Abstract
The 22-nt c-kit87 promoter sequence is unique within the human genome. Its fold and tertiary structure have recently been determined by NMR methods [Phan,A.T., Kuryavyi,V., Burge,S., Neidle,S. and Patel,D.J. (2007) Structure of an unprecedented G-quadruplex scaffold in the c-kit promoter. J. Am. Chem. Soc., 129, 4386–4392], and does not have precedent among known DNA quadruplexes. We show here using bioinformatics and molecular dynamics simulations methods that (i) none of the closely related sequences (encompassing all nucleotides not involved in the maintenance of structural integrity) occur immediately upstream (<100 nt) of a transcription start site, and (ii) that all of these sequences correspond to the same stable tertiary structure. It is concluded that the c-kit87 tertiary structure may also be formed in a very small number of other loci in the human genome, but the likelihood of these playing a significant role in the expression of particular genes is very low. The c-kit87 quadruplex thus fulfils a fundamental criterion of a ‘good’ drug target, in that it possesses distinctive three-dimensional structural features that are only present in at most a handful of other genes.
INTRODUCTION
The proto-oncogene c-kit encodes for a 145–160 kDa tyrosine kinase receptor, which is especially expressed in mast cells, melanocytes and hematopoetic stem cells (1,2). The tyrosine kinase domain of c-kit has become an important molecular target for the treatment of gastrointestinal stromal tumors (GIST), and the small molecule kinase inhibitor Gleevec has become the most significant therapy for GIST, where it has made a major difference to survival rates (3–6). Over-expression and/or mutation of c-kit may also play a significant role in several other cancers, including some leukaemias (7) and testicular cancers (8). However, resistance to Gleevec occurs as a result of deactivating mutations in the kinase active site (6,9–11). These diminish binding and rapidly reduce the clinical effectiveness of the drug. Several 2nd-generation c-kit kinase inhibitors are currently being developed to overcome this resistance (12–15), although it is possible that they in turn may produce new patterns of resistance mutations in the kinase active site.
Selective gene regulation at the transcriptional level is a potential alternative to targeting a protein, the product of gene expression. One way in which this can be achieved is by the induction of higher-order G-quadruplex DNA structures (16) in a G-rich region such as a promoter sequence (17–20) by a small-molecule ligand. This has been demonstrated for the c-myc oncogene at the nuclease hypersensitivity element (NHE) III1 that is responsible for up to 90% of c-myc transcription (21,22). G-quadruplexes, which may have transient stability by themselves when embedded within the double-stranded DNA of a eukaryotic gene, may thus be stabilized further by a small-molecule ligand. The structure and topology of two c-myc DNA quadruplexes have been determined by NMR spectroscopy (23,24), as well as that of a ligand (TMPyP4) complex (25). These are structurally complex parallel-stranded quadruplexes, with several strand-reversal loops and base-pair platforms.
Two discrete G-rich quadruplex-forming sequences have been identified (26,27) in the human c-kit core promoter region (28–30). These are within the nuclease hypersensitive region of the promoter, suggesting that they are not involved in a chromatin complex. Biophysical and 1-D NMR studies have shown that these individual sequences can both form G-quadruplex structures (26,27). One sequence, d(AGGGAGGGCGCTGGGAGGAGGG), which occurs 87-nt upstream of the transcription start site, forms a single G-quadruplex species in solution (26). The occurrence of four tracts of three consecutive guanines (underlined), separated by linkers of either one or four residues initially suggested that the sequence forms a G-quadruplex structure with these G-tracts forming the G-tetrad core, and the linker sequences forming loops, analogous to the parallel-stranded structure of the human intramolecular telomeric quadruplex (31). However, this proposed model was unable to explain the dramatic quadruplex destabilizing effect caused by mutations in the linker sequences (26).
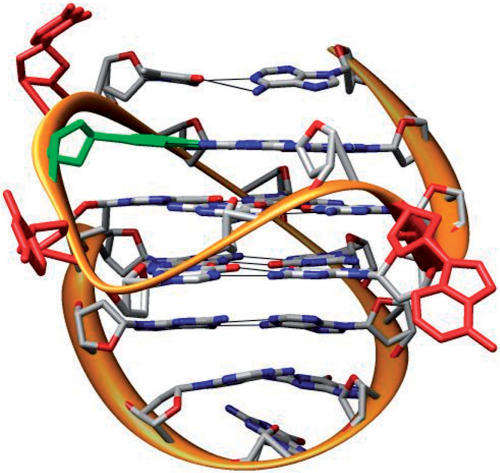
The NMR-based solution structure of the G-quadruplex formed by this precise sequence in K+ solution has now been determined (32), and shows that c-kit87 has an unprecedented G-quadruplex folding topology that involves 18 of the 22 nt in tertiary interactions (Figure 1), and providing rationales for the mutant data (26,32). These four non-essential nucleotides are in bold in the above sequence. One of the ‘loop’ guanine bases is directly involved in G-tetrad core formation, contrary to expectations and despite the presence of four three-guanine tracts. There are also four loops; two single-nucleotide double-chain-reversal loops, a two-residue loop, and a five-residue d(AGGAG) stem-loop. The net result is a tertiary quadruplex structure with complex features absent in simpler quadruplexes such as the human telomeric parallel and antiparallel arrangements. In particular, the presence of two well-defined clefts in the structure that are defined by the stem-loop and the two-residue loop strongly suggest that the c-kit87 quadruplex could be a target for the design of selective small molecules that would serve to stabilize the structure within the context of the core promoter sequence, and thus down-regulate c-kit expression. The structure allows for straightforward continuation of a DNA sequence in both 5′ and 3′ directions, suggesting that it could be formed within the promoter region without undue steric constraint.
Figure 1.
One of the NMR structures of the c-kit87 quadruplex, taken from the PDB entry 2O3M, and drawn with the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (Pettersen,E.F., Goddard,T.D., Huang,C.C., Couch,G.S., Greenblatt,D.M., Meng,E.C., and Ferrin,T.E. UCSF chimera - a visualization system for exploratory research and analysis. (2004). J. Comput. Chem., 25, 1605–1612). Nucleotides A5, C9 and C11 are coloured red, and G18 is in green.
The potential of c-kit87 as a therapeutic target raises the question of the degree of its sequence and structural uniqueness. This issue is addressed here using a combined bioinformatics, circular dichroism (CD) and molecular dynamics simulation approach.
