Abstract
Although adjustable transgene expression systems are considered essential for future therapeutic and biopharmaceutical manufacturing applications, the currently available transcription control modalities all require side-effect-prone inducers such as immunosupressants, hormones and antibiotics for fine-tuning. We have designed a novel mammalian transcription-control system, which is reversibly fine-tuned by non-toxic vitamin H (also referred to as biotin). Ligation of vitamin H, by engineered Escherichia coli biotin ligase (BirA), to a synthetic biotinylation signal fused to the tetracycline-dependent transactivator (tTA), enables heterodimerization of tTA to a streptavidin-linked transrepressor domain (KRAB), thereby abolishing tTA-mediated transactivation of specific target promoters. As heterodimerization of tTA to KRAB is ultimately conditional upon the presence of vitamin H, the system is vitamin H responsive. Transgenic Chinese hamster ovary cells, engineered for vitamin H-responsive gene expression, showed high-level, adjustable and reversible production of a human model glycoprotein in bench-scale culture systems, bioreactor-based biopharmaceutical manufacturing scenarios, and after implantation into mice. The vitamin H-responsive expression systems showed unique band pass filter-like regulation features characterized by high-level expression at low (0–2 nM biotin), maximum repression at intermediate (100–1000 nM biotin), and high-level expression at increased (>100 000 nM biotin) biotin concentrations. Sequential ON-to-OFF-to-ON, ON-to-OFF and OFF-to-ON expression profiles with graded expression transitions can all be achieved by simply increasing the level of a single inducer molecule without exchanging the culture medium. These novel expression characteristics mediated by an FDA-licensed inducer may foster advances in therapeutic cell engineering and manufacturing of difficult-to-produce protein therapeutics.
INTRODUCTION
Precise transgene expression dosing in mammalian cells has become a cornerstone for synthetic biology (1–4), (pre)clinical gene therapy studies (5–8), drug discovery (9,10), biopharmaceutical manufacturing (11–13) as well as for numerous applications in functional genomic research (14–16).
Research and development of inducible mammalian transgene expression systems has focused on clinically licensed small-molecule inducers to minimize pleiotropic side effects in cell culture, animal models or ultimately in clinical trials (17). Several antibiotics (18–22), steroid hormone analogs (23–25) and immunosuppressive substances (26,27) have been successfully used to modulate transgene expression in vitro and in vivo. Immunosuppresive drugs such as rapamycin can trigger transgene expression by conditional heterodimerization of FKBP, fused to a DNA-binding domain, and FRB, fused to a transactivation domain, which form a chimeric transactivor inducing transcription from specific promoters (26). Steroid hormone-based systems capitalize on the cytosolic sequestration of nuclear hormone receptors engineered to contain heterologous DNA binding and transactivation domains by endogenous heat-shock protein 90 [Hsp90; (28)]. Addition of steroid hormone analogs (for example, tamoxifen) displaces Hsp90 which results in nuclear translocation of the transactivator and induction of cognate promoters (29,30). Antibiotic-dependent expression systems [tetracyclines (19), macrolides (21), streptogramins (18)] take advantage of engineered prokaryotic repressors which activate or repress synthetic target promoters in an antibiotic-responsive manner (17).
The prototype tool for heterologous mammalian gene regulation is the tetracycline-responsive expression system (known as the TET system) which consists of an Escherichia coli-derived tetracycline-dependent transactivator (tTA, a fusion of the TetR repressor and the Herpes simplex VP16 transactivation domain) that binds and activates tetracycline-responsive promoters (PhCMV*−1, heptameric TetR-specific operator sites [tetO7] linked to a minimal version of the human cytomegalovirus immediate early promoter) (19). In the absence of tetracycline antibiotics, tTA binds tetO7 and triggers PhCMV*−1-driven transgene expression. In the presence of tetracycline, tTA-tetO7 binding is abolished and transcription remains shut down (19). Despite solid regulation characteristics, ongoing concerns about long-term side effects of antibiotics (31,32), as well as the rapid emergence of antibiotic resistance in bacteria exposed to subclinical environmental antibiotic concentrations (33), have prevented the use of the TET systems in gene therapy and biopharmaceutical manufacturing scenarios. Despite advantages of using clinically licensed drugs for therapeutic transgene expression fine-tuning, there are ongoing concerns about side effects associated with their prolonged administration in a clinical setting (28,32,34). For example, sustained administration of immunosuppressive drugs increases the susceptibility to bacterial infections (35), persistent intake of steroid hormone analogs may trigger different side effects including pancreatitis (36,37) and continued ingestion of antibiotics compromises the intestinal flora (38) and increases the risk for the emergence of multidrug-resistant pathogenic bacteria (33). Therefore, the design of novel transgene control modalities responsive to side-effect-free trigger molecules remains a current priority (17,39).
In E. coli synthesis of activated vitamin H (biotin) as well as biotinylation of target proteins is catalyzed by the dual-function biotin ligase BirA: (i) BirA activates biotin (biotinyl-5′-adenylate) and transfers the biotin moiety to an acceptor lysine within a specific signal sequence (40). (ii) Furthermore, intracellular biotin concentrations are feedback-controlled by biotinyl-5′-adenylate which triggers binding of BirA to a specific 40 bp operator site, thereby inhibiting transcription of the divergently oriented bioA-BFCD operon and preventing de novo synthesis of biotin (40). Several synthetic biotinylation signals have been recently described [Avitag signal, GLNDIFEAQKIEWHE, target lysine in bold print, (41)] and used in combination with E. coli BirA for in vivo and in vitro purification of biotinylated target proteins (42), chromatin immunoprecipitation, as well as for detection of protein–DNA complexes (43,44).
Capitalizing on the high affinity of biotin to (strept)avidin [10−15 M; (45)], we have designed a chimeric transrepressor, which is assembled from tTA-Avitag and streptavidin-KRAB (krueppel-associated box protein of the human kox-1 gene) components by biotin-triggered heterodimerization, and capable of tTA-mediated induction of PhCMV*−1 in a vitamin H-adjustable manner. Precise transgene expression fine-tuning using a non-toxic vitamin may represent an important advancement for the gene therapy and biopharmaceutical manufacturing communities.
MATERIAL AND METHODS
Vector construction
pWW938 (PSV40-tTA-AT-pA) encoding the tetracycline-dependent transactivator [tTA; (19)]—Avitag (AT) fusion (tTA-AT) under the control of the simian virus 40 promoter (PSV40) was constructed by PCR-mediated amplification of the Herpes simplex VP16 transactivation domain from pWW35 (21) using oligonucleotides OWW18 (5′-GTACGAATTCCCACCatgccccgccccaagctcaa-3′, annealing sequence in lower case) and OWW428 (5′-GGATCAAGCTTGCGGCCGCTTATTCGTGCCATTCGATTTTCTGAGCCGA AGATGTCGTTCAGACCcccaccgtactcgtcaattcc-3′, Avitag sequence underlined, annealing sequence in lower case, HindIII restriction site in italics) and subsequent ligation of the VP16-AT module (BssHII/HindIII) into pSAM200 (18). pWW944 (PSV40-SA-KRAB-pA) encoding streptavidin (SA) fused to the krueppel-associated box protein of the human kox-1 gene (KRAB) was constructed by PCR-mediated amplification of streptavidin from pWW801 [PSV40-TetR-SA-pA, PSV40, simian virus 40 promoter; TetR, tetracycline-dependent repressor; (39)] using oligonucleotides OWW434 (5′-GGATCGAATTCCACCatggctagcatgactggtggac-3′, annealing sequence in lower case, EcoRI restriction site in italics) and OWW433 (5′-GGATCATGCGCGCGGCTGTACGCGGActgctgaacggcgtcgagc-3′, annealing in lower case, BssHII restriction site in italics) and subsequent ligation of the SA module (EcoRI/BssHII) into pWW43 (21). pWW732 (PhCMV*−1-IFN-β-pA) harboring beta interferon (IFN-β) under the control of the tetracycline-responsive promoter (PhCMV*−1) was constructed by excising IFN-β (EcoRI/HindIII) from pWW430 (46) and ligating it into pMF111 [PhCMV*−1-SEAP-pA, (18)], thereby replacing SEAP (human placental secreted alkaline phosphatase). pWW804 (PhCMV-BirA-pA-PSV40-neoR-pA, PhCMV, human cytomegalovirus immediate early promoter; PSV40, simian virus 40 promoter; neoR, neomycin resistance gene) encoding the E. coli biotin ligase BirA has been described previously (39), in brief, BirA was excised (EcoRI/SpeI) from pGEM-SD2 [Strouboulis,J., unpublished data, (42)] and ligated (EcoRI/XbaI) into pMF150 [PhCMV-PIP-pA-PSV40-neoR-pA (18)]. BirA harbors an N-terminal hemagluttinin (HA) tag. The triple-transcript vector pTT-Bio [pWW1091, PSV40-SA-KRAB-PGTX-tTA-AT-PGTX-BirA-pA, PGTX, PGTX, synthetic promoter (47)] was constructed in a three-step procedure: (i) SA-KRAB excised from pWW944 (EcoRI/HindIII) was ligated into the corresponding sites of the first multiple cloning site (MCS) of pCF263 [PSV40-MCSI-PGTX-MCSII-PGTX-MCSIII-pA, (47,48)] resulting in plasmid pWW1089. (ii) BirA excised from pWW804 (SpeI/PmeI) was ligated (SpeI/SwaI) into the third MCS of pWW1089 resulting in plasmid pWW1090. (iii) Finally, tTA-AT excised from pWW938 (NotI) was ligated in sense orientation (NotI) into the second MCS of pWW1090 thereby resulting in pTT-Bio.
