Abstract
Ancient DNA (aDNA) research has long depended on the power of PCR to amplify trace amounts of surviving genetic material from preserved specimens. While PCR permits specific loci to be targeted and amplified, in many ways it can be intrinsically unsuited to damaged and degraded aDNA templates. PCR amplification of aDNA can produce highly-skewed distributions with significant contributions from miscoding lesion damage and non-authentic sequence artefacts. As traditional PCR-based approaches have been unable to fully resolve the molecular nature of aDNA damage over many years, we have developed a novel single primer extension (SPEX)-based approach to generate more accurate sequence information. SPEX targets selected template strands at defined loci and can generate a quantifiable redundancy of coverage; providing new insights into the molecular nature of aDNA damage and fragmentation. SPEX sequence data reveals inherent limitations in both traditional and metagenomic PCR-based approaches to aDNA, which can make current damage analyses and correct genotyping of ancient specimens problematic. In contrast to previous aDNA studies, SPEX provides strong quantitative evidence that C > U-type base modifications are the sole cause of authentic endogenous damage-derived miscoding lesions. This new approach could allow ancient specimens to be genotyped with unprecedented accuracy.
INTRODUCTION
Traces of genetic material preserved within ancient specimens can provide a unique and important real-time record of the past (e.g. 1–4). However, this record is compromized because ancient DNA (aDNA) is invariably damaged and degraded to some extent, initially by endogenous nucleases and microorganisms after death, and subsequently by hydrolysis and oxidation reactions that can fragment the DNA backbone and chemically modify bases (5,6). Spontaneous base-loss events, creating non-coding abasic sites (6,7), and certain base modifications (8,9) can block the amplification of aDNA templates by halting DNA polymerase-mediated primer extension. In contrast, other base modifications can create damage-derived miscoding lesions (DDMLs) which do permit polymerase extension but which have altered base-pairing properties, leading to altered sequences in newly amplified DNA (7).
Almost all aDNA studies to date have been PCR-based, as this method can generate sequence data from the trace amounts of DNA preserved in ancient specimens. However, PCR can generate incorrect sequence data from aDNA for a number of reasons. In addition to an intrinsic background rate of polymerase misincorporation errors, the altered base-pairing properties of endogenous DDMLs can cause considerable amounts of sequence variation in PCR-amplified aDNA (10,11). ‘Jumping-PCR’, where partially extended primers switch between different damaged and degraded aDNA template strands during the early cycles of PCR amplification, has been shown to create non-authentic, recombinant, sequences (12–14). ‘Jumping-PCR’ artefacts may also be compounded by the tendency of many DNA polymerases to add a single nucleotide to the 3′-end of primer extensions in a non-template directed fashion (10,14–19). PCR can also generate additional, non-endogenous, sequence artefacts such as so-called ‘Type 1 damage’ (20–22). PCR amplification of low copy number templates is known to create products with highly skewed representations (13,23). This means that sequence artefacts can easily come to dominate the products of PCR-amplified aDNA (10,11,13,14,23). Sequence accuracy is therefore a major issue in aDNA research.
These factors have been recognized to varying degrees. The overlapping ‘best-of-three’ PCR amplification and cloning strategy currently used when key ancient samples are amplified by standard or multiplex PCR (10,24,25), explicitly accepts the inherent shortcomings of PCR-generated aDNA sequences and significantly increases the chances of correctly inferring the original endogenous DNA sequence. However, there are two essential prerequisites for a quantitative investigation into the molecular nature of aDNA damage and its effects on sequence accuracy. First, authentic endogenous DDMLs must be disentangled from other non-endogenous, PCR-generated, forms of sequence variation. Secondly, due to the complementary double-stranded nature of DNA, the template strand-of-origin of particular DDML base modification events must somehow be unambiguously specified. Exponential amplification from both strands of a DNA template is intrinsic to PCR and this prevents the strand-of-origin of particular base changes from being determined (20). Together with the demonstrated potential for the generation of additional non-authentic sequence variation, these limitations of PCR-based methods have prevented full resolution of the molecular nature of DDMLs in aDNA.
Although there has always been strong theoretical and biochemical evidence that C > U-type DDMLs are a major cause of Type 2 ‘damage’ (C > T/G > A) transitions in PCR-amplified aDNA sequences (e.g. 5,6,10), there has also been considerable debate about the existence, or otherwise, of a genuine biochemical basis for Type 1 ‘damage’ (T > C/A > G) transitions. However, it has recently become clear that so-called Type 1 ‘damage’, observed at significant levels by some traditional PCR-based studies (e.g. 20,26–28), disappears once alternative techniques are employed (21,22; this study), and this is now recognized as a non-endogenous, PCR-generated, phenomenon (22).
The potential role(s) of aDNA templates that are shorter than the target region in PCR amplifications is an issue that requires closer examination. Following the first cycle, only those initial primer extensions long enough to cover the entire target region could be utilized directly by both PCR primers. As we demonstrate however, as the PCR target length increases so does the proportion of shorter, abortive, primer extensions. These have the potential to contribute to the creation of recombinant and other non-authentic sequence artefacts in subsequent cycles. These findings raise questions about the widespread use of quantitative PCR (qPCR) methods to estimate the numbers of aDNA templates contributing to the products of PCR amplification reactions. qPCR results give no information about the number of templates below the target size that end up contributing to amplification products via ‘jumping-PCR’ and other PCR-generated mechanisms. PCR amplification from ancient extracts with template copy numbers estimated by qPCR to be in the tens-of-thousands have been shown to produce significant levels of non-endogenous ‘Type 1 damage’ artefacts (e.g. 27). Therefore the widespread assumption that given a sufficiently high estimated starting number, endogenous DDML sequence diversity in aDNA templates will necessarily be reflected by the sequence variation within PCR-generated products simply cannot be sustained.
As traditional PCR-based approaches have proven incapable of fully resolving the molecular nature of DDMLs, we have developed a novel SPEX-based approach (Figure 1) to generate detailed information about post mortem DDML base modifications in aDNA. In direct contrast to PCR, SPEX is an amplification methodology that specifically targets only one of the aDNA template strands at a locus-of-interest and imposes no predefined target length. This allows the production of first-generation copies of aDNA template molecules, with quantifiable (up to 40-fold or more) coverage from a single reaction. SPEX is shown to be capable of producing sequence data of unprecedented accuracy from aDNA, without the generation of additional, non-endogenous, sequence artefacts over and above a background rate of misincorporation errors common to polymerase-based methodologies.
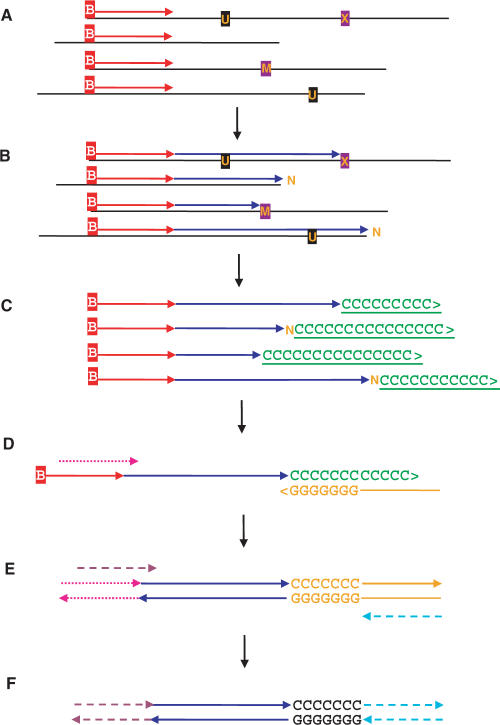
Figure 1.