METHODS
Informatics
The Ensembl human genome core database (33) version 38 (NCBI build 36) was searched for sequences of the patterns:
AGGGwGGGwGwTGGGAGGAGGG
AGGGwGGGwGwTGGGAGwAGGG
TGGGwGGGwGwAGGGAGwAGGG
CGGGwGGGwGwGGGGAGwAGGG
GGGGwGGGwGwCGGGAGwAGGG
where w represents any base, at positions 5, 9, 11 and 18 that are not involved in tertiary interactions in the structure.
The search software was that developed for earlier quadruplex searches (17). The positions of each hit within the chromosome and its relation to the surrounding genes, or the gene within which it occurred, was recorded and compiled into a mySQL database. This database was then queried so that the results could be ordered and grouped as desired. Where sequences occurred upstream relative to the transcription direction of a gene, the distance between the gene and the transcription start site was retrieved and in the case where one of the hit sequences occurred within a gene, it was noted in which intron, exon or untranslated region the sequence occurred.
Searches for the mutated c-kit sequences which were previously examined (26) and shown not to form quadruplex structures, were also carried out, in the same way as described above. These sequences are:
d(AGGGAGGGAGGAGGGAGGAGGG)
d(AGGGAGGGCGCTGGGCGCTGGG)
d(AGGGAGGGCGCTGGGCGGCGGG)
Additional variations of these mutated sequences were also investigated. The human genome was searched for the following sequences with systematic variations at the 5, 9, 11 and 18 positions:
d(AGGGwGGGwGwAGGGAGwAGGG) sequences 1m
d(AGGGwGGGwGwTGGGCGwTGGG) sequences 2m
d(AGGGwGGGwGwTGGGCGwCGGG) sequences 3m
The c-kit upstream regions from various different species were obtained from the Ensembl web site www.ensembl.org (using Ensembl release 43). Upstream regions for the orthologues to the human c-kit sequence were found for macaque, rat, mouse, cow, opossum chicken and zebrafish. A multiple sequence alignment was carried out on these sequences using the CLUSTAL software package (34).
CD studies
The c-kit87 and the ten mutant sequences were synthesized and hplc purified (Eurogentec), and were then used in this study:
[A5G] d(AGGGGGGGCGCTGGGAGGAGGG)
[A5C] d(AGGGCGGGCGCTGGGAGGAGGG)
[A5T] d(AGGGTGGGCGCTGGGAGGAGGG)
[C9G] d(AGGGAGGGGGCTGGGAGGAGGG)
[C9A] d(AGGGAGGGAGCTGGGAGGAGGG)
[C9T] d(AGGGAGGGTGCTGGGAGGAGGG)
[C11G] d(AGGGAGGGCGGTGGGAGGAGGG)
[C11A] d(AGGGAGGGCGATGGGAGGAGGG)
[C11T] d(AGGGAGGGCGTTGGGAGGAGGG)
[G18T] d(AGGGAGGGCGCTGGGAGTAGGG)
c-kit87 native d(AGGGAGGGCGCTGGGAGGAGGG)
CD spectra for them were acquired on a Chirascan spectrometer (Applied Photophysics Ltd) at King's College London. All samples were prepared at 100 μM in 50 mM potassium chloride and heated to 95°C and slowly annealed overnight to room temperature. The samples were further diluted, with buffer to 1 optical density unit prior to data collection. UV absorbance and CD spectra were measured between 360 and 200 nm in a 10 mm path-length cell. Spectra were recorded with a 0.5 nm step size, a 1.5 s time-per-point and a spectral bandwidth of 1 nm. All spectra were acquired at room temperature and buffer baseline corrected. The concentrations of the above oligonucleotides were determined by using the absorbance value at 260 nm and the Beer–Lambert law.
Molecular dynamics simulations
One of the experimental c-kit87 NMR structures (PDB accession code 2O3M) was arbitrarily chosen and used as a starting point for all calculations. Mutants occurring with high frequency were identified using the bioinformatics techniques outlined above. Structural modifications were made to the native c-kit87 model to generate 3D models from these mutant sequences, changing only the base; backbone conformations were not altered at all. This was carried out using the Insight suite of programs (www.accelrys.com). In all, ten mutants were constructed and are listed in Table 1.
Table 1.
List of c-kit87 structures examined by molecular dynamics simulations
| Position 5 | Position 9 | Position 11 | RMSD (Å) | |
|---|---|---|---|---|
| Native | A(2) | C(2) | C(6) | 1.6 |
| Mutant 1: A5G | G(0) | C | C | 2 |
| Mutant 2: A5C | C(0) | C | C | 1.8 |
| Mutant 3: A5T | T(59) | C | C | 1.8 |
| Mutant 4: C9G | A | G(36) | C | 1.8 |
| Mutant 5: C9A | A | A(17) | C | 2.2 |
| Mutant 6: C9T | A | T(6) | C | 1.6 |
| Mutant 7: C11G | A | C | G(42) | 1.9 |
| Mutant 8: C11A | A | C | A(6) | 2 |
| Mutant 9: C11T | A | C | T(7) | 2.1 |
| Mutant 10: AGTAG | A | C | C | 1.8 |
The numbers in parentheses are the frequency of occurrence of that residue in a particular position, as found in this work. The right-hand column lists the RMSD values arising from the simulations.
Molecular dynamics simulations were carried out using the ff99 forcefield in the AMBER v9.0 package (36). Each system was equilibrated with explicit solvent molecules (TIP3P) using 1000 steps of minimization and 20 ps of molecular dynamics at 300 K. The entire systems were kept constrained, while allowing the ions and the solvent molecules to equilibrate. The systems were then subjected to a series of dynamics calculations in which the constraints were gradually relaxed, until no constraints at all were applied. The final production run was performed without any restrain on the complex for 10 ns and co-ordinates were saved after every 10 ps for analysis of their trajectories. The simulation protocols were consistent for all of the systems. Periodic boundary conditions were applied, with the particle-mesh Ewald (PME) method (37) used to treat the long-range electrostatic interactions. The solute was first solvated in a TIP3P water box (38), the boundaries of which were at least at a distance of 10 Å from any solute atoms. Additional positively charged K+ counter-ions were included in the system to neutralize the charge on the DNA backbone. The counter-ions were automatically placed by the LEAP program throughout the water box at grid points of negative Coulombic potential. The final system had net zero charge.
All calculations were carried out using the SANDER module, trajectories were analysed using the PTRAJ module from the AMBER9.0 suite and viewed using the VMD program (39).
