Abstract
In an effort to improve the knowledge about the rules which direct the effect of the early ORF sequences on translation efficiency, we have analyzed the effect of pairs of the six arginine codons at the second and third positions on the expression of lacZ variants. Whereas the pairs of identical AGA or AGG codons were favorable for the gene expression, identical pairs of each of the four CGN codons were very inefficient. This result was unexpected because tandems of AGA or AGG codons located in more internal gene positions provoke deficient expression whilst internally located CGU and CGC are the most abundant and efficiently translated arginine codons. The mixed combinations of AGA and each of the CGN codons usually resulted in efficient rates of lacZ expression independently of the peptidyl-tRNA propensity to dissociate from the ribosome. Thus, the variant harboring the pair of AGA codons was expressed as efficiently as the variant carrying a pair of AAA codons in the same positions, a configuration reported as one of the most common and efficient for gene expression. We explain these results assuming that the presence of adenines in these early positions enhance gene expression. As expected, specific mRNA levels correlated with the intensity of lacZ expression for each variant. However, the induction of lacZ AGA AGA gene in pth cells accumulated peptidyl-tRNAArg4 as well as a short 5′-proximal lacZ mRNA fragment suggesting ribosome stalling due to depletion of aminoacylated-tRNAArg4.
INTRODUCTION
The early steps after translation initiation are critical for the efficiency of gene expression. In addition to the initiation codon in the mRNA, recognized by the initiator tRNA, the flanking sequences affect the rate of translation (1). Upstream from the initiation codon there is usually a four to six nucleotide tract, named Shine–Dalgarno sequence (2) or SD, that pairs with a complementary, or anti-SD sequence, close to the 3′-end of the 16S RNA of the 30S ribosomal subunit. The SD region directs the ribosome to the initiation codon during translation initiation (3). The downstream region (DR), the nucleotide sequence following the initiation codon also affects the efficiency of translation (4–7). It is unlikely that the DR acts by pairing with a complementary sequence in the 16S rRNA as SD does (8,9). It has been suggested that the effect of the DR on gene expression stems from the base sequence in mRNA rather than from the encoded amino acid sequence in the protein. Comparison of DRs containing different iso-codons, thus generating an identical protein, could give significant differences of gene expression (10).
Particularly critical for gene expression is the nature of the codon next to the initiation triplet, the +2 position. Codon changes in +2 can affect gene expression by 15- to 20-fold (7,11). Also, the effect of the +2 codon on the gene expression can be modulated by the subsequent triplets (7,10). In general, a high adenine content of the +2 codon is associated with high gene expression (7). The lysine codon AAA is the most common codon at +2 to +5 positions in Escherichia coli reading frames and it usually determines efficient gene expression (7,11,12). Indeed, the changes of gene expression due to variations in the content of adenines downstream of the initiation codon, correlates with changes in in vitro ribosome-binding strength, an association probably mediated by protein molecules (13).
Tandems of the low usage arginine codons AGA or AGG at different positions in the reading frame inhibit gene expression (14–17). The longer and closer to the initiation codon is the tandem the stronger is the inhibition; starting up at the +10 codon, the closest assayed position (14). The effect of low-usage codon tandems located farther downstream from the initiation codon is observed under conditions which favor tRNA limitation. For example, translation of a lacZ variant harboring contiguous AGA codons in the positions +352 and +353 is arrested at these codons upon the concurrent expression of a minigene that sequestered the cognate tRNAArg4 as peptidyl-tRNAArg4 (pep-tRNAArg4) in a host defective for peptidyl-tRNA hydrolase (pth) (18). Also, depleting the pool of tRNAArg4 stops the ribosome movement and enhances tagging at tandem of AGA codons by the SsrA system (19).The naturally occurring AGA and AGG codons in the positions +3 and +4 of the phage lambda int gene modulate the expression by tRNA sequestration (20,21). It is likely that the expression of int in pth+ cells may be limited by the elevated drop-off rate of pep-tRNAArg4 that overwhelms the Pth activity in the cell.
We have investigated the effect of the substitutions of arginine codons at positions +2 and +3 on the expression of a reporter gene. Unexpectedly, CGU and CGC, the arginine codons more frequently used in bacteria, were deficient in supporting the gene expression whilst AGA and AGG, two of the less frequent arginine codons, were the most effective in wild-type bacteria. In spite these results, a variant substituted for the AGA AGA codons in a pth mutant strain was deficient in gene expression due to ribosome stalling at these codons. This indicates that the efficiency of translation does not necessarily correlate with the propensity of the pep-tRNAs to dissociate from the ribosomes. The nucleotide composition of the codons at +2 and +3 is a dominant factor in translation efficiency. Therefore, the deficiency of the CGU and CGC codons located at these positions, is due to the unfavorable base composition for translation rather than to an increased rate of abortive pep-tRNA dissociation from the ribosome.
MATERIALS AND METHODS
Bacterial strains, plasmids and growth conditions
We carried experimental procedures on the E. coli K-12 strains P90C [ara Δ(lac-pro) thi] and its pth mutant P90C rap [P90C pth(rap) zch::Tn10]. This pth mutation just expresses 10% of the normal Pth activity (22).
The plasmids used were ampicillin-resistant derivatives of pKQV4 (23) containing lacZ gene variants in the second and third codons of the ORF (Figure 1). We also employed pDC952 which carries argU, the gene for tRNAArg4, cognate to the AGA codon (24) and pGREC, containing the pth+ gene of E. coli (20,25). Both constructs are chloramphenicol-resistant pACYC184-based derivatives. The cell cultures of the strains harboring the lacZ variant plasmids were grown at 37°C in Luria–Bertani (LB) medium containing ampicillin 100 µg/ml (Amp). The strains co-transformed with the lacZ variant plasmids and pDC952 or pGREC were grown in LB-Amp medium plus chloramphenicol 50 µg/ml (Cm).
Figure 1.
