Abstract
Pif1p is the prototypical member of the PIF1 family of DNA helicases, a subfamily of SFI helicases conserved from yeast to humans. Baker's yeast Pif1p is involved in the maintenance of mitochondrial, ribosomal and telomeric DNA and may also have a general role in chromosomal replication by affecting Okazaki fragment maturation. Here we investigate the substrate preferences for Pif1p. The enzyme was preferentially active on RNA–DNA hybrids, as seen by faster unwinding rates on RNA–DNA hybrids compared to DNA–DNA hybrids. When using forked substrates, which have been shown previously to stimulate the enzyme, Pif1p demonstrated a preference for RNA–DNA hybrids. This preferential unwinding could not be correlated to preferential binding of Pif1p to the substrates that were the most readily unwound. Although the addition of the single-strand DNA-binding protein replication protein A (RPA) stimulated the helicase reaction on all substrates, it did not diminish the preference of Pif1p for RNA–DNA substrates. Thus, forked RNA–DNA substrates are the favored substrates for Pif1p in vitro. We discuss these findings in terms of the known biological roles of the enzyme.
INTRODUCTION
Helicases are ubiquitous enzymes that harness the energy of nucleotide hydrolysis to translocate on a nucleic acid strand. Helicases participate in most transactions involving nucleic acids, including DNA replication, recombination, repair, transcription, protein synthesis and ribonucleoprotein assembly and remodeling (1,2). Saccharomyces cerevisiae Pif1p is the founding member of a DNA helicase family conserved from yeast to humans (3,4). The helicase was originally discovered by its effects on maintenance of yeast mitochondrial DNA (5,6). In the absence of Pif1p, yeast cells lose mitochondrial DNA at high rates, which generates respiratory-deficient (petite) cells.
Pif1p was rediscovered by its nuclear role in the synthesis of telomeric DNA by telomerase (7,8), the specialized reverse transcriptase that maintains chromosome ends in most eukaryotes (9). The nuclear and mitochondrial isoforms of Pif1p, which originate from the same mRNA, are generated by differential codon start usage (7,8). The effects of the nuclear isoform of Pif1p on telomerase have been extensively studied, both in vivo and in vitro. In vivo, Pif1p inhibits telomerase-mediated telomere lengthening and de novo telomere addition at double-strand breaks (7,8,10,11). De novo addition is rare in wild-type cells, but is increased 600–1000-fold in pif1Δ cells (7,10). Telomere length is inversely related to Pif1p levels: reduced Pif1p results in long telomeres, and Pif1p over-expression results in modest telomere shortening (7,8). The effects of Pif1p on telomere length and de novo telomere addition are telomerase dependent (7,8,10,11), suggesting that Pif1p affects telomerase directly. In support of this possibility, in vitro assays show that Pif1p removes telomerase from DNA ends, which limits telomerase nucleotide addition processivity (12).
The role of Pif1p in the nucleus is not limited to telomeres. PIF1 deletion has a modest effect on ribosomal DNA (rDNA) replication by inhibiting fork movement past the replication fork barrier (RFB) (13). Pif1p may have a more general role in chromosomal replication as its deletion suppresses the lethality associated with loss of the helicase/endonuclease Dna2p, which functions in Okazaki fragment maturation (14). Pif1p may also function to resolve recombination intermediates as Pif1p over-expression suppresses the Sgs1p-induced damage that occurs in the absence of the topoisomerase Top3p (15).
Understanding how Pif1p functions in the cell requires a detailed analysis of its enzymological properties. Several characteristics of the enzyme have already been reported. Using an enzyme purified from yeast mitochondria or a recombinant protein produced in insect or bacterial cells, Pif1p was shown to be a distributive 5'–3' DNA helicase (6,8,12,16), whose activity is stimulated by forked substrates (16). Pif1p is also able to unwind RNA–DNA hybrids if the substrates contain a 5' single-stranded DNA overhang but does not unwind RNA–RNA hybrids (12). Here we show that Pif1p preferentially unwinds RNA–DNA hybrids, and that the enzyme is synergistically stimulated by the presence of a forked structure.
MATERIALS AND METHODS
Proteins
Recombinant Yeast RPA purified from Escherichia coli was a generous gift from P. Sung (17). The nuclear form of yeast Pif1p fused at its N-terminus to a 6-histidine tag was purified essentially as described (12). Briefly, the DNA coding for the nuclear form of Pif1p (amino acids 40–859) was cloned into vector pET28 (Novagen) and transformed into E. coli Rosetta strain, which is a derivative of the BL21(DE3) strain (Novagen). Protein expression was induced at 23°C by addition of 1 mM IPTG to a mid-log phase culture in rich media, and the culture was further grown for 16 h. Pif1p was purified by affinity chromatography to the 6-histidine tag, followed by cation exchange chromatography on a resource 15S column (GE Healthcare). Fractions containing pure Pif1p as assessed by Coomassie staining were pooled, concentrated and stored at –80°C in 25 mM HEPES pH 7.0, 100 mM NaCl, 25 mM (NH4)2SO4, 25 mM Mg(OAc), 1 mM DTT, 50% glycerol.
Oligonucleotide and helicase substrates
Oligonucleotides substrates were synthesized and purified by high pressure liquid chromatography by Integrated DNA Technology (IDT). These oligonucleotides were designed as random sequences containing a high GC content and unable to form stable secondary structures. For formation of the oligonucleotide substrate, the top (short) strand was labeled with γ-32P-ATP using polynucleotide kinase (New England Biolabs), according to standard protocols. Top and bottom strand oligos were mixed at equimolar ratios and annealed by slow cooling from 95 to 25°C in 10 mM MgSO4, 10 mM Tris pH 7.5. Substrates were subsequently gel purified and stored in 10 mM Tris pH 8.0 at −20°C. Concentration of each substrate was calculated based on the specific activity of the labeled oligonucleotide.
UV spectroscopy and melting profiles
Duplexes were prepared as described above for helicase substrates but in the following buffer: 0.15 M NaCl, 5 mM MgCl2, 10 mM cacodylate pH 7.0. Thermal melting profiles were determined in a computer-driven AVIV 14DS spectrophotometer equipped with a thermoelectrically controlled cell holder for cells of 1 cm path length. Melting profiles were obtained by measuring absorbance at 260 nm every 0.5°C over the indicated temperature range. Tm values (±0.5°C) were determined from the midpoint of the transition (inflexion point). All of the melting profiles were measured against a solvent blank.