Single primer extension (SPEX) amplification. (A) Denaturation and hybridization of a single biotinylated primer to one target strand at the locus-of-interest. (B) Primer extension by a thermostable DNA polymerase until halted at the physical end of an aDNA template molecule; at a polymerase-blocking modified base [M]; or at an abasic site, some other non-coding lesion, or some kind of physical block [X]. Miscoding lesions [U] do not block primer extension, but result in altered sequences. Polymerases can catalyze the non-directed addition of a single 3′-terminal nucleotide [N] following primer extension to either the physical end of a fragmented aDNA template, or to an abasic site or other non-coding lesion. Single or multiple cycles of SPEX primer extension can be used. Biotinylated molecules were then bound to Streptavidin-coated beads and stringent washes removed everything else (e.g. aDNA template molecules, enzymes, buffer, etc). (C) Biotinylated primers and extended primers (with single-stranded, direct first-generation copies of individual aDNA template molecules) were then polyC-tailed using terminal transferase (TdT), followed again by bead-wash removal of TdT and buffer. (D) Locus-specific, primer-extended, polyC-tailed ssDNA molecules were then selectively amplified by PCR; using a partially-nested, locus-specific, SPEX-2 forward primer (Tables S1 and S2) and a polyG-based, 5′ adapter-tagged, reverse primer (Tables S1 and S2). (E) These products were amplified a final time by PCR using a further partially nested, locus-specific, SPEX-3 forward primer (Tables S1 and S2) and a 5′-adapter reverse primer (Tables S1 and S2). (F) The final products of SPEX amplification underwent restriction digestion, directional cloning and sequencing.
Recently, massively parallel metagenomic sequencing approaches have also been used to investigate aDNA damage. By inferring the sequences of individual single-stranded DNA (ssDNA) templates generated from aDNA via the 454-methodology (29), independent studies concluded that in addition to C > U-type DDMLs, a substantial proportion of Type 2 transitions were due to modification(s) of G residues (by an unknown biochemical process) that caused them to be read as A by polymerases (21,22,30). Here, we use SPEX to overcome limitations inherent to both traditional PCR- and current 454-based approaches. The ability of SPEX to rigorously distinguish between authentic aDNA and first-generation copied sequences, whilst simultaneously quantitatively generating highly accurate sequence data from designated loci, has enabled the molecular nature of DDMLs to be fully revealed for the first time.
MATERIAL AND METHODS
Samples
The main body of this study analyses data from SPEX amplification experiments on fifteen aDNA extracts, performed in a dedicated aDNA laboratory at the Henry Wellcome Ancient Biomolecules Centre, University of Oxford. DNA had previously been extracted and analysed for each specimen using well-established aDNA methods (as for 4,31). Each SPEX analysis focussed on sections of the mitochondrial control region where diagnostic SNPs or sequence ‘fingerprints’ were known to characterize individual specimens. Samples from three mammalian species (bison, human, Eurasian cave lion) were selected to cover a broad range of ages, environments, regions and types of site (Table S2). SPEX experiments using synthetic oligonucleotide templates were carried out independently at the Australian Centre for Ancient DNA (ACAD) in Adelaide.
Design of SPEX amplification
The SPEX strategy for amplification of aDNA is shown schematically in Figure 1. All polymerase-based methodologies introduce a background level of polymerase misincorporation errors. However, the use of a single primer extension, followed by homopolymer tailing, avoided the potential creation of additional PCR-generated artefacts. Any deviations from the known underlying primary aDNA sequences in the first-generation copies of aDNA were quantitatively analysed. Partially nested SPEX primer sets are shown in Tables S1 and S2. SPEX can access genetic information from highly fragmented and damaged aDNA templates since, unlike PCR, it does not have a pre-defined target size based on the primer pair. SPEX primer extension continues until halted by aDNA template fragmentation or a polymerase-blocking lesion. After the primer extension stage, all aDNA templates were completely removed by Strepavidin bead-washes and the remaining, single-stranded, first-generation copies of aDNA target strands were then permanently ‘trapped’ by polyC-tailing. Nested PCR amplification was then used to amplify what were now effectively ‘modern’ ssDNA template molecules, with minimal risk therefore of subsequent ‘jumping-PCR’ events. A multi-cycle extension variant of SPEX was also examined. Sequence data was generated using SPEX from the following sequence positions for the bison, human and cave lion samples. Bison: equivalent position to 16 178 (single-cycle SPEX) or 16 175 (multi-cycle SPEX) upwards on the Bos taurus mitochondrial genome [Genbank V00654; (32)]. Humans: from three parts of the human mitochondrial control region (according to the revised Cambridge Reference Sequence (33), Genbank J01415.2); 16 223 upwards; 16 364 downwards and 16 267 downwards. Cave lions: from the equivalent to position 95 onwards on the Panthera leo spelaea partial mitochondrial sequence (Genbank DQ899910).
Single-cycle primer extension
Reactions were performed in 50 μl volumes with 1–5 μl of aDNA extract added to reactions comprising: 1 mg/ml rabbit serum albumin (RSA; Sigma) to help overcome polymerase inhibitors; 1× High Fidelity PCR buffer; 2 mM magnesium sulphate; 200 μM of each dNTP; 1.5 Units (U) Platinum Taq DNA Polymerase High Fidelity (Invitrogen) and 0.2 μM of the appropriate 5′-biotinylated SPEX-1 primer (Tables S1 and S2). Synthetic oligonucleotide templates (Figure S3) were used at 0.25 μM. The thermocycling profile was: 95°C for 3°min, 53–57°C (depending on primer) for 1 min, 68°C for 10 min; then 4°C until bead washing.
Multi-cycle primer extension
Reactions were performed in 50 μl volumes with 1.0–5.0 μl of aDNA extract added to reactions comprising: 1 mg/ml bovine serum albumin (BSA; Sigma) for non-bison extracts (RSA for bison extracts) to help overcome polymerase inhibtors; 1× AmpliTaq Gold buffer II; 2.5 mM magnesium chloride; 200 μM of each dNTP; 1.5 U AmpliTaq Gold (Perkin Elmer) and 0.2 μM of the appropriate 5′-biotinylated SPEX-1 primer (Tables S1 and S2). The thermocycling profiles were: 95°C for 5 min; followed by 50 cycles of 30 s at 95°C, 30 s at 54–60°C and 1 min at 72°C; then 4°C until bead washing.
Bead washing
Aliquots of 20 μl of Streptavidin magnetic beads (New England Biolabs, S1420S) were pre-washed three times with 2× BW buffer (34); resuspended in 50 μl 2× BW; mixed with the 50 μl SPEX primer extension reaction and rotated at room temperature for 30 min to immobilize biotinylated molecules to the beads; then a series of washes with 2× BW, 0.15 M NaOH and 1× Tris/EDTA (TE, pH 7.5) were carried out as described by Chen et al. (34) to remove everything but biotinylated molecules. The beads were resuspended to 14 μl with 0.1× Qiagen buffer EB (10 mM Tris·Cl, pH 8.5).
PolyC-tailing
PolyC-tailing was performed for 1 h at 37°C in 20 μl reactions comprising: 15 U of Terminal Deoxynucleotidyl Transferase, Recombinant (rTdT) (Invitrogen, 10 533–065); 1× TdT reaction buffer; 500 μM dCTP; and the 14 μl of resuspended beads. Washes with 1× TE (pH 7.5) were carried out to remove everything but polyC-tailed, biotinylated molecules. The beads were resuspended to 15 μl with 0.1× Qiagen buffer EB.
First partially nested PCR amplification
Resuspended beads were used in 50 μl reactions with: 1× High Fidelity PCR buffer; 2 mM magnesium sulphate; 200 μM of each dNTP; 1.5 U Platinum Taq DNA Polymerase High Fidelity and 0.2 μM of the appropriate SPEX-2 (F & R) primers (Tables S1 and S2). The thermocycling profiles were: 2 min at 95°C; followed by 50 cycles of 30 s at 95°C, 30 s at 48–54°C and 1 min at 68°C; with a final extension of 10 min at 68°C. Excess primers and nucleotides were removed with QIAprep PCR purification columns (Qiagen).
Second partially nested PCR amplification
In order to ensure complete specificity prior to the extensive cloning and sequencing of SPEX amplicons required for quantitative aDNA damage analyses, a second round of partially nested PCR amplifications were performed. However, this additional step could be omitted for general SPEX experiments. Reactions were performed in 25 μl volumes comprising: a 100-fold dilution of the cleaned-up first-round products; 1× High Fidelity PCR buffer; 2 mM magnesium sulphate; 200 μM of each dNTP; 0.75 U Taq; and 0.2 μM of the appropriate SPEX-3 (F & R) primers (Tables S1 and S2). The thermocycling profiles were: 2 min at 95°C; followed by 35 cycles of 20 s at 95°C, 20 s at 54–57°C, and 1 min at 68°C; with a final extension of 10 min at 68°C. Excess primers and nucleotides were removed as above.