Cell culture
Wild-type Chinese hamster ovary cells (CHO-K1; ATCC CCL-61) were cultivated either in biotin-containing or biotin-free ChoMaster® HTS medium (Cell Culture Technologies, Gravesano, Switzerland) supplemented with 5% fetal calf serum (FCS, PAN Biotech GmbH, Aidenbach, Germany, Cat. No. 3302, Lot. No. P231902) or 10% biotin-free serum replacement (KOSR, Invitrogen, Carlsbad, CA, USA, cat. no. 10828-028), respectively, and 1% of a penicillin/streptomycin solution (Sigma, St. Louis, MO, USA, Cat. No. 4458). Human embryonic kidney cells [HEK293-T (49)] were cultivated in Dulbecco's modified Eagle's medium (DMEM) medium supplemented with 10% FCS or 10% serum replacement and 1% penicillin/streptomycin solution.
Transfection
All transfection protocols were optimized for a well of a 24-well plate. CHO-K1: 30 000 CHO-K1 were seeded 12 h before transfection (0.5 ml biotin-free HTS medium supplemented with 5% FCS). An aliquot of 12 µl 0.5 M CaCl2 solution containing 1.2 µg total plasmid DNA (for co-transfections, equal amounts of individual plasmids were used) were mixed with 12 µl phosphate solution (50 mM HEPES, pH 7.05, 280 mM NaCl, 1.5 mM Na2HPO4), vortexed for 5 s, incubated for 25 s at room temperature to allow formation of the transfection precipitate and transferred to 0.4 ml ChoMaster® HTS medium containing 2% FCS. The culture medium was replaced by the transfection mixture and the plates were centrifuged for 5 min at 1200 × g prior to a 90 min incubation period and a 30 s glycerol shock (0.5 ml 15% glycerol in ChoMaster® HTS medium containing 2% FCS). After washing once in 0.5 ml biotin-free ChoMaster® HTS medium, the cells were cultivated in biotin-free or biotin-containing ChoMaster® HTS medium supplemented with 10% KOSR serum replacement or 5% FCS as indicated. HEK293-T: 40 000 HEK293-T were seeded 12 h before transfection (0.5 ml DMEM, 10% FCS). Twenty microliter of 0.25 M CaCl2 containing 0.6 µg total DNA (for co-transfections, equal amounts of individual plasmids were used) were mixed with 20 µl 2× HBS solution (100 mM HEPES, 280 mM NaCl, 1.5 mM Na2HPO4, pH 7.1), incubated for 20 min at room temperature to allow formation of the transfection precipitate, which was transferred dropwise to the cell culture and centrifuged onto the cells (5 min at 1200 × g). Transfected cells were incubated for 90 min at 37°C before they were washed once in FCS-free DMEM and then cultivated in DMEM containing 10% biotin-free KOSR.
Construction of the stable cell line BioCHO-SEAP
CHO-K1 were first co-transfected with plasmids pWW938 (PSV40-tTA-AT-pA), pWW944 (PSV40-SA-KRAB-pA) and pWW804 (PhCMV-BirA-pA, also carrying a constitutive expression cassette conferring resistance to G418) at a ratio of 15:15:1, selected for 12 days in ChoMaster® HTS medium containing 5% FCS and 400 µg/ml G418 and subjected to single-cell cloning. Individual clones, which functionally express pWW938- and pWW944-encoded genes, were then co-transfected with pMF111 (PhCMV*−1-SEAP-pA), pWW804 (PhCMV-BirA-pA) and pPUR (conferring constitutive resistance to puromycin; Clontech, Palo Alto, CA, USA) at a ratio of 15:15:1 followed by selection for 10 days in ChoMaster® HTS medium containing 5% FCS, 400 µg/ml G418 and 15 µg/ml puromycin and single-cell cloning. Individual cell clones were assessed for biotin-triggered SEAP production and BioCHO-SEAP1 was chosen for further studies.
Analytic assays
BirA (HA-tagged) and tTA-AT (TetR-VP16-Avitag) were quantified by resolving cleared cell lysates on a 10% SDS–polyacrylamide gel followed by western blot analysis using anti-HA-tag (Santa Cruz Biotechnology, Santa Cruz, CA, USA, Cat. No. sc-805) or anti-VP16 (Santa Cruz, Cat. No. sc-7545) as primary antibodies for detection and horseradish peroxidase-couple anti-IgG antibodies for chemiluminescence-based visualization (ECL plus, GE Healthcare, Piscataway, NJ, USA, Cat. No RPN2132) with a Chemilux CCD camera (Intas, Göttingen, Germany). Chemiluminescence signals were indicated as integrated optical density (IOD). Streptavidin-KRAB was quantified by incubating cleared cell lysate (40 µl) with 1 µg/ml FITC-biotin (60 µl) (Fluka, Buchs, Switzerland, Cat. No. 53608) for 30 min before measuring fluorescence intensity. Fluorescence of FITC-Biotin bound to streptavidin is quenched and can be used to quantify streptavidin (50). Fluorescence quenching is indicated in relative fluorescence units (RFU). SEAP production was quantified as detailed by Schlatter and coworkers (51) In brief, the cell culture supernatant, collected 48 h after seeding/transfection, was heated to 65°C for 30 min to inactivate endogenous phosphatases and then centrifuged for 2 min at 14 000 × g to remove cell debris. An aliquot of 80 µl supernatant were mixed with 100 µl SEAP buffer (20 mM homoarginine, 1 mM MgCl2, 21% (v/v) diethanolamine, pH 9.8) and the reaction was started by addition of 20 µl pNPP (120 mM p-nitrophenylphosphate in SEAP buffer) and the light absorbance timecourse scored at 405 nm. SEAP levels were calculated according to Lambert-Beer's law using the slope of the timecourse and the specific absorption coefficient of the reaction product p-nitrophenolate (εpNP = 18 600 cm−1 M−1). IFN-β production was profiled by ELISA (PBL Biomedical Laboratories, Piscataway, NJ, USA, Cat. No. 41400-1A) and the cell number was determined either by a Casy™ Counter TTC (Schärfe System GmbH, Reutlingen, Germany) or by using a colorimetric WST-1-based assay (Roche Applied Science, Rotkreuz, Switzerland, Cat. No. 11644807001). All experimental data represent average values derived from three independent experiments with SDs indicated by the error bars.
Bioreactor operation
Cells were cultivated in a BioWave® 50SPS bioreactor (Wave Biotech AG®, Tagelswangen, Switzerland) equipped with Wave Bag® 2LOpt for optical pH and DO control of the 1 l culture. The bioreactor was operated at a rocking rate of 16 min−1, a rocking angle of 5° and an aeration rate of 50 ml/min with inlet gas humidification to prevent evaporation of the medium (HumiCare® 200, Gruendler Medical, Freudenstadt, Germany). Two milliliter samples were withdrawn at the indicated points in time. Samples for assessment of SEAP production were centrifuged (2 min, 14 000 × g) and the supernatant was stored at −20°C until analysis. Cell density was quantified using the colorimetric WST-1 kit (Roche Applied Science, Rotkreuz, Switzerland, Cat. No. 11644807001).