RESULTS
Informatics
The c-kit87 experimental structure contains three looped-out bases (A5, C9 and C11), which visual inspection (Figure 1) shows do not play a role in maintaining structural integrity, since they do not interact directly with any part of the folded structure. Other notable features of the structure are (i) a Watson–Crick base pair between A1 and T12 and (ii) the AGGAG stem loop, which contains two A … G base pairs. The middle guanine, G18 in this loop sequence, stacks on the end of the loop and is not involved in any hydrogen bonding with other residues. Changing A5 and C9 to thymines has been found (32) to produce a structure with the same topology as the native; changing C11 to thymine produces a mixture of structures with a similar topology but where one structure contains an A1–T11 base pair and the other an A1–T12 base pair, as in the native structure. Modification of G18 to T18 also maintains the topology.
We have therefore searched through the human genome for all possible sequence variations at these four positions, in order to assess their implications for the uniqueness of the sequence and the structure. All variations have been examined, at positions 5, 9, 11 and 18 and also for all four alternative Watson–Crick base pairings between positions 1 and 12. These searches were done in stages, as outlined below. An initial search revealed that the native 22-mer c-kit87 sequence has only a single occurrence in the human genome.
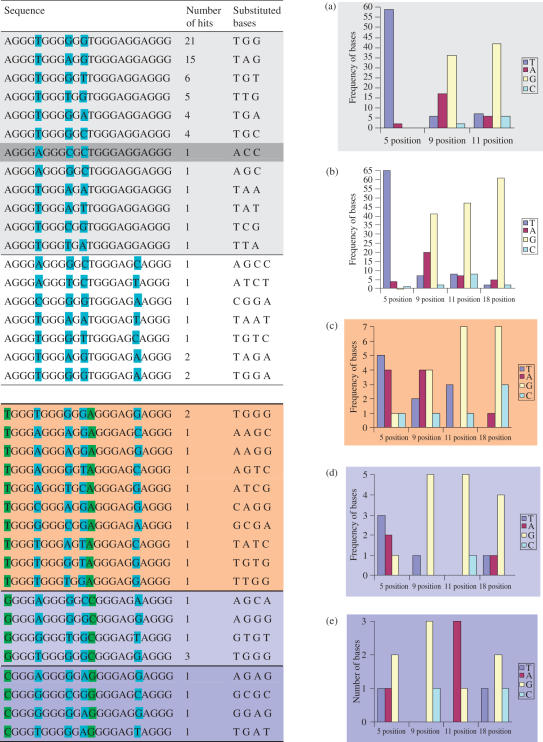
There are 64 possible combinations for the three 'flipped out' bases at the 5, 9 and 11 positions. A total of 61 sequence occurrences were found, corresponding to just 12 unique sequences (Figure 2a). The relative frequencies with which different bases occur are not random, with sequences that have T, G and G substitutions at the 5, 9 and 11 positions being the most common type, of which 21 were found. The thymine substitution at the 5 position occurs in ca. 97% of the sequence hits and the 9 and 11 positions were most frequently guanines. Sequences closely similar to the c-kit87 sequence itself are exceptionally rare. Only one other sequence has an adenine at the 5 position, only one other sequence has a cytosine at the 9 position and of the five other sequences which have a cytosine at the 11 position only one has another of the substituted bases in common with the c-kit87 native sequence.
Figure 2.
(a) Variations at the 5, 9 and 11 positions, Figure 2 (b) variations at the 5, 9, 11 and 18 positions (c) variants at the 5, 9, 11 and 18 position when A1 and T12 are transposed to T1 and A12, (d) variants at the 5, 9, 11 and 18 positions with G1 and C12 and (e) variants at the 5, 9, 11 and 18 positions with C1 and G12.
Examination of substitutions at position 18 in addition to those at the 5, 9 and 11 positions, showed that although there are a further 192 possible sequence combinations, only nine more actually occur (Figure 2b). Again none of these nine additional sequences have more than two of the substituted bases in common with the c-kit87 sequence, and only two sequences have two bases in common, and the remaining seven have none in common. Our previous analysis of quadruplex loop occurrences in the human genome (17) found that loops of sequence AGGA, and therefore sequences containing AGGAG, are highly over-represented. Out of the many thousands of loop sequences which were found when searching for potential quadruplex sequences, AGGA was the 14th most frequently found loop.
Searches for alternative Watson–Crick base-pairing combinations between the A1 and T12 positions yielded only 21 further hits (from a possible 768 more sequences), the majority of which have T at the 1st position and an A at the 12th position (Figure 2c). Again there were no other sequences which differ from the native c-kit87 in only the alternative 1–12 pairing, although one sequence differed in only the 1–12 pairing together with the 9 position. Figure 2d and e shows that the alternative 1–12 base pairings G-C and C-G have even fewer sequence hits, with just six and four sequences found respectively. Again these were dissimilar to the native c-kit87 sequence.
A search for occurrences of the mutated c-kit sequences used in the initial study (26) (none of which form a stable quadruplex structure), found no hits for two of the sequences examined, (AGGGAGGGCGCTGGGCGCTGGG and AGGGAGGGCGCTGGGCGGCGGG). There are seven occurrences of the third sequence, AGGGAGGGAGGAGGGAGGAGGG (Table 2) in the human genome. Two instances of this high purine-content sequence occurred in a potential promoter region, two instances were close together within an intron, and the other two were not near any regions of biological importance as far as is known.
Table 2.