Cloning, sequencing and sequence analysis
All steps followed standard protocols, according to manufacturer's instructions where appropriate, and are described in detail as Supplementary Data.
Behaviour of platinum Taq polymerase mix following primer extension
To examine the behaviour of the Platinum Taq DNA Polymerase High Fidelity mix (commonly used in aDNA research) upon completing primer extension to either the physical end of a template molecule, or to an internal abasic site, simplified systems of HPLC-purified synthetic oligonucleotide templates were amplified by single-cycle SPEX and cloned and sequenced as above. These constructs used the same SPEX primers as the single cycle SPEX amplification of bison aDNA to allow a direct comparison between otherwise equivalent regions of ancient and non-ancient DNA.
Distribution of Type 2 aDNA damage amongst 454-derived molecules
We re-analysed 1449 Mammuthus primigenius mitochondrial sequences from a data set of ssDNA molecules produced by the GS 20 Sequencing System (454 Life Sciences, Branford, CT) as previously described (22,35) to investigate whether Type 2 transitions are randomly distributed across the molecules. A Kolmogorov–Smirnov one sample test was used to test the hypothesis that the positions of 514 C > T and 231 G > A transitions were each distributed according to a uniform distribution along the length of the sequence traces. A t-test and Wilcoxon rank sum test were also performed to compare the positions of the C > T and G > A transitions relative to one another, along each trace. These tests allowed us to accept or reject the null hypothesis that, respectively, the mean and median relative positions of the two types of mutation are the same. The summary data from this analysis was compiled and represented using a box-plot (36).
Distribution of C > U-type damage-derived miscoding lesion events
The distribution of C > U-type DDML events (observed as complementary G > A transitions on SPEX-derived first-generation copied aDNA sequences) was investigated for each of the six ancient bison extracts amplified with single-cycle SPEX using a generalized linear model (Poisson family, log link function) in the R statistical package (http://www.r-project.org/): testing for a random distribution of C > U-type events whilst taking into account the lengths of each molecule. We calculated the lack of fit to this model by treating the residual deviance as a χ2 random deviate with the residual number of degrees of freedom. We calculated (1-P) where P is the probability of obtaining a random deviate as large, or larger, than that observed. Small values of (1-P) suggest a lack of fit of the model because of over-dispersion—a departure from a Poisson distribution such that damaged sites are clustering on particular strands within a sample, even when differences in length are taken into account. Samples that displayed a trend towards over-dispersion were parametrically modelled using the negative-binomial family version of the Generalized Linear Model (37).
RESULTS
Single primer extension amplification of aDNA
Sections of the mitochondrial control region were amplified by single-cycle SPEX from six bison extracts covering a wide range of ages and environments (Table S2). A ‘total cloned’ data set (TCDS) contained all sequences obtained. A ‘conservative’ data set (CDS; Figure S1) comprised inserts with discrete lengths and/or primary sequences. Differences in SPEX length resulted from differences in the sites of aDNA template fragmentation or polymerase-blocking lesions. Differences in primary sequence reflected contributions from endogenous DDMLs, ‘non-directed’ polymerase activity at the 3′-end following primer extension and polymerase misincorporation errors. Many polymerases can add a single nucleotide in a non-directed manner when primer extension stalls at a non-coding lesion, such as an abasic site (17–19), or halts after reaching the physical end of a template molecule (15,16). Fifty-two percent of 3′-terminal bases in the CDS did not match the known underlying template sequence (Figures S1 and S2), meaning at least 69% (Table S3) of SPEX events must have undergone non-directed nucleotide addition (NDNA) at the 3′-end. For this reason, bases at the 3′-terminal position (immediately 5′ to the polyC-tail) were excluded from all SPEX aDNA damage analyses.
The single-cycle SPEX TCDS covered 10 644 bases from 548 amplicons, while the CDS covered 7654 bases from 337 discrete sequences (Table 1). Most CDS sequences were distinguishable on the basis of length (Figure S1). Of those with identical length, the vast majority of these differed due to G > A transitions (i.e. largely from endogenous C > U-type DDMLs on the template strand) and/or NDNA at the 3′-end. Only 5/337 of the CDS sequences differed from one another due solely to a non-C > U-derived transition. Therefore, we estimate that at least 98% of the SPEX amplicon sequences in the CDS were ultimately derived from discrete SPEX events on discrete aDNA template molecules (with <2% reflecting polymerase errors). Single-cycle SPEX amplification appears to be highly robust since, despite subsequent partially nested amplification steps, 548 sequenced clones did not produce a single example of cross-contamination between any of the six individual bison specimens (Figure S1). Multi-cycle SPEX gave essentially indistinguishable results to single-cycle SPEX (Table 2).
Table 1.
Transitions and transversions observed after single-cyclea SPEX amplification of six ancient bison extracts
| A | Type 1 | Type 2 | ||||||
| Observed transitionsb | A > G | T > C | G > A | C > T | ||||
| aDNA DDMLs? c | T > C? | A > HX | C > U | G > X | ||||
| Conservative data setd | 9 | 3 | 210 | 2 | ||||
| Total cloned data sete | 10 | 3 | 279 | 2 | ||||
| B | ||||||||
| Observed transversions | C > A | C > G | G > C | G > T | T > G | T > A | A > T | A > C |
| Conservative data set | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 |
| Total cloned data set | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 5 |
aEach SPEX primer extension reaction initially comprising a single cycle of primer annealing and extension using Platinum Taq DNA Polymerase High Fidelity (Invitrogen), followed by bead clean-up and polyC-tailing. bEvery SPEX-amplified sequence is derived from an initial direct first-generation copy of an aDNA template strand, so the observed transitions represent the complement of DDML events that actually existed on the original aDNA templates. The base composition amongst those nucleotides common to all specimens (i.e. non-‘fingerprinting’ positions; Figure S1) across the first 56 bp of this locus (completely encompassing 94% of observed primer extension events) was: G = 13 (27%); A = 10 (20%); T = 8 (16%); C = 18 (37%). cA > HX, C > U and G > X are base modifications that have previously been suggested as the causes of particular DDMLs (10,21,39). dThe conservative data set only considered discrete sequences derived from primer extension events which differed in length, primary sequence, or 3′-terminal NDNA and covered 7654 nucleotides from 337 discrete SPEX-derived amplicons.eThe total cloned data set represents every sequence obtained after extensive cloning and covered 10 644 nucleotides from 548 SPEX-derived amplicons.
Table 2.
Transitions and transversions observed after multi-cyclea SPEX amplification of five ancient bison, four ancient human and three ancient Eurasian cave lion extracts
| Type 1 | Type 2 | Transversions | |||
|---|---|---|---|---|---|
| Observed transitionsb | A > G | T > C | G > A | C > T | |
| aDNA DDMLs?c | T > C? | A > HX | C > U | G > X | |
| Five ancient bison extracts; one locus | |||||
| Conservative data setd | 2 | 0 | 91 | 1 | 5 |
| Total cloned data sete | 2 | 0 | 111 | 1 | 5 |
| Four ancient human extracts; three loci | |||||
| Conservative data setf | 0 | 1 | 50 | 1 | 1 |
| Total cloned data setg | 0 | 1 | 57 | 1 | 1 |
| Three ancient cave lion extracts; one locus | |||||
| Conservative data seth | 2 | 1 | 25 | 0 | 0 |
| Total cloned data seti | 2 | 1 | 26 | 0 | 0 |
| Combined multi-cycle SPEX data | |||||
| Conservative data setj | 4 | 2 | 166 | 2 | 6 |
| Total cloned data setk | 4 | 2 | 194 | 2 | 6 |
aEach SPEX primer extension reaction initially comprising 50 cycles of single primer annealing and extension using AmpliTaq Gold (Perkin Elmer), followed by bead clean-up and polyC-tailing. b,cObserved transitions and proposed miscoding lesions are explained in Table 1. d4187 nucleotides from 171 discrete SPEX-derived amplicons. e4934 nucleotides from 219 SPEX-derived amplicons. f3304 nucleotides from 164 discrete SPEX-derived amplicons. g4647 nucleotides from 271 SPEX-derived amplicons. h4086 nucleotides from 111 discrete SPEX-derived amplicons. i4470 nucleotides from 141 SPEX-derived amplicons. j11 577 nucleotides from 446 discrete SPEX-derived amplicons. k14 051 nucleotides from 1121 SPEX-derived amplicons.