Animal studies
CHO-K1 transgenic for pWW938, pWW944, pWW804 and pMF111 were encapsulated in 400 µm alginate-PLL (poly-l-lysine)-alginate beads using the Inotech Encapsulator Research IE-50R (Inotech Biotechnologies Ltd., Basel, Switzerland) according to the manufacturer's protocol and at the following specific settings: 0.2 mm nozzle, 20 ml syringe at a flow rate of 315 units, nozzle vibration frequency 1108 s−1, voltage for bead dispersion 800 V. Female OF1 (oncins france souche 1) mice were obtained from Iffa-Credo and kept on a biotin-free diet (Société SAFE, Augy, France) for two weeks, whereas the control mice were fed the same diet supplemented with 4 mg/kg biotin. Biotin deficiency was monitored using the forced swim test (52). 700 microliter of ChoMaster® HTS medium containing 50% capsules (2 × 106 cells, 200 cells/capsule) were injected intraperitoneally into the mice. When indicated, biotin was dissolved in 0.9% NaCl and administered by intraperitoneal injection 1 h after capsule implantation. Blood samples were collected retroorbitally and serum was produced using microtainer SST tubes (Beckton Dickinson, Franklin Lakes, NJ, USA, Cat. No. 365968). Serum of control mice (implanted with capsules and kept with or without biotin supplementation) did not show any detectable SEAP levels. All experiments involving animals were conducted according to European Community legislation (86/609/EEC), and have been approved by the French Republic (No. 69266310) and performed by M.D-E. at the Institut Universitaire de Technologie, IUTA, F-69622 Villeurbanne Cedex, France.
Inducers
D(+)-biotin (Acros Organics, Geel, Belgium, Cat. No. 23009) was prepared as a 10 µM stock solution in H2O and was typically used at a final concentration of 100 nM unless stated otherwise. At concentrations above 10 µM, biotin was dissolved as stock solution in DMSO. Avidin (Pierce, Rockford, IL, USA, Cat. No. 21121) was prepared as a 100× stock solution in phosphate-buffered saline (PBS; 20 mM NaH2PO4, 150 mM NaCl, pH 7.2). The concentration of avidin was expressed as biotin-binding capacity. Tetracycline (Sigma, Cat. No. T7660) was prepared as a 1 mg/ml stock solution in H2O and used at a final concentration of 2 µg/ml. For determination of biotin stability, ChoMaster® containing 100 µM biotin and 5% FCS was incubated for one week at 37°C and biotin samples were quantified daily using a HABA (2-(4′hydroxyazobenzene) benzoic acid)-based biotin quantification kit (EZ biotin quantification kit, Pierce, Rockford, IL, USA Cat. No. 28005). No significant decrease in biotin concentration could be observed. In pigs, biotin shows a half-life time of up to 22 h (53).
RESULTS
Design of a vitamin H-responsive mammalian expression system
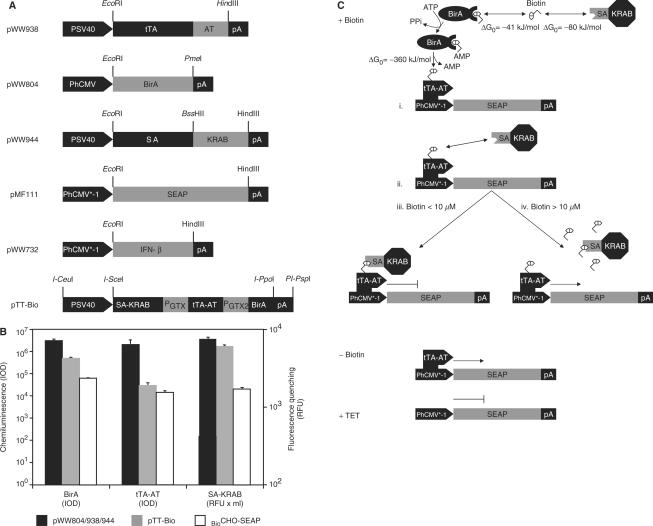
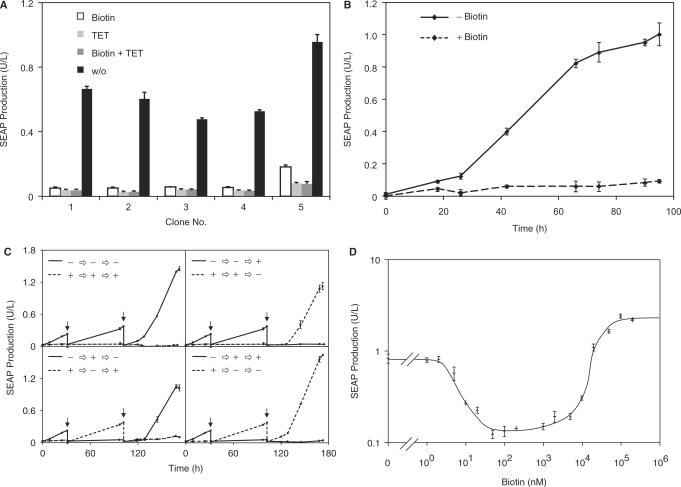
In order to enable precise expression dosing by vitamin H, we have converted proven tetracycline-responsive expression system into a vitamin H-adjustable transcription control modality (Figure 1A). In the presence of biotin (+Biotin) constitutive expression of the Escherichia coli biotin ligase BirA [pWW804; PhCMV-BirA-pA; (42,54)] biotinylates tetracycline- [tTA, TetR-VP16; (19)] dependent transactivators, which have been fused to the synthetic biotinylation signal (Avitag, AT; (41); pWW938, PSV40-tTA-AT-pA, Figure 1C-i) thereby promoting biotin (B)-dependent heterodimerization with the krueppel-associated box protein of the human kox-1 gene (KRAB; SA-KRAB; pWW944, PSV40-SA-KRAB-pA) which has been fused to streptavidin (SA) (Figures 1A and C-ii). Since KRAB-mediated silencing overrides the VP16-based transactivation capacity of tTA-AT, the chimeric transrepressor (tTA-TA-B-SA-KRAB) binds and represses the established tetraycline- (PhCMV*−1; pMF111, PhCMV*−1-SEAP-pA) responsive promoter (Figure 1C-iii). At high biotin concentrations (Figure 1C-iv), all avitag and streptavidin sites are expected to be saturated by free biotin thereby competitively inhibiting SA-AT heterodimerization and de-repressing target gene expression. Biotin deprivation (Figure 1C, −Biotin) prevents or abolishes heterodimerization of chimeric transrepressors thereby enabling classic tTA-mediated transgene expression which can be repressed by addition of regulating tetracycline antibiotics (Figure 1C, +TET) independent of biotin availability. Expression of all biotin-responsive regulatory genes has been validated by western blot analysis or fluorescence quenching (Figure 1B).
Figure 1.
Vitamin H-responsive gene expression in mammalian cells. (A) Expression vectors for vitamin H-responsive gene expression. AT, synthetic BirA-specific biotinylation signal (Avitag, AT); BirA, Escherichia coli biotin ligase; IFN-β, human beta interferon; PGTX, synthetic promoter; KRAB, krueppel-associated box protein of the human kox-1 gene; pA, simian virus 40-derived polyadenylation signal; PhCMV*−1, tTA-specific tetracycline-responsive promoter; PhCMV, human cytomegalovirus immediate early promoter; PSV40, simian virus 40 promoter; SA, streptavidin; SEAP, human placental secreted alkaline phosphatase; tTA, tetracycline-dependent transactivator. (B) Quantification of BirA, tTA-AT and SA-KRAB expression levels. Cells were transfected with either all three regulatory vectors (pWW804, pWW938 and pWW944) or pTT-Bio alone followed by quantification of the expression levels by chemiluminiscence-based western blot analysis (BirA, tTA-AT) or by incubating SA-KRAB-containing cell lysates with FITC-biotin, the fluorescence of which is quenched upon binding to streptavidin. The expression levels of the three proteins were also quantified in the stable cell line BioCHO-SEAP. IOD, integrated optical density of chemiluminescence signal; RFU, relative fluorescence units. (C) Mode of function. In the presence of biotin (+Biotin), the avitagged tTA (tTA-AT) is biotinylated by BirA (i) thereby triggering binding of streptavidin-KRAB [SA-KRAB, (ii)] and silencing of tTA-AT-mediated PhCMV*−1 activation (iii). At high biotin concentrations all tTA-AT and SA-KRAB-binding sites are saturated thereby preventing heterodimerization and de-repression of the target gene (iv). In the absence of biotin (−Biotin) heterodimerization of tTA-AT and SA-KRAB is prevented and tTA-AT binding to PhCMV*−1 induces SEAP expression. In the presence of tetracycline (+TET), tTA-AT-binding to PhCMV*−1 is prevented regardless of whether or not it has heterodimerized with SA-KRAB, and SEAP expression remains silent. Free standard binding enthalpies (ΔG0) for the covalent biotin-avitag bond as well as for the non-covalent biotin-streptavidin interaction are indicated.