Map not to scale of the pKQV4-based constructs used in this work. Upon addition of the gratuitous inducer IPTG the Lac repressor, encoded by lacIq (open arrow) dissociates from the operator region Olac (gray box) and transcription initiates at promoter Ptac (bold arrow). Transcription terminates at the transcription terminator Trrnb (gray box). The transcription initiated at Ptac drives the expression of the lacZ gene (large open arrow) cloned between the EcoRI and HindIII sites (underlined). The used lacZ variants carried different codons in the +2 and +3 positions. Translation initiation occurs, thorough association of the Shine-Dalgarno (SD, bold) sequence in the mRNAs and the ribosomes, at the translation initiation codon ATG. The constructs were selected in transformed cells resistant to ampicillin conferred by the gene bla (open arrow), which encodes β-lactamase. The relative position of the plasmid replication origin (ori) is indicated. The segments in bold indicate other vector DNA sequences.
Construction of lacZ variant plasmids
The lacZ gene from pLEX/lacZ plasmid (Invitrogen) was amplified by PCR and the final product cloned between the EcoRI and HindIII restriction sites of pKQV4 (Figure 1). The second and third codons of the lacZ ORF were replaced by identical or combined pairs of all six arginine (AGA, AGG, CGA, CGG, CGC and CGT), leucine (CTA and CTC) and lysine (AAA) codons. The lacZ constructs were obtained by site-directed mutagenesis using a Quick@Change mutagenesis kit (Stratagene) and pairs of complementary oligonucleotides with the common sequence 5′-CAGAATTCATGNNNNNNCCCGTCGTTTTACAACG-3′ and 5′-CGTTGTAAAACGACGGGNNNNNNCATGAATTCTG-3′ where the tracts of Ns represent the above-mentioned codon substitutions and their complementary sequences.
β-Galactosidase activity
Fresh cell cultures at an OD600 of 0.3 were diluted to an OD600 of 0.1 in the same fresh medium preheated at 37°C. After 10 min, 1 mM of IPTG was added to induce β-galactosidase (β-Gal) synthesis from the lacZ variant plasmids. Samples were extracted at different times and the β-Gal activity was determined by employing a modified procedure of the Miller protocol (26). The cellular density of the samples was measured at OD600 and immediately, 0.5 ml of each sample was mixed with 0.5 ml of Z buffer containing 30 µl of chloroform and 15 µl of 0.1% SDS. The mixes were vortexed for 30 s and then incubated at room temperature for at least 10 min. To start the reaction, 200 µl of ONPG solution (4 mg/ml) in Z buffer was added to the samples. The reaction was stopped by mixing 0.5 ml of 1 M sodium carbonate (the reaction time fluctuated depending on the velocity of β-Gal synthesis). All the samples were centrifuged at 10 000 r.p.m. for 10 min to sediment the cell debris and to measure the OD420 of the supernatants. With these data, we calculated the β-Gal activity in Miller units as previously indicated (26). Reaction rates (Miller units/min) were calculated by linear regression from the β-Gal synthesis obtained at different incubation times for each lacZ variant.
Peptidyl-tRNA levels
Pep-tRNA levels were measured by northern blot assays as previously described (27). Briefly, cultures of the pth mutant transformed with lacZ variant plasmids were grown at 37°C to an OD600 of 0.3 in LB-Amp. Then, 1 mM IPTG was added and the cultures incubated for 40 min more. The cells were harvested at 4°C and the total tRNA was isolated under acidic conditions (27). To estimate the fraction of pep-tRNA relative to total tRNA, aminoacyl-tRNA was hydrolyzed with copper sulfate in one of two aliquots. Four microgram of RNA from each sample were resolved overnight by acid/urea PAGE, transferred to Hybond-N+ nylon membranes (Amersham Biosciences) and hybridized to 5′-32P end-labeled oligonucleotides. The radioactive signals were quantified using a Typhoon Scan (Amersham Biosciences). The amount of pep-tRNAs in the samples was estimated using the following formula:% pep-tRNA = c.p.m. of pep-tRNA × 100/ c.p.m. of uncharged-tRNA + c.p.m. of aminoacylated-tRNA + c.p.m. of pep-tRNA. The tRNA-specific oligodeoxyribonucleotide probes: 5′- CCTGCGGCCCACGACTTAG-3′, for tRNAArg4; 5′-CCTGCAATTAGCCCTTAGG-3′, for tRNAArg5; 5′-CCTCCGACCGCTCGGTTCG-3′, for tRNAArg2; 5′-CCTGAGACCTCTGCCTCCGGA-3′, for tRNAArg3; 5′-CCTGCGACCAATTGATTAAA-3′, for tRNALys; 5′-CACCTTGCGGCGCCAGAA-3′, for tRNALeu3; 5′-CCCGCACAGCGCGAACGCCG-3′, for tRNALeu5 were chemically synthesized according to the sequences reported by Dong et al. (28).
lacZ mRNA detection
Cultures induced with 1 mM IPTG were grown to an OD600 of 0.4 and harvested by centrifugation. Total RNA was extracted with hot phenol (65°C) from a 10 ml culture as described by Aiba et al. (29). About 30 μg of RNA was denatured in 40% formamide plus 5 μg/ml ethidium bromide solution at 65°C for 10 min. The RNA species were resolved by electrophoresis through 1.5% denaturing agarose gel containing 2.2 M formaldehyde and transferred to a nylon membrane (Hybond-N, Amersham Pharmacia Biotech). Hybridization was carried out at 42°C in 5× SSPE, 0.5% SDS, 100 μg/ml salmon sperm DNA, 0.1% bovine serum albumin, 0.1% Ficoll and 0.1% polyvinyl pyrrolidone and 5′-32P end-labeled antisense oligonucleotide: 5′-CGTTGTAAAACGACGGGTCTTCTCATGAATTCTG-3′, for lacZ AGA2 AGA3 variant; 5′-CGTTGTAAAACGACGGGCCGCCGCATGAATTCTG-3′, for lacZ CGG2 CGG3 variant; or 5′-CGTTGTAAAACGACGGGTAGTAGCATGAATTCTG-3′, for lacZ CTA2 CTA3 variant. After 16 h incubation the membranes were rinsed twice at room temperature with 2× SSPE, 0.1% SDS, dried and exposed to X-ray film to develop the signal.