SPEX can access highly damaged and fragmented aDNA template molecules
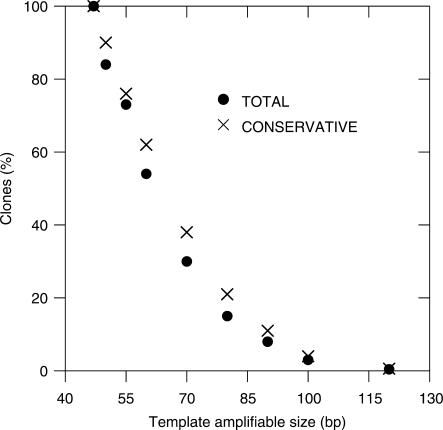
As single-cycle SPEX primer extension is functionally equivalent to the first cycle of PCR, the observed lengths of first-generation copied aDNA were analysed to estimate what proportion would, or would not, have been available for subsequent exponential PCR amplification with a reverse PCR primer placed at defined distances away. When given the opportunity, PCR is known to preferentially amplify shorter targets (38), meaning that there is likely to have been at least some drift towards SPEX amplicons representing shorter primer extension events. Nevertheless, a plot of apparent ‘template amplifiable size’ by PCR (i.e. the maximal SPEX-amplified aDNA product length) versus the percentage of all clones for each product length (Figure 2) clearly shows that as the size of the target PCR amplicon increased, the production of directly PCR-amplifiable primer extensions would dramatically decrease compared to the production of shorter, abortive, primer extensions.
Figure 2.
Graph showing the percentage of clones of the single-cycle SPEX CDS and TCDS (Figure S1; Table 1) versus ‘template amplifiable size’ (in bp) by PCR. The best-fit function that describes the data is an exponential decay (ExpGauss) with formula y = exp(a+b*x+c*x2); where: a = 3.966; b = 0.019 and c = 0.050. The best-fit line was obtained by non-linear least-square curve fitting algorithms, and the curve shows the relationship between SPEX-amplified aDNA product length (x) and the percentage of all clones for each product length (y). Template amplifiable mean: 83.5 bp and SD: 24.9 bp. The minimum size considered was 47 bp, which corresponds to SPEX primer extension events that originally extended 1 base past the 3′-end of the final SPEX-3 partially nested forward PCR primer (Tables S1 and S2).
By analogy to the single-cycle SPEX data, the kinds of sizes targeted in most PCR-based aDNA studies so far e.g. (4,10,20,26–28,39,40) should also have led to the production of many times more abortive primer extensions than directly PCR-amplifiable ones during the initial PCR cycles. Abortive primer extensions like these would be available to contribute to the creation of recombinant ‘jumping-PCR’ artefacts in subsequent cycles. Moreover, the majority of these abortive primer extension events could be expected to undergo NDNA at their 3′-end. Therefore the potential contribution of molecular events like these to the generation of non-endogenous sequence artefacts in subsequent cycles during traditional PCR-based aDNA studies is an area that needs further investigation.
Physical fragmentation or non-coding lesions, like abasic sites, halt most primer extension events on aDNA templates
Many polymerases can catalyse the non-templated addition of a single overhanging nucleotide (overwhelmingly A) to blunt-ended double-stranded DNA (dsDNA), following full primer extension to the physical end of a template molecule (15,16). The requirement for blunt-ended dsDNA is absolute as NDNA does not occur on ssDNA (15). On the other hand, most known polymerase-blocking base modifications do not result in non-authentic nucleotides at the 3′-terminal position (8). Physical blocks to polymerase extension (e.g. inter-strand crosslinks) should also lead to correctly-paired 3′ nucleotides.
The high proportion of positions exhibiting a non-authentic 3′-terminal A (Figures S1 and S2; Table S3) might suggest full primer extension and NDNA following a fairly random and extensive fragmentation of aDNA template molecules. However at abasic sites, polymerases can also ‘non-instructionally’ add A as well as lower levels of G, T or C, nucleotides (17–19). Purine (A or G) bases are known to be released from aDNA at a higher rate than pyrimidines (C or T) to create abasic sites (6). If abasic sites played a significant role in NDNA, we would expect to find higher levels of non-authentic 3′-terminal bases opposite A and G sites on the complementary strand. All six bison specimens shared an identical sequence over the first 17 bases of single-cycle SPEX primer extension (Figure S1). Over this region, 62 non-authentic 3′-terminal A, G or T bases were observed opposite the sites of the eight purines on the complementary strand, while only 24 were observed opposite the sites of the nine pyrimidines (Figure S2). (The use of polyC-tailing prevents an analysis of NDNA events involving C.) These findings point towards a significant contribution from abasic sites. A less likely alternative, given the significant levels of non-authentic G and T bases at the 3′-terminal position, is that there is an elevated rate of strand breakage immediately 3′ to purine bases, due to some unknown mechanism.
Single-cycle SPEX was used to amplify synthetic oligonucleotide templates (Figure S3) in a detailed analysis of the behaviour of the Platinum Taq DNA Polymerase High Fidelity mix; both at an abasic site and when primer extension reached the physical end of a template. The level of NDNA on aDNA templates (69%) lies between the levels observed at an abasic site (94%) and following full primer extension (50%) in the test system (Table S3). Overall, the NDNA data is therefore consistent with the great majority of primer extension events on aDNA templates being halted due to either template fragmentation or an abasic site, with these being the major factors limiting the effective amplifiable size of aDNA. This evidence contradicts other studies that have argued that crosslinks play the major role (41). However, the elevated levels of G and T at the 3′-terminal position with aDNA templates (Figure S2) suggests that the detailed picture may be more complicated than the test system and that other non-coding lesions, as well as abasic sites, may also play a role.
SPEX strongly suggests post mortem C > U-type base modification events are solely responsible for damage-derived miscoding lesions in aDNA
The single-cycle SPEX sequences provided no evidence for so-called Type 1 (T > C/A > G) ‘damage’ events (Figure 3; Table 1). If the previously proposed A > HX (hypoxanthine) lesion (39) plays a role in aDNA, then significant levels of complementary T > C transitions should have been observed on first-generation copied SPEX sequences. These results concur with the findings of recent large-scale 454-based aDNA studies, which also precluded the possibility of ‘jumping-PCR’-type events, and similarly produced no evidence of Type 1 transition artefacts (21,22,30).
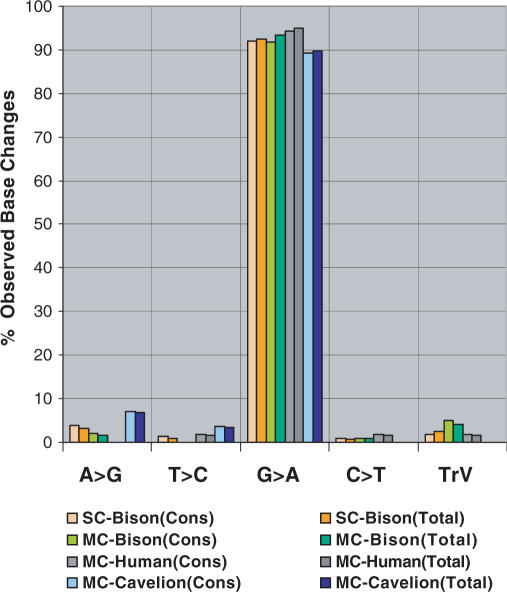
Figure 3.
The percentage of the total observed base changes corresponding to each different transition and all transversions (grouped) following the single- or multi-cycle SPEX amplification of various ancient extracts. SC = Single-cycle SPEX. MC = Multi-cycle SPEX. Cons = CDS. Total = TCDS. TrV = transversions. SC-Bison = 6 ancient bison extracts: (Cons) 7,654 bp, 337 discrete sequences; (Total) 10,644 bp, 548 independent amplicons. MC-Bison = 5 ancient bison extracts: (Cons) 4,187 bp, 171 discrete sequences; (Total) 4,934 bp, 219 independent amplicons. MC-Human = 4 ancient human extracts (at 3 loci): (Cons) 3304 nucleotides, 164 discrete sequences; (Total) 4,647 bp, 271 independent amplicons. MC-Cavelion = 3 ancient Eurasian cave lion extracts: (Cons) 4,086 bp, 111 discrete sequences; (Total) 4,470 bp, 141 independent amplicons.
