 in Rad9. This exchange might increase the entropic cost of Rad9 binding to the histones.
What role for the recognition of DNA by Rad9 tudor region?
Human 53BP1 interacts with chromatin in vivo at DSBs following X-irradiation but not after treatment with UV or hydroxyurea (33). DNAse or phosphatase treatment abolishes this interaction. Blot experiments using a double-stranded 10 bp oligonucleotide have shown that the only region capable of DNA binding in 53BP1 is comprised between residues 1052 and 1709. This region contains the minimal 53BP1 fragment necessary for foci formation at DSBs, which corresponds to the tandem tudor domain. EMSA experiments have shown that the fragments 1319–1480 (containing the RG-rich stretch) and 1480–1616 (containing the tudor region) of human 53BP1 interact with single-stranded and double-stranded DNA in vitro (33). We have previously tested the interaction of the mouse tandem tudor region Mm53BP1[1463–1617] with the already cited 10 bp oligonucleotide using NMR (15). We observed that the conserved aromatic patch of 53BP1 tudor domain mediates a millimolar affinity interaction with this DNA fragment. However, using EMSA experiments, we could not visualize an interaction of higher affinity with a longer double-stranded DNA fragment. Thus, we suggest that the tudor region of 53BP1 may participate to chromatin binding, but needs to be associated to another 53BP1 region as its RG-rich region to increase its affinity for DNA up to a level detectable by EMSA.
In the case of Rad9, the tudor region ScRad9[754–947] is clearly sufficient to bind DNA with a micromolar affinity. Moreover, this region possesses a positively charged patch in the second tudor fold which mediates DNA binding (Figure 6C). This patch is conserved neither in 53BP1, nor in Crb2. However, it is present in several other yeast Rad9 analogs (Figure 1E). Thus, DNA binding through the positive surface of the second tudor could be a property common to a large subset of yeast Rad9 analogs. The shorter ScRad9[754–931] also binds to the 35 bp double-stranded DNA with a micromolar affinity, whereas the ScRad9[754–931,C789A,C812S,C853S] mutant does not recognize DNA anymore. Comparison of the chemical shifts between ScRad9[754–931] and ScRad9[754–931,C789A,C812S,C853S] shows that the two proteins share the same global fold (Supplementary Figure S1A and B). Chemical shifts are perturbed around the mutated cysteines, but can still be assigned, except in loop β2′β3′ (i.e. residues 854–869, close to C853S) where major changes are observed: the 3D structure of this loop is probably largely modified by the mutations. Interestingly, loop β2′β3′ contains R854, whose side-chain amide proton signal is shifted after addition of DNA, and R862, whose side chain is close to that of R854 but its amide proton is not assigned (Figure 6B). These arginines are relatively conserved in yeast: position 854 is mainly positively charged and position 862 is essentially occupied by an arginine or a threonine. They are not conserved in Crb2. Finally, in metazoan, position 854 is never positively charged and position 862 is absent (it is in a deleted loop). We suggest that the positioning of loop β2′β3′, and in particular of R854 and R862 side chains, is crucial for DNA binding in a large subset of Rad9 analogs.
We looked on the DNA side which type of structure was recognized by Rad9 tudor region. We observed that this region does not preferentially bind to nick, gap or mismatch. Furthermore, its apparent affinity for DNA is not modified whether the DNA consists of 35, 146, 211 and 357 bp. As these fragments do not possess a common DNA sequence of more than 4 nucleotides, we suggest that Rad9 binding is not sequence specific, but instead is based on the recognition of DNA ends. Consistently, blockage of one of the DNA ends by streptavidin divides the affinity of ScRad9[754–947] for DNA by a factor 2, and blockage of both ends largely modify the gel shift pattern.
CONCLUSION
We have shown that Rad9, as Crb2 and 53BP1, possesses a tandem tudor domain upstream from its BRCT region. This domain exhibits additional N-terminal β-strand and loops when compared to the published structure of 53BP1 and Crb2 tudor regions, and is highly dynamic both on a fast ns to ps timescale and on a slower μs to ms timescale. Per se, it is not capable of recognizing the methylated histone peptides reported to bind 53BP1 tudor region. However, it possesses a large and positively charged loop β3′β4′ which recognizes DNA with a micromolar affinity and is conserved in most identified yeast Rad9 analogs. Thus, both the tudor regions of 53BP1 and Rad9 seem to recognize chromatin, however through different interactions: 53BP1 binds with a micromolar affinity to H4 dimethylated on K20 through a five-residue binding cage located on the first tudor fold, whereas Rad9 binds with a similar affinity to DNA through a positively charged region situated on the second tudor fold. The two binding regions are located on the same side of the tandem tudor domain; they are distinct but contiguous. In the case of Crb2, Botuyan et al. (14) reported that the affinity of the tudor region for H4K20me2 is only in the millimolar range, but proposed that in vivo, the effective affinity is higher if one considers that Crb2 not only interacts with histones but also with DNA in the context of nucleosomes. Consistently, they noticed some affinity of the Crb2 tudor region for nucleic acids. Rad9 and 53BP1 tudor regions could similarly be involved in such multi contact interaction with chromatin.