RESULTS
The expression efficiency of the lacZ variants harboring identical pairs of codons in the positions +2 and +3
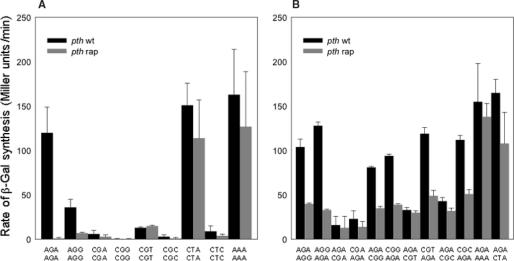
It has been noticed that the presence of low usage AGA triplets in lacZ +2 and +3 codon positions (AGA2 AGA3) induces a robust expression of the lacZ gene (18). This was so in spite of reports that contiguous low usage codons near the initiation codon were deleterious for gene expression (14,16,17,30–33). In order to further examine this apparent paradox we analyzed the effects of AGA2 AGA3 and other pairs of identical arginine codons substituted in these positions of the lacZ ORF. By maintaining the N-terminal amino acid composition of the protein, variations in the β-Gal activity due to different N-terminal protein composition were eliminated. We generated the appropriate lacZ variants by site-directed mutagenesis on the construct shown in Figure 1. Then, P90C a strain lacking in the lac operon, was transformed with the different constructs bearing each of the double substituted variants. The lacZ expression was induced with IPTG and the rates of β-Gal synthesis calculated (see Materials and Methods section). The results (Figure 2A) showed that the AGA2 AGA3 lacZ variant generated the highest rate of β-Gal synthesis in the wild-type strain. The rate of β-Gal synthesis measured for the AGA2 AGA3 variant was nearly as high as that observed for a variant which contained a pair of AAA codons in +2 and +3 positions (Figure 2A). This last pair of lysine codons has been reported as one of the most efficient pairs for gene expression (7,12) and one of the most frequently located at these positions (12,34). The variant harboring low-usage AGG codon, AGG2 AGG3, followed expressing lacZ about one-third as fast as the AGA2 AGA3 construct. The variants with pairs of other identical low usage (CGA, and CGG) or common (CGC or CGU) arginine codons generated low levels of β-Gal activity. A similar result was observed using leucine codons. A pair of CUA leucine low-usage codons in these positions promoted lacZ expression comparable to that expressed by the AAA2 AAA3 and AGA2 AGA3 pairs (Figure 2A). However, a variant substituted with the leucine common codons CUC2 CUC3, was defective to promote gene expression. Thus, the differences of gene expression due to changes in the second and third positions of lacZ ORF cannot be assigned to the codon usage or to the relative abundance of the tRNA isoacceptors.
Figure 2.
The effect of base composition of the codons located at the positions +2 and +3 on the expression of lacZ gene in wild-type and pth strains. Each pair of columns correspond to the rates in P90C and P90C pth(rap) as indicated. Results represent the mean ± SD of at least two, and up to nine independent experiments. The pairs of codons of each of the lacZ variants, one (+2) above the other (+3), are indicated below each pair of columns. (A) identical codon substitutions; (B) mixed codon substitutions. (see Materials and Methods section for details).
The accumulation of pep-tRNA upon expression of the lacZ variants in the pth(rap) strain
We investigated whether the abortive translation process of pep-tRNA drop-off played a role in the scant lacZ expression observed for some of the variants. The appropriate constructs were transformed into the pth(rap) strain. The rates of β-Gal synthesis and the levels of cognate pep-tRNA accumulation were determined upon IPTG induction (Figure 2A and Table 1). The AGA2 AGA3 variant, which expressed β-Gal very efficiently in the wild-type cells, promoted poor β-Gal synthesis and high accumulation (70%) of pep-tRNAArg4 in the pth strain (see Figures 2A, 4B, 4C and Table 1). On the other hand, the AGG2 AGG3 variant, which expressed β-Gal moderately in wild-type cells, mediated 5-fold less expression in the pth cells and accumulated intermediate level (36%) of pep-tRNAArg5 (Figure 2A and Table 1). The high rate of the lacZ gene expression, especially that of the AGA2 AGA3 variant in the wild-type cells, is not incompatible with the high level of pep-tRNAArg4 drop-off under limiting Pth activity. Rather, this observation suggests either that drop-off does not occur in the wild-type cells or that, if it does, the level of Pth activity present readily hydrolyzes the released pep-tRNAs preventing starvation for free tRNAs. Associated to the lacZ expression inhibition, the lacZ AGA2 AGA3 variant also affected the grow rate of the pth mutant (Figure 4D, pACYC). No comparable levels of accumulation of the corresponding pep-tRNAs were observed for the other variants assayed (Table 1). Unlike the AGA2 AGA3 and AGG2 AGG3 lacZ variants, the efficient expression of the AAA2 AAA3 and CTA2 CTA3 variants was only somewhat reduced in the pth strain (Figure 2A) in agreement with the low levels of the pep-tRNA accumulated upon induction of these variants (Table 1). Therefore, it appears that there is no correlation between the high rates of protein synthesis in the wild-type cells and the accumulation of pep-tRNA in the pth cells. On the other hand, the poor β-Gal expression mediated by other lacZ variants (CGG2 CGG3, CGC2 CGC3, CGA2 CGA3, CGT2 CGT3 or CTC2 CTC3) in the wild-type cells was also accompanied by inefficient β-Gal synthesis and modest accumulation of the cognate pep-tRNA in the pth strain (Figure 2A and Table 1). Thus, for this last group of variants, the inefficient synthesis of β-Gal protein in wild-type cells may result from defective translation of the codons located in the +2 and +3 positions or defective interaction of the mRNA with the ribosome, but not from abortive translation.
Table 1.