G > A transitions (from C > U-type DDMLs on the original aDNA template strand) account for >90% of observed base changes in the single-cycle SPEX sequence data (Figures 3 and S1; Table 1). The remaining <10% are distributed between the other 11 possible transitions and transversions with an overall level of ∼2.5 × 10−5 base changes per nucleotide per cycle - an error rate comparable to that observed with Platinum Taq High Fidelity on non-aDNA templates (42,43). A comparison between the percentage of base changes per site over the first 17 bases of primer extension, for both bison control region aDNA and an equivalent synthetic oligonucleotide, also strongly suggests that aside from C > U-type DDMLs, base changes in aDNA are at background levels consistent with polymerase misincorporation errors (Figure S4). The multi-fold coverage of first-generation copies from a known strand of origin provided by SPEX clearly suggests C > U-type base modification events are the only significant cause of authentic endogenous DDMLs in aDNA.
Comparison between single-cycle SPEX with Platinum Taq High Fidelity and multi-cycle SPEX with AmpliTaq Gold
aDNA damage studies using traditional PCR with either Platinum Taq High Fidelity or AmpliTaq Gold polymerase systems have often produced strikingly different findings (cf. 10,11,20–22,26–28,39). Multi-cycle SPEX was used with AmpliTaq Gold to perform repeated cycles of single primer annealing and extension with: five bison extracts (at one mitochondrial locus); four human extracts (at three mitochondrial loci—each with key identifying SNPs to monitor modern contamination) and three Eurasian cave lion extracts (at one mitochondrial locus). Figure 3 and Table 2 show the results for all three groups of samples (11 577 nucleotides from 446 discrete sequences for the CDS and 14 051 nucleotides from 1121 independently cloned sequences for the TCDS). All species and loci examined again showed ∼90% of observed base changes were G > A transitions (due to C > U-type DDMLs in template aDNA), with an overall spectrum of base changes from multi-cycle SPEX using AmpliTaq Gold essentially indistinguishable from single-cycle SPEX using Platinum Taq High Fidelity.
Distribution of C > U-type base modification events
The spectrum of observed G > A transitions on discrete single-cycle SPEX first-generation copied strands (Figure S1) strongly suggests that particular individual aDNA templates had undergone multiple, clustered, C > U-type DDML event ‘hits’. Statistical analyses confirm that the distribution of G > A transitions is non-random, with three of the four most highly damaged extracts (BS143; BS477; BS569) exhibiting clustering (over-dispersion) of hits onto certain strands independent of sequence length (P = 0.02, 0.09 and 0.01, respectively). The dispersion parameter [θ] of the negative binomial distribution fitted by GLM (37) reported for all three samples shows low values of θ and relatively narrow standard errors (0.89, SE 0.54; 1.49, SE 1.06; 0.83, SE 0.37, respectively), indicating a better fit than to a Poisson model (which is a special case of the negative binomial, when θ tends to infinity). Further investigation into possible post mortem mechanisms for the creation of this intra-molecular clustering of C > U-type DDML base modification events is required.
Post mortem G > A-type base modification events inferred from 454-derived data
The conclusions about aDNA damage processes reached from the SPEX data directly contradict those reached by two recent large-scale 454-based aDNA damage analyses (21,22). High-throughput pyrosequencing on the 454 platform can generate sequence data from thousands of individual ssDNA molecules derived from ‘enzymatically polished’, adapter-tagged, DNA (29). In contrast to SPEX, 454-based aDNA damage analyses generated both C > T and G > A Type 2 transitions (21,22). Since the sequence data was generated from ssDNA templates, both studies independently concluded that in addition to C > U-type events, distinct DDMLs must also be causing some G residues to be read as A. To further investigate this apparent contradiction over the existence of endogenous ‘G > A’ DDMLs, we examined whether an inability of the 454 approach to clearly distinguish between regions of authentic aDNA sequence and regions of sequence derived from first-generation copied aDNA might be an issue. The specific combination of the conditions used in the pre-PCR ‘polishing’ steps of 454-based studies so far and the damaged, fragmented, nature of the aDNA template molecules suggested a potential mechanism (Figure S5).
A significant proportion of ssDNA starting templates would originally have been double-stranded aDNA templates with 3′ recessed ends, filled in by T4 DNA polymerase (30) or both T4 DNA polymerase and the Klenow fragment of E. coli DNA polymerase I (21,35). Due to its strong strand displacement activity, the Klenow enzyme would also extend from any single-stranded breaks (SSBs) or nicks with 3′-OH ends within dsDNA (44), displacing ‘downstream’ 3′ regions of endogenous aDNA and replacing these with a newly synthesized first-generation copy of the complementary aDNA template strand (Figure S5B). The subsequent ‘nick repair’ step (21,29,30,35) by the strand-displacing Bst DNA polymerase would similarly replace 3′ regions downstream of suitable SSB sites in cases where Klenow had not been used (30).
As seen with SPEX-derived sequence data, first-generation copied aDNA naturally produces high levels of G > A transitions derived from authentic C > U-type DDML events on the template strand (Figures 3, S1 and S4). If these mechanisms were responsible for creating the G > A transitions observed in 454-based aDNA studies, then there are several explicit, testable, predictions. First, under the model in Figure S5, the 5′ > 3′ direction of DNA synthesis should strongly skew G > A transitions towards the 3′ ends of individual 454-derived ssDNA templates in a highly non-random distribution. Secondly, since authentic aDNA should always be 5′ to newly synthesized first-generation copies of aDNA in enzymatically modified molecules, then all damage-derived C > T transitions should be 5′ to all G > A transitions in any sequence which contained both. On the other hand, genuine DDMLs of G residues should produce a distribution of G > A transitions wholly independent of the distribution of C > T miscoding lesions.
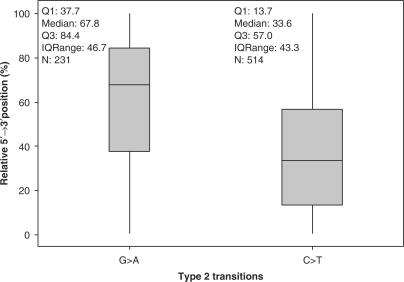
Kolmogorov–Smirnov one sample tests on the relative positions of 514 C > T and 231 G > A transitions from 1449 454-derived Mammuthus primigenius mitochondrial sequences (Table S4) allowed us to reject the null hypothesis that their relative positions are uniformly distributed along the DNA strand (C > T, D = 0.1904, P < 0.001; G > A, D = 0.2014, P < 0.001). The two sided t-test and Wilcoxon rank sum test performed on the relative 5′ > 3′ positions of both C > T and G > A transitions also allowed us to reject the null hypothesis that the mean and median relative positions of the two types of mutation are the same (t = −10.96, nCT = 515, nGA = 231, P < 0.001; W = 32337, nCT = 515, nGA = 231, P < 0.001, Wilcoxon test). G > A transitions are skewed towards the 3′ end with a median location of 67.8% from the 5′ end (Figure 4). As this effect evidently occurred in enough molecules to also similarly skew the distribution of authentic endogenous C > U-type DDMLs (resulting C > T transitions have a median location of 33.6% from the 5′ end; Figure 4), a significant proportion of double-stranded aDNA templates must originally have possessed extendable recessed 3′-ends and/or SSBs (Figure S5). An analogous reciprocal skewing of Type 2 transitions is also observed with 454-derived sequences from a Neanderthal specimen (30).
Figure 4.
Box-plot showing the 5′ to 3′ distribution of Type 2 (C > T/G > A) transitions following the PCR amplification of 454-generated ssDNA molecules (514 C > T and 231 G > A). The plot comprises a box and whiskers. A line is drawn across the box to represent the median; the bottom of the box is the first quartile (Q1) and the top is the third quartile (Q3). The lower whisker extends to the lowest value within the lower limit, whilst the upper whisker extends to the highest value within the upper limit. The limits are defined by: Q1 − 1.5(Q3 − Q1) (lower limit) and Q3 + 1.5(Q3 − Q1) (upper limit). The exact position of each type of damage was normalized to allow comparison between molecules of different lengths where 0% corresponds to the 5′-terminal base and 100% the 3′-terminal base. The horizontal line represents the median of the relative position of the damage and the box represents the middle 50% (the inter-quartile) of the relative position of each type of damage from all ssDNA molecules combined.
