Abstract
Homing endonuclease genes (HEGs) are ‘selfish’ genetic elements that combine the capability to selectively disrupt specific gene sequences with the ability to rapidly spread from a few individuals to an entire population through homologous recombination repair events. Because of these properties, HEGs are regarded as promising candidates to transfer genetic modifications from engineered laboratory mosquitoes to wild-type populations including Anopheles gambiae the vector of human malaria. Here we show that I-SceI and I-PpoI homing endonucleases cleave their recognition sites with high efficiency in A. gambiae cells and embryos and we demonstrate HEG-induced homologous and non-homologous repair events in a variety of functional assays. We also propose a gene drive system for mosquitoes that is based on our finding that I-PpoI cuts genomic rDNA located on the X chromosome in A. gambiae, which could be used to selectively incapacitate X-carrying spermatozoa thereby imposing a severe male-biased sex ratio.
INTRODUCTION
Mosquito species of the Anopheles gambiae complex represent the major vectors of human malaria and they pose an enormous burden on global health and economies. Every year 300–500 million people are infected by malaria and over a million people die as consequence of Plasmodium parasite infections (1). While many insect pests have long been successful targeted with population control measures such as insecticides or release of sterile males (2), for others, including A. gambiae, classical control measures have largely failed to deliver long-term solutions. Disease endemic countries often do not have the economic resources and the logistics to sustain control efforts like the massive and prolonged use of insecticides. New control strategies that are affordable, easy to implement and sustainable are desperately needed.
This global health problem has prompted an unprecedented effort aimed at generating new molecular tools and a better understanding of the biology and the genetics of Anopheline mosquitoes that culminated in the sequencing of the A. gambiae genome (3) and development of gene transfer technology for a series of vectors species (4,5). These molecular advances have made it possible to express genes that can block the transmission of Plasmodium in model systems (6–8) or express traits facilitating the implementation of sterile insect techniques for vector control (9).
The translation of these achievements in suitable control measures still represents a major scientific and technical challenge. Genetically modified mosquitoes carrying a desired trait such as malaria refractoriness would need to be released on a gigantic scale given the vast numbers of these insects and the wide areas that are inhabited by vectors of human tropical diseases. Therefore, a mechanism must be developed to spread the desired genetic modification from a few laboratory-reared mosquitoes to a large fraction of the wild-type vector population (10,11). Naturally occurring ‘selfish’ genetic elements that have non-mendelian inheritance mechanisms and spread through populations even when they provide no benefit to the host organism (12) have been proposed to transform wild-type mosquito populations.
Homing endonuclease genes (HEGs) are highly specific DNA endonucleases found in some viruses, bacteria and eukaryotes. The endonuclease promotes the movement of its encoding DNA from one allele to the other by creating a double-strand break (DSB) at a specific, long (15–40 bp) target site in an allele that lacks the HEG. Homologous DNA repair then copies the HEG to the cut chromosome in a process called ‘gene conversion’ (13,14).
The observation that HEGs can be engineered to cleave novel DNA sequences (15–18) offers a multitude of opportunities to utilize these elements for mosquito control. For example, HEGs could be used to disrupt genes regulating the ability of Anopheles mosquitoes to function as efficient vectors for Plasmodium parasites, or to drive recombinant refractory genes through a mosquito population, rendering them unable to transmit malaria. Alternatively, HEGs designed to target an essential mosquito gene or a gene required for female fertility could be utilized to introduce a genetic load on the population leading to population size reduction or collapse (19). More recently, it has been suggested that a harmful selfish element put under the control of a promoter which is active in individuals susceptible to Plasmodium infection but inactive in refractory individuals should drive alleles causing refractoriness through the population (20). Finally, HEGs could be used to bias the sex ratio towards males, using an endonuclease that targets X-linked sequences and is expressed during male spermatogenesis from the Y chromosome (19).