Vitamin H-controlled transgene expression in mammalian cell lines
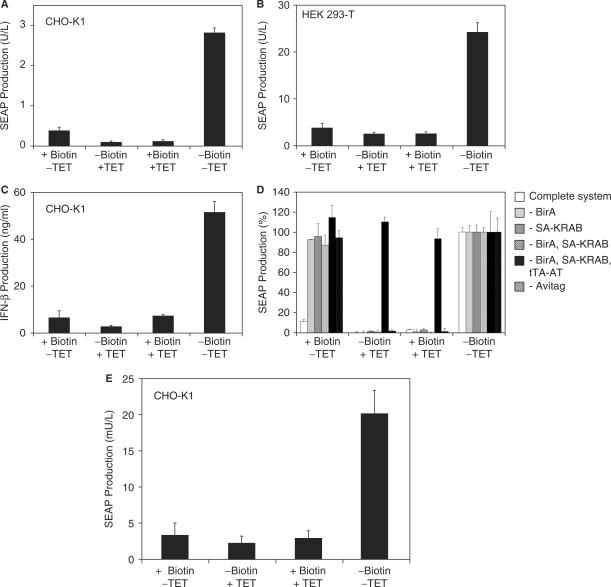
Vitamin H-controlled transgene expression was validated by co-transfection of CHO-K1 with pWW804 (PhCMV-BirA-pA), pWW944 (PSV40-SA-KRAB-pA), pWW938 (PSV40-tTA-AT-pA) and pMF111 (PhCMV*−1-SEAP-pA) followed by SEAP quantification 48 h after cultivation in the presence or absence of biotin and the regulating antibiotic tetracycline (Figure 2A). Cells exhibited high-level SEAP expression in the absence of regulating agents and repressed transgene expression in the presence of biotin or tetracycline. The biotin-regulated gene expression system also mediated conditional SEAP expression in human embryonic kidney cells (HEK293-T) (Figure 2B) and both vitamin H and tetracycline could be used to control the production of the multiple sclerosis therapeutic beta interferon (IFN-β; pWW732, PhCMV*−1-IFN-β-pA) (Figure 2C).
Figure 2.
Functional validation of vitamin H-responsive expression technology. (A) 40 000 CHO-K1 cells were co-transfected with plasmids pWW944, pWW938, pMF111 and pWW804 (see Figure 1A) and cultivated in biotin-free medium supplemented with (+) or without (−) biotin (100 nM) or tetracycline (TET; 2 µg/ml) for 48 h prior to quantification of SEAP production. (B) 30 000 HEK293-T cells were co-transfected with plasmids pWW944, pWW938, pMF111 and pWW804 (see Figure 1A) and cultivated in biotin-free medium in the presence (+) or absence (−) of exogenous biotin (100 nM) or TET (2 µg/ml) for 48 h prior to quantification of SEAP production. (C) 30 000 CHO-K1 cells were co-transfected with plasmids pWW944, pWW938, pWW732 and pWW804 (see Figure 1A) and cultivated in biotin-free medium in the presence (+) or absence (−) of exogenous biotin (100 nM) or TET (2 µg/ml) for 48 h prior to quantification of beta interferon (IFN-β) production. (D) Specificity of biotin-regulated gene expression. All components of the biotin-responsive expression system [tTA-AT (pWW938), SA-KRAB (pWW944), BirA (pWW804)] were co-transfected with the reporter plasmid pMF111 into 30 000 CHO-K1 which were cultivated for 48 h in the presence (+) or absence (−) of biotin (100 nM) or tetracycline (2 µg/ml) before SEAP production was quantified. In parallel control experiments single components/plasmids were omitted. In order to better visualize the effect of each component, SEAP production was normalized to the inducer-free condition (−Biotin. −TET). (E) Compact genetic design for biotin-regulated gene expression. A total of 30 000 CHO-K1 cells were transfected with plasmids pTT-Bio and pMF111 (see Figure 1A for genotype) and cultivated in the presence (+) or absence (−) of biotin (100 nM) or tetracycline (2 µg/ml) for 48 h prior to quantification of SEAP production.
Specificity of the biotin-controlled expression system was assessed by selective omission individual components including (i) the biotin ligase BirA (pWW804), (ii) SA-KRAB (pWW944), (iii) BirA and SA-KRAB (pWW804, pWW944), (iv) BirA, SA-KRAB, tTA-AT (pWW804, pWW944, pWW938) or (v) by replacing the avitag-containing tTA-AT (pWW938) by the original tTA (pSAM200, PSV40-tTA-pA). Exclusion of either BirA, SA-KRAB or Avitag abolished biotin-responsive transgene repression while tetracycline-responsive expression control remained intact (Figure 2D). In the absence of either tTA-AT or tTA no SEAP expression could be observed indicating that all components are essential and sufficient for biotin-controlled gene expression (Figure 2D). In all of these experiments, SEAP production reached identical maximum levels in the absence of biotin and tetracycline, indicating that the AVITAG fusion does not impact tTA function (tTA-AT, pWW938, 2.9 ± 0.3 U/l; tTA, pSAM200, 2.7 ± 0.1 U/l).
To provide biotin-controlled gene regulation in a easy-to-handle two-vector format (19), we have cloned all components (SA-KRAB, tTA-AT, BirA) into a triple-transcript expression format, in which the first cistron (SA-KRAB) is transcribed from a simian virus 40 promoter (PSV40) while subsequent cistrons two (tTA-AT) and three (BirA) are under control of compact synthetic promoters (PGTX) (47,48) (pTT-Bio; PSV40-SA-KRAB-PGTX-tTA-AT-PGTX-BirA-pA). Co-transfection of pTT-Bio with the response vector pMF111 into CHO-K1 cells resulted in high-level SEAP expression in the absence of biotin while addition of biotin and/or tetracycline shut off SEAP production (Figure 2E).
Adjustability and induction kinetics of the biotin control system
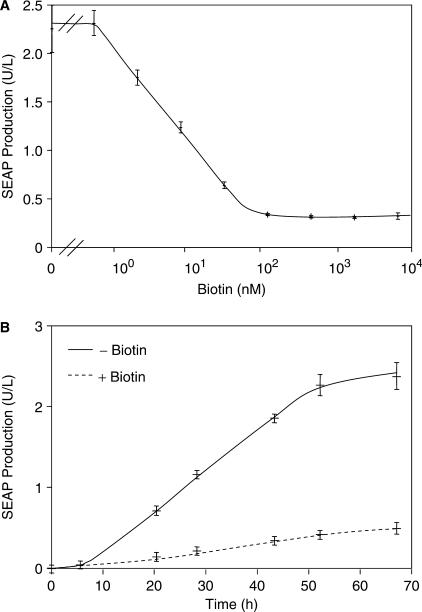
Exposure of CHO-K1 containing components mediating biotin-controlled SEAP expression (pWW938, pWW944, pWW804 and pMF111) to increasing vitamin H concentrations followed by scoring of SEAP production 48 h after transfection revealed adjustability over 2 logs of biotin concentrations. Maximum SEAP production occurred below 1 nM of biotin and SEAP repression to background levels occurred at 100 nM of biotin (Figure 3A). Biotin-dependent expression kinetics were assessed by cultivating CHO-K1 cells transfected with plasmids pWW938, pWW944, pWW804 and pMF111 for 68 h in the presence (100 nM) or absence of biotin which resulted in linear induction profiles between 5 and 53 h after transfection (Figure 3B).
Figure 3.
Analysis of vitamin H-responsive transgene expression. (A) Dose-response characteristics. A total of 30 000 CHO-K1 cells were co-transfected with plasmids pWW944, pWW938, pMF111 and pWW804 and cultivated for 48 h in biotin-free medium supplemented with increasing biotin concentrations prior to scoring SEAP production. (B) Expression kinetics. A total of 30 000 CHO-K1 cells were co-transfected with plasmids pWW944, pWW938, pMF111 and pWW804 and SEAP production was profiled for 68 h in the presence (+) or absence (−) of biotin (100 nM).
Validation of biotin-controlled product protein secretion in mice
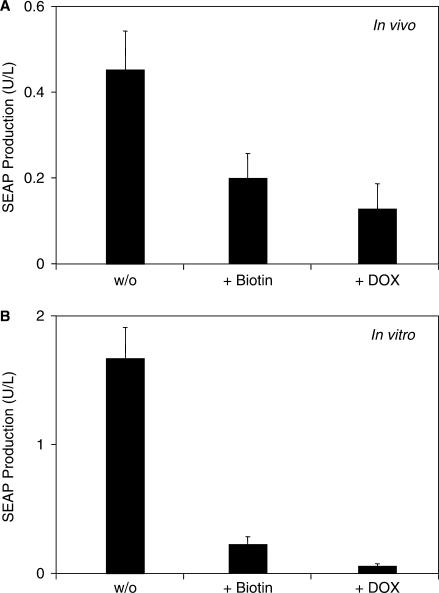
In order to validate biotin-responsive expression technology in a prototype gene therapy scenario, we microencapsulated CHO-K1 engineered for biotin-controlled production of the human model glycoprotein SEAP into coherent alginate-poly-l-lysine-alginate capsules and injected them intraperitoneally into mice kept on a biotin-free diet. After implantation, the mice were injected with either biotin (100 µg/ml) or doxycycline (100 mg/kg) and the serum levels of SEAP were profiled after 48 h. SEAP profiles confirmed maximum product protein production in the absence of biotin and repressed protein production in the presence of biotin or doxycycline (Figure 4A). Relative in vivo regulation profiles were similar compared to identical subcultures maintained in vitro suggesting that the biotin control system retains its regulation characteristics in vivo (Figure 4B). The higher basal expression in the presence of doxycycline or biotin in vivo (Figure 4A) compared to in vitro (Figure 4B) may result from lower bioavailability of the inducer in vivo. However, since biotin- and doxycycline-mediated in vitro and in vivo repression profiles are comparable, the regulation performance of both systems may be considered equivalent.