To graphically illustrate this effect, Figure S6 shows all 17 sequence reads which had both C > T and G > A transitions, but otherwise had a 100% match to the Mammuthus primigenius mitochondrial consensus sequence (i.e. no other transitions, transversions, indels, etc). In 16 of these 17 reads, all C > T transitions are 5' to all G > A ones. This result is also significantly non-random: G = 45.73, df = 1, P < 0.001. We assume that the single exception to the predicted pattern (1/24 G > A transitions) is due to a polymerase misincorporation error, although this cannot be proven.
This re-analysis of 454-derived aDNA sequence data strongly supports both the general conclusions on the molecular nature of DDMLs from the SPEX data, and the hypothesis that 454-derived G > A transitions are actually generated opposite authentic endogenous C > U-type DDML events on the complementary aDNA strand by polymerase activity. These findings have serious implications for aDNA damage analyses using current 454-based approaches.
DISCUSSION
SPEX versus traditional PCR-based approaches to aDNA damage
The well-characterized samples and loci chosen for this study provided a stringent test for SPEX, as any generation of either endogenous DDML or artefactual sequence changes could easily be identified and quantified. Aside from C > U-type DDML events (Figures 3 and S4), SPEX amplification of aDNA produced both a spectrum and level of sequence differences typical of the background level of polymerase misincorporation errors on non-ancient specimens. SPEX-amplified sequences also provided a simple means to estimate the minimum number of aDNA templates contributing to product sequences, thereby permitting the molecular nature of miscoding and other lesions to be assessed on a quantifiable basis.
With standard PCR methods, the lengths of target amplicons are pre-defined by the primer pair. In order to gain as much data as possible, most phylogenetic and aDNA damage studies so far (e.g. 4,10,20,26–28,39,40) have tended towards the analysis of amplicons that are larger, sometimes significantly larger, than the smallest possible amplifiable fragments from given extracts (but that are nevertheless ‘reliable’ according to currently accepted aDNA criteria; e.g. 11,45). Figure 2 makes it clear that unless the PCR primer pair directly abutted one another, then during the initial cycles of PCR-based aDNA studies like these the numbers of primer extensions of directly PCR-amplifiable length would generally be greatly exceeded by the numbers of short, abortive, primer extensions. As Figure S1 and Table S3 demonstrate, the majority of primer extensions also undergo the addition of non-authentic 3′-terminal bases and any of these products could serve as potential protagonists in ‘jumping-PCR’ events in subsequent PCR amplification cycles. Whether these processes play a role in creating PCR-generated sequence artefacts, like so-called ‘Type 1 damage’ (observed at significant levels in several PCR-based studies of aDNA damage; e.g. 20,26–28,39), is currently unclear and requires further analysis. However, this class of artefact is strikingly absent from SPEX- or 454-derived sequences, where ‘jumping-PCR’ should not be an issue.
It has long been observed that analysing the products of a single PCR amplification from aDNA can lead to wholly incorrect inferences about the underlying endogenous sequence (e.g. 10,11,40). One explanation of this phenomenon might be that the absolute number of initial primer extensions of directly PCR-amplifiable length was small or zero (for the particular target size) in amplifications like these. Primer extension steps that created only one or a small number of molecules that traversed both PCR primer binding sites, perhaps containing authentic endogenous DDMLs or polymerase-generated/‘jumping-PCR’ artefacts, could then undergo a form of positive selection and come to dominate the exponentially amplified products (cf. 13,23). However, aDNA extracts with high estimated copy numbers (according to qPCR) can still generate significant levels of non-endogenous, PCR-generated, ‘Type 1 damage’ (e.g. 27). Therefore, perhaps the relative proportions of intact, directly PCR-amplifiable, templates versus fragmented, damaged, templates may be key. Further investigation is required.
Until now, a 3-fold redundancy PCR amplification and cloning strategy has been employed to attempt to generate credible consensus sequences from key ancient samples (e.g. 25,46), but this approach is both labour and sample intensive and has been shown to be fallible even with high-quality, frozen, aDNA templates (24). Overall, traditional and multiplex PCR can probably be relied upon to produce correct consensus sequences over the great majority of nucleotide positions in non-human samples by the ‘best-of-three’ strategy, provided that enough suitable aDNA templates are available and appropriate care is taken (e.g. 11,24,25,45,46). However, despite many years of effort, the inherent features of the methodology discussed above have meant that no PCR-based study has been able to fully resolve the molecular nature of DDMLs.
Unlike PCR, single-cycle SPEX synthesizes a first-generation copy from only one of the aDNA template strands, thereby precluding any ‘jumping-PCR’-type mechanisms. Multi-cycle SPEX also did not exhibit any obvious indications of these kinds of artefacts (e.g. repeated characteristic DDML motifs in SPEX amplicons of different lengths or enhanced levels of base changes not due to endogenous DDMLs). Multi-cycle SPEX produced a spectrum of transitions and transversions indistinguishable from single-cycle SPEX (Figure 3). The linear mode of multi-cycle SPEX primer extension amplification (as opposed to the exponential mode of PCR amplification) meant that the single SPEX-1 primer should have remained at vast molar excess to aDNA targets and extended primers throughout the reaction. The absence of a reverse PCR primer in the multi-cycle SPEX primer extension stage meant there was no potential for the positive selection of ‘jumping-PCR’-type events (13).
Another potential source of non-authentic sequence diversity is the cloning of heteroduplexes. When 50 or 60 PCR cycles are used (e.g. 10,21), high levels of exponentially amplified product can drastically reduce primer-to-template ratios during the final cycles, favouring self-annealing of complementary PCR-amplified strands over productive primer-template binding (47). With heterologous starting sequences, the subsequent cloning of heteroduplexes has been shown to allow the Escherichia coli mismatch repair system to generate further non-authentic sequence microvariation (48–50). As PCR-amplified aDNA is known to have extensive sequence variation, due to both endogenous DDMLs and PCR-generated artefacts, this issue should not be neglected by PCR-based studies. SPEX minimizes potential hetroduplex formation prior to cloning by amplifying a wide range of insert sizes for only 35 cycles.
Damage-derived miscoding lesions
Quantitative damage analyses on both the CDS and TCDS for both single- and multi-cycle SPEX amplified sequences from three separate species support the same two conclusions. First, that Type 1 (T > C/A > G) ‘damage’ transitions are non-endogenous, PCR-generated, sequence artefacts. Secondly, C > U-type base modification events appear to be the only DDMLs present at significant levels in ancient DNA. Comparing SPEX-amplified sequences from bison aDNA and an equivalent synthetic oligonucleotide template also emphasizes that C > U-type DDMLs occur at a remarkably consistent level (∼11–12% per site), regardless of local sequence context (Figure S4). Therefore, the increased accuracy of this quantitative SPEX sequence data provides no support for the DDML ‘hotspots’ inferred by some traditional PCR-based aDNA studies (e.g. 28,39).
Recent 454-based aDNA studies (21,22,30) argued that a currently unknown DDML must be causing some G residues to be read as A during PCR amplification from individual ssDNA templates. The quantitative demonstration of the predicted highly non-random distribution of G > A transitions towards the 3′ ends of 454-derived sequences, coupled with the skewing of C > T transitions towards the 5′ ends, strongly supports the hypothesis that G > A transitions are generated during the pre-PCR ‘enzymatic polishing’ and/or subsequent ‘nick repair’ steps, from C > U-type DDML events on the complementary aDNA strand (Figure S5). Re-interpreted in this way, 454-generated metagenomic sequence data supports the central finding from the SPEX aDNA studies that post mortem C > U-type base modification events are effectively the sole cause of authentic DDMLs in aDNA.