Occurrences in the human genome of the non-quadruplex forming sequences examined by Rankin et al. (26)
| 1 | AGGGAGGGAGGAGGGAGGAGGG | In the middle of chromosome 3: nearest known gene is 82008 bases away |
| 2 | CCCTCCTCCCTCCTCCCTCCCT | 2694 bases upstream of ENSG00000004866 (suppression of tumorigenicity 7) |
| 3 | CCCTCCTCCCTCCTCCCTCCCT | Within the third intron of ENSG00000133195 (solute carrier family 39 (metal ion transporter, member 11) |
| 4 | CCCTCCTCCCTCCTCCCTCCCT | Within the third intron of ENSG00000133195 (solute carrier family 39 (metal ion transporter, member 11) |
| 5 | CCCTCCTCCCTCCTCCCTCCCT | 3143 bases upstream of ENSG00000183019 (function seems to be unknown) |
| 6 | CCCTCCTCCCTCCTCCCTCCCT | 228 bases upstream of ENSG00000185985 SLIT and NTRK-like family, member 2 |
| 7 | CCCTCCTCCCTCCTCCCTCCCT | 210 bases upstream of ENSG00000185985 SLIT and NTRK-like family, member 2 |
The distances to putative transcription start sites (TSS) were then examined for all of these sequence variants (Tables 3 and 4). The majority do not occur within genes, but are distributed in non-coding regions. The c-kit87 native sequence, which is 34 bases upstream of the TSS, is by far the closest to its TSS. The next closest is ENSG00000185245 (coding for Platelet glycoprotein Ib alpha chain precursor) which appears 147 bases upstream of its transcription start site. The sequence which bears the greatest similarity to the c-kit87 sequence, differing only by a C in the 9th position, occurs upstream of the transcription start site of the gene ENSG00000136213 (coding for the protein carbohydrate sulfotransferase 12); however this sequence is far upstream, at ∼18.6 kb from the TSS). The remaining sequences are also, in general, located in quite remote positions.
Table 3.
Sequences which are upstream relative to known genes. Those with genes on either side are in the upstream regions of both genes
| Gene ensembl ID | Number of bases from TSS | Sequence | Number of bases from TSS | Gene ensembl ID |
|---|---|---|---|---|
| AGGGAGGGCGCTGGGAGGAGGG | 34 | ENSG00000157404 | ||
| ENSG00000205709 | 9359 | AGGGTGGGAGATGGGAGTAGGG | 147 | ENSG00000185245 |
| ENSG00000092148 | 401 | CGGGAGGGGGAGGGGAGGAGGG | 37043 | ENSG00000202402 |
| ENSG00000195330 | 1024 | AGGGTGGGGGTTGGGAGGAGGG | ||
| ENSG00000195067 | 4844 | AGGGTGGGGGTTGGGAGGAGGG | 1051 | ENSG00000194918 |
| ENSG00000169892 | 1340 | AGGGTGGGAGGTGGGAGGAGGG | ||
| ENSG00000109956 | 126892 | AGGGAGGGGGCTGGGAGCAGGG | 196763 | ENSG00000204236 |
| AGGGAGGGGGCTGGGAGGAGGG | 18651 | ENSG00000136213 | ||
| ENSG00000190177 | 512336 | AGGGCGGGGGGTGGGAGAAGGG | ||
| ENSG00000111716 | 77565 | AGGGTGGGAGATGGGAGGAGGG | ||
| ENSG00000179862 | 87893 | AGGGTGGGAGGTGGGAGAAGGG | 29076 | ENSG00000171793 |
| AGGGTGGGAGGTGGGAGAAGGG | 116715 | ENSG00000166035 | ||
| AGGGTGGGAGGTGGGAGGAGGG | 54970 | ENSG00000199297 | ||
| ENSG00000193578 | 3626 | AGGGTGGGAGGTGGGAGGAGGG | ||
| AGGGTGGGAGGTGGGAGGAGGG | 51108 | ENSG00000182824 | ||
| ENSG00000128185 | 32965 | AGGGTGGGAGGTGGGAGGAGGG | ||
| ENSG00000187979 | 50633 | AGGGTGGGAGGTGGGAGGAGGG | ||
| ENSG00000181250 | 1371257 | AGGGTGGGAGGTGGGAGGAGGG | 372140 | ENSG00000199778 |
| ENSG00000190535 | 622126 | AGGGTGGGAGGTGGGAGGAGGG | ||
| ENSG00000199652 | 615165 | AGGGTGGGAGGTGGGAGGAGGG | 747692 | ENSG00000193275 |
| ENSG00000193660 | 216206 | AGGGTGGGAGGTGGGAGGAGGG | ||
| ENSG00000163492 | 155574 | AGGGTGGGAGGTGGGAGGAGGG | ||
| ENSG00000113504 | 82344 | AGGGTGGGCGGTGGGAGGAGGG | 7270 | ENSG00000174358 |
| AGGGTGGGGGATGGGAGGAGGG | 118987 | ENSG00000205666 | ||
| ENSG00000123243 | 22080 | AGGGTGGGGGATGGGAGGAGGG | 14218 | ENSG00000151655 |
| AGGGTGGGGGCTGGGAGGAGGG | 144423 | ENSG00000199222 | ||
| AGGGTGGGGGGTGGGAGAAGGG | 3151 | ENSG00000187806 | ||
| ENSG00000191596 | 21490 | AGGGTGGGGGGTGGGAGAAGGG | ||
| ENSG00000136149 | 2368405 | AGGGTGGGGGGTGGGAGGAGGG | ||
| ENSG00000177138 | 212606 | AGGGTGGGGGGTGGGAGGAGGG | 24604 | ENSG00000194029 |
| AGGGTGGGGGGTGGGAGGAGGG | 236887 | ENSG00000192765 | ||
| ENSG00000185555 | 2200 | AGGGTGGGGGGTGGGAGGAGGG | ||
| AGGGTGGGGGGTGGGAGGAGGG | 219635 | ENSG00000018236 | ||
| ENSG00000197445 | 191931 | AGGGTGGGGGGTGGGAGGAGGG | ||
| AGGGTGGGGGGTGGGAGGAGGG | 22768 | ENSG00000185744 | ||
| ENSG00000189981 | 187334 | AGGGTGGGGGGTGGGAGGAGGG | ||
| AGGGTGGGGGGTGGGAGGAGGG | 455385 | ENSG00000154478 | ||
| AGGGTGGGGGGTGGGAGGAGGG | 34226 | ENSG00000154478 | ||
| ENSG00000133424 | 734086 | AGGGTGGGGGTTGGGAGCAGGG | 411717 | ENSG00000175329 |
| AGGGTGGGGGTTGGGAGGAGGG | 18610 | ENSG00000100739 | ||
| ENSG00000186964 | 5596 | AGGGTGGGTGATGGGAGGAGGG | ||
| AGGGTGGGTGGTGGGAGGAGGG | 199573 | ENSG00000102290 | ||
| ENSG00000201475 | 645873 | AGGGTGGGTGGTGGGAGGAGGG | 179263 | ENSG00000099715 |
| AGGGTGGGTGGTGGGAGGAGGG | 58391 | ENSG00000088538 | ||
| AGGGTGGGTGGTGGGAGGAGGG | 13880 | ENSG00000193070 | ||
| ENSG00000204966 | 127294 | GGGGGGGGTGGCGGGAGTAGGG | 120459 | ENSG00000189221 |
| ENSG00000196406 | 100265 | GGGGTGGGGGGCGGGAGGAGGG | 38941 | ENSG00000165509 |
| ENSG00000071564 | 103462 | TGGGAGGGAGGAGGGAGGAGGG | 19364 | ENSG00000205922 |
| ENSG00000192873 | 551362 | TGGGTGGGGGGAGGGAGGAGGG | 242860 | ENSG00000200960 |
| ENSG00000190169 | 169776 | TGGGTGGGGGGAGGGAGGAGGG | 1955948 | ENSG00000202478 |
| ENSG00000118487 | 471843 | TGGGTGGGGGTAGGGAGGAGGG |
Table 4.