Helicase assays
Helicase assays were carried out by incubating various amounts of Pif1p and 1 nM nucleic acid substrate at 25 or 35°C, as indicated. Unless otherwise noted, standard reaction buffer was 20 mM Tris pH 7.5, 50 mM NaCl, 100 µg/ml bovine serum albumin, 2 mM DTT, 5 mM Mg2+ and 4 mM ATP. For kinetic studies, reactions were started by addition of ATP. 5 µl aliquots were withdrawn at indicated times and the reactions stopped by addition of 2 µl quenching/loading buffer (6% Ficoll, 13 mM EDTA pH 8.0, 300 nM unlabeled top strand, 0.05% bromophenol blue and 0.05% xylene cyanol). Reaction products were loaded on a 12% polyacrylamide (20:1 acry:bis-acrylamide ratio) non-denaturing gel and resolved by electrophoresis at 4°C and 10 V/cm in TBE 1× buffer. Gels were dried and scanned with a storm PhosphorImager (Molecular Dynamics) and quantified using ImageQuant software (GE Healthcare). Data were fitted using PRISM software (Graphpad Software) to the single order exponential law A(t) = A(1 − e−kt), where A describes the amplitude of the reaction and k the apparent turnover rate of the reaction. For single cycle kinetics, the reaction was started with ATP and 500 nM single-stranded 36 bp trap DNA oligonucleotide of the following sequence: 5′-CGTAATCATGGTCATAGCTGTTTCCTGTGTGAAATT.
Gel retardation assay
Pif1p was incubated at various concentrations (ranging from 5 nM to 1 µM) in the presence of 50 pM of the indicated γ-32P labeled substrate at 25°C in the following buffer: 20 mM Tris pH 7.5, 10 mM NaCl, 100 µg/ml bovine serum albumin, 2 mM DTT and 5 mM Mg2+, ±4 mM AMPPNP (Sigma) or ATP-γS (Fluka). After 30 min incubation at room temperature, binding reactions were supplemented with loading buffer (6% ficoll, 0.05% bromophenol blue and 0.05% xylene cyanol) and resolved by electrophoresis at 4°C on a 6% (40:1 acry:bis-acrylamide ratio) non-denaturing gel run at 10 V/cm. Gels were dried, scanned and quantified as described above. Percentage of binding was calculated as the ratio between bound and (bound+unbound) radiolabeled substrate.
Centrifugation analysis of Pif1p
Five micromolar of Pif1p was incubated at 4°C for 3 h in a buffer containing 10 mM Tris pH 7.5, 50 mM NaCl, 4 mM MgCl2 and 5% glycerol. Aggregated protein was removed by centrifugation at 23 000g for 30 min. The supernatant was loaded on a 5 ml 15–40% glycerol gradient equilibrated with 10 mM Tris pH 7.5, 50 mM NaCl, 4 mM MgCl2 and centrifuged for 12 h at 151 693 g in a sw55ti rotor (Beckman Coulter). 250 μl fractions were collected and analyzed for the presence of Pif1p by western blot using an anti-His monoclonal antibody (Novagen). Molecular weight markers (Sigma) were resolved on an identical gradient run at the same time. Linearity of the gradient was verified by running a blank gradient and measuring diffraction index of each 250 µl fraction with a Bausch & Lomb Abbe-3L Refractometer.
RESULTS
RNA–DNA and DNA–DNA substrates unwinding by Pif1p at various salt concentrations
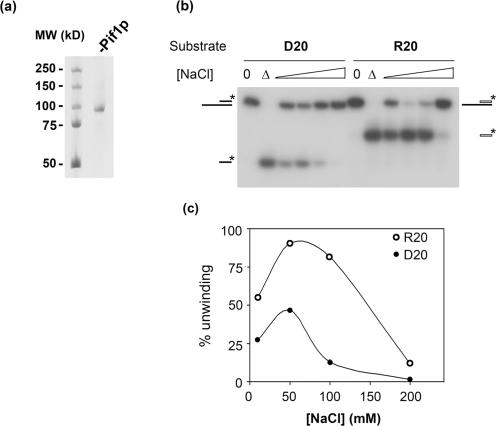
Pif1p was overexpressed in bacteria and purified to near homogeneity using a two-step procedure involving a cobalt affinity column and cation exchange chromatography (12) (Figure 1a). In a recent study, we showed that Pif1p is able to displace short DNA or RNA strands, provided that the loading strand is made of DNA. This result, along with our demonstration that Pif1p inhibits telomerase in vivo and in vitro (8,12) raised the possibility that Pif1p inhibits yeast telomerase by unwinding the RNA–DNA hybrid formed between the telomerase RNA, TLC1 and the end of the TG1–3 telomeric DNA. In order to compare the activity of Pif1p on RNA–DNA hybrids versus DNA–DNA substrates, we first determined if Pif1p unwinding of these two types of substrates had the same optimal salt requirements. For this experiment, we used as a substrate a 40-mer DNA oligonucleotide to which was annealed a complementary 20-mer oligonucleotide made of either DNA or RNA, leaving a 20-nt 5′ single-stranded overhang (substrates D20 and R20, Table 1 and Figure 2a). The experiments were conducted in conditions in which the enzyme was in excess, as significant unwinding was not observed if the substrates were present in excess or in the presence of a DNA trap (data not shown). Both DNA–DNA (closed circles) and RNA–DNA (open circles) hybrids were optimally unwound at 50 mM NaCl, and no activity was detected at 200 mM NaCl (Figure 1b and c). Therefore, the optimal salt concentration for unwinding both RNA–DNA and DNA–DNA hybrids is 50 mM NaCl. At all salt concentrations, a larger fraction of the RNA–DNA substrate compared to the DNA–DNA substrate was unwound.
Figure 1.
Pif1p unwinding activity as a function of NaCl concentration. (a) Coomassie gel staining of the purified recombinant nuclear isoform of Pif1p. (b) Unwinding of DNA–DNA (D20, left) and RNA–DNA (R20, right) hybrids at increasing salt concentrations. A total of 60 nM Pif1p was incubated with 1 nM D20 or R20 substrates at 35°C for 15 min. Products of the reaction were analyzed as described in the Materials and Methods section. (0) indicates zero time point, (▵) indicates heat denatured sample. (c) Quantification of the unwinding of RNA–DNA (R20, open circles) and DNA–DNA (D20, closed circles) substrates shown in (b). This experiment was done several times with similar results.
Table 1.