Figure 4.
In vivo validation of vitamin H-responsive gene expression. (A) CHO-K1 cells were co-transfected with plasmids pWW944, pWW938, pMF111 and pWW804, encapsulated in coherent alginate-poly-l-lysine-alginate capsules and intraperitoneally injected into biotin-deficient mice (2 × 106 cells/mouse). Mice further received biotin (+Biotin, 100 µg/kg) or doxycycline (+DOX, 100 mg/kg) injections and were subsequently kept for 48 h prior to quantification of SEAP production in the serum. (B) Parallel in vitro validation. Encapsulated CHO-K1 cells co-transfected with plasmids pWW944, pWW938, pMF111 and pWW804 were cultivated in vitro (2 × 106 cells/20 ml biotin-free medium) in the presence of exogenous biotin (+Biotin, 100 nM) or doxycycline (+DOX, 2 µg/ml) for 48 h prior to quantification of SEAP production.
Stable CHO-K1 cell derivatives tetra-transgenic for biotin-responsive transcription-control components
Extensive characterization of the biotin-responsive transcription control system for biopharmaceutical manufacturing requires availability of a stable cell line for long-term studies. We have therefore constructed a stable CHO-K1-derived cell line, which is tetratransgenic for pWW938 (PSV40-tTA-AT-pA), pWW944 (PSV40-SA-KRAB-pA), pWW804 (PhCMV-BirA-pA) and pMF111 (PhCMV*−1-SEAP-pA) and produces SEAP in a biotin and tetracycline-responsive manner. Five out of 40 stable cell clones were cultivated for 48 h in biotin- and/or tetracycline-containing medium and profiled for SEAP production. All cell clones showed high SEAP production when grown in the absence of regulating molecules (−biotin, −tetracycline), while either addition of biotin or tetracycline abolished SEAP production (Figure 5A). The different expression levels observed for individual cell clones may result from random chromosomal integration and transcription-modulating impact of adjacent genetic elements. Likewise, transgene expression differences also exist between stable and transiently transfected populations, which produce the product protein from expression vector episomes (55,56). Since BioCHO-SEAP clone no. 1 (BioCHO-SEAP1) showed the best regulation profile it was chosen for further characterization. Expression stability of BioCHO-SEAP1 was confirmed by continuous cultivation of the cell line for 90 days, which did not reveal any statistically significant decrease in SEAP production (day 0, 0.67 ± 0.5 U/l; day 90, 0.65 ± 0.6 U/l). Biotin-responsive SEAP production kinetics for which BioCHO-SEAP1 was seeded in biotin-free or biotin-containing medium (t = 0) revealed a steady increase over time when cultured in the absence of an inducer, while biotin addition resulted in background SEAP levels only (Figure 5B). Reversibility of biotin-responsive gene expression induction was assessed by cultivating initial BioCHO-SEAP1 populations in either biotin-free or biotin-containing medium. After 31 and 100 h of cultivation, the cell population was split and the biotin status maintained or reversed as indicated. Sequential induction and repression of SEAP production was followed over a one-week period (Figure 5C). Biotin-controlled SEAP expression was reversible and the induction or repression kinetics were not influenced by the biotin cultivation history of the BioCHO-SEAP population (Figure 5C). The dose-response profiles recorded over 7 biotin concentration logs revealed unique expression characteristics: (i) gradual ON-to-OFF expression profiles between 1 and 100 nM, reflecting increasing heterodimerization of transsilencer and transsilencer-mediated repression of tTA-AT-driven PhCMV*−1 transcription. (ii) Full OFF state at biotin concentrations ranging from 100 to 1000 nM, reflecting maximum transsilencing capacity. (iii) graded OFF-to-ON expression at biotin levels above 100 µM, suggesting that free biotin levels exceeding the ligation capacity of BirA may bind and sequestrate SA-KRAB thereby competitively inhibiting its heterodimerization with biotinylated tTA-AT (Figure 5D).
Figure 5.
Characterization of the transgenic cell line BioCHO-SEAP engineered for vitamin H-responsive SEAP expression. (A) CHO-K1 cells were stably transfected with plasmids pWW944, pWW938, pMF111 and pWW804 (BioCHO-SEAP) and subjected to single-cell cloning. Five representative clones were cultivated (60 000 cells/ml) in biotin-free medium in the presence or absence of biotin (100 nM) or tetracycline (2 µg/ml) for 48 h prior to quantification of SEAP expression. (B) Expression kinetics of BioCHO-SEAP1. SEAP production from BioCHO-SEAP1 cells was monitored when grown in biotin-free medium for 96 h in the presence (+, 100 nM) or absence (−) of biotin. At t = 0 the cells were seeded in the respective media. (C) Reversibility of SEAP expression in BioCHO-SEAP1 cells. SEAP expression from BioCHO-SEAP1 cells was followed during cultivation in biotin-free medium for 170 h in the presence (+) or absence (−) of 100 nM biotin. At the indicated times (arrows) each culture was split into two and cultivated in fresh medium with the same or an alternating −/+ biotin status. (D) Dose-response characteristics of BioCHO-SEAP1 cells. BioCHO-SEAP1 was cultivated (60 000 cells/ml) in biotin-free medium supplemented with increasing biotin concentrations for 48 h prior to quantification of SEAP production.
Biotin-inducible expression in biotin-containing environments
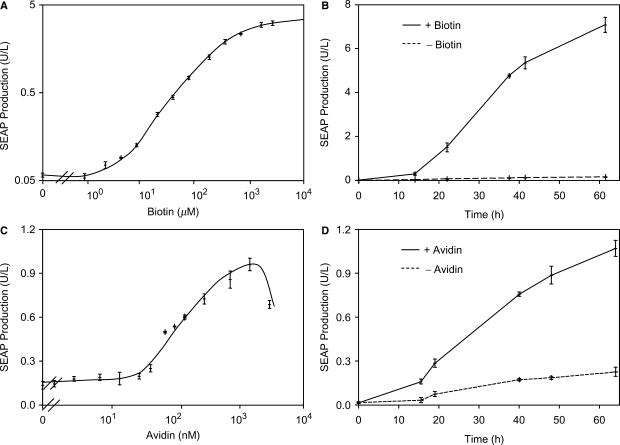
While all aforementioned experiments were conducted in biotin-free medium, standard cell culture media and serum additives contain biotin at an average level of 50 nM which fully represses the biotin control system (Figure 5D). Dose-response characteristics of BioCHO-SEAP1 cultivated in standard serum-containing cell culture medium and exposed to increasing biotin concentrations resulted in a gradual increase in SEAP production, confirming that the biotin control modality operates as an inducible expression system under standard culture conditions (Figure 6A). Furthermore, expression kinetics in the presence of 2000 µM biotin increased steadily, whereas the absence of additional biotin resulted in background expression only (Figure 6B).
Figure 6.
Vitamin H-responsive transgene expression in standard culture environments containing natural biotin levels. (A) Dose-response characteristics of BioCHO-SEAP1 cultivated in standard serum (biotin)-containing medium. BioCHO-SEAP1 (30 000 cells/ml) was cultivated in standard serum (biotin)-containing medium in the presence of increasing biotin concentrations for 48 h prior to scoring SEAP production. (B) Expression kinetics. SEAP production from BioCHO-SEAP1 (60 000 cells/ml) was monitored for 65 h in the presence (+, 2 mM) or absence (−) of biotin supplementation. (C) Regulated SEAP expression by avidin-mediated biotin sequestration. BioCHO-SEAP1 (60 000 cells/ml) was cultivated in the presence of increasing avidin concentrations (expressed as biotin-binding capacity) for 48 h prior to assessing SEAP production. (D) Avidin-controlled induction kinetics of vitamin H-regulated SEAP expression. SEAP production from BioCHO-SEAP1 cells (60 000 cells/ml) was monitored during cultivation for 65 h in the presence (+, 1.5 µM) or absence (−) of avidin.