Description of highlighted genes in Table 3
| Ensembl ID | Description |
|---|---|
| ENSG00000185245 | (GP1BA) glycoprotein Ib (platelet) alpha polypeptide |
| ENSG00000092148 | (HECTD1) HECT domain containing 1 |
| ENSG00000195330 | tRNA pseudogene |
| ENSG00000194918 | tRNA pseudogene |
| ENSG00000169892 | CDNA FLJ46366 fis, clone TESTI4051388 |
We have also examined the phylogenetic features of the c-kit87 sequence. Table 5 shows the results of the multiple sequence alignment between the upstream sequences of several c-kit87 orthologues. The two non-mammalian sequences gave very dissimilar alignments to the rest of the species, however the mammalian sequences were similar enough to identify the relevant, orthologous upstream regions. The opossum and macaque sequences were identical to the human while the cow sequence differed by only one base, where a cytosine appears instead of guanine at position 21. The mouse and rat sequences are identical. However, they have an adenine inserted at the 2nd position and a deletion at 9 and 15 which seem to make it impossible for them to form quadruplex structures with the same topology as the human c-kit sequence. They remain guanine-rich however, so it is not impossible that they can fold into an alternative quadruplex topology.
Table 5.
Multiple sequence alignment of the human c-kit87 and surrounding region with orthologous regions from various species
| HUMAN | TGGCCGGCGCG-CAGAGGGAGGGCGCTGGGAGGAGGGGCTGCT - - - - - - - GCTCGCC- |
| MACAQUE | TGGCCGGCGCG-CAGAGGGAGGGCGCTGGGAGGAGGGGCTGCT - - - - - - - GCTCGGC- |
| MOUSE | TGGCCA-CGAG-CTGGGAGGAGG-GCTGG-AGGAGGGGCTGTC - - - - - - - GCGCGCC- |
| RAT | TGGCCA-CGCG-CTGGGAGGAGG-GCTGG-AGGAGGGGCTGTC - - - - - - - GCGCGCC- |
| COW | TGGCCGCCGCT-CAGGGGGAGGGCGCTGGGAGGAGCGGCCGCG - - - - - - - GCTTGGC- |
| OPOSSUM | TGGCCGGCGTGGCAAGGGGAGGGCGCTGGGAGGAGGGGCTGCTCTCCTTTGCTAGCCT |
| CHICKEN | GGCCGGCAGTACTCCGC-AGCCTCCCGCG–GGGTTCGGGCATATATGCGCGCCGGGT |
| ZEBRAFISH | TGTTGATGTTGTTACCTCCCTGTCCCCGCCCAGGCTCGCTCGTCGTTC–CGCATGAC |
As a check on the sequence occurrences we have compared search results for different G-rich sequences, using the non-quadruplex-forming c-kit87 mutants 1 m, 2 m and 3 m. In total there were 38 hits for sequence 1m, none for sequence 2 m and two for sequence 3 m (Tables 6 and 7). One of the sequences appears 71 bases upstream of ensembl gene ENSG00000133466 (HGNC name: C1q tumor necrosis factor-related protein 6) and one occurs 228 bases upstream of the transcription start site of ensembl gene ENSG00000185985 (HGNC name: SLIT and NTRK-like family, member 2).
Table 6.
Variants of non-quadruplex forming sequence 1m which occur upstream of known genes
| Gene ensembl ID <- - - - - - - | | Number of bases from TSS | Sequence | Number of bases from TSS | Gene ensembl ID |- - - - - - - - -> |
|---|---|---|---|---|
| ENSG00000004866 | 2694 | AGGGAGGGAGGAGGGAGGAGGG | 6219 | ENSG00000195520 |
| ENSG00000199778 | 125965 | AGGGTGGGGGGAGGGAGCAGGG | 483712 | ENSG00000181250 |
| ENSG00000162825 | 2997 | AGGGTGGGGGGAGGGAGGAGGG | ||
| ENSG00000120370 | 161143 | AGGGTGGGGGGAGGGAGGAGGG | ||
| ENSG00000199285 | 179234 | AGGGTGGGGGTAGGGAGGAGGG | 552024 | ENSG00000176435 |
| AGGGAGGGTGGAGGGAGAAGGG | 127190 | ENSG00000176769 | ||
| ENSG00000205866 | 1042 | AGGGAGGGGGGAGGGAGGAGGG | ||
| ENSG00000205864 | 10129 | AGGGAGGGGGGAGGGAGGAGGG | 1886 | ENSG00000205865 |
| AGGGAGGGAGCAGGGAGGAGGG | 71 | ENSG00000133466 | ||
| ENSG00000144810 | 403994 | AGGGTGGGAGGAGGGAGAAGGG | ||
| ENSG00000185985 | 228 | AGGGAGGGAGGAGGGAGGAGGGA GGGAGGAGGGAGGAGGG | ||
| ENSG00000183019 | 3143 | AGGGAGGGAGGAGGGAGGAGGG | ||
| ENSG00000176783 | 37266 | AGGGAGGGGGGAGGGAGGAGGG | 21147 | ENSG00000202120 |
Table 7.