DNA–DNA substrates used in this studya
| Substrate | |
|---|---|
| D13 | CGTGAGCCTAGTG-5′ |
| 5′-CACTGGCCGTCTTACGGTCGGCACTCGGATCAC | |
| D20 | CGTGAGCCTAGTGGTACCGC-5′ |
| 5’-CACTGGCCGTCTTACGGTCGGCACTCGGATCACCATGGCG | |
| D40 | CGTGAGCCTAGTGGTACCGCCTGAGAGACGAGAGCACGAG-5’ |
| 5′-CACTGGCCGTCTTACGGTCGGCACTCGGATCACCATGGCGGACTCTCTGCTCTCGTGCTC | |
| fD20 | AAAAAAAAAACGTGAGCCTAGTGGTACCGC-5′ |
| 5′-CACTGGCCGTCTTACGGTCGGCACTCGGATCACCATGGCG |
aRNA–DNA hybrid counterparts of the above DNA–DNA substrates share the same bottom strand, but the top strand is made of ribonucleotides. RNA–DNA substrates counterparts are called R13, R20, R40 and fR20, respectively. The region of the forked substrate where the two strands are not complementary is underlined.
Figure 2.
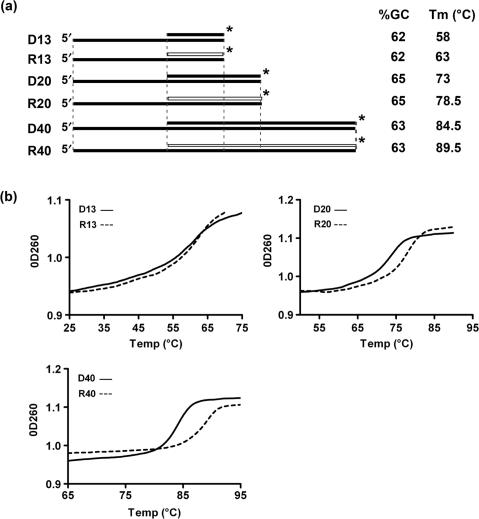
Structure and stability of helicase substrates. (a) Schematic of the structures of substrates used for helicase assays. Black bars represent DNA strands; white bars are RNA strands. Vertical dotted lines indicate regions of identical sequence among the various substrates. The asterisk indicates the position of the 32P label. GC content of the substrates in the hybrid region and the experimentally determined Tm (±0.5°C, as in b) are also indicated for each substrate. (b) Thermal denaturation profiles of the helicase substrates depicted in (a). The experiment was done at least twice for each substrate with identical results. A representative experiment is shown.
Pif1p activity on oligonucleotide substrates of various sizes
From the previous experiment, it seemed that Pif1p was more potent at unwinding the RNA–DNA substrate than the DNA–DNA substrate (Figure 1). To characterize this preference more quantitatively, we designed a family of RNA–DNA and DNA–DNA substrates of various sizes. These substrates contain a 13-, 20- or 40-nt-long duplex region with the same 5' single-stranded 20-nt overhang (Table 1, Figure 2a). The different substrates were designed to have high G+C content but an otherwise random sequence and predicted not to form stable secondary structures at 35°C. Not only was the overhang identical on all of the substrates, but in addition, longer substrates contained the sequence of the shorter substrates at the 5′ end of the duplex region (Figure 2a). To ensure that the differences in unwinding rates were not due to differential stability of the substrates, we measured the thermal melting curves of all the substrates. RNA–DNA hybrids of high GC content are predicted to be more stable in solution than their DNA–DNA counterparts (18). As expected, RNA–DNA hybrids were consistently as stable as or more stable than their DNA counterpart (Figure 2b).
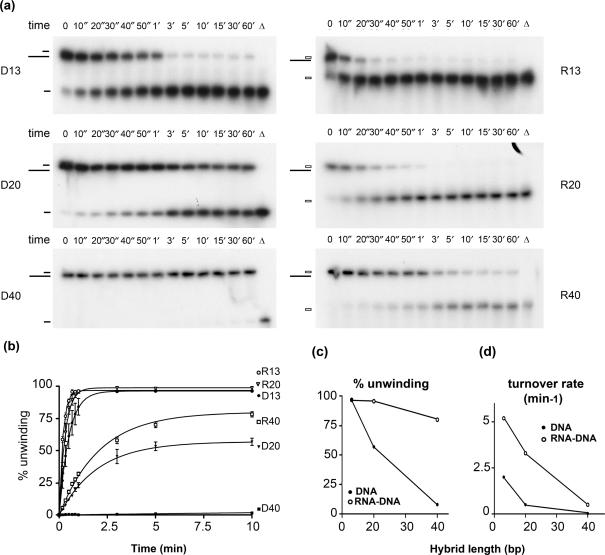
The six substrates were incubated with ATP, 50 mM NaCl, and excess Pif1p (60:1 enzyme/substrate ratio), which corresponded to saturating amounts of the enzyme for these substrates (data not shown). The displacement of the 32P-labeled top (short) strand of the substrate was monitored by electrophoresis in a non-denaturing polyacrylamide gel and quantified by phosphorImager analysis of the dried gels (Figure 3a). Efficient unwinding was observed for each RNA–DNA substrate. Even the longest RNA–DNA substrate, R40, was 60% unwound by the 3-min time point (Figure 3a and b). In contrast, only the 13-mer DNA substrates were fully unwound during the 60-min time course (Figure 3a and b; D13). In each case, Pif1p unwound the RNA–DNA hybrid more efficiently than its DNA–DNA counterpart, and the difference in the amplitude of the reaction and in the turnover rate between the two types of substrates increased with the size of the hybrid (Figure 3c and d). Indeed, the difference in the turnover rates between the RNA–DNA and DNA–DNA substrates was respectively 2-fold for the 13-mer, 13-fold for the 20-mer and 75-fold for the 40-mer. Pif1p was almost completely unable to unwind the 40-mer DNA substrate in these conditions even after 60 min (Figure 3a and b; D40). When the reactions were started in the presence of a single-strand DNA trap, none of the substrates was unwound at a quantifiable level, which made it impossible to measure single cycle kinetic rates on these substrates. Thus, although the activity of Pif1p is strongly stimulated by RNA–DNA substrates, the enzyme is poorly processive on both DNA–DNA and RNA–DNA substrates.
Figure 3.