A further strategy to induce the biotin control system is to sequester repressing biotin in standard medium by addition of avidin. Cultivation of BioCHO-SEAP1 in the presence of increasing avidin concentrations resulted in a gradual increase in SEAP production, thereby enabling adjustable transgene expression by administration of an exogenous protein (Figure 6C). Expression kinetics in the presence or absence of avidin increased steadily over time (Figure 6D). However, the overall regulation performance using avidin as an inducer was lower compared to biotin-triggered expression control (Figure 6A and C), suggesting that avidin is unable to fully compete with the thermodynamically favored enzymatic biotinylation of tTA-AT.
Biotin-inducible protein production in a prototype biopharmaceutical manufacturing scenario
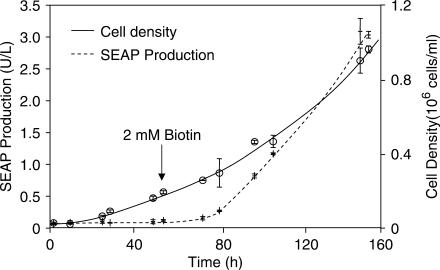
Despite intensive research no significant side effects have been discovered for biotin, neither following oral uptake nor after injection (57). Therefore, this FDA-licensed molecule represents an ideal inducer for any bioprocess application requiring precisely timed expression control (11,12,58). We have therefore evaluated the compatibility of vitamin H-responsive gene expression with standard bioreactor operation by cultivating BioCHO-SEAP1 for 53 h in a BioWave® bioreactor using 1 l of standard biotin-containing (non-supplemented) culture medium before adding 2 mM biotin to trigger SEAP production. While BioCHO-SEAP1 grew exponentially during the entire period, SEAP expression remained silent until shortly after addition of biotin, thereby validating the suitability for timely controlled biotin-responsive gene expression in a prototype biopharmaceutical manufacturing scenario (Figure 7).
Figure 7.
Performance of vitamin H-responsive gene expression during cultivation in bioreactors. BioCHO-SEAP1 was cultivated in a BioWave® bioreactor in 1 l culture volume using serum- (biotin) containing standard medium. SEAP expression and cell density were monitored over time. After 53 h (arrow) SEAP expression was induced by adding 2 mM biotin.
DISCUSSION
Adjustable transgene expression technology is fundamental to prototype gene therapy scenarios (6,7,59–61), functional genomic research (14,16), synthetic biology (1–3,62) and forefront biopharmaceutical manufacturing strategies (11,12,58,63). The novel vitamin H-responsive expression technology exhibits several advantages regarding standard operation and regulation performance compared to established systems which are typically triggered by side-effect-prone immunosuppressants, hormones or antibiotics (32,34,35,37,38): Vitamin H (biotin) is the first inducing agent with no or only low known side effects (e.g. to our knowledge, no LD50 has ever been described). As biotin is a natural component of cell culture media, its removal or supplementation to native levels is not expected to require additional validation or licensing efforts for biopharmaceutical manufacturing as would be the case for pharmacologically active inducers such as hormones, immunosupressors and antibiotics.
When comparing biotin-adjustable regulation characteristics to classic expression systems, biotin-adjustable transgene control exhibits unique band pass filter-like expression characteristics using a single inducer molecule: (i) within the low biotin concentration window (0–2 nM), transcription is fully induced, (ii) it is maximally repressed between 100 and 1000 nM biotin and (iii) again entirely turned on beyond 100 µM. This exclusive feature enables sequential induction, repression and re-induction using increasing doses of vitamin H, an expression pattern which has so far only been achieved in bacteria using a sophisticated gene network (64). The unique biotin-dependent band pass filter characteristic could be explained by a competition of tTA-AT and SA-KRAB for free biotin. At low biotin concentrations (0–100 nM), binding of biotin to tTA-AT [via the intermediate biotin–BirA complex, (65)] is thermodynamically favored [ΔG0 = −360 kJ/mol for an amide bond (in trans) versus ΔG0 = −80 kJ/mol for the non-covalent binding of biotin to SA-KRAB (66,67)], resulting in biotinylated tTA-AT and subsequent transcriptional shutdown after binding to SA-KRAB. At high biotin concentrations (>100 µM) free biotin is expected to saturate all tTA-AT and SA-KRAB biotin-binding sites thereby preventing dimerization with biotinylated tTA-AT. While basic research scientists may appreciate the extended regulation bandwidth emulating developmental expression patterns (64,68–70), the biopharmaceutical manufacturing could welcome the first opportunity to reverse the expression state of difficult-to-produce protein therapeutics without media exchanges and filtration operations.
Owing to the use of TetR as the DNA-binding component, the biotin-responsive gene expression technology is compatible with standard tetracycline-responsive promoters, established cell lines and mouse strains (15,19,20,59) and is unique in integrating two input signals: biotin and tetracycline. This dual-input sensitivity permits override of the biotin-controlled transgene expression by administration of tetracycline which may represent a useful safety feature in future cell engineering strategies. Since biotin is a physiological component the synthetic control modality may be used to convert endogenous biotin concentrations into a therapeutic transgene response. Scenarios to treat biotinidase-associated metabolic disorders by triggering therapeutic biotinidase expression at low pathologic biotin concentrations and shutting it off when physiologic biotin concentrations are reached, could now become a reality (71–73). Despite the ubiquitous presence of biotin in living systems, interference-free transgene control can still be achieved by (i) addition of biotin-sequestrating avidin or by (ii) administration of excess biotin (>1000 nM) thereby taking advantage of the high-biotin regulation window of the aforementioned band-pass filter.
Considering all of the design and performance characteristics in mammalian cells, mice as well as during standard bioreactor operation, we are convinced that biotin-controlled transgene expression will foster advances in basic research, gene therapy scenarios as well as in biopharmaceutical manufacturing.
ACKNOWLEDGEMENTS
We thank Martine Gilet and Mattia Hamberger for skilled technical assistance as well as Marcia Schoenberg and David Greber for critical comments on the manuscript. This work was supported by the Swiss National Science Foundation (grant no. 3100A0-112549), the Swiss State Secretariat for Education and Research within EC Framework 6 and Cistronics Cell Technology GmbH, P.O.B. 145, CH-8093 Zurich, Switzerland. Funding to pay the Open Access publication charges for this article was provided by ETH Zurich.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kramer BP, Fischer C, Fussenegger M. BioLogic gates enable logical transcription control in mammalian cells. Biotechnol. Bioeng. 2004;87:478–484. doi: 10.1002/bit.20142. [DOI] [PubMed] [Google Scholar]
- 2.Kramer BP, Fussenegger M. Hysteresis in a synthetic mammalian gene network. Proc. Natl Acad. Sci. USA. 2005;102:9517–9522. doi: 10.1073/pnas.0500345102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kramer BP, Viretta AU, Daoud-El-Baba M, Aubel D, Weber W, Fussenegger M. An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol. 2004;22:867–870. doi: 10.1038/nbt980. [DOI] [PubMed] [Google Scholar]
- 4.Kramer BP, Weber W, Fussenegger M. Artificial regulatory networks and cascades for discrete multilevel transgene control in mammalian cells. Biotechnol. Bioeng. 2003;83:810–820. doi: 10.1002/bit.10731. [DOI] [PubMed] [Google Scholar]