Variants of non-quadruplex forming sequence 3m which occur upstream of known genes
| Gene ensembl ID <- - - - - - - | | Number of bases from TSS | Sequence | Number of bases from TSS | Gene ensembl ID |- - - - - - - - -> |
|---|---|---|---|---|
| ENSG00000160145 | 61150 | AGGGCGGGCGTTGGGCGGCGGG | 53107 | ENSG00000065371 |
| ENSG00000202265 | 28176 | AGGGTGGGGGCTGGGCGGCGGG | 134617 | ENSG00000195069 |
CD studies
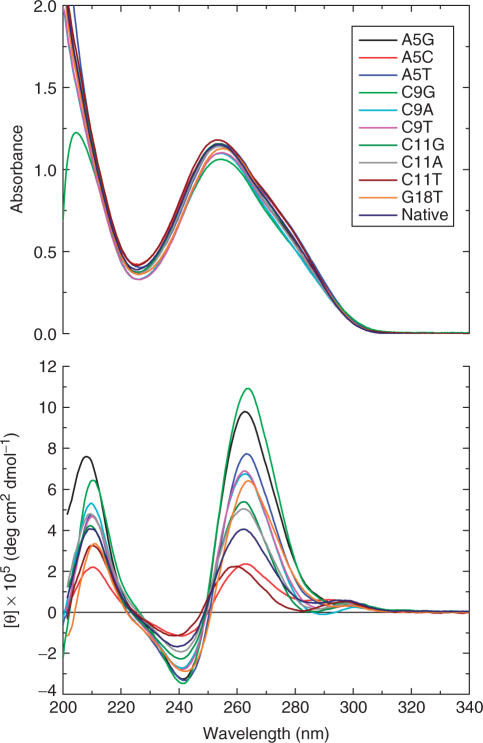
The UV and CD spectra for the c-kit87 sequence and the ten mutants are shown in Figure 3. All of the UV spectra are identical. The CD spectra all show the same pattern of minimum at 240 nm and maximum at 262 nm, although there are significant differences in peak heights.
Figure 3.
UV and CD spectra of the c-kit87 native sequence and the ten mutant sequences, taken in 50 mM KCl solution with a 1 cm path-length cell.
Simulations
We have undertaken molecular dynamics simulations on the native c-kit87 structure and ten mutants, as detailed above and in Table 1. The root mean-square deviation (RMSD) over the course of a molecular dynamics simulation was used as a measure of the conformational stability of a structure or model during that simulation. The native c-kit87 NMR model and the mutant models examined here are extremely stable structures, as is evident from the stable and small RMSDs over the timescales of 10 ns simulations, starting from the initial structure. The maximum variance ranged between 1.6 and 2.2 Å for the native and mutant 5 respectively and is shown in Table 1.
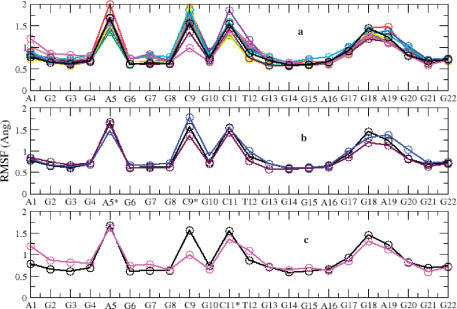
A more detailed picture of differences in residue mobility within and between simulations was obtained from graphs of the root mean-square fluctuation (RMSF) of residues relative to the average structure. The RMSF profiles of all the mutants are somewhat similar to that observed for the native structure. In particular, the peaks in the RMSF profile correspond to residues 5, 9 and 11 (Figure 4a). The NMR structure shows that these three bases do not interact directly with any other part of the structure and hence do not play any role in stabilizing it. This is fully confirmed by the simulation of the native structure and of the 5, 9 and 11 mutants.
Figure 4.
Root mean square fluctuation (Å) plotted versus residue, compared from all c-kit87 and mutant simulations (a). The plots highlight the flexibility of residues at positions 5, 9, 11 and 18; the flexible nature of the unpaired residues shows as sharp peaks, which are interspersed by the other 18 residues that contribute to the stability of the structure. Mutant 3 (A5T; coloured blue) and mutant 9 (C11T; coloured maroon) exhibit flexibility patterns similar to that observed in the native (black) structure (b), whereas residues A1, C9 and T12 exhibit a pattern of flexibility that is different from the native structure (c).
The guanine bases which contribute to quartet formation are extremely stable, whereas the AGGAG loop (and nucleotide G18 in particular) shows significant flexibility. Interestingly, as predicted by our bioinformatics results, mutation of G18 to T18 results in the retention of the same topology as the native sequence. This can be explained by the overall flexible nature of the AGGAG loop, which would allow the G18T modification to be adopted into a similar folding topology. The G17–G18 stacking in the loop is similar to that found in the T4-T5 stacking adopted in the loop region of the crystal structure of the Oxytricha telomeric sequence G4T4G4 (40: PDB id 1JPQ).
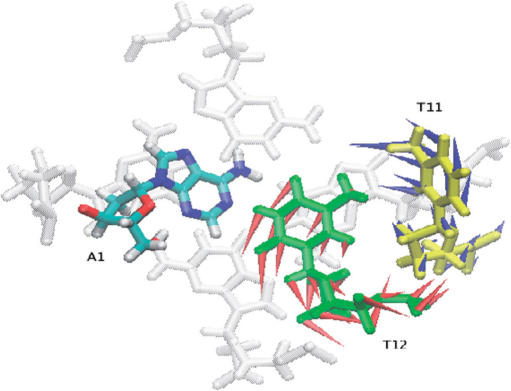
Mutants 3 (A5T) and 6 (C9T) also exhibit patterns of flexibility that are very similar to the native structure (Figure 4b). This is in accord with the NMR studies where again these modifications produced a structure with same topology as the native. However, modification of C11 to T11 was found to produce a mixture of structures with A1–T12 base pairing (in the native structure) and A1–T11 base pairing (in the mutant). Examining the RMSF profile for the mutant-9 (C11T) simulation (Figure 4c), we see that residue A1 has increased flexibility compared to the flexibility of residue A1 in the native structure simulation. Furthermore, the flexibility of residue C9 in mutant 9 is considerably reduced. A slight increase in flexibility of residue T12 is also observed, suggesting that some minor structural changes may have occurred during the course of the simulations. To investigate the dominant motions, principal components analysis was performed. By calculating the eigenvectors from the covariance matrix of a simulation and then filtering the trajectories along each of the different eigenvectors, it is possible to identify the dominant motions observed during a simulation, by visual inspection. Plotting the start and the end points of eigenvectors as arrows, highlights the direction of motion for a particular atom. Application of such an analysis to these simulations enabled us to identify the structural changes occurring between A1–T2 and T11. In order for the A1–T11 base pair to form, the A1–T12 base pair needs to be broken and T12 has to move out and pave the way for T11 to occupy its place (Figure 5). This is clearly observed in the PCA analysis; however the timescale of the simulations are too short for these entire processes to be fully simulated.