Kinetics of unwinding by Pif1p of RNA–DNA and DNA–DNA hybrids of various sizes. (a) Unwinding by Pif1p of the substrates D13, D20, D40 and their RNA–DNA hybrid counterparts during a 1-h time course. For these experiments, 60 nM Pif1p was incubated with 1 nM substrate. Aliquots were removed at the indicated time points and analyzed on non-denaturing PAGE gels as described in the Materials and Methods section. (b) Quantification of RNA–DNA and DNA–DNA substrate unwinding by Pif1p. Errors bars indicate SD. Amplitude (% unwinding) of the reaction and apparent turnover rate for the different substrates are as follows A(D13) = 96 ± 2%; k(D13) = 2 ± 0.15 min−1; A(R13) = 97 ± 0.4%; k(R13) = 5 ± 0.15 min−1; A(D20) = 57 ± 1%; k(D20) = 0.5 ± 0.03 min−1; A(R20) = 99 ± 1%; k(R20) = 3 ± 0.2 min−1; A(D40) = 8 ± 1%; k(D40) = 0.02 ± 0.005 min−1; A(R40) = 80 ± 1%; k(R40) = 0.5 ± 0.02 min−1. (c) % unwinding and (d) turnover rate of the helicase reactions as a function of the hybrid length, derived from the three independent experiments analyzed in (b). SD are displayed but are too small to be visible at this scale.
Unwinding of forked substrates
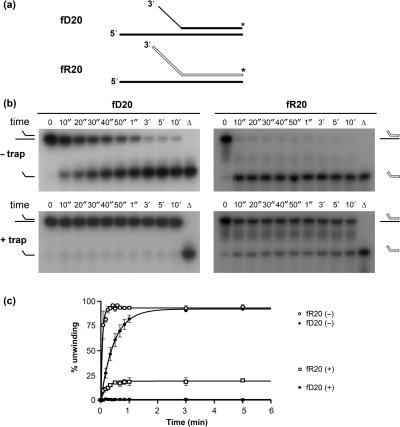
The activity of Pif1p purified from yeast mitochondria is stimulated by forked substrates (16). Here we determine if this stimulation is also observed with RNA–DNA substrates. The substrates used for this experiment are based on the 20-mer hybrids D20 and R20 (Table 1, Figure 2a). To generate forked substrates, a 10-mer poly(dA) segment was added to the 3' end of the labeled (top) strand of the D20 substrate to generate fD20. Similarly, a 10-mer poly(rA) was added to the 3' end of the top strand of the R20 substrate to generate fR20 (Table 1, Figure 4a). At 35°C, fD20 and fR20 were rapidly and completely unwound (<20 s, data not shown). This result confirmed that Pif1p activity is stimulated by forked substrates, but in these conditions the unwinding of the substrates was too fast to allow quantitative analyses. Lowering the temperature to 25°C slowed the reaction kinetics and allowed quantitative comparison of the unwinding of the RNA–DNA and DNA–DNA forked substrates in presence of saturating amounts of Pif1p.
Figure 4.
Activity of Pif1p on forked substrates at 25°C. (a) Structure of the forked substrates, using the same conventions as in Figure 2a. (b) Unwinding of fD20 and fR20 forked substrates in multiple cycle (– trap) and single cycle conditions (+ trap). (c) Quantification of three independent unwinding reactions of the fD20 and fR20 forked substrates. (+) and (–) indicate, respectively, reactions done in the presence and absence of a single-strand DNA trap. Amplitude and apparent turnover rates are as follows. Multiple cycle conditions: A(fD20) = 92 ± 2%, k(fD20) = 2.1 ± 0.3 min−1; A(fR20) = 93 ± 1%, k(fR20) = 18.3 ± 2 min−1. For single cycle kinetics, no unwinding was observed for the fD20 substrate; values for fR20 were A(fR20) = 19 ± 1%, k(fR20) = 5.2 ± 0.7 min−1.
At 25°C, both fD20 and fR20 were readily unwound (Figure 4b). However, the RNA–DNA hybrid was unwound at a faster rate [18.3 versus 2.1 min−1, compare fR20 (−) to fD20 (−), Figure 4c]. We then tested if the increased unwinding rate for forked substrates was accompanied by a change in the ability of the enzyme to unwind these substrates under single cycle condition. In the presence of a 500-fold excess of a 36-nt-long single-strand DNA trap, which prevents the enzyme from re-associating if it dissociates from the substrate during the course of the unwinding reaction, no unwinding of fD20 was observed (Figure 4b, fD20, left panel). In contrast, ∼20% of fR20 was unwound under these conditions (Figure 4b, fR20, right panel; Figure 4c). This result provides two mechanistic insights. First it indicates that, in contrast to what is observed with DNA–DNA substrates, the enzyme is able to unwind the 20-mer forked RNA–DNA hybrid without dissociating. Since no unwinding of the unforked substrates was observed in single turnover experiments (data not shown), this result also suggests that the presence of a RNA–DNA fork helps stabilize the enzyme–substrate complex upon binding and/or during the initial steps of unwinding.
Binding of Pif1p to DNA–DNA and RNA–DNA substrates
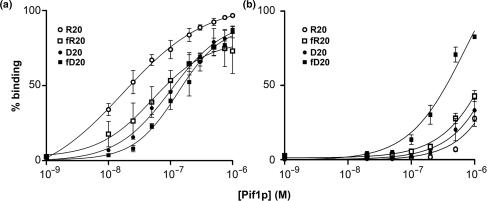
A possible explanation for the increased activity of Pif1p on RNA–DNA hybrids and on forked substrates could be that Pif1p binds preferentially to the substrates that are more easily unwound. Using a quantitative gel retardation assay, we compared the binding of Pif1p to forked and unforked substrates comprised of either DNA–DNA or RNA–DNA hybrids in absence of nucleotide, in presence of AMP–PNP, a nonhydrolyzable ATP analog (Figure 5) or in presence of the poorly hydrolyzable ATP analog ATPγS. In absence of nucleotide, Pif1p bound about as well to the fR20, fD20 and D20 substrates. The Kd for binding to these substrates was, respectively, 1.0 × 10−7 M (D20), 1.3 × 10−7 M (fD20) and 5.2 × 10−8 M (fR20). Pif1p bound most efficiently to the unforked RNA–DNA R20 substrate (Kd = 1.7 × 10−8 M), although this was not the most readily unwound substrate. These data indicate that preferential binding in absence of nucleotide does not account for the preferential unwinding of forked substrates, at least not in a way that is measurable by gel retardation (compare squares to circles, Figure 5a). However, Pif1p does show preferential binding to RNA–DNA substrates compared to their DNA–DNA counterparts, although this 2- to 5-fold preference is relatively small (Figure 5a, compare circles to circles and squares to squares). In presence of saturating amount of AMPPNP (Figure 5b) or in presence of ATPγS (data not shown), the binding affinity decreased significantly by more than 10-fold for all substrates except for the forked DNA substrate fD20. Thus, under conditions containing nucleotide analogs, which are the most similar to conditions used for unwinding assays, a DNA–DNA substrate had the highest binding affinity rather than the RNA–DNA substrates that were more readily unwound.