All these results underline that a common mechanism exists in the biological functions of Rad9, Crb2 and 53BP1: they bind chromatin through their similar tudor domains. As proposed by Kron and co-workers (1), these transient interactions may allow the initial targeting of Rad9 to DSBs. Then, phosphorylation of H2A by Tel1 may induce a stabilization of the Rad9/chromatin interaction and promote Rad9 hyperphosphorylation and Rad53 transautophosphorylation by the Ddc2/Mec1 complex. However, the mode of chromatin recognition by Rad9, Crb2 and 53BP1 is different, which implies that the corresponding interactions might be differently regulated. It was proposed by several authors that Rad9 tandem tudor region interacts directly with H3 dimethylated on K79 (16,32). We show in this paper that our Rad9 fragment does not bind to such modified histone in vitro. Methylation of H3 K79 may play a more general role in coordinating various repair processes (34). More recently, it was proposed that dimethylation of H4 K20 plays a critical role in the direct binding of yeast Rad9 homologs to chromatin (14). This hypothesis is not supported by our study. However, Rad9 tandem tudor domain might recognize another region of H4, or the multimeric functional state of Rad9 might be critical for histone binding. Lastly, the targeting of yeast Rad9 homologs to DSBs might depend on indirect interactions with methylated histones. More quantitative binding studies and 3D structural information are needed in order to understand how Rad9 cooperates with other BRCT proteins in order to recognize chromatin at the vicinity of DSBs and thus prevent accumulation of DSBs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Alain Lecoq at the SIMOPRO (CEA Saclay, France) for H4K20 and H4K20-long peptide chemical syntheses, Steven Dubois and Robert Thai at SIMOPRO (CEA Saclay, France) for amino acid composition and peptide sequence analyses, Nathalie Nozerand and Alain Lorphelin (CEA Marcoule, France) for their help in protein expression and purification and Cédric Laguri at the IBS (CEA Grenoble, France) for the annealing of the different 35 bp oligonucleotides (double strand, nick, mismatch and gap). We are grateful to François Bontems at Ecole Polytechnique (Palaiseau, France) and Laure Guilhaudis at the University of Rouen (France) who kindly lent us their 600 MHz spectrometers. The 800 MHz HSQC-NOESY spectra were recorded at the IBS (CEA Grenoble, France) with the help of Adrien Favier. This work was supported by grant number 4524 from Association pour la Recherche sur le Cancer. Funding to pay the Open Access publication charges for this article was provided by CEA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Javaheri A, Wysocki R, Jobin-Robitaille O, Altaf M, Cote J, Kron SJ. Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling. Proc. Natl Acad. Sci. USA. 2006;103:13771–13776. doi: 10.1073/pnas.0511192103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vialard JE, Gilbert CS, Green CM, Lowndes NF. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol. Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert CS, Green CM, Lowndes NF. Budding yeast Rad9 is an ATP-dependent Rad53 activating machine. Mol. Cell. 2001;8:129–136. doi: 10.1016/s1097-2765(01)00267-2. [DOI] [PubMed] [Google Scholar]
- 5.Sun Z, Hsiao J, Fay DS, Stern DF. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 6.Du LL, Nakamura TM, Moser BA, Russell P. Retention but not recruitment of Crb2 at double-strand breaks requires Rad1 and Rad3 complexes. Mol. Cell. Biol. 2003;23:6150–6158. doi: 10.1128/MCB.23.17.6150-6158.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saka Y, Esashi F, Matsusaka T, Mochida S, Yanagida M. Damage and replication checkpoint control in fission yeast is ensured by interactions of Crb2, a protein with BRCT motif, with Cut5 and Chk1. Genes Dev. 1997;11:3387–3400. doi: 10.1101/gad.11.24.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia Z, Morales JC, Dunphy WG, Carpenter PB. Negative cell cycle regulation and DNA damage-inducible phosphorylation of the BRCT protein 53BP1. J. Biol. Chem. 2001;276:2708–2718. doi: 10.1074/jbc.M007665200. [DOI] [PubMed] [Google Scholar]
- 9.Rappold I, Iwabuchi K, Date T, Chen J. Tumor suppressor p53 binding protein 1 (53BP1) is involved in DNA damage-signaling pathways. J. Cell. Biol. 2001;153:613–620. doi: 10.1083/jcb.153.3.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson L, Henderson C, Adachi Y. Phosphorylation and rapid relocalization of 53BP1 to nuclear foci upon DNA damage. Mol. Cell. Biol. 2001;21:1719–1729. doi: 10.1128/MCB.21.5.1719-1729.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz LB, Chehab NH, Malikzay A, Halazonetis TD. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks. J. Cell. Biol. 2000;151:1381–1390. doi: 10.1083/jcb.151.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Callebaut I, Mornon JP. From BRCA1 to RAP1: a widespread BRCT module closely associated with DNA repair. FEBS Lett. 1997;400:25–30. doi: 10.1016/s0014-5793(96)01312-9. [DOI] [PubMed] [Google Scholar]