To investigate the feasibility of using HEGs as driving genetic element in mosquitoes, we have analysed the activity of ectopic HEGs in both A. gambiae cells and embryos, using experimental systems that are highly predictive of in vivo behaviour of mobile genetic elements (21,22). We determined the activity of two well-characterized HEGs, I-PpoI (a member of the His-Cys box family of endonucleases from the slime mold Physarum polycephalum) (23–25) and I-SceI (a LAGLIDADG class endonuclease originally isolated from Saccharomyces cerevisiae mitochondria) (26–28), both of which have been used in a variety of organisms (including Drosophila) to induce DNA DSBs (29–32). We systematically analysed the nature of HEG-mediated integration and recombination events in A. gambiae and the effect of expressing these endonucleases on cell proliferation.
MATERIALS AND METHODS
Plasmids and cloning
The target plasmids pBC/SacRB S1/S2/S3 and pBC/SacRB P1 were constructed as follows: pBC/SacRB (21) was cut with SalI and PstI, ends filled in with T4 DNA polymerase and religated to remove a redundant EcoRI site. Linkers were created by annealing 5′ phosphorylated oligonucleotides (S1: AATTCATTACCCTGTTATCCTAG and AATTCTAGGGATAACAGGGTAATG; S2: AATTCGATAGGGATAACAGGGTAATTG and AATTCAATTACCCTGTTATCCCTATCG; S3: AATTCAATTACCCTGTTATCCCTACCG and AATTCGGTAGGGATAACAGGGTAATTG; P1: AATTCCGCTACCTTAAGAGAGTCG and AATTCGACTCTCTTAAGGTAGCGG), which were cloned into the now unique EcoRI site in the SacRB CDS. Selection against SacRB was performed in LB agar lacking NaCl containing 15% sucrose (w/v) and chloramphenicol (25 μg/ml).
To create pSL-SacRBTet, the minimal 1.5 kb tetracycline (Tet) resistance cassette from pBR322 was amplified with the primers TTCAAGAATTCTCATGTTTGACAG and ATGAATTCTGCTAACCAGTAAGGCAACC and cloned into pBC/SacRB with EcoRI. The Tet cassette replaces the I-SceI site and disrupts the SacRB gene. The 3.4 kb SacRBTet cassette was moved to pSLfa1180fa (33) using XhoI/XbaI to create pSL-SacRBTet.
To create pDR-CMV-GFP, the CMV promoter from pEGFP-Ppo was amplified with the primers AAAGGGCCCTAGTTATTAATAGTAATCAATTACGGGGTCATTAG and AAAGAATTCGATCTGACGGTTCACTAAACCAGCTC and cut with ApaI and EcoRI. The fragment was ligated into partially ApaI and EcorI digested pDR-GFP (34). This replaces the 3′ part of the chicken β-actin promoter with CMV. To remove the remaining sequences of the chicken β-actin promoter, the resulting vector was cut with SnaBI and religated.
PSL-Act-EGFP was constructed in pSLFa1180fa to contain the 2.5 kb Drosophila Actin5C promoter driving EGFP (BamHI/XbaI fragment) and the Drosophila Hsp70 terminator.
Cell culture and transfections
Cells from the stable anchorage-dependent A. gambiae cell line, Suakoko 4 (Sua 4.0) (Müller,H.M. et al., 1999) were cultured in Schneider's Drosophila medium (Invitrogen) supplemented with 10% FCS (Invitrogen) and 200 U/ml penicillin and 200 μg/ml streptomycin sulphate (Invitrogen) in a cooled incubator at 27°C.
In vivo HEG activity assays were performed by lipid-mediated transient transfections (Effecten, Qiagen) of 1–3 × 105 cells with 2 μg/ml (culture volume) recipient plasmid and 4 μg/ml donor plasmid. When necessary, cells were heat-shocked for 1 h at 41°C, 24 h post-transfection to induce expression of I-SceI from the Drosophila Hsp70 promoter on the pP[v+, 70 I-SceI] plasmid. Total DNA was extracted 48 h post-transfection (Promega Wizard Genomic DNA purification Kit) and re-suspended at 25–40 μl. This preparation was used to transform Escherichia coli DH5α strain.