- 5.McLoughlin JM, McCarty TM, Cunningham C, Clark V, Senzer N, Nemunaitis J, Kuhn JA. TNFerade, an adenovector carrying the transgene for human tumor necrosis factor alpha, for patients with advanced solid tumors: surgical experience and long-term follow-up. Ann. Surg. Oncol. 2005;12:825–830. doi: 10.1245/ASO.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 6.Regulier E, Pereira de Almeida L, Sommer B, Aebischer P, Deglon N. Dose-dependent neuroprotective effect of ciliary neurotrophic factor delivered via tetracycline-regulated lentiviral vectors in the quinolinic acid rat model of Huntington's disease. Hum. Gene. Ther. 2002;13:1981–1990. doi: 10.1089/10430340260355383. [DOI] [PubMed] [Google Scholar]
- 7.Regulier E, Trottier Y, Perrin V, Aebischer P, Deglon N. Early and reversible neuropathology induced by tetracycline-regulated lentiviral overexpression of mutant huntingtin in rat striatum. Hum. Mol. Genet. 2003;12:2827–2836. doi: 10.1093/hmg/ddg305. [DOI] [PubMed] [Google Scholar]
- 8.Weber W, Fussenegger M. Approaches for trigger-inducible viral transgene regulation in gene-based tissue engineering. Curr. Opin. Biotechnol. 2004;15:383–391. doi: 10.1016/j.copbio.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 9.Aubel D, Morris R, Lennon B, Rimann M, Kaufmann H, Folcher M, Bailey JE, Thompson CJ, Fussenegger M. Design of a novel mammalian screening system for the detection of bioavailable, non-cytotoxic streptogramin antibiotics. J. Antibiot. (Tokyo) 2001;54:44–55. doi: 10.7164/antibiotics.54.44. [DOI] [PubMed] [Google Scholar]
- 10.Weber CC, Link N, Fux C, Zisch AH, Weber W, Fussenegger M. Broad-spectrum protein biosensors for class-specific detection of antibiotics. Biotechnol. Bioeng. 2005;89:9–17. doi: 10.1002/bit.20224. [DOI] [PubMed] [Google Scholar]
- 11.Boorsma M, Nieba L, Koller D, Bachmann MF, Bailey JE, Renner WA. A temperature-regulated replicon-based DNA expression system. Nat. Biotechnol. 2000;18:429–432. doi: 10.1038/74493. [DOI] [PubMed] [Google Scholar]
- 12.Fussenegger M, Schlatter S, Datwyler D, Mazur X, Bailey JE. Controlled proliferation by multigene metabolic engineering enhances the productivity of Chinese hamster ovary cells. Nat. Biotechnol. 1998;16:468–472. doi: 10.1038/nbt0598-468. [DOI] [PubMed] [Google Scholar]
- 13.Umana P, Jean-Mairet J, Bailey JE. Tetracycline-regulated overexpression of glycosyltransferases in Chinese hamster ovary cells. Biotechnol. Bioeng. 1999;65:542–549. [PubMed] [Google Scholar]
- 14.Malleret G, Haditsch U, Genoux D, Jones MW, Bliss TV, Vanhoose AM, Weitlauf C, Kandel ER, Winder DG, et al. Inducible and reversible enhancement of learning, memory, and long-term potentiation by genetic inhibition of calcineurin. Cell. 2001;104:675–686. doi: 10.1016/s0092-8674(01)00264-1. [DOI] [PubMed] [Google Scholar]
- 15.Mansuy IM, Bujard H. Tetracycline-regulated gene expression in the brain. Curr. Opin. Neurobiol. 2000;10:593–596. doi: 10.1016/s0959-4388(00)00127-6. [DOI] [PubMed] [Google Scholar]
- 16.Gossen M, Bujard H. Studying gene function in eukaryotes by conditional gene inactivation. Annu. Rev. Genet. 2002;36:153–173. doi: 10.1146/annurev.genet.36.041002.120114. [DOI] [PubMed] [Google Scholar]
- 17.Weber W, Fussenegger M. Pharmacologic transgene control systems for gene therapy. J. Gene. Med. 2006;8:535–556. doi: 10.1002/jgm.903. [DOI] [PubMed] [Google Scholar]
- 18.Fussenegger M, Morris RP, Fux C, Rimann M, von Stockar B, Thompson CJ, Bailey JE. Streptogramin-based gene regulation systems for mammalian cells. Nat. Biotechnol. 2000;18:1203–1208. doi: 10.1038/81208. [DOI] [PubMed] [Google Scholar]
- 19.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl Acad. Sci. USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gossen M, Freundlieb S, Bender G, Muller G, Hillen W, Bujard H. Transcriptional activation by tetracyclines in mammalian cells. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 21.Weber W, Fux C, Daoud-el Baba M, Keller B, Weber CC, Kramer BP, Heinzen C, Aubel D, Bailey JE, et al. Macrolide-based transgene control in mammalian cells and mice. Nat. Biotechnol. 2002;20:901–907. doi: 10.1038/nbt731. [DOI] [PubMed] [Google Scholar]
- 22.Zhao HF, Boyd J, Jolicoeur N, Shen SH. A coumermycin/novobiocin-regulated gene expression system. Hum. Gene. Ther. 2003;14:1619–1629. doi: 10.1089/104303403322542266. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, O’Malley B.W., Jr., Tsai SY, O’Malley BW. A regulatory system for use in gene transfer. Proc. Natl Acad. Sci. USA. 1994;91:8180–8184. doi: 10.1073/pnas.91.17.8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Palli SR, Kapitskaya MZ, Kumar MB, Cress DE. Improved ecdysone receptor-based inducible gene regulation system. Eur. J. Biochem. 2003;270:1308–1315. doi: 10.1046/j.1432-1033.2003.03501.x. [DOI] [PubMed] [Google Scholar]
- 25.Serguera C, Bohl D, Rolland E, Prevost P, Heard JM. Control of erythropoietin secretion by doxycycline or mifepristone in mice bearing polymer-encapsulated engineered cells. Hum. Gene. Ther. 1999;10:375–383. doi: 10.1089/10430349950018823. [DOI] [PubMed] [Google Scholar]
- 26.Rivera VM, Clackson T, Natesan S, Pollock R, Amara JF, Keenan T, Magari SR, Phillips T, Courage NL, et al. A humanized system for pharmacologic control of gene expression. Nat. Med. 1996;2:1028–1032. doi: 10.1038/nm0996-1028. [DOI] [PubMed] [Google Scholar]
- 27.Rollins CT, Rivera VM, Woolfson DN, Keenan T, Hatada M, Adams SE, Andrade LJ, Yaeger D, van Schravendijk MR, et al. A ligand-reversible dimerization system for controlling protein-protein interactions. Proc. Natl Acad. Sci. USA. 2000;97:7096–7101. doi: 10.1073/pnas.100101997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fussenegger M. The impact of mammalian gene regulation concepts on functional genomic research, metabolic engineering, and advanced gene therapies. Biotechnol. Prog. 2001;17:1–51. doi: 10.1021/bp000129c. [DOI] [PubMed] [Google Scholar]
- 29.Roscilli G, Rinaudo CD, Cimino M, Sporeno E, Lamartina S, Ciliberto G, Toniatti C. Long-term and tight control of gene expression in mouse skeletal muscle by a new hybrid human transcription factor. Mol. Ther. 2002;6:653–663. [PubMed] [Google Scholar]
- 30.Hanazono Y, Nagashima T, Takatoku M, Shibata H, Ageyama N, Asano T, Ueda Y, Dunbar CE, Kume A, et al. In vivo selective expansion of gene-modified hematopoietic cells in a nonhuman primate model. Gene. Ther. 2002;9:1055–1064. doi: 10.1038/sj.gt.3301781. [DOI] [PubMed] [Google Scholar]
- 31.Cohlan SQ. Tetracycline staining of teeth. Teratology. 1977;15:127–129. doi: 10.1002/tera.1420150117. [DOI] [PubMed] [Google Scholar]
- 32.Kapunisk-Uner JE, Sande MA, Chambers HFS. Tetracycline, Chloramphenicol, Erythromycin and Miscellaneous Antibacterial Agents. 9th. New York: McGraw-Hill; 1996. [Google Scholar]
- 33.Aarestrup FM. Veterinary drug usage and antimicrobial resistance in bacteria of animal origin. Basic Clin. Pharmacol. Toxicol. 2005;96:271–281. doi: 10.1111/j.1742-7843.2005.pto960401.x. [DOI] [PubMed] [Google Scholar]
- 34.Rang A, Will H. The tetracycline-responsive promoter contains functional interferon-inducible response elements. Nucleic Acids Res. 2000;28:1120–1125. doi: 10.1093/nar/28.5.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gee I, Trull AK, Charman SC, Alexander GJ. Sirolimus inhibits oxidative burst activity in transplant recipients. Transplantation. 2003;76:1766–1768. doi: 10.1097/01.TP.0000093995.08240.49. [DOI] [PubMed] [Google Scholar]
- 36.Elisaf MS, Nakou K, Liamis G, Pavlidis NA. Tamoxifen-induced severe hypertriglyceridemia and pancreatitis. Ann. Oncol. 2000;11:1067–1069. doi: 10.1023/a:1008309613082. [DOI] [PubMed] [Google Scholar]
- 37.Grunberg SM, Weiss MH, Russell CA, Spitz IM, Ahmadi J, Sadun A, Sitruk-Ware R. Long-term administration of mifepristone (RU486): clinical tolerance during extended treatment of meningioma. Cancer Invest. 2006;24:727–733. doi: 10.1080/07357900601062339. [DOI] [PubMed] [Google Scholar]
- 38.Sullivan A, Edlund C, Nord CE. Effect of antimicrobial agents on the ecological balance of human microflora. Lancet Infect. Dis. 2001;1:101–114. doi: 10.1016/S1473-3099(01)00066-4. [DOI] [PubMed] [Google Scholar]