- 13.Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- 14.Botuyan MV, Lee J, Ward IM, Kim JE, Thompson JR, Chen J, Mer G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–1373. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Charier G, Couprie J, Alpha-Bazin B, Meyer V, Quemeneur E, Guerois R, Callebaut I, Gilquin B, Zinn-Justin S. The Tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding. Structure. 2004;12:1551–1562. doi: 10.1016/j.str.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 16.Huyen Y, Zgheib O, Ditullio R.A., Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, et al. Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature. 2004;432:406–411. doi: 10.1038/nature03114. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Daniel J, Espejo A, Lake A, Krishna M, Xia L, Zhang Y, Bedford MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanders SL, Portoso M, Mata J, Bahler J, Allshire RC, Kouzarides T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell. 2004;119:603–614. doi: 10.1016/j.cell.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 19.Wysocki R, Javaheri A, Allard S, Sha F, Cote J, Kron SJ. Role of Dot1-dependent histone H3 methylation in G1 and S phase DNA damage checkpoint functions of Rad9. Mol. Cell. Biol. 2005;25:8430–8443. doi: 10.1128/MCB.25.19.8430-8443.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Du LL, Moser BA, Russell P. Homo-oligomerization is the essential function of the tandem BRCT domains in the checkpoint protein Crb2. J. Biol. Chem. 2004;279:38409–38414. doi: 10.1074/jbc.M403326200. [DOI] [PubMed] [Google Scholar]
- 21.Ward I, Kim JE, Minn K, Chini CC, Mer G, Chen J. The tandem BRCT domain of 53BP1 is not required for its repair function. J. Biol. Chem. 2006;281:38472–38477. doi: 10.1074/jbc.M607577200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clemenson C, Marsolier-Kergoat MC. The spindle assembly checkpoint regulates the phosphorylation state of a subset of DNA checkpoint proteins in Saccharomyces cerevisiae. Mol. Cell. Biol. 2006;26:9149–9161. doi: 10.1128/MCB.00310-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alpha-Bazin B, Lorphelin A, Nozerand N, Charier G, Marchetti C, Berenguer F, Couprie J, Gilquin B, Zinn-Justin S, et al. Boundaries and physical characterization of a new domain shared between mammalian 53BP1 and yeast Rad9 checkpoint proteins. Protein Sci. 2005;14:1827–1839. doi: 10.1110/ps.041305205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marsolier MC, Roussel P, Leroy C, Mann C. Involvement of the PP2C-like phosphatase Ptc2p in the DNA checkpoint pathways of Saccharomyces cerevisiae. Genetics. 2000;154:1523–1532. doi: 10.1093/genetics/154.4.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 26.Duband-Goulet I, Courvalin JC. Inner nuclear membrane protein LBR preferentially interacts with DNA secondary structures and nucleosomal linker. Biochemistry. 2000;39:6483–6488. doi: 10.1021/bi992908b. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. NY, USA: Cold Spring Harbor Laboratory Press; 1989. edn. [Google Scholar]
- 28.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 29.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 30.Du LL, Nakamura TM, Russell P. Histone modification-dependent and -independent pathways for recruitment of checkpoint protein Crb2 to double-strand breaks. Genes Dev. 2006;20:1583–1596. doi: 10.1101/gad.1422606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charier G, Alpha-Bazin B, Couprie J, Callebaut I, Berenguer F, Quemeneur E, Gilquin B, Zinn-Justin S. 1H, 13C and 15N resonance assignments of the region 1463–1617 of the mouse p53 binding protein 1 (53BP1) J. Biomol. NMR. 2004;28:303–304. doi: 10.1023/B:JNMR.0000013688.79211.8c. [DOI] [PubMed] [Google Scholar]
- 32.Grenon M, Costelloe T, Jimeno S, O'Shaughnessy A, Fitzgerald J, Zgheib O, Degerth L, Lowndes NF. Docking onto chromatin via the Saccharomyces cerevisiae Rad9 Tudor domain. Yeast. 2007;24:105–119. doi: 10.1002/yea.1441. [DOI] [PubMed] [Google Scholar]
- 33.Iwabuchi K, Basu BP, Kysela B, Kurihara T, Shibata M, Guan D, Cao Y, Hamada T, Imamura K, et al. Potential role for 53BP1 in DNA end-joining repair through direct interaction with DNA. J. Biol. Chem. 2003;278:36487–36495. doi: 10.1074/jbc.M304066200. [DOI] [PubMed] [Google Scholar]
- 34.Bostelman LJ, Keller AM, Albrecht AM, Arat A, Thompson JS. Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair (Amst.) 2007;6:383–395. doi: 10.1016/j.dnarep.2006.12.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.