Embryo microinjections and preparation of low molecular weight DNA
Anopheles gambiae adult females (G3 strain) were allowed to deposit their embryos 72 h after a blood meal on a moist filter paper. Embryos were injected 60–120 min after oviposition, essentially as described (35). Embryos were injected with a mixture of I-SceI donor plasmid pP[v+, 70 I-SceI] (400 μg/ml), Tet donor plasmid pSL-SacRBTet (500 μg/ml) and HEG target plasmid pBC/SacRB S1 (500 μg/ml). Between 150 and 200 embryos were microinjected and 12 h later embryos were heat-shocked for 1 h at 41°C and left to recover for 2 h. Low molecular weight DNA was extracted (21), re-suspended in 20 μl and 150 ng of the recovered DNA was used to transform E. coli DH5α strain.
Southern blot and primer extension
Genomic DNA was digested with ClaI in the presence and absence of I-PpoI. As a probe we used a 2 kb rDNA PCR fragment amplified from genomic DNA using the primers GCCGAAGCAATTAGCCCTTAAAATGGATG and CACCAGTAGGGTAAAACTAACCTGTCTCACG. The probe was P32 labelled using the High Prime DNA labelling kit (Roche) and purified with ProbeQuant™ G-50 columns (GE Healthcare). For primer extension, genomic DNA was digested with HincII (a.k.a. HindII). The reaction was performed essentially as described (36) using the 5′ P32-labelled primer rPrex GTTAATCCATTCATGCGCGTCACTAATTAG and vent (exo-) polymerase (New England Biolabs). The reaction products were resolved on a 6% denaturing polyacrylamide gel. Results for both experiments were visualized using a FUJIFILM-FLA-5000 Phosphoimager (Fuji Photo Film Co. Ltd, Stamford, CT, USA). For in vitro digestions, we used commercially available I-PpoI (Promega) and I-SceI (New England Biolabs) enzymes.
Cell proliferation assay
Sua 4.0 cells were transfected with either of the two endonuclease plasmids (4 μg/ml) together with pIB/V5-His (2 μg/ml), conferring resistance to blasticidin (Invitrogen). Forty-eight hours post-transfection, blasticidin was supplemented to complete medium at 50 μg/ml. Cells were incubated in blasticidin for an initial proliferation period of 5 days, at which point they were harvested and re-seeded at 1.5 × 105 cells/ml and grown for a further 5 days. Transfections were performed in triplicates and cell numbers for each were counted in four replicates.
GFP gene conversion assay
Sua 4.0 cells were transfected with pDR-CMV-GFP (2 μg/ml) in the presence or absence of donor plasmid pP[v+, 70 I-SceI] (4 μg/ml). Twenty-four hours post-transfection, cells were heat-shocked for 1 h at 41°C. Gene conversion was measured by fluorescence activated cell sorting (FACS) 72 h post-transfection using a Beckman FACS Calibur; 50 000 size-dependent gated events were analysed for GFP fluorescence. Data were analysed using the FlowJo software package. Transfection efficiency was assessed by parallel co-transfections with an Actin5C-DsRed plasmid.
Fluorescence microscopy
Anopheles gambiae Sua 4.0 cells were transfected with pEGFP-Ppo, pEGFP-Ppo H98A and the control pSL-Act-GFP, and 48 h later cells were fixed for 5 min in 4% paraformaldehyde and permiabilized for 10 min with 0.1% Triton X-100. Nuclei were stained with DAPI (2 ng/μl) and actin filaments with Alexa546-phalloidin (1 U/ml, Invitrogen). Cell micrographs were taken at ×40 magnification using a Zeiss widefield microscope.