- 39.Weber W, Stelling J, Rimann M, Keller B, Daoud-El Baba M, Weber CC, Aubel D, Fussenegger M. A synthetic time-delay circuit in mammalian cells and mice. Proc. Natl Acad. Sci. USA. 2007;104:2643–2648. doi: 10.1073/pnas.0606398104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin KC, Campbell A, Shiuan D. Binding characteristics of Escherichia coli biotin repressor-operator complex. Biochim. Biophys. Acta. 1991;1090:317–325. doi: 10.1016/0167-4781(91)90196-s. [DOI] [PubMed] [Google Scholar]
- 41.Schatz PJ. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology (NY) 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- 42.de Boer E, Rodriguez P, Bonte E, Krijgsveld J, Katsantoni E, Heck A, Grosveld F, Strouboulis J. Efficient biotinylation and single-step purification of tagged transcription factors in mammalian cells and transgenic mice. Proc. Natl Acad. Sci. USA. 2003;100:7480–7485. doi: 10.1073/pnas.1332608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Werven FJ, Timmers HT. The use of biotin tagging in Saccharomyces cerevisiae improves the sensitivity of chromatin immunoprecipitation. Nucleic Acids Res. 2006;34:e33. doi: 10.1093/nar/gkl003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viens A, Mechold U, Lehrmann H, Harel-Bellan A, Ogryzko V. Use of protein biotinylation in vivo for chromatin immunoprecipitation. Anal. Biochem. 2004;325:68–76. doi: 10.1016/j.ab.2003.10.015. [DOI] [PubMed] [Google Scholar]
- 45.Kohanski RA, Lane MD. Monovalent avidin affinity columns. Methods Enzymol. 1990;184:194–200. doi: 10.1016/0076-6879(90)84274-k. [DOI] [PubMed] [Google Scholar]
- 46.Weber W, Rimann M, Spielmann M, Keller B, Daoud-El Baba M, Aubel D, Weber CC, Fussenegger M. Gas-inducible transgene expression in mammalian cells and mice. Nat. Biotechnol. 2004;22:1440–1444. doi: 10.1038/nbt1021. [DOI] [PubMed] [Google Scholar]
- 47.Hartenbach S, Fussenegger M. A novel synthetic mammalian promoter derived from an internal ribosome entry site. Biotechnol. Bioeng. 2006;95:547–559. doi: 10.1002/bit.21174. [DOI] [PubMed] [Google Scholar]
- 48.Fux C, Langer D, Kelm JM, Weber W, Fussenegger M. New-generation multicistronic expression platform: pTRIDENT vectors containing size-optimized IRES elements enable homing endonuclease-based cistron swapping into lentiviral expression vectors. Biotechnol. Bioeng. 2004;86:174–187. doi: 10.1002/bit.20028. [DOI] [PubMed] [Google Scholar]
- 49.Mitta B, Rimann M, Ehrengruber MU, Ehrbar M, Djonov V, Kelm J, Fussenegger M. Advanced modular self-inactivating lentiviral expression vectors for multigene interventions in mammalian cells and in vivo transduction. Nucleic Acids Res. 2002;30:e113. doi: 10.1093/nar/gnf112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kada G, Falk H, Gruber HJ. Accurate measurement of avidin and streptavidin in crude biofluids with a new, optimized biotin-fluorescein conjugate. Biochim. Biophys. Acta. 1999;1427:33–43. doi: 10.1016/s0304-4165(98)00178-0. [DOI] [PubMed] [Google Scholar]
- 51.Schlatter S, Rimann M, Kelm J, Fussenegger M. SAMY, a novel mammalian reporter gene derived from Bacillus stearothermophilus alpha-amylase. Gene. 2002;282:19–31. doi: 10.1016/s0378-1119(01)00824-1. [DOI] [PubMed] [Google Scholar]
- 52.Osada K, Komai M, Sugiyama K, Urayama N, Furukawa Y. Experimental study of fatigue provoked by biotin deficiency in mice. Int. J. Vitam. Nutr. Res. 2004;74:334–340. doi: 10.1024/0300-9831.74.5.334. [DOI] [PubMed] [Google Scholar]
- 53.Wang KS, Kearns GL, Mock DM. The clearance and metabolism of biotin administered intravenously to pigs in tracer and physiologic amounts is much more rapid than previously appreciated. J. Nutr. 2001;131:1271–1278. doi: 10.1093/jn/131.4.1271. [DOI] [PubMed] [Google Scholar]
- 54.Chapman-Smith A, Mulhern TD, Whelan F, Cronan J.E., Jr The C-terminal domain of biotin protein ligase from E. coli is required for catalytic activity. Protein Sci. 2001;10:2608–2617. doi: 10.1110/ps.22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gopalkrishnan RV, Christiansen KA, Goldstein NI, DePinho RA, Fisher PB. Use of the human EF-1alpha promoter for expression can significantly increase success in establishing stable cell lines with consistent expression: a study using the tetracycline-inducible system in human cancer cells. Nucleic Acids Res. 1999;27:4775–4782. doi: 10.1093/nar/27.24.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wurm FM. Production of recombinant protein therapeutics in cultivated mammalian cells. Nat. Biotechnol. 2004;22:1393–1398. doi: 10.1038/nbt1026. [DOI] [PubMed] [Google Scholar]
- 57.Fiume MZ. Final report on the safety assessment of biotin. Int. J. Toxicol. 2001;20(Suppl. 4):1–12. [PubMed] [Google Scholar]
- 58.Boorsma M, Hoenke S, Marrero A, Fischer R, Bailey JE, Renner WA, Bachmann MF. Bioprocess applications of a Sindbis virus-based temperature-inducible expression system. Biotechnol. Bioeng. 2002;79:602–609. doi: 10.1002/bit.10311. [DOI] [PubMed] [Google Scholar]
- 59.Bohl D, Heard JM. Modulation of erythropoietin delivery from engineered muscles in mice. Hum. Gene. Ther. 1997;8:195–204. doi: 10.1089/hum.1997.8.2-195. [DOI] [PubMed] [Google Scholar]
- 60.Rivera VM, Gao GP, Grant RL, Schnell MA, Zoltick PW, Rozamus LW, Clackson T, Wilson JM. Long-term pharmacologically regulated expression of erythropoietin in primates following AAV-mediated gene transfer. Blood. 2005;105:1424–1430. doi: 10.1182/blood-2004-06-2501. [DOI] [PubMed] [Google Scholar]
- 61.Rivera VM, Ye X, Courage NL, Sachar J, Cerasoli F., Jr., Wilson JM, Gilman M. Long-term regulated expression of growth hormone in mice after intramuscular gene transfer. Proc. Natl Acad. Sci. USA. 1999;96:8657–8662. doi: 10.1073/pnas.96.15.8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kramer BP, Fischer M, Fussenegger M. Semi-synthetic mammalian gene regulatory networks. Metab. Eng. 2005;7:241–250. doi: 10.1016/j.ymben.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 63.Weber W, Rimann M, de Glutz FN, Weber E, Memmert K, Fussenegger M. Gas-inducible product gene expression in bioreactors. Metab. Eng. 2005;7:174–181. doi: 10.1016/j.ymben.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 64.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 65.Kwon K, Streaker ED, Beckett D. Binding specificity and the ligand dissociation process in the E. coli biotin holoenzyme synthetase. Protein Sci. 2002;11:558–570. doi: 10.1110/ps.33502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sanderson RT. Chemical Bonds and Bond Energy. New York: Academic Press; 1971. [Google Scholar]
- 67.Weber PC, Ohlendorf DH, Wendoloski JJ, Salemme FR. Structural origins of high-affinity biotin binding to streptavidin. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 68.Crick F. Diffusion in embryogenesis. Nature. 1970;225:420–422. doi: 10.1038/225420a0. [DOI] [PubMed] [Google Scholar]
- 69.Roth S, Stein D, Nusslein-Volhard C. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell. 1989;59:1189–1202. doi: 10.1016/0092-8674(89)90774-5. [DOI] [PubMed] [Google Scholar]
- 70.Stein D, Roth S, Vogelsang E, Nusslein-Volhard C. The polarity of the dorsoventral axis in the Drosophila embryo is defined by an extracellular signal. Cell. 1991;65:725–735. doi: 10.1016/0092-8674(91)90381-8. [DOI] [PubMed] [Google Scholar]
- 71.Mock DM. Skin manifestations of biotin deficiency. Semin. Dermatol. 1991;10:296–302. [PubMed] [Google Scholar]
- 72.Pacheco-Alvarez D, Solorzano-Vargas RS, Del Rio AL. Biotin in metabolism and its relationship to human disease. Arch. Med. Res. 2002;33:439–447. doi: 10.1016/s0188-4409(02)00399-5. [DOI] [PubMed] [Google Scholar]
- 73.Velazquez A, Zamudio S, Baez A, Murguia-Corral R, Rangel-Peniche B, Carrasco A. Indicators of biotin status: a study of patients on prolonged total parenteral nutrition. Eur. J. Clin. Nutr. 1990;44:11–16. [PubMed] [Google Scholar]