Figure 5.
Pif1p binding to forked and unforked substrates. Increasing amounts of Pif1p were incubated at 25°C with 50 pM nucleic acid substrate, in absence (a) or presence (b) of the nonhydrolyzable nucleotide analog AMPPNP, and analyzed on 12% non-denaturing gels as described in the Materials and Methods section. Data from three different experiments were averaged and plotted. Error bars represent the SD between the three experiments. Binding parameters are as follows; in the absence of a nucleotide, Kd(D20) = 1.0 × 10−7 M; Kd(R20) = 1.7 × 10−8 M; Kd(fD20) = 1.3 × 10−7 M; Kd(fR20) = 5.2 × 10−8 M; in the presence of AMPPNP, Kd(D20) = 2.0 × 10−6 M; Kd(R20) = 3.2 × 10−6 M; Kd(fD20) = 3.2 × 10−7 M; Kd(fR20) = 1.4 × 10−6 M.
Yeast RPA stimulates Pif1p activity but does not alleviate its preference for RNA–DNA substrates
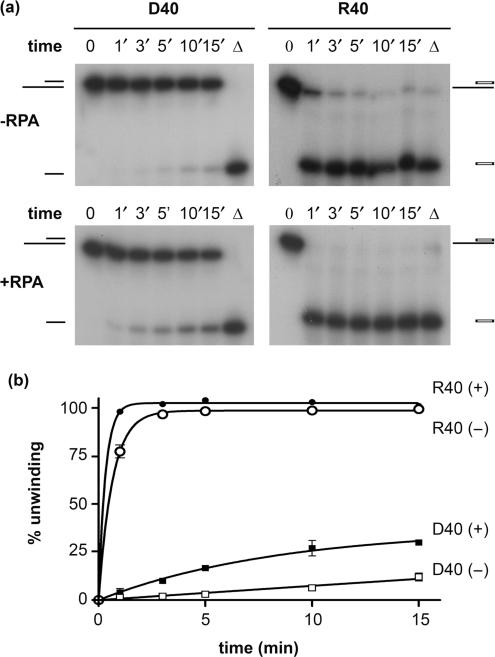
It has been shown for a number of helicases, including the yeast Srs2p and the human RecQ helicases, RECQ1, WRN and BLM, that addition of a sequence non-specific single-strand DNA-binding protein, such as E. coli SSB or the eukaryotic replication protein A (RPA), significantly enhances their ability to unwind longer substrates (1,19–22). The mechanistic explanation for this effect is that SSB or RPA prevents the re-annealing of partially unwound substrates by trapping the unwound region of the substrate in single-strand form. If, during the unwinding reaction, the DNA strand of the DNA–DNA substrate had a higher tendency than the RNA oligonucleotide to re-anneal to the opposite DNA strand, it would lead to an apparent slower unwinding rate for DNA–DNA substrates. We carried out the unwinding reaction of the 40-mer substrate (1 nM) in the presence or absence of saturating amounts of yeast RPA (5 μM) at 30°C and 100 nM Pif1p. As seen for other helicases, Pif1p unwinding activity was stimulated by RPA (Figure 6). RPA stimulation of Pif1p was detected with both DNA–DNA and RNA–DNA substrates, although stimulation of unwinding of the RNA–DNA substrate was relatively modest (Figure 6a and b). However, RPA did not alleviate the preference for RNA–DNA unwinding. Therefore, the relative inefficiency of Pif1p for unwinding DNA–DNA substrates is not due to a preferential re-annealing of the displaced DNA strand, but is due to an intrinsic lower processivity of the enzyme when unwinding DNA.
Figure 6.
Pif1p activity is stimulated by yeast RPA. (a) Unwinding of the D40 and R40 substrates in presence or absence of yeast RPA. 100 nM Pif1p was incubated with 1 nM nucleic acid substrate at 25°C, in the presence or absence of 50 ng/µl recombinant yeast RPA (generous gift from P. Sung). (b) Quantification of three independent experiments as shown in (a). Bars indicate SDs, which are present on every point. Amplitude and turnover rates are as follows. In the presence of RPA, A(D40) = 37 ± 5%, k(D40) = 0.1 ± 0.02 min−1; A(R40) = 100%, k(R40) = 3 ± 0.2 min−1; in the absence of RPA, A(R40) = 99 ± 1%, k(R40) = 1.5 ± 0.07 min−1. Reaction parameters for D40 were not determined.
Pif1p exists in solution as a monomer
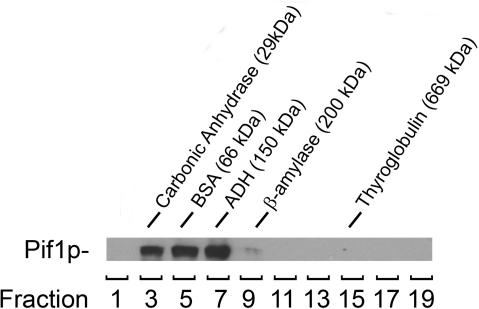
Pif1p purified from mitochondria has been shown to exist as a monomer in solution (16). We analyzed the nuclear recombinant Pif1p by glycerol gradient sedimentation. Pif1p was incubated in the standard reaction buffer at high concentration (5 µM) before being sedimented in a 15–40% glycerol gradient. The position of Pif1p was revealed by western blotting using an anti-Histidine tag monoclonal antibody (Figure 7). Pif1p eluted between carbonic anhydrase (29 kDa) and alcohol dehydrogenase (ADH, 150 kDa). The predicted molecular weight for His-tagged nuclear Pif1p is 95 kDa. Although Pif1p fractionated over a broad range in this assay, none of the protein could be detected above 150 kDa. This result suggests that the recombinant nuclear form of Pif1p exists as a monomer in solution, similar to what has been observed for mitochondrial Pif1p or for Pfh1p, the Schizosaccharomyces pombe homolog of Pif1p (16,23).
Figure 7.