Effect of the pairs of identical codons in lacZ on the accumulation of pep-tRNA in P90C pth(rap)
| lacZ ORF codona | % of specific pep-tRNA accumulatedb | |
|---|---|---|
| +2 | +3 | |
| AGA | AGA | 71 ± 6 (4) |
| AGG | AGG | 36 ± 6 (3) |
| CGG | CGG | 9 |
| CGT | CGT | 7 ± 2 (2) |
| CGA | CGA | 12 ± 4 (2) |
| CGC | CGC | 6 |
| CTA | CTA | 21 ± 2 (2) |
| AAA | AAA | 17 |
aThe lacZ variants contained the indicated codons at the second (+2) and third (+3) codon positions.
bThe percentages of pep-tRNAs were calculated relative to the total concentrations of specific tRNA isoacceptors (see Materials and Methods section).
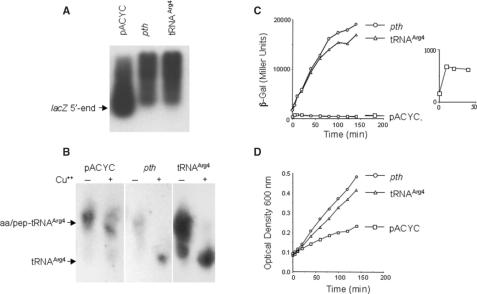
Figure 4.
Analysis of β-Gal synthesis, lacZ mRNA levels, pep-tRNAArg4 accumulation and cellular growth during the expression of the lacZ AGA2 AGA3 variant in pth cells. Cultures of P90C pth(rap) co-transformed with the plasmid construction harboring the lacZ AGA2 AGA3 variant and pACYC (vector), pDC952 (a tRNAArg4 overproducer) or pGREC (a Pth overproducer) were induced for lacZ expression by the addition of 1 mM IPTG. (A) Analysis of lacZ mRNA. Total RNA was extracted after 40 min of the induction and a northern blot assay was performed with a specific 5′-end lacZ oligonucleotide probe to detect the lacZ mRNAs. (B) Estimation of accumulated pep-tRNAArg4. Total RNA was extracted as indicated above. The samples were halved and each aliquot treated (+) or not (−) with a copper salt solution to hydrolyze aminoacyl-tRNA and therefore, unmask the pep-tRNA. A northern blot assay was performed to reveal the accumulated pep-tRNAArg4 using a radioactively labeled oligonucleotide probe specific for tRNAArg4. (C) Time course of β-Gal activities determined in culture samples drawn at the indicated times after induction. The inset is an enlargement of the β-Gal activity generated by the cells co-transformed with the empty vector. (D) Growth curves measured as optical density of the P90C pth(rap) strain transformed with the indicated overproducing constructs or with the empty vector (pACYC) after induction with IPTG. For experimental details see Material and Methods section.
Expression efficiency of the lacZ variants containing combinations of AGA and other codons located in the positions +2 and +3
We investigated whether the favorable effect of AGA codons on the lacZ expression was associated with its location in the second, the third or both codon positions. lacZ variants containing combinations of the AGA codon in the +2 or +3 positions and each of the other five arginine codons placed in the reciprocal +3 or +2 positions were assayed. The appropriate constructs were transformed into the P90C strain and the rates of β-Gal synthesis determined upon IPTG induction (see Materials and Methods section). The results, shown in Figure 2B, indicated that the different variants containing the combinations of AGA and the other arginine codons expressed broadly different levels of β-Gal activity which spanned almost a 10-fold range. But in all cases, the variants harboring the AGA codon in position +3 expressed the lacZ gene at higher rates than the corresponding variants harboring the AGA codon in position +2 (compare wild-type columns in Figure 2B). The variants containing an AGA codon, either in positions +2 or +3, and any of the other five arginine codons in the alternative position, enhanced the rate of lacZ expression relative to those variants harboring identical pairs of non-AGA arginine codons (compare Figure 2A and B). The variants carrying combinations of the AGA codon in position +2 and other favorable codons such as the CUA or AAA codons in location +3, promoted as high rates of lacZ expression in the wild-type cells as those mediated by lacZ variants carrying the pairs AGA2 AGA3, CUA2 CUA3 and AAA2 AAA3 (compare Figure 2A and B). These non-arginine favorable codons may share with AGA the enhancing effect with the CGN codons.
The β-Gal expression efficiency and pep-tRNA accumulation of the lacZ variants containing AGA and other codons in +2 and +3 in the pth(rap) cells
From the results in Table 1, it appears that AGA and AGG are the arginine codons that mediate the highest accumulation of pep-tRNAs in the pth strain during expression of corresponding lacZ variants. We asked whether the position of the AGA codon, either in the positions +2 or +3 induces drop-off of the involved pep-tRNAs. The expression of lacZ variants containing alternative combinations between AGA and other arginine codons in positions +2 and +3 were assayed in the pth(rap) mutant. The results (Figure 2B) showed, in general, that the rate of lacZ expressed by a variant in the wild-type cells was higher than the rate promoted by the same variant in the pth strain. These results are compatible with the notion that, under limiting Pth activity, the AGA codon and/or the accompanying arginine codon of the pair have a propensity to drop-off. Because the pth cells are defective in the regeneration of aminoacylable tRNAs from pep-tRNAs, they would be limited in their capacity to synthesize β-Gal. Then, the relative accumulation of pep-tRNAArg4 and the pep-tRNAs specific for the accompanying arginine codons was measured (Table 2, see Materials and Methods section). With the exception of the AGA2 CGT3 variant, all the lacZ variants carrying the AGA codon accumulated pep-tRNAArg4 at higher levels than 20%. The pep-tRNA specific for the accompanying non-AGA arginine codon in the pair was accumulated by the variants where AGA resided in the +3 codon position, but not in the +2 location (Table 2, compare lines 1 and 2, 3 and 4, etc.). These data argue that the AGA codon promoted translation of the accompanying arginine codon and that the pep-tRNAs of these codons accumulated in response to the drop-off at the subsequent AGA codon. Therefore, AGA in the positions +2 and/or +3 promoted translation of the associated arginine codon and facilitated the dissociation of pep-tRNA from the ribosome affecting the rate of lacZ expression under limiting Pth activity.
Table 2.