RESULTS
Development of a reporter system for HEG activity
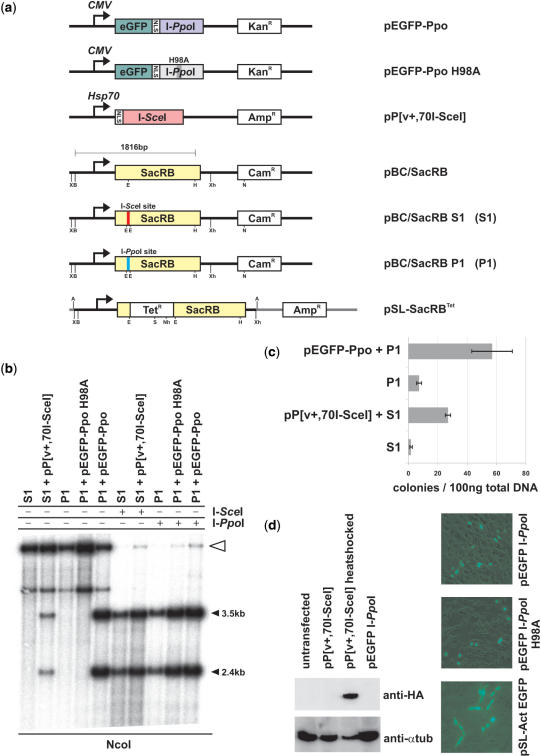
To assess the functionality of HEGs in Anopheles cells, we developed a rapid and efficient reporter system to score HEG-mediated activity (site specific cleavage and homologous recombination events) in both A. gambiae cells and embryos. This assay, based on the interplasmid transposition assay utilized in insect cells and embryos to assess the activity of transposable elements (21), utilizes two sets of plasmids (Figure 1a). A donor plasmid directs the production of either I-SceI [pP(v+,70I-SceI)] or I-PpoI (pEGFP-PpoI), and the target plasmid (pBC/SacRB) contains the bacterial suicidal gene SacRB, coding for levansucrase, engineered to contain either the I-SceI or the I-PpoI recognition sequences. SacRB catalyses the hydrolysis of sucrose and the synthesis of levans, high-molecular-weight fructose polymers that accumulate in the periplasmic space, and are toxic to gram negative bacteria (37,38). HEG cleavage of its recognition sequence results in the inactivation of SacRB genes, which is then detected in bacteria transformed with plasmid DNA recovered from transiently transfected cells and injected embryos. Negative selection against the functional SacRB gene in medium containing sucrose is accompanied by positive selection for the chloramphenicol (Cam) resistance marker, which is also present on pBC/SacRB (Figure 1).
Figure 1.
I-SceI and I-PpoI expression constructs and their activity in A. gambiae cells. (a) Maps of HEG expression and target vectors used in the interplasmid activity assay. CMV, cytomegalovirus promoter; Hsp70, Drosophila heatshock protein 70 promoter; eGFP enhanced green fluorescent protein; SacRB, levansucrase gene; X, XbaI; B, BamHI; E, EcoRI; H, HindIII; Xh, XhoI; N, NcoI; A, AscI; S, SalI; Nh, NheI; KanR, kanamycin resistance cassette; CamR, chloramphenicol resistance cassette; TetR, tetracycline resistance cassette; AmpR, ampicilin resistance cassette; NLS, nuclear localization signal. (b) Analysis of I-SceI and I-PpoI activity in Sua4.0 cells by Southern blot. Cells were co-transfected with I-SceI or I-PpoI expression and target plasmids. Total DNA from these cells was digested with NcoI and hybridized with the 1.8 kb BamHI/HindIII fragment of pBC/SacRB as a probe (Lanes 1–5). In lanes 6–10, DNA was also digested with I-SceI or I-PpoI in vitro. The white arrow marks linearized full-length plasmids (lanes 1–5) and plasmids resistant to in vitro endonuclease cleavage. (c) Number of Cam/Suc-resistant colonies after bacterial transformation of plasmid DNA isolated from transfected Sua4.0 cells. Co-transfection of the target plasmids together with I-SceI and I-PpoI expression vectors increases the number of colonies. (d) Expression of I-SceI and I-PpoI in A. gambiae Sua 4.0 cells. Western blot of transfected cells using anti-hemagglutinin (anti-HA) and anti-α tubulin (left). The I-PpoI-GFP fusion proteins show the expected nuclear localization unlike the Actin5C-driven GFP control (right).