Pif1p exists as a monomer in solution. Pif1p was subjected to centrifugation through a 15–40% glycerol gradient. The protein markers used as molecular weight standards were Carbonic Anhydrase (29 kDa; s20,w = 2.8 S), BSA (66 kDa, s20,w = 4.41 S), ADH (150 kDa, s20,w = 4.8 S), β-Amylase (200 kDa, s20,w = 8.9 S) and Thyroglobulin (669kDa, s20,w = 19.4 S). Two hundred and fifty micro liter fractions from the 5 ml gradient were analyzed for protein content by Coomassie gel staining to determine elution peak of molecular weight markers, and by anti-His western blotting for the detection of Pif1p.
DISCUSSION
In this study, we show that Pif1p has a preference for unwinding RNA–DNA hybrids. This preference was manifest at several levels. First, the rate of unwinding of a given RNA–DNA substrate was always faster than the rate of unwinding of the comparable DNA–DNA substrate. In the case of the linear 40-mer substrates, the rate of unwinding of the RNA–DNA substrate was almost two orders of magnitude higher than for the DNA–DNA 40-mer substrate (Figure 3). Second, the fraction of RNA–DNA substrate that was unwound was consistently higher. Indeed, Pif1p was able to unwind efficiently a 40-mer RNA–DNA substrate, while <5% of a 40-mer DNA–DNA substrate was unwound even in the presence of a 60-fold excess of Pif1p (Figure 3). Third, even though the unwinding reactions for RNA–DNA and DNA–DNA substrates shared the same optimal salt concentration, the RNA–DNA unwinding reaction was more resistant to increasing ionic strength in the physiological range (Figure 1).
The ability to unwind RNA–DNA hybrids has been reported for several DNA and RNA helicases, such as the E. coli DNA repair helicase UvrD (24), the RNA helicase NPH-II (25) and several replicative DNA helicases, including E. coli DnaB, the Methanothermobacter thermotrophicus MCM protein and the S. pombe MCM4, 6, 7 complex (26). However, of these helicases, only UvrD also exhibits preferential unwinding of RNA–DNA hybrids (24). Therefore, the preference for unwinding this type of substrate does not appear to be a common property of helicases. Indeed, Pif1p's preference for RNA–DNA hybrids contrasts with the emerging view on the mechanism of unwinding for several helicases that suggests that most helicases are not particularly sensitive to the chemical identity of the displaced strand. For example, studies with the bacteriophage T4 Dda helicase, a 5′–3′ SFI superfamily member, show that the rate-limiting step for unwinding is relatively insensitive to the chemical nature of the displaced strand (27). Similar conclusions have been reached for NPH-II, a 3′–5′ SFII RNA helicase (25). Therefore, the mechanism of nucleic acids recognition and/or unwinding by Pif1p are likely different from that of these prototypical SF1 and SF2 superfamily helicases.
Pif1p is also stimulated by forked substrates (16). Here we show that the stimulatory effect of the fork structure occurs with both DNA–DNA and RNA–DNA substrates (Figure 4). At 35°C, both the forked RNA–DNA hybrids and the forked DNA–DNA substrates were unwound very rapidly, in <20 s. In comparison, the unwinding of R20 was complete in 1 min under identical experimental conditions (Figure 3). Therefore, we can estimate that the presence of the fork increases the apparent kinetic rate of unwinding by at least 5-fold. Since Pif1p binding to forked substrates was not significantly higher than its binding to conventional tailed substrates, both in absence of nucleotide and in presence of the nucleotide analog AMPPNP (Figure 5) or ATPγS, this substrate specificity is probably not established by differential binding. However, it is possible that the fork structure increases the stability of the helicase–substrate complex and/or triggers the transition between the binding form of the enzyme and the translocating form upon ATP binding. A detailed analysis of the effects of nucleotide binding on nucleic acids binding by Pif1p will be needed to address this question.
When the unwinding of the two forked substrates was done at 25°C, the unwinding of both forked substrates was slower, but the unwinding of fR20 was still nine times faster than unwinding of fD20. The stimulatory effect of the fork was observed with both DNA–DNA and RNA–DNA substrates (Figure 4). This result indicates that the stimulation provided by the forked structure is additive to the stimulation provided by the top strand being made of RNA. The combined stimulatory effect of the forked structure and the RNA strand on Pif1p activity has an important consequence for Pif1p helicase activity as Pif1p was able to unwind the forked 20-mer RNA–DNA hybrid in a single turnover reaction (Figure 4). Interestingly, the calculated rate of unwinding for the single turnover reaction was ∼3 times lower that the calculated rate for multiple cycle conditions (5.2 min−1 versus 18.3 min−1; Figure 4), which indicates that the rate-limiting step is likely to be different in the two reaction conditions. Pif1p is a poorly processive helicase in vitro. An intriguing possibility is that enzyme stalling and/or dissociation from substrates accounts for the low rate of unwinding in single cycle conditions. In multiple cycle conditions, alignment of multiple Pif1p monomers ‘pushing’ each other forward along the substrate could have a stimulatory effect on the overall kinetic rate of the reaction. Increased activity when multiple helicases are bound to a substrate has been shown with the Dda helicase, another monomeric helicase, in a different experimental setting (28). A detailed study of Pif1p kinetic mechanism is necessary to resolve this question.
Since Pif1p does not bind RNA (12), an interesting possibility is that during the unwinding of DNA–DNA substrates, Pif1p interacts with both strands of the duplex. If the interaction with the upper DNA strand counteracts the unwinding reaction, whether it is achieved by translocation of the enzyme or another mechanism that remains to be identified, this interaction could result in lower unwinding rates for DNA–DNA hybrids. This scenario could arise if the active form of Pif1p is a dimer with each monomer attached to a different DNA strand. However, we did not detect self-association of the protein in vitro even when the enzyme was incubated at 50 times the concentration that was used in helicase assays (Figure 6), suggesting that nuclear Pif1p is active as a monomer. If Pif1p is active as a monomer and dissociates from the bottom strand, it could potentially re-associate with either the lower or the partially unwound upper strand if it is made of DNA, which would also slow down unwinding of the DNA–DNA substrate.