Effect of the pairs of codon combinations in lacZ on the accumulation of pep-tRNA in P90C pth(rap)
| lacZ ORF codona | % of specific pep-tRNA accumulatedb | ||
|---|---|---|---|
| +2 | +3 | +2 | +3 |
| AGA | AGG | 29 ± 5 (2) | 12 ± 3 (2) |
| AGG | AGA | 38 ± 6 (3) | 24 ± 1 (2) |
| AGA | CGA | 44 ± 2 (4) | 6 ± 1 (3) |
| CGA | AGA | 32 ± 4 (3) | 28 ± 3 (4) |
| AGA | CGG | 21 ± 13 (2) | 10 |
| CGG | AGA | 25 | 20 ± 3 (2) |
| AGA | CGC | 25 | 8 |
| CGC | AGA | 18 | 22 |
| AGA | CGT | 12 ± 2 (2) | 9 ± 1 (2) |
| CGT | AGA | 27 ± 4 (2) | 27 ± 3 (2) |
| AGA | AAA | 10 | – |
aThe lacZ variants contained the indicated codons at the second (+2) and third (+3) codon positions.
bThe percentages of pep-tRNAs were calculated relative to the total concentrations of specific tRNA isoacceptors (see Materials and Methods section). The two percentage columns correspond to the pep-tRNAs specific for the codons located at the +2 and +3 position in each case. The percentages are the averages of the number of experiments indicated in parentheses.
The variants harboring AAA2 AAA3 or CTA2 CTA3 expressed lacZ efficiently in the wild-type cells and only slightly less well in the pth strain (Figure 2A). This was observed also with variants bearing the codon combinations AGA2 AAA3 and AGA2 CUA3 (Figure 2B). Thus, combinations of codons that express lacZ variants efficiently as identical pairs placed in +2 and +3 are also efficient in mixed combinations. In addition the levels of expression of these variants in the pth cells, suggest that they do not promote the accumulation of pep-tRNAs. Thus, it seems that the pep-tRNAArg4 accumulates in conditions where the codon subsequent to AGA is difficult to translate.
Correlation between drop-off and accumulation of a short 5′-proximal lacZ mRNA by the AGA2 AGA3 variant
It is assumed that starvation for an aminoacylated-tRNA due to the accumulation of pep-tRNA in pth cells induces ribosome stalling at the ‘hungry’ codons in the mRNA. The stalled ribosomes protect the associated mRNA against the ribonucleolytic activities of the cell (27,35). Therefore, it is expected that ribosomes stalled in the AGA codons, located next to the initiation codon, would protect a 5′-proximal segment of the mRNA. To test this prediction the lacZ variant harboring AGA codons in the positions +2 and +3 was expressed in the pth strain. Total RNA was extracted and submitted to northern blot analysis using an oligonucleotide-probe complementary to the 5′-end lacZ mRNA (see Materials and Methods section). The results showed that the expression of lacZ AGA2 AGA3 variant generated a short transcript containing the 5′-end sequences of lacZ mRNA (Figure 3B). The expression of the same variant in the wild-type strain, that expressed the lacZ gene efficiently, yielded lacZ transcripts of a wide size range, mostly larger than the 5′-end fragment (Figure 3A). The CTA2 CTA3 variant, that expresses the lacZ gene nearly as efficiently in the pth mutant as it does in wild-type cells, produced the wide size range pattern of lacZ transcripts in both strains. On the other hand, the lacZ CGG2 CGG3 variant, that was very ineffective to express lacZ (Figure 2A), yielded no lacZ mRNA signals at all (Figure 3). Likewise, no evidence of lacZ mRNA signals were observed for the low expressed variants carrying codons such as CGA2 CGA3, CGC2 CGC3, CGU2 CGU3, CGA2 AGA3 or AGA2 CGA3 (data not shown). We suggest that these pairs of codons in positions +2 and +3 are defective in translation either because they fail to interact properly with the ribosome or because these codons are difficult to read by the specific tRNA.
Figure 3.
lacZ mRNA detection during the expression of the lacZ variants. Cultures of the strains P90C (A) and P90C pth(rap) (B) transformed with the indicated lacZ +2, +3 variants were induced with 1 mM IPTG for 40 min. Total RNA was extracted, resolved by gel electrophoresis and transferred to nylon membranes. Upper panels: northern blot assays performed with 5′-end labeled oligonucleotide probes to detect the lacZ mRNAs molecules. Heterogeneous lacZ mRNA and the 5′-proximal lacZ mRNA segment are indicated. As a RNA loading control, lower panels, it is shown the ethidium bromide stained rRNAs transferred to the same membrane which was used for the northern blot assay.
The presence of the 5′-end lacZ mRNA segment in the pth cells expressing lacZ AGA2 AGA3 variants (Figure 3B and Figure 4A, pACYC) correlated with accumulation of pep-tRNAArg4 (Figure 4B, pACYC), deficiency of lacZ expression (Figure 4C, pACYC) and reduction of the rate of cellular growth (Figure 4D, pACYC). Cells supplemented with an excess of Pth protein or tRNAArg4 showed longer species of lacZ mRNA (Figure 4A, lanes 2 and 3), did not accumulate pep-tRNAArg4 (Figure 4B, lanes 4 and 6), restored β-Gal activity and rescued the cellular growth (Figure 4C and D). These results support the notion that, under limited Pth activity, the production of β-Gal protein from the lacZ mRNA containing the AGA2 AGA3 codons was limited by the starvation for charged tRNAArg4 and the ribosome pause at the AGA codons in the lacZ mRNA. The presence of the 5′-proximal short mRNA was not observed in a preparation of the lacZ CTA2 CTA3 variant (Figure 3). Instead, they produced the pattern of long lacZ transcripts associated with the high rates of lacZ expression (Figure 3). In general, the presence of large-size lacZ mRNAs correlated with high rates of β-Gal synthesis, but low rates of β-Gal synthesis were compatible either with high levels of truncated lacZ mRNA, as in the AGA2 AGA3 variant in the pth cells, or no lacZ mRNA at all.