Figure 5.
Porcupine plots of the first eigenvector for simulation of mutant 9 (C11T). Residues A1, T12 (green) and T11 (yellow) are viewed sitting on a G-tetrad (coloured grey). The arrows attached to each atom in T11 and T12 indicate the direction of the eigenvector and the magnitude of the corresponding eigenvalue. The arrows summarize the direction of motion. In order for the A1–T11 base pair to form, A1–T12 base pair has to be broken and T12 subsequently moved to allow T11 to take its place.
CONCLUSIONS
The 22-nt c-kit87 promoter sequence is unique within the human genome. Its fold and tertiary structure does not have precedent among known DNA quadruplexes. The present theoretical and experimental studies have shown that (i) none of the closely related sequences (encompassing all nucleotides not involved in the maintenance of structural integrity) occur immediately upstream (<100 nt) of a transcription start site, and (ii) that all of these sequences correspond to the same stable tertiary structure. The identity of the CD spectral maxima and minima indicate that all the ten related mutant sequences adopt the same overall fold as the native c-kit87 sequence; the differences in peak height can be ascribed to the sequence differences, although a detailed analysis is beyond the scope of this article. It is concluded that the c-kit87 tertiary structure may also be formed in a small number of other loci in the human genome, but the likelihood of these playing a significant role in the expression of particular genes is small. The c-kit87 quadruplex thus fulfils a fundamental criterion of a ‘good’ drug target, of possessing distinctive 3D structural features that are only present in at most a handful of other genes, with only one, that for platelet glycoprotein Ib alpha chain precursor (ENSG00000185245) also being in a likely core promoter region. The genome searches with mutant c-kit87 sequences that are known not to form quadruplexes, found a number of hits; two are close to transcription start sites, demonstrating the importance of knowledge of the folding behaviour.
DNA is normally considered as a structurally homogeneous molecule, defined in its flexibility by the constraints of the double helix. The possibility of DNA forming higher order structures is not new, and triplexes and quadruplexes have long been postulated, especially in regulatory sequences. However, until now even these features have not been considered to possess a high degree of complexity and variation (though the structures of the c-myc quadruplexes do show features that are absent in previous quadruplex structures). The c-kit87 structure, involving 18 out of 22 nt in tertiary interactions, shows that non-duplex DNA sequences can adopt highly stable and complex arrangements. We are as yet far from knowing the rules governing these folds or the extent to which they may occur.
Searches for potential quadruplex sequences in non-telomeric DNAs have always used a template pattern based on known quadruplex sequences and their topologies (17,20,41,42), in which four runs of guanine bases are separated by three distinct loop regions: Gm Xn Gm Xo Gm Xp Gm where m = 3–5 and n,o,p = 1–7. In lieu of structural data providing evidence that additional sequence patterns are valid, we suggest that this remains a reasonable assumption. Important caveats are (i) that a particular topology cannot be assumed purely on the basis of the sequence alone, and (ii) that the occurrence of a sequence per se does not necessarily mean that it corresponds to a stable or potentially stable quadruplex—as is the case with a number of the c-kit87 mutants (26). The distinctly non-random distribution of particular bases at the non-essential 5, 9, 11 and 18 positions of the c-kit87 sequence is a surprising observation, which is being further examined experimentally and theoretically.
ACKNOWLEDGEMENTS
Funding to pay the Open Access publication charges for this article was provided by Cancer Research UK (programme grant to S. N.).
Conflict of interest statement. None declared.
REFERENCES
- 1.Yarden Y, Kuang WJ, Yang-Feng T, Coussens L, Munemitsu S, Dull TJ, Chen E, Schlessinger J, Francke U, et al. Human proto-oncogene C-Kit - a new cell-surface receptor tyrosine kinase for an unidentified ligand. EMBO J. 1987;6:3341–3351. doi: 10.1002/j.1460-2075.1987.tb02655.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roskoski R. Structure and regulation of Kit protein-tyrosine kinase–the stem cell factor receptor. Biochem. Biophys. Res. Commun. 2005;337:1307–1315. doi: 10.1016/j.bbrc.2005.09.150. [DOI] [PubMed] [Google Scholar]
- 3.Hirota S, Isozaki K, Moriyama Y, Hashimoto K, Nishida T, Ishiquro S, Kawano K, Hanada M, Kurata A, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science. 1998;279:577–580. doi: 10.1126/science.279.5350.577. [DOI] [PubMed] [Google Scholar]
- 4.Taniguchi M, Nishida T, Hirota S, Isozaki K, Ito T, Nomura T, Matsuda H, Kitamura Y. Effect of c-kit mutation on prognosis of gastrointestinal stromal tumors. Cancer Res. 1999;59:4297–4300. [PubMed] [Google Scholar]
- 5.Tarn C, Godwin AK. Molecular research directions in the management of gastrointestinal stromal tumors. Curr. Treat. Options Oncol. 2005;6:473–486. doi: 10.1007/s11864-005-0026-x. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher JA, Rubin BP. KIT mutations in GIST. Curr. Opin. Genet. Dev. 2007;17:3–7. doi: 10.1016/j.gde.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 7.Wang YY, Zhou GB, Yin T, Chen B, Shi JY, Liang WX, Jin XL, You JH, Yang G, et al. AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia: implication in stepwise leukemogenesis and response to Gleevec. Proc. Natl Acad. Sci. USA. 2005;102:1104–1109. doi: 10.1073/pnas.0408831102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Looijenga LH, de Leeuw H, van Oorschot M, van Gurp RJ, Stop H, Gillis A, de Gouveia Brazao CA, Weber RE, Kirkels WJ, et al. Stem cell factor receptor (c-KIT) codon 816 mutations predict development of bilateral testicular germ-cell tumors. Cancer Res. 2003;63:7674–7678. [PubMed] [Google Scholar]