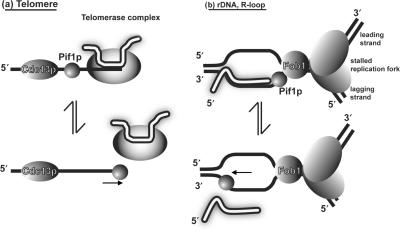
Taken together, our results indicate that Pif1p is sensitive to both the structure of the duplex region of the substrate, preferring forked to linear duplexes, and to the chemical nature of the top strand, preferring to displace a RNA strand. How do these data inform our understanding of Pif1p function in vivo? The best studied function of Pif1p is its inhibition of telomerase lengthening of existing telomeres and telomere addition to double-strand breaks (8,10–12,29). The preference of Pif1p for unwinding forked RNA–DNA hybrids in vitro supports the hypothesis that Pif1p inhibits telomerase by unwinding the RNA–DNA hybrid formed between telomerase RNA and the telomeric DNA end. Indeed, the TLC1-RNA annealed to the telomeric single-stranded DNA resembles a forked RNA–DNA hybrid (Figure 8a) (12).
Figure 8.
Hypothetical RNA–DNA hybrid substrates for Pif1p. Two possible RNA–DNA hybrids are shown. RNA strands are shown in white, DNA strands in black (a) Pif1p acts catalytically to remove telomerase from DNA ends, possibly by unwinding the RNA–DNA hybrid formed by TLC1 and the end of the telomere (7,8,12). (b) Pif1p could be involved in removing stable RNA–DNA hybrids in front of the replication fork. At the rDNA locus (pictured), Pif1p could help remove rDNA transcripts that approach the RFB established by Fob1p (36). Elsewhere, the R-loop could originate from a transcribed gene or an incompletely processed Okazaki fragment RNA primer (37,38). An alternative hypothesis has been proposed for the role of Pif1p in Okazaki fragment processing that does not involve RNA–DNA hybrid unwinding, in which Pif1p extends the DNA flap in front of the lagging strand polymerase (14).
In addition to its telomeric functions, nuclear Pif1p helps maintain the RFB in the rDNA (13). Given its preference for forked RNA–DNA substrates (Figure 4) and its association with rDNA in vivo (13), Pif1p might sit at a fork stalled at the RFB and act on the rRNA transcript in a manner that prevents the transcript from running into the RFB (Figure 8b). Nuclear Pif1p also acts in cooperation with the helicase/endonuclease Dna2p. Dna2p is a helicase/nuclease that is required for Okazaki fragment maturation (30). While dna2Δ cells are dead, pif1Δ dna2Δ are viable (although not at high temperatures) and have improved repair and replication capabilities. It has been proposed that Pif1p helps extend the DNA flap generated during Okazaki fragment maturation (14), a proposal consistent with Pif1p's preference for forked substrates. Pif1p is also required in cells lacking topoisomerase III activity, and this requirement is dependent upon an active Sgs1p helicase and homologous recombination (15). Again, this role can be explained by Pif1p acting on the non-linear DNA–DNA hybrids that arise during recombinational repair. Finally, Pif1p is also important for maintenance of mitochondrial DNA (5,31), especially in the presence of oxidative damage or the intercalating agent ethidium-bromide (32–34). The increased mitochondrial DNA breakage detected in ethidium bromide treated pif1 cells occurs at discrete sites, suggesting that Pif1p action is required only at specific loci in the mitochondrial genome (34).
Pif1p might carry out its role at non-telomeric sites by unwinding forked DNA–DNA hybrids. However, it is tempting to speculate from its in vitro substrate preferences that in vivo Pif1p acts primarily or even exclusively on RNA–DNA hybrids. One interesting possibility is that Pif1p participates in the removal of the RNA–DNA hybrids that form during transcription and which, if not removed, lead to the formation of R-loops, which can trigger replication arrest, fork breakage and recombination (35). Because replicative helicases from various organisms are able to unwind RNA–DNA hybrids in vitro (26), in most situations, replicative helicases are likely able to remove most RNA–DNA hybrids that are present in front of the replication fork. Nonetheless, it is possible that particular sequences in the genome are prone to the formation of stable RNA–DNA hybrids that require a dedicated helicase like Pif1p for efficient processing. This model would reconcile the observation that Pif1p acts at discrete loci of the genome (3,34) with its preference for RNA–DNA hybrids (this study). The fact that the enzyme is involved in maintenance of mitochondrial and rDNA, two loci that contain highly transcribed genes, is compatible with this hypothesis. Given the evolutionary conservation of the PIF1 family helicases (3,4,8), it will be interesting to determine if other members of the family also act preferentially on RNA–DNA hybrids, as a first step to identify their in vivo substrates.
ACKNOWLEDGEMENTS
We thank P. Sung for reagents, Olga Amosova for help with melting profiles measurements and Sarah Aubert, Gary LeRoy, Maria Mateyak, Stefan Pinter and Yun Wu for critical reading of the manuscript. This work was supported by NIH grant R37GM 026938. J.-B.B. was supported in part by a fellowship from the Association pour la Recherche contre le Cancer and a fellowship from the New Jersey Commission on Cancer Research. Funding to pay the Open Access publication charges for this article was provided by NIH grant R37GM 026938.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sharma S, Doherty KM, Brosh R.M., Jr Mechanisms of RecQ helicases in pathways of DNA metabolism and maintenance of genomic stability. Biochem. J. 2006;398:319–337. doi: 10.1042/BJ20060450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delagoutte E, von Hippel PH. Helicase mechanisms and the coupling of helicases within macromolecular machines. Part II: integration of helicases into cellular processes. Q. Rev. Biophys. 2003;36:1–69. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]
- 3.Bessler JB, Torres JZ, Zakian VA. The Pif1p subfamily of helicases: region-specific DNA helicases? Trends Cell. Biol. 2001;11:60–65. doi: 10.1016/s0962-8924(00)01877-8. [DOI] [PubMed] [Google Scholar]