DISCUSSION
The objective of the current study was to understand how the sequences of the early codons in ORFs affect gene expression in bacteria. A recent study shows that the low-usage AGA triplets in early codon positions induce a robust expression of the lacZ gene (18). In order to know how these codons affect mRNA translation, we have analyzed their effect on the efficiency of lacZ expression, induction of pep-tRNA drop-off and lacZ mRNA concentration. The AGA2 AGA3 lacZ variant promoted the highest rate of β-Gal synthesis, the AGG2 AGG3 variant was fairly efficient and all the variants carrying identical pairs of CGN codons (CGU, CGC, CGA or CGG) were rather defective (Figure 2A). The combination of one AGA codon, in either position +2 or +3, with any other arginine codon, in the alternate +3 and +2 positions, promoted lacZ expression efficiently (Figure 2B). Interestingly, the high levels of lacZ expression induced by AGA codons in wild-type cells were compatible with the high rate of pep-tRNA drop-off in pth bacteria (Tables 1 and 2). On the other hand, it was difficult to assess whether the low rate of lacZ expression promoted by the CGN codons was accompanied by pep-tRNA drop-off (Tables 1 and 2). Therefore, in the analyzed cases, there is no correlation between gene expression and pep-tRNA accumulation. As expected, the level of lacZ expression corresponded with the concentration of lacZ mRNA accumulated (Figure 3). These findings support and extend the notion that the sequence downstream the initiation codon affects the efficiency of gene expression. Possibly, the AGA2 AGA3, unlike CGN2 CGN3, facilitates the interaction of the ribosome with the mRNA (see subsequently).
The efficiency of gene expression is affected by multiple factors relative to the early codon composition: secondary structure of mRNA, efficiency of mRNA association with the ribosomes, degree of ribosome pausing at specific codons, propensity of the different pep-tRNAs to drop-off, rates of codon reading by the tRNAs and codon context itself. Here, it is shown that the variant that harbors AGA2 AGA3 promote quite an efficient expression of the lacZ gene in bacteria. This expression is comparable to that attained by the lacZ variant that carries AAA2 AAA3 (Figure 2A), a codon configuration frequently found in the highly expressed genes of E. coli (7,10–12). The efficient expression of the AGA2 AGA3 variant in wild-type cells occurred in spite of elevated rates of pep-tRNAArg4 drop-off. This was revealed by the high levels of pep-tRNAArg4 accumulated upon expression of the variant in a mutant deficient in Pth activity (Table 1) (36). It seems that the mechanism involved in enhancing translation efficiency by the AGA codons overwhelms the negative effect of the pep-tRNA drop-off that occurs at these codons. It is likely that in the pth mutant, the expression of lacZ leveled off soon after an initial burst of β-Gal synthesis due to starvation for the aminoacylable tRNAArg4 which was sequestered as pep-tRNAArg4 (Figure 4C, inset). Accordingly, the overproduction of Pth and tRNAArg4, conditions that increase the pool of aminoacylable tRNAArg4 in the cell, mitigated the defect in β-Gal synthesis (Figure 4C). The propensity of pep-tRNAArg4 to drop-off did not depend on the presence of two contiguous AGA codons because the expression of variants carrying the AGA codon in combinations with the other arginine codons also resulted in the accumulation of pep-tRNAArg4 (Table 2).
We did not observe any correlation between the rate of pep-tRNA drop-off (Tables 1 and 2) or the frequency of codon usage and the efficiency of lacZ expression (Figure 2A and B) consistent with previous findings relative to codons located next to the initiation codon (37). Instead, our results conform to the correlation between gene expression and adenine content of the early ORF sequences that has been proposed for different genes (13,38–40). Accordingly, the AGA2 AGA3 variant showed the highest rate of lacZ expression, the AGG2 AGG3 variant expressed lacZ at an intermediate rate and the CGN2 CGN3 variants (where N is not adenine, see subsequently) expressed lacZ the poorest. It has been proposed that the early adenines enhance translation by increasing the rate of association of mRNA and ribosomes. Rather than a direct interaction between mRNA and 16S rRNA by sequence complementation, the association could be mediated by ribosomal proteins (13). One could argue that the CGA2 CGA3 lacZ variant, expressed poorly in spite the fact that it contained as many adenines as the AGG2 AGG3 variant. However, the CGA variant may be a special case as CGA has been reported as a codon difficult to translate by its correlative tRNAArg2 (41). As the defective expression of the CGN2 CGN3 variants was not related to the accumulation of pep-tRNA specific to these codons under limiting Pth activity (Table 1), we assume that the mRNAs from these variants are deficient in binding to ribosomes or in the formation of ternary complexes with aminoacyl-tRNAs. The possibility that these codons, per se, are difficult to translate when located in the positions +2 or +3 is unlikely because when they are next to an AGA codon they express lacZ efficiently (Figure 2B). Again the exception was codon CGA.
We considered the possibility that the different degrees of expression of the lacZ variants were explained by other mechanisms: translation reinitiation, formation of internal secondary structures in the mRNA, and the AG-rich arginine codons acting as secondary SD sequences. In wild-type cells, it is unlikely that re-initiation of translation downstream of the AGA tandem would explain lacZ expression because the released pep-tRNA would be readily hydrolyzed by Pth and ribosomal stalling would not be expected to occur. Under limiting Pth, however, stalling does occur as a consequence of pep-tRNA accumulation and reduction in the pool of charged tRNA. However, if reinitiation takes place in pth(rap) cells, it should be negligible because the β-Gal activity synthesized was very scant (Figures 2A and 4C).
To assess the formation of secondary structures between the different codons in +2 and +3 positions and the neighboring nucleotide sequences in the mRNAs of the variants, an informational search using an appropriate program was used (42). The data did not reveal a consistent correlation between the degree of lacZ expression and the proposed stability of the generated structures (data not shown). Then, it was examined whether the efficient gene expression of the lacZ variants carrying the codon pairs AGA2 AGA3 and AGG2 AGG3 was due to these pairs acting as secondary SD regions (43). However, if this assumption was true, the facts indicate that it did not correspond to a simple scheme because first, the lacZ variant carrying the codon sequence AGG2 AGG3, which is a near consensus SD, expresses the gene less efficiently than the codon sequence AGA2 AGA3, a less SD-like sequence and second, unlike authentic SD sequences that anchor the mRNA to the 16S rRNA, the pairs of codons AGA2 AGA3 and AGG2 AGG3 in the lacZ mRNAs are translated robustly as shown by the accumulation of the respective pep-tRNAs in cells defective for Pth activity (Table 1). Furthermore, the levels of the pep-tRNAs accumulated in the pth(rap) mutant upon expression of the variants, correlated with the efficiency of lacZ expression in the wild-type cells. Thus the AGA and AGG codons seem to affect the rate of lacZ expression mainly by their contribution to the translation rate of the lacZ mRNA rather than by acting as secondary SD regions. However, a mechanism invoking the transient association of the ribosome with the secondary SD region represented by these codons, followed by sliding back to the original SD sequence to start translation (44,45) cannot be ruled out by our results.