The I-SceI or I-PpoI recognition sites were inserted in the SacRB sequence after Glu67, downstream of the signal peptide required for secretion (39), at a position predicted not to be essential (40). We assessed the effect of inserting several recognition site variants in different frames on SacRB activity. None of the inserted amino acid variants (pBC/SacRB S1: ITLLSL, pBC/SacRB S2: DRDNRVI, pBC/SacRB S3: NYPVIPI for I-SceI; pBC/SacRB P1: ATLRE for I-PpoI) interfered with SacRB function as inferred by the continued inability of bacteria transformed with these vectors to grow on Cam supplemented with 15% sucrose (data not shown). We therefore concluded that the SacRB gene is tolerant to amino acid insertions at this position and a variety of recognition sequences of natural and reengineered HEGs could be inserted and tested using this approach.
I-SceI and I-PpoI homing endonuclease activity in A. gambiae cell lines
We transfected the plasmids pP[v+,70I-SceI] and pEGFP-Ppo together with their corresponding target pBC/SacRB S1 and pBC/SacRB P1 into A. gambiae Sua4.0 cell lines (Figure 1a). The plasmid pP[v+,70I-SceI] (30) expresses the I-SceI ORF with an N-terminal SV40 NLS and hemagglutinin (HA) tag under the control of the Drosophila Hsp70 promoter, which directs a significant inducible expression of I-SceI in Sua4.0 cells (Figure 1d, lanes 2 and 3). The plasmid pEGFP-Ppo expresses the EGFP-I-PpoI fusion protein containing a SV40 NLS (Figure 1a and d) under the control of the CMV promoter. An inactive variant pEGFP-Ppo H98A (Raymond Monnat personal communication) was used as a control. Cells were heat-shocked for 1 h at 41°C 24 h post transfection and DNA extracted after an additional 24 h. Under these experimental conditions, we observed that the target plasmids were cut in vivo by the corresponding endonuclease (Figure 1b). Digestion of DNA extracted from transfected cells with purified endonucleases in vitro revealed the presence of a fraction of endonuclease resistant plasmids (Figure 1b, lanes 6–10, white arrow), possibly generated by non-homologous religation of HEG-mediated cleavage events.
To recover and analyse non-homologous end joining (NHEJ) events generated in mosquito cells, total DNA (which includes plasmid DNA) extracted from transfected cells was used to transform bacterial cells plated on Cam or Cam/Suc selective media. Compared to control experiments (target plasmid only), transfection of mosquito cells with I-SceI- or I-PpoI expressing plasmids increases the number of Cam/Suc resistant clones by 15- and 8-fold, respectively (Figure 1c). By comparing the colony numbers on Cam and Cam/Suc plates of bacteria transformed with recovered DNA from A. gambiae cells, we found that ∼0.5–2% of recovered plasmids allowed growth on Cam/Suc as compared to Cam alone. This indicates that although HEG cleavage is efficient (Figure 1b) only a small number of these plasmids are subsequently religated.