- 4.Boule JB, Zakian VA. Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res. 2006;34:4147–4153. doi: 10.1093/nar/gkl561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foury F, Kolodynski J. pif mutation blocks recombination between mitochondrial rho+ and rho- genomes having tandemly arrayed repeat units in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1983;80:5345–5349. doi: 10.1073/pnas.80.17.5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lahaye A, Stahl H, Thines-Sempoux D, Foury F. PIF1: a DNA helicase in yeast mitochondria. EMBO J. 1991;10:997–1007. doi: 10.1002/j.1460-2075.1991.tb08034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schulz VP, Zakian VA. The saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- 8.Zhou J, Monson EK, Teng SC, Schulz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- 9.Autexier C, Lue NF. The structure and function of telomerase reverse transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 10.Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- 11.Mangahas JL, Alexander MK, Sandell LL, Zakian VA. Repair of chromosome ends after telomere loss in Saccharomyces. Mol. Biol. Cell. 2001;12:4078–4089. doi: 10.1091/mbc.12.12.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boule JB, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- 13.Ivessa AS, Zhou JQ, Zakian VA. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell. 2000;100:479–489. doi: 10.1016/s0092-8674(00)80683-2. [DOI] [PubMed] [Google Scholar]
- 14.Budd ME, Reis CC, Smith S, Myung K, Campbell JL. Evidence suggesting that Pif1 helicase functions in DNA replication with the Dna2 helicase/nuclease and DNA polymerase delta. Mol. Cell. Biol. 2006;26:2490–2500. doi: 10.1128/MCB.26.7.2490-2500.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner M, Price G, Rothstein R. The absence of Top3 reveals an interaction between the Sgs1 and Pif1 DNA helicases in Saccharomyces cerevisiae. Genetics. 2006;174:555–573. doi: 10.1534/genetics.104.036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lahaye A, Leterme S, Foury F. PIF1 DNA helicase from Saccharomyces cerevisiae. Biochemical characterization of the enzyme. J. Biol. Chem. 1993;268:26155–26161. [PubMed] [Google Scholar]
- 17.Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- 18.Sugimoto N, Nakano S.-I, Katoh M, Matsumura A, Nakamuta H, Ohmichi T, Yoneyama M, Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- 19.Brosh R.M., Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 20.Van Komen S, Reddy MS, Krejci L, Klein H, Sung P. ATPase and DNA helicase activities of the Saccharomyces cerevisiae anti-recombinase Srs2. J. Biol. Chem. 2003;278:44331–44337. doi: 10.1074/jbc.M307256200. [DOI] [PubMed] [Google Scholar]
- 21.Cui S, Arosio D, Doherty KM, Brosh R.M., Jr, Falaschi A, Vindigni A. Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 2004;32:2158–2170. doi: 10.1093/nar/gkh540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doherty KM, Sommers JA, Gray MD, Lee JW, von Kobbe C, Thoma NH, Kureekattil RP, Kenny MK, Brosh R.M., Jr Physical and functional mapping of the replication protein a interaction domain of the werner and bloom syndrome helicases. J. Biol. Chem. 2005;280:29494–29505. doi: 10.1074/jbc.M500653200. [DOI] [PubMed] [Google Scholar]
- 23.Ryu G.-H, Tanaka H, Kim D.-H, Kim J.-H, Bae S.-H, Kwon Y.-N, Rhee JS, MacNeill SA, Seo Y.-S. Genetic and biochemical analyses of Pfh1 DNA helicase function in fission yeast. Nucleic Acids Res. 2004;32:4205–4216. doi: 10.1093/nar/gkh720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matson SW. Escherichia coli DNA helicase iI (uvrD gene product) catalyzes the unwinding of RNA:DNA Hybrids in vitro. Proc. Natl Acad. Sci. USA. 1989;86:4430–4434. doi: 10.1073/pnas.86.12.4430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kawaoka J, Pyle AM. Choosing between DNA and RNA: the polymer specificity of RNA helicase NPH-II. Nucleic Acids Res. 2005;33:644–649. doi: 10.1093/nar/gki208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin J.-H, Kelman Z. The replicative helicases of Bacteria, Archaea, and Eukarya can unwind RNA-DNA hybrid substrates. J. Biol. Chem. 2006;281:26914–26921. doi: 10.1074/jbc.M605518200. [DOI] [PubMed] [Google Scholar]
- 27.Tackett AJ, Morris PD, Dennis R, Goodwin TE, Raney KD. Unwinding of unnatural substrates by a DNA helicase. Biochemistry. 2001;40:543–548. doi: 10.1021/bi002122+. [DOI] [PubMed] [Google Scholar]
- 28.Byrd AK, Raney KD. Protein displacement by an assembly of helicase molecules aligned along single-stranded DNA. Nat. Struct. Mol. Biol. 2004;11:531–538. doi: 10.1038/nsmb774. [DOI] [PubMed] [Google Scholar]
- 29.Eugster A, Lanzuolo C, Bonneton M, Luciano P, Pollice A, Pulitzer JF, Stegberg E, Berthiau AS, Forstemann K, et al. The finger subdomain of yeast telomerase cooperates with Pif1p to limit telomere elongation. Nat. Struct. Mol. Biol. 2006;13:734–739. doi: 10.1038/nsmb1126. [DOI] [PubMed] [Google Scholar]
- 30.Hui IK, Robert AB. The protein components and mechanism of eukaryotic Okazaki fragment maturation. Crit. Rev. Biochem. Mol. Biol. 2003;38:433–452. doi: 10.1080/10409230390259382. [DOI] [PubMed] [Google Scholar]
- 31.Foury F, Dyck EV. A PIF-dependent recombinogenic signal in the mitochondrial DNA of yeast. EMBO J. 1985;4:3525–3530. doi: 10.1002/j.1460-2075.1985.tb04112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doudican NA, Song B, Shadel GS, Doetsch PW. Oxidative DNA damage causes mitochondrial genomic instability in Saccharomyces cerevisiae. Mol. Cell. Biol. 2005;25:5196–5204. doi: 10.1128/MCB.25.12.5196-5204.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Rourke TW, Doudican NA, Mackereth MD, Doetsch PW, Shadel GS. Mitochondrial dysfunction due to oxidative mitochondrial DNA damage is reduced through cooperative actions of diverse proteins. Mol. Cell. Biol. 2002;22:4086–4093. doi: 10.1128/MCB.22.12.4086-4093.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng X, Dunaway S, Ivessa AS. The role of Pif1p, a DNA helicase in Saccharomyces cerevisiae, in maintaining mitochondrial DNA. Mitochondrion. 2007;7:211–222. doi: 10.1016/j.mito.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 35.Prado F, Aguilera A. Impairment of replication fork progression mediates RNA polII transcription-associated recombination. EMBO J. 2005;24:1267–1276. doi: 10.1038/sj.emboj.7600602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguilera A. The connection between transcription and genomic instability. EMBO J. 2002;21:195–201. doi: 10.1093/emboj/21.3.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arudchandran A, Cerritelli S, Narimatsu S, Itaya M, Shin DY, Shimada Y, Crouch RJ. The absence of ribonuclease H1 or H2 alters the sensitivity of Saccharomyces cerevisiae to hydroxyurea, caffeine and ethyl methanesulphonate: implications for roles of RNases H in DNA replication and repair. Genes Cells. 2000;5:789–802. doi: 10.1046/j.1365-2443.2000.00373.x. [DOI] [PubMed] [Google Scholar]