ACKNOWLEDGEMENTS
The authors wish to thank Guadalupe Aguilar González and Eva Jacinto Loeza for skilful technical support, to Guillermina Rosas for the pGREC construct, to Emanuel Goldman for careful reading and useful suggestions to the manuscript. The Consejo Nacional de Ciencia y Tecnología, México (28401N, 3775 N) and The Consejo del Sistema Nacional de Educación Tecnológica, México (402003247MP). Funding to pay the Open Access publication charges for this article was provided by Centro de Investigación y de Estudios Avanzados.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ringquist S, Shinedling S, Barrick D, Green L, Binkley J, Stormo GD, Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol. Microbiol. 1992;6:1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- 2.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc. Natl Acad. Sci. USA. 1974;71:1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarthy JE, Brimacombe R. Prokaryotic translation: the interactive pathway leading to initiation. Trends Genet. 1994;10:402–407. doi: 10.1016/0168-9525(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 4.Etchegaray JP, Inouye M. A sequence downstream of the initiation codon is essential for cold shock induction of cspB of Escherichia coli. J. Bacteriol. 1999;181:5852–5854. doi: 10.1128/jb.181.18.5852-5854.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Faxen M, Plumbridge J, Isaksson LA. Codon choice and potential complementarity between mRNA downstream of the initiation codon and bases 1471-1480 in 16S ribosomal RNA affects expression of glnS. Nucleic Acids Res. 1991;19:5247–5251. doi: 10.1093/nar/19.19.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sprengart ML, Fatscher HP, Fuchs E. The initiation of translation in E. coli: apparent base pairing between the 16srRNA and downstream sequences of the mRNA. Nucleic Acids Res. 1990;18:1719–1723. doi: 10.1093/nar/18.7.1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stenstrom CM, Holmgren E, Isaksson LA. Cooperative effects by the initiation codon and its flanking regions on translation initiation. Gene. 2001;273:259–265. doi: 10.1016/s0378-1119(01)00584-4. [DOI] [PubMed] [Google Scholar]
- 8.Moll I, Huber M, Grill S, Sairafi P, Mueller F, Brimacombe R, Londei P, Blasi U. Evidence against an interaction between the mRNA downstream box and 16S rRNA in translation initiation. J. Bacteriol. 2001;183:3499–3505. doi: 10.1128/JB.183.11.3499-3505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connor M, Asai T, Squires CL, Dahlberg AE. Enhancement of translation by the downstream box does not involve base pairing of mRNA with the penultimate stem sequence of 16S rRNA. Proc. Natl Acad. Sci. USA. 1999;96:8973–8978. doi: 10.1073/pnas.96.16.8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stenstrom CM, Isaksson LA. Influences on translation initiation and early elongation by the messenger RNA region flanking the initiation codon at the 3' side. Gene. 2002;288:1–8. doi: 10.1016/s0378-1119(02)00501-2. [DOI] [PubMed] [Google Scholar]
- 11.Looman AC, Bodlaender J, Comstock LJ, Eaton D, Jhurani P, de Boer HA, van Knippenberg PH. Influence of the codon following the AUG initiation codon on the expression of a modified lacZ gene in Escherichia coli. EMBO J. 1987;6:2489–2492. doi: 10.1002/j.1460-2075.1987.tb02530.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sato T, Terabe M, Watanabe H, Gojobori T, Hori-Takemoto C, Miura K. Codon and base biases after the initiation codon of the open reading frames in the Escherichia coli genome and their influence on the translation efficiency. J. Biochem., (Tokyo) 2001;129:851–860. doi: 10.1093/oxfordjournals.jbchem.a002929. [DOI] [PubMed] [Google Scholar]
- 13.Brock JE, Paz RL, Cottle P, Janssen GR. Naturally occurring adenines within mRNA coding sequences affect ribosome binding and expression in Escherichia coli. J. Bacteriol. 2007;189:501–510. doi: 10.1128/JB.01356-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen GF, Inouye M. Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res. 1990;18:1465–1473. doi: 10.1093/nar/18.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen GT, Inouye M. Role of the AGA/AGG codons, the rarest codons in global gene expression in Escherichia coli. Genes Dev. 1994;8:2641–2652. doi: 10.1101/gad.8.21.2641. [DOI] [PubMed] [Google Scholar]
- 16.Gurskii Ia G, Marimont N, Bibilashvili R. The effect of intracellular concentrations of tRNA, corresponding to the rare arginine codons AGG and AGA, on the gene expression in Escherichia coli. Mol. Biol. (Mosk) 1992;26:1080–1087. [PubMed] [Google Scholar]
- 17.Gurskii Ia G, Marimont N, Shevelev A, Iuzhakov AA, Bibilashvili R. Rare codons and gene expression in Escherichia coli. Mol. Biol. (Mosk) 1992;26:1063–1079. [PubMed] [Google Scholar]
- 18.Delgado-Olivares L, Zamora-Romo E, Guarneros G, Hernandez-Sanchez J. Codon-specific and general inhibition of protein synthesis by the tRNA-sequestering minigenes. Biochimie. 2006;88:793–800. doi: 10.1016/j.biochi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Roche ED, Sauer RT. SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 1999;18:4579–4589. doi: 10.1093/emboj/18.16.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olivares-Trejo JJ, Bueno-Martinez JG, Guarneros G, Hernandez-Sanchez J. The pair of arginine codons AGA AGG close to the initiation codon of the lambda int gene inhibits cell growth and protein synthesis by accumulating peptidyl-tRNAArg4. Mol. Microbiol. 2003;49:1043–1049. doi: 10.1046/j.1365-2958.2003.03611.x. [DOI] [PubMed] [Google Scholar]