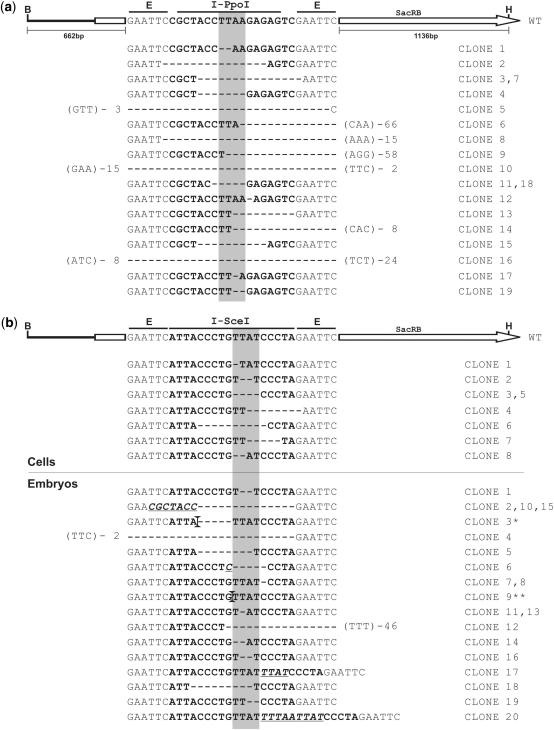
Plasmids from Cam/Suc-resistant bacterial cells were isolated and digested with BamHI and HindIII endonucleases. Only plasmids showing a 1.8 kb band, indicating the presence of an intact SacRB gene (Figure 1a), were analysed by sequencing (Figure 2). This step was undertaken to ensure that sucrose resistance is due to HEG-mediated disruption of SacRB, as we occasionally observed the growth of Cam/Suc-resistant bacteria in the absence of a HEG expression vector (Figure 1c). Digestion of plasmids with BamHI/HindIII in the presence of I-SceI or I-PpoI showed that all recovered clones were also resistant to endonuclease cleavage in vitro (data not shown). The sequence of the regions surrounding the HEG cleavage sites was analysed in plasmids recovered from Cam/Suc-resistant bacteria (Figure 2). All 27 sequenced clones showed deletion events of variable sizes, ranging from 1 to 80 bp, close to the predicted HEG cleavage site. We did not observe any nucleotide insertions, contrary to reports of NHEJ repair products in human cell lines (41,42).
Figure 2.
DNA sequence analysis of clones created by HEG cleavage and non-homologous repair. (a) Clones of pBC/SacRB P1 isolated from Sua4.0 cells after co-transfection with I-PpoI expression vector pEGFP-Ppo. (b) Clones of pBC/SacRB S1 isolated from Sua4.0 cells (top) and G3 embryos (bottom) after co-transfection/injection with I-SceI expression vector pP[v+,70I-SceI]. The native 15 bp minimal I-PpoI and 18-bp I-SceI recognition sites are shown on top of the isolated clones in the context of the SacRB gene. Deleted nucleotides are indicated by dashes (-), inserted nucleotides are underlined. If deletions extend beyond the EcoRI sites flanking the HEG recognition sites the number of base pairs deleted is indicated and the first 3 bp after the deletion are shown in brackets. The shaded area indicates the 4 bp between the HEG cleavage positions on both DNA strands. Larger insertions are marked by vertical bars: (*) Insertion of 66 bp partially homologous to A. gambiae genome. (**) Insertion of 43 bp. B, BamHI; E, EcoRI; H, HindIII.
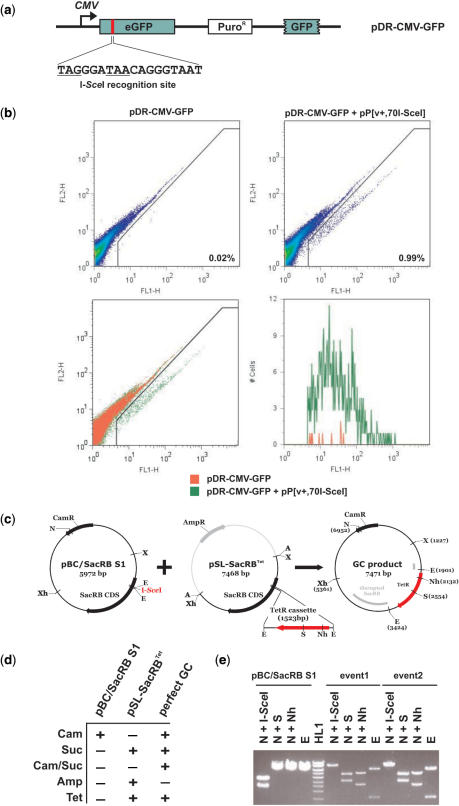
I-SceI expression induces homologous repair events in A. gambiae cells
Copying of HEGs from one allelic site to the other requires that cleavage of the target site is followed by gene conversion events using the HEG-containing allele as a repair template. We therefore adapted our reporter system to test for the occurrence HEG-induced gene conversion events in mosquito cells. We used plasmid pDR-CMV-GFP (34) (Figure 3a), which contains the CMV promoter 5′ of a non-functional eGFP gene. In this gene, 11 bp of the GFP CDS have been deleted and replaced by an I-SceI site which also introduces two stop codons. The plasmid also carries a second promoterless and inactive GFP locus, which lacks 220 bp of the GFP C terminus, functioning as a repair template. Cells were transfected with pDR-CMV-GFP or co-transfected with pDR-CMV-GFP and pP[v+,70I-SceI]. As expected, transfection of mosquito cells with pDR-CMV-GFP alone did not result in a high rate of spontaneous repair of the GFP gene as shown by the low frequency (0.02%) of fluorescent cells (Figure 3b), whereas we found that co-transfection with pP[v+,70I-SceI] increased the number of GFP+ cells 50-fold (0.99%). Adjusted for transfection efficiency, GFP positive cells represent 0.15% (pDR-CMV-GFP) and 10.9% (pDR-CMV-GFP + pP[v+,70I-SceI]) of transfected cells. Co-transfection with the I-SceI expressing plasmid increases the average and peak intensity of fluorescence of GFP+ cells (Figure 3b, lower right panel and data not shown), thus suggesting the presence of a higher number of repaired plasmids per cell in these experiments.
Figure 3.
Analysis of I-SceI-induced homologous repair and interplasmid gene conversion events from A. gambiae cells and embryos. (a) Reporter plasmid pDR-CMV-GFP. In frame stop codons introduced by I-SceI recognition site are underlined. (b) Fluorescence activated cell sorting (FACS) of transfected cells performed 72 h post-transfection. Two-colour fluorescence analysis of cell line Sua4.0 transfected with pDR-CMV-GFP in the presence and absence of pP[v+,70I-SceI]. A gate using SSC-H (complexity) versus FSC-H (size) was used for cell size analysis (data not shown). The percentage of green fluorescent cells falling below the diagonal for each transfection are indicated (upper panels). The panel at the lower left side shows an overlay of both transfections (pDR-CMV-GFP transfected cells shown in red, pDR-CMV-GFP + pP[v+,70I-SceI] transfected cells shown in green) and the panel on the lower right side shows a histogram of all GFP+ events. FL1-H, green fluorescence; FL2-H, red autofluorescence. (c) Detailed maps of pBC/SacRB S1, pSL-SacRBTet and the predicted product plasmid created by a perfect gene conversion event. (d) Resistance properties of pBC/SacRB S1, pSL-SacRBTet and the predicted product plasmid created by a perfect gene conversion event. (e) Restriction analysis of pBC/SacRB S1 and 2 clones isolated from microinjected embryos. X, XbaI; B, BamHI; E, EcoRI; H, HindIII; Xh, XhoI; N, NcoI; A, AscI; S, SalI; Nh, NheI; HL1, Hyperladder 1 (1.5, 2, 2.5, 3, 4, 5, 6 and 10 kb).
I-SceI is expressed and active in A. gambiae embryos
We also investigated HEG activity in A. gambiae embryos. Briefly, I-SceI donor plasmid pP[v+,70I-SceI] and pBC SacRB S1 were co-injected into preblastoderm embryos. I-PpoI was not used in these experiments due to its toxic effect on insect cells (see subsequently). One day post-injection embryos were heat-shocked for 1 h at 41°C and low molecular weight DNA was extracted 2 h later. Selection for target plasmids modified by NHEJ was performed as described above for cells. As previously observed in mosquito cells, co-injection of the plasmid pP[v+,70I-SceI] increased the number of Cam/Suc-resistant bacteria colonies ∼10-fold: 0.8% of the recovered pBC/SacRB S1 pool were able to grow on Cam/Suc, a similar frequency to that obtained from cells. We analysed the sequence of the I-SceI recognition site in 20 plasmids recovered from injected embryos (Figure 2b). In contrast to what we observed in mosquito cells, both deletion and insertion events were found. One of the recovered plasmids contained a 66 bp insertion homologous to an A. gambiae genomic sequence.