- 9.Heinrich MC, Corless CL, Demetri GD, Blanke CD, von Mehren M, Joensuu H, McGreevey LS, Chen CJ, Van den Abbeele AD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J. Clin. Oncol. 2003;21:4342–4349. doi: 10.1200/JCO.2003.04.190. [DOI] [PubMed] [Google Scholar]
- 10.Mol CD, Dougan DR, Schneider TR, Skene RJ, Kraus ML, Scheibe DN, Snell GP, Zou H, Sang BC, et al. Structural basis for the autoinhibition and STI-571 inhibition of c-Kit tyrosine kinase. J. Biol. Chem. 2004;279:31655–31663. doi: 10.1074/jbc.M403319200. [DOI] [PubMed] [Google Scholar]
- 11.Heinrich MC, Corless CL, Blanke CD, Demetri GD, Joensuu H, Roberts P J, Eisenberg BL, von Mehren M, Fletcher CD, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J. Clin. Oncol. 2006;24:4764–4774. doi: 10.1200/JCO.2006.06.2265. [DOI] [PubMed] [Google Scholar]
- 12.Corbin AS, Griswold IJ, La Rosee P, Yee KW, Heinrich MC, Reimer CL, Druker BL, Deininger MW. Sensitivity of oncogenic KIT mutants to the kinase inhibitors MLN518 and PD180970. Blood. 2004;104:3754–3757. doi: 10.1182/blood-2004-06-2189. [DOI] [PubMed] [Google Scholar]
- 13.Schittenhelm MM, Shiraga S, Schroeder A, Corbin AS, Lee FY, Bokemeyer C, Deininger MW, Druker BJ, Heinrich MC. Dasatinib (BMS-354825), a dual src/abl kinase inhibitor, inhibits the kinase activity of wild-type, juxtamembrane, and activation loop mutant kit isoforms associated with human malignancies. Cancer Res. 2006;66:473–481. doi: 10.1158/0008-5472.CAN-05-2050. [DOI] [PubMed] [Google Scholar]
- 14.Debiec-Rychter M, Cools J, Dumez H, Sciot R, Stul M, Mentens N, Vranckx H, Wasag B, Prenen H, et al. Mechanisms of resistance to imatinib mesylate in gastrointestinal stromal tumors and activity of the PKC412 inhibitor against imatinib-resistant mutants. Gastroenterology. 2005;128:270–279. doi: 10.1053/j.gastro.2004.11.020. [DOI] [PubMed] [Google Scholar]
- 15.Prenen H, Cools J, Mentens N, Folens C, Sciot R, Schoffski P, Van Oosterom A, Marynen P, Debiec-Rychter M. Efficacy of the kinase inhibitor SU11248 against gastrointestinal stromal tumor mutants refractory to imatinib mesylate. Clin. Cancer Res. 2006;12:2622–2627. doi: 10.1158/1078-0432.CCR-05-2275. [DOI] [PubMed] [Google Scholar]
- 16.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 20.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hurley LH, Von Hoff DD, Siddiqui-Jain A, Yang D. Drug targeting of the c-MYC promoter to repress gene expression via a G-quadruplex silencer element. Seminars Oncol. 2006;33:498–512. doi: 10.1053/j.seminoncol.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Phan AT, Modi YS, Patel DJ. Propeller-type parallel-stranded G-quadruplexes in the human c-myc promoter. J. Am. Chem. Soc. 2004;126:8710–8716. doi: 10.1021/ja048805k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ambrus A, Chen D, Dai J, Jones RA, Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-myc promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. doi: 10.1021/bi048242p. [DOI] [PubMed] [Google Scholar]
- 25.Phan AT, Kuryavyi V, Gaw HY, Patel DJ. Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter. Nat. Chem. Biol. 2005;1:167–173. doi: 10.1038/nchembio723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fernando H, Reszka AP, Huppert J, Ladame S, Rankin S, Venkitaraman AR, Neidle S, Balasubramanian S. A conserved quadruplex motif located in a transcription activation site of the human c-kit oncogene. Biochemistry. 2006;45:7854–7860. doi: 10.1021/bi0601510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamamoto K, Tojo A, Aoki N, Shibuya M. Characterization of the promoter region of the human c-kit proto-oncogene. Jpn. J. Cancer Res. 1993;84:1136–1144. doi: 10.1111/j.1349-7006.1993.tb02813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park GH, Plummer HK, Krystal GW. Selective Sp1 binding is critical for maximal activity of the human c-kit promoter. Blood. 1998;92:4138–4149. [PubMed] [Google Scholar]
- 30.Cairns LA, Moroni E, Levantini E, Giorgetti A, Klinger FG, Ronzoni S, Tatangelo L, Tiveron C, De Felici M, et al. Kit regulatory elements required for expression in developing hematopoietic and germ cell lineages. Blood. 2003;102:3954–3962. doi: 10.1182/blood-2003-04-1296. [DOI] [PubMed] [Google Scholar]
- 31.Parkinson GN, Lee MP, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 32.Phan AT, Kuryavyi V, Burge S, Neidle S, Patel DJ. Structure of an unprecedented G-quadruplex scaffold in the c-kit promoter. J. Am. Chem. Soc. 2007;129:4386–4392. doi: 10.1021/ja068739h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higgins D, Thompson J, Gibson T. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Durbin R, Eddy S, Krogh A, Mitchison G. Biological Sequence Analysis. Probabilistic Models of Proteins and Nucleic Acids. Cambridge: Cambridge University Press; 1998. [Google Scholar]
- 36.Case DA, Cheatham T.E., III, Darden T, Gohlke H, Luo R, Merz K.MJr, Onufriev A, Simmerling C, Wang B, et al. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darden T, Perera L, Li L, Pedersen L. New tricks for modelers from the crystallography toolkit: the particle-mesh Ewald algorithm and its use in nucleic acid simulations. Structure. 1999;7:R55–R60. doi: 10.1016/s0969-2126(99)80033-1. [DOI] [PubMed] [Google Scholar]
- 38.Price DJ, Brooks CL. A modified TIP3P water potential for simulation with Ewald summation. J. Chem. Phys. 2004;121:10096–10103. doi: 10.1063/1.1808117. [DOI] [PubMed] [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD - visual molecular dynamics. J. Molec. Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 40.Haider SM, Parkinson GN, Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 41.Kostadinov R, Malhotra N, Viotti M, Shine R, D’Antonio L, Bagga P. GRSDB: a database of quadruplex forming G-rich sequences in alternatively processed mammalian pre-mRNA sequences. Nucleic Acids Res. 2006;34:D119–124. doi: 10.1093/nar/gkj073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rawal P, Kummaraseitti VB, Ravindran J, Kumar N, Halder K, Sharma R, Mukerji M, Das SK, Chowdhury S. Genome-wide prediction of G4 DNA as regulatory motifs: role in Escherichia coli global regulation. Genome Res. 2006;16:644–655. doi: 10.1101/gr.4508806. [DOI] [PMC free article] [PubMed] [Google Scholar]