- 21.Zahn K, Landy A. Modulation of lambda integrase synthesis by rare arginine tRNA. Mol. Microbiol. 1996;21:69–76. doi: 10.1046/j.1365-2958.1996.6201335.x. [DOI] [PubMed] [Google Scholar]
- 22.Cruz-Vera LR, Toledo I, Hernandez-Sanchez J, Guarneros G. Molecular basis for the temperature sensitivity of Escherichia coli pth(Ts) J. Bacteriol. 2000;182:1523–1528. doi: 10.1128/jb.182.6.1523-1528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strauch MA, Spiegelman GB, Perego M, Johnson WC, Burbulys D, Hoch JA. The transition state transcription regulator abrB of Bacillus subtilis is a DNA binding protein. EMBO J. 1989;8:1615–1621. doi: 10.1002/j.1460-2075.1989.tb03546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Del Tito BJ, Ward JM, Hodgson J, Gershater CJ, Edwards H, Wysocki LA, Watson FA, Sathe G, Kane JF. Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J. Bacteriol. 1995;177:7086–7091. doi: 10.1128/jb.177.24.7086-7091.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindsley D, Gallant J, Guarneros G. Ribosome bypassing elicited by tRNA depletion. Mol. Microbiol. 2003;48:1267–1274. doi: 10.1046/j.1365-2958.2003.03514.x. [DOI] [PubMed] [Google Scholar]
- 26.Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 27.Cruz-Vera LR, Hernandez-Ramon E, Perez-Zamorano B, Guarneros G. The rate of peptidyl-tRNA dissociation from the ribosome during minigene expression depends on the nature of the last decoding interaction. J. Biol. Chem. 2003;278:26065–26070. doi: 10.1074/jbc.M301129200. [DOI] [PubMed] [Google Scholar]
- 28.Dong H, Nilsson L, Kurland CG. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 1996;260:649–663. doi: 10.1006/jmbi.1996.0428. [DOI] [PubMed] [Google Scholar]
- 29.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J. Biol. Chem. 1981;256:11905–11910. [PubMed] [Google Scholar]
- 30.Gao W, Tyagi S, Kramer FR, Goldman E. Messenger RNA release from ribosomes during 5′-translational blockage by consecutive low-usage arginine but not leucine codons in Escherichia coli. Mol. Microbiol. 1997;25:707–716. doi: 10.1046/j.1365-2958.1997.5081871.x. [DOI] [PubMed] [Google Scholar]
- 31.Goldman E, Rosenberg AH, Zubay G, Studier FW. Consecutive low-usage leucine codons block translation only when near the 5′ end of a message in Escherichia coli. J. Mol. Biol. 1995;245:467–473. doi: 10.1006/jmbi.1994.0038. [DOI] [PubMed] [Google Scholar]
- 32.Rosenberg AH, Goldman E, Dunn JJ, Studier FW, Zubay G. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J. Bacteriol. 1993;175:716–722. doi: 10.1128/jb.175.3.716-722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shu P, Dai H, Gao W, Goldman E. Inhibition of translation by consecutive rare leucine codons in E. coli: absence of effect of varying mRNA stability. Gene Expr. 2006;13:97–106. doi: 10.3727/000000006783991881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rudd KE, Schneider TD. Compilation of E. coli ribosome binding sites. In: Miller J, editor. A Short Course in Bacterial Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1992. pp. 17.19–17.46. [Google Scholar]
- 35.Valadez JG, Hernandez-Sanchez J, Magos MA, Ontiveros C, Guarneros G. Increased bar minigene mRNA stability during cell growth inhibition. Mol. Microbiol. 2001;39:361–369. doi: 10.1046/j.1365-2958.2001.02214.x. [DOI] [PubMed] [Google Scholar]
- 36.Cruz-Vera LR, Magos-Castro MA, Zamora-Romo E, Guarneros G. Ribosome stalling and peptidyl-tRNA drop-off during translational delay at AGA codons. Nucleic Acids Res. 2004;32:4462–4468. doi: 10.1093/nar/gkh784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gonzalez de Valdivia EI, Isaksson LA. A codon window in mRNA downstream of the initiation codon where NGG codons give strongly reduced gene expression in Escherichia coli. Nucleic Acids Res. 2004;32:5198–5205. doi: 10.1093/nar/gkh857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H, Pomeroy-Cloney L, Bjerknes M, Tam J, Jay E. The influence of adenine-rich motifs in the 3′ portion of the ribosome binding site on human IFN-gamma gene expression in Escherichia coli. J. Mol. Biol. 1994;240:20–27. doi: 10.1006/jmbi.1994.1414. [DOI] [PubMed] [Google Scholar]
- 39.Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J. Mol. Biol. 1988;204:79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- 40.Martin-Farmer J, Janssen GR. A downstream CA repeat sequence increases translation from leadered and unleadered mRNA in Escherichia coli. Mol. Microbiol. 1999;31:1025–1038. doi: 10.1046/j.1365-2958.1999.01228.x. [DOI] [PubMed] [Google Scholar]
- 41.Curran JF. Decoding with the A:I wobble pair is inefficient. Nucleic Acids Res. 1995;23:683–688. doi: 10.1093/nar/23.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin H, Zhao Q, Gonzalez de Valdivia EI, Ardell DH, Stenstrom M, Isaksson LA. Influences on gene expression in vivo by a Shine-Dalgarno sequence. Mol. Microbiol. 2006;60:480–492. doi: 10.1111/j.1365-2958.2006.05110.x. [DOI] [PubMed] [Google Scholar]
- 44.Weiss RB, Dunn DM, Atkins JF, Gesteland RF. Slippery runs, shifty stops, backward steps, and forward hops: -2, -1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- 45.Weiss RB, Dunn DM, Dahlberg AE, Atkins JF, Gesteland RF. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli. EMBO J. 1988;7:1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]