I-SceI expression induces inter-plasmid gene conversion events in A. gambiae embryos
In order to test for HEG-induced gene conversion events in A. gambiae embryos, we used the plasmid pSL-SacRBTet. This plasmid contains a Tet resistance cassette inserted into the SacRB gene on a pSL backbone (Figures 1a and 3c). Therefore, this plasmid shares two regions of homology with the pBC target plasmids which flank the Tet resistance cassette but contains a different backbone (Figure 3c). As shown in Figure 3c and d, the outcome of a perfect gene conversion event would generate a plasmid that contains both Cam and Tet resistance cassettes but a non-functional SacRB gene. Plasmids pBC/SacRB S1, pSL-SacRBTet and pP[v+,70I-SceI] were co-injected into pre-blastoderm stage embryos and DNA extracted as described above. As a control a second set of injections was performed excluding the pP[v+,70I-SceI] plasmid. In control experiments, where only the HEG target plasmid was injected we obtained six Cam/Suc bacterial colonies and none of these were able to grow when plated on Tet. To demonstrate that gene conversion cannot be achieved by the bacteria alone, bacteria were transformed with the injection mixture and plated on Cam, Cam/Suc, Tet as well as Cam/Suc/Tet. Whereas bacteria transformed with injection mix can grow on Cam or Tet, no colonies were obtained on Cam/Suc or Cam/Suc/Tet plates.
From injection experiments performed with all three plasmids (pBC/SacRB S1, pSL-SacRBTet and pP[v+,70I-SceI]) we obtained 98 bacterial colonies growing on Cam/Suc in the initial selection round. Of these 11 were also able to grow on Tet (11.2%). Further analysis revealed that two of them were unable to grow on ampicillin (Amp) (the Amp resistance gene is found in the pSL-SacRBTet but not in the pBC/SacRB S1 backbone). We analysed these two colonies by restriction endonuclease digestion (Figure 3e) and sequencing. The sequence analysis revealed the occurrence of perfect gene conversion events, in which the Tet cassette had been inserted into pBC/SacRB S1 as predicted. The nine clones that were able to grow on Amp were resistant to cleavage with I-SceI but they appeared to be larger than the expected product. All nine plasmids were cleavable by AscI, a rare cutting enzyme that flanks the SacRBTet cassette (Figures 1a and 3c), indicating that larger parts of pSL-SacRBTet had been transferred during the process of homologous recombination.
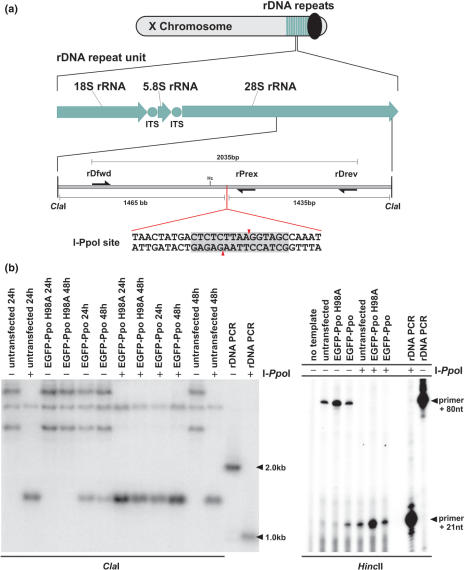
The A. gambiae 28S rDNA gene contains a conserved I-PpoI recognition site
I-PpoI mediates homing of intron 3 (Pp LSU 3) in the extrachromosomal nuclear rDNA of the acellular slime mold Physarum polycephalum (23). This region of the 28S rDNA is highly conserved in eukaryotes and I-PpoI has been shown to cleave human rDNA repeats in vivo (42). In order to determine whether an I-PpoI site is present in A. gambiae rDNA, the 28