This identifies significant methodological issues for 454-based aDNA studies, as 454-derived sequence variation does not reflect the authentic underlying pattern of DDMLs in an aDNA extract. The 454 sequence traces contain DDML sequence variation from both of the original aDNA template strands (in the form of variable and unquantifiable proportions of 5′ regions of authentic, endogenous, aDNA and 3′ segments of first-generation copied DNA derived from the complementary strand). Until input ssDNA templates can be unequivocally produced from single strands-of-origin in 454-based studies this will remain a key issue. Theoretically, multiple overlapping traces could allow correct consensus sequences to be inferred, enabling Type 2 miscoding lesion transitions (whether observed as C > T or G > A) to be clearly discounted. However, with most aDNA specimens, current 454-based methodologies appear unlikely to regularly generate a sufficient depth-of-coverage to allow the accurate SNP-typing of key sites in this way.
CONCLUSION
SPEX has shown why almost 20 years of PCR-based approaches have not been able to fully resolve the molecular basis of DDMLs. Traditional PCR and current 454-based aDNA studies cannot unambiguously resolve the template strand-of-origin for any particular endogenous Type 2 DDML. Moreover, the production of significant levels of non-endogenous PCR-generated sequence artefacts, such as so-called ‘Type 1 damage’ in some PCR-based investigations (e.g. 20,26–28,39), clearly demonstrates that any firm inferences and conclusions about authentic endogenous DDMLs from these studies are now questionable. In contrast, PCR-based strategies using the ‘best-of-three’ approach are likely to yield correct consensus sequences most of the time, particularly in studies of well-preserved ancient specimens with reasonably high template copy numbers.
The development of the SPEX approach to aDNA has allowed the processes of post mortem aDNA damage to be disentangled from PCR-generated sequence artefacts, and revealed the molecular nature of DDMLs. Although much work remains to be done before SPEX could be more widely used in high-throughput situations, a far greater and quantifiable, depth-of-coverage could potentially be achieved compared to other current aDNA methodologies. Sequence data of unprecedented accuracy can be produced from single aDNA target strands with only a single aliquot of extract, a simple system and no specialized equipment. By allowing post mortem C > U-type base modification events to be unambiguously identified as the sole significant cause of DDMLs in ancient specimens, SPEX also shows that potential miscoding lesions at key sites could be avoided altogether in future SNP-typing studies by simply targeting the appropriate aDNA strand. This could reduce SNP-typing errors in aDNA studies down towards the theoretical limit of the background rate of polymerase misincorporation errors and, at the same time, introduce quantifiable genotyping from many other kinds of low copy number, damaged, DNA such as forensic, environmental or fixed clinical samples.
Note added after completion
During the review process of this paper, we became aware of a study submitted after ours by the research group of one of our referees. This study presents an equivalent model for the skewing of C > T and G > A transitions in 454-based data and infers the molecular nature of aDNA miscoding lesion damage from statistical evidence (51).
SUPPLEMENTARY DATA
Supplementary data is available at NAR Online.
CONTRIBUTIONS OF AUTHORS
P.B. designed and developed the SPEX approach to aDNA damage and performed the aDNA experiments with support from A.C. The SPEX-derived sequence data was analysed by P.B., P.E., J.J.S. and A.C. P.B. proposed the mechanism for the origin of 454-derived G>A transitions. P.E. provided ancient human extracts, sequence data and appropriate target SNPs. J.J.S., P.E. and M.B. carried out statistical analyses on SPEX and 454-derived data. R.B. provided ancient Eurasian cave lion extracts and sequence data. J.A. generated SPEX data from synthetic oligonucleotide templates. P.B., A.C., P.E. J.J.S. and M.B wrote the manuscript.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank: Richard Harington, Canadian Museum of Nature; Andrei Sher, Paleontological Institute, Moscow; Jim Burns, Royal Alberta Museum; Larry Martin, University of Kansas Natural History Museum; Beth Shapiro (for assistance with the various ancient bison extracts) as well as other members of the Henry Wellcome Ancient Biomolecules Centre, University of Oxford and the Australian Centre for Ancient DNA in Adelaide. Many thanks to Chris Allinson and Susan Wallace for helpful discussions and correspondence. The work was supported by grants to A.C. from the UK NERC, Wellcome and Leverhulme Trusts, and the Australian Research Council. J.J. Sanchez was supported by a grant from ‘Plan Nacional I+D+I’ (MEC,18.08.463B.780). Funding to pay the Open Access publication charges for this article was provided by the Australian Research Council (ARC), via the University of Adelaide.
Conflict of interest statement. None declared.
REFERENCES
- 1.Endicott P, Gilbert MT, Stringer C, Lalueza-Fox C, Willerslev E, Hansen AJ, Cooper A. The genetic origins of the Andaman Islanders. Am. J. Hum. Genet. 2003;72:178–184. doi: 10.1086/345487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haak W, Forster P, Bramanti B, Matsumura S, Brandt G, Tanzer M, Villems R, Renfrew C, Gronenborn D, et al. Ancient DNA from the first European farmers in 7500-year-old Neolithic sites. Science. 2005;310:1016–1018. doi: 10.1126/science.1118725. [DOI] [PubMed] [Google Scholar]
- 3.Ritchie PA, Millar CD, Gibb GC, Baroni C, Lambert DM. Ancient DNA enables timing of the pleistocene origin and holocene expansion of two adelie penguin lineages in antarctica. Mol. Biol. Evol. 2004;21:240–248. doi: 10.1093/molbev/msh012. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro B, Drummond AJ, Rambaut A, Wilson MC, Matheus PE, Sher AV, Pybus OG, Gilbert MT, Barnes I, et al. Rise and fall of the Beringian steppe bison. Science. 2004;306:1561–1565. doi: 10.1126/science.1101074. [DOI] [PubMed] [Google Scholar]
- 5.Pääbo S. Ancient DNA: extraction, characterization, molecular cloning and enzymatic amplification. Proc. Natl Acad. Sci. USA. 1989;86:1939–1943. doi: 10.1073/pnas.86.6.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 7.Friedberg E, Walker G, Siede W. DNA repair and mutagenesis. Washington, DC: ASM Press; 1995. [Google Scholar]
- 8.Wallace SS. Biological consequences of free radical-damaged DNA bases. Free Radic Biol Med. 2002;33:1–14. doi: 10.1016/s0891-5849(02)00827-4. [DOI] [PubMed] [Google Scholar]
- 9.Höss M, Jaruga P, Zastawny TH, Dizdaroglu M, Pääbo S. DNA damage and DNA sequence retrieval from ancient tissues. Nucleic Acids Res. 1996;24:1304–1307. doi: 10.1093/nar/24.7.1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Pääbo S. DNA sequences from multiple amplifications reveal artefacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29:4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pääbo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, et al. Genetic analyses from ancient DNA. Annu. Rev. Genet. 2004;38:645–679. doi: 10.1146/annurev.genet.37.110801.143214. [DOI] [PubMed] [Google Scholar]
- 12.Bandelt HJ. Mosaics of ancient mitochondrial DNA: positive indicators of nonauthenticity. Eur. J. Hum. Genet. 2005;13:1106–1112. doi: 10.1038/sj.ejhg.5201476. [DOI] [PubMed] [Google Scholar]
- 13.Ruano G, Brash DE, Kidd KK. PCR: the first few cycles. Amplifications. 1991;7:1–4. [Google Scholar]
- 14.Pääbo S, Irwin DM, Wilson AC. DNA damage promotes jumping between templates during enzymatic amplification. J Biol Chem. 1990;265:4718–4721. [PubMed] [Google Scholar]
- 15.Clark JM. Novel non-templated nucleotide addition reactions catalyzed by prokaryotic and eukaryotic DNA polymerases. Nucleic Acids Res. 1988;16:9677–9686. doi: 10.1093/nar/16.20.9677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brownstein MJ, Carpten JD, Smith JR. Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modifications that facilitate genotyping. Biotechniques. 1996;20:1004–1010. doi: 10.2144/96206st01. [DOI] [PubMed] [Google Scholar]
- 17.Strauss BS. The ‘A rule’ of mutagen specificity: a consequence of DNA polymerase bypass of non-instructional lesions? Bioessays. 1991;13:79–84. doi: 10.1002/bies.950130206. [DOI] [PubMed] [Google Scholar]
- 18.Shibutani S, Takeshita M, Grollman AP. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the ‘A rule’. J. Biol. Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 19.Berthet N, Roupioz Y, Constant JF, Kotera M, Lhomme J. Translesional synthesis on DNA templates containing the 2′-deoxyribonolactone lesion. Nucleic Acids Res. 2001;29:2725–2732. doi: 10.1093/nar/29.13.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen A, Willerslev E, Wiuf C, Mourier T, Arctander P. Statistical evidence for miscoding lesions in ancient DNA templates. Mol. Biol. Evol. 2001;18:262–265. doi: 10.1093/oxfordjournals.molbev.a003800. [DOI] [PubMed] [Google Scholar]
- 21.Stiller M, Green RE, Ronan M, Simons JF, Du L, He W, Egholm M, Rothberg JM, Keates SG, et al. Patterns of nucleotide misincorporations during enzymatic amplification and direct large-scale sequencing of ancient DNA. Proc. Natl Acad. Sci. USA. 2006;103:13578–13584. doi: 10.1073/pnas.0605327103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert MT, Binladen J, Miller W, Wiuf C, Willerslev E, Poinar H, Carlson JE, Leebens-Mack JH, Schuster SC. Recharacterization of ancient DNA miscoding lesions: insights in the era of sequencing-by-synthesis. Nucleic Acids Res. 2007;35:1–10. doi: 10.1093/nar/gkl483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner A, Blackstone N, Cartwright P, Dick M, Misof B, Snow P, Wagner GP, Bartels J, Murtha M, et al. Surveys of gene families using polymerase chain reaction: PCR selection and PCR drift. Syst. Biol. 1994;43:250–261. [Google Scholar]
- 24.Krause J, Dear PH, Pollack JL, Slatkin M, Spriggs H, Barnes I, Lister AM, Ebersberger I, Pääbo S, et al. Multiplex amplification of the mammoth mitochondrial genome and the evolution of Elephantidae. Nature. 2006;439:724–727. doi: 10.1038/nature04432. [DOI] [PubMed] [Google Scholar]
- 25.Krings M, Stone A, Schmitz RW, Krainitzki H, Stoneking M, Pääbo S. Neandertal DNA sequences and the origin of modern humans. Cell. 1997;90:19–30. doi: 10.1016/s0092-8674(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert MTP, Willerslev E, Hansen AJ, Barnes I, Rudbeck L, Lynnerup N, Cooper A. Distribution patterns of post-mortem damage in human mitochondrial DNA. Am. J. Hum. Genet. 2003a;72:32–47. doi: 10.1086/345378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Binladen J, Wiuf C, Gilbert MTP, Bunce M, Larson G, Barnett R, Hansen AJ, Willerslev E. Comparing miscoding lesion damage in mitochondrial and nuclear ancient DNA. Genetics. 2006;172:733–741. doi: 10.1534/genetics.105.049718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert MTP, Shapiro BA, Drummond A, Cooper A. Post mortem DNA damage hotspots in Bison (Bison bison and B. bonasus) provide supporting evidence for mutational hotspots in human mitochondria. J. Archaeol. Sci. 2005;32:1053–1060. [Google Scholar]
- 29.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green RE, Krause J, Ptak SE, Briggs AW, Ronan MT, Simons JF, Du L, Egholm M, Rothberg JM, et al. Analysis of one million base pairs of Neanderthal DNA. Nature. 2006;444:330–336. doi: 10.1038/nature05336. [DOI] [PubMed] [Google Scholar]
- 31.Endicott P, Metspalu M, Stringer C, Macaulay V, Cooper A, Sanchez JJ. Multiplexed SNP Typing of Ancient DNA Clarifies the Origin of Andaman mtDNA Haplogroups amongst South Asian Tribal Populations. PLoS ONE. 2006;1:e81. doi: 10.1371/journal.pone.0000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson S, de Bruijn MH, Coulson AR, Eperon IC, Sanger F, Young IG. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J. Mol. Biol. 1982;156:683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- 33.Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 34.Chen HH, Kontaraki J, Bonifer C, Riggs AD. Terminal Transferase-Dependent PCR (TDPCR) for in vivo UV photofootprinting of vertebrate cells. Sci. STKE. 2001 doi: 10.1126/stke.2001.77.pl1. 77, PL1. [DOI] [PubMed] [Google Scholar]
- 35.Poinar HN, Schwarz C, Qi J, Shapiro B, Macphee RD, Buigues B, Tikhonov A, Huson DH, Tomsho LP, et al. Metagenomics to paleogenomics: large-scale sequencing of mammoth DNA. Science. 2006;311:392–394. doi: 10.1126/science.1123360. [DOI] [PubMed] [Google Scholar]
- 36.Tukey J. Exploratory Data Analysis. Addison-Wesley Publishing Co, London; 1977. [Google Scholar]
- 37.Venables WN, Ripley BD. Modern Applied Statistics with S. 4th. New York: Springer-Verlag; 2002. [Google Scholar]
- 38.Walsh PS, Erlich HA, Higuchi R. Preferential PCR amplification of alleles: mechanisms and solutions. PCR Methods Appl. 1992;1:241–250. doi: 10.1101/gr.1.4.241. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert MTP, Hansen AJ, Willerslev E, Rudbeck L, Barnes I, Lynnerup N, Cooper A. Characterization of genetic miscoding lesions caused by post-mortem damage. Am. J. Hum. Genet. 2003b;72:48–61. doi: 10.1086/345379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Handt O, Krings M, Ward RH, Pääbo S. The retrieval of ancient human DNA sequences. Am. J. Hum. Genet. 1996;59:368–376. [PMC free article] [PubMed] [Google Scholar]
- 41.Hansen AJ, Mitchell DL, Wiuf C, Paniker L, Brand TB, Binladen J, Gilichinsky DA, Ronn R, Willerslev E. Crosslinks rather than strand breaks determine access to ancient DNA sequences from frozen sediments. Genetics. 2006;173:1175–1179. doi: 10.1534/genetics.106.057349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Belanger AE, Lai A, Brackman MA, Le Blanc DJ. PCR-based ordered genomic libraries: a new approach to drug target identification for Streptococcus pneumoniae. Antimicrob. Agents Chemother. 2002;46:2507–2512. doi: 10.1128/AAC.46.8.2507-2512.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rual JF, Hirozane-Kishikawa T, Hao T, Bertin N, Li S, Dricot A, Li N, Rosenberg J, Lamesch P, et al. Human ORFeome version 1.1: a platform for reverse proteomics. Genome Res. 2004;14:2128–2135. doi: 10.1101/gr.2973604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Masamune Y, Richardson CC. Strand displacement during deoxyribonucleic acid synthesis at single strand breaks. J. Biol. Chem. 1971;246:2692–26701. [PubMed] [Google Scholar]
- 45.Cooper A, Poinar HN. Ancient DNA: do it right or not at all. Science. 2000;289:1139. doi: 10.1126/science.289.5482.1139b. [DOI] [PubMed] [Google Scholar]
- 46.Krings M, Geisert H, Schmitz RW, Krainitzki H, Pääbo S. DNA sequence of the mitochondrial hypervariable region II from the Neandertal type specimen. Proc. Natl Acad. Sci. USA. 1999;96:5581–5585. doi: 10.1073/pnas.96.10.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruano G, Kidd KK. Modeling of heteroduplex formation during PCR from mixtures of DNA templates. PCR Methods Appl. 1992;2:112–116. doi: 10.1101/gr.2.2.112. [DOI] [PubMed] [Google Scholar]
- 48.Suzuki MT, Giovannoni SJ. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 1996;62:625–630. doi: 10.1128/aem.62.2.625-630.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu X, Wu L, Huang H, McDonel PE, Palumbo AV, Tiedje JM, Zhou J. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 2001;67:880–887. doi: 10.1128/AEM.67.2.880-887.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Speksnijder AG, Kowalchuk GA, De Jong S, Kline E, Stephen JR, Laanbroek HJ. Microvariation artefacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 2001;67:469–472. doi: 10.1128/AEM.67.1.469-472.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Briggs AW, Stenzel U, Johnson PLF, Green RE, Kelso J, Prüfer K, Meyer M, Krause J, Ronan MT, et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl Acad. Sci. USA. 2007;104 doi: 10.1073/pnas.0704665104. 10.1073/pnas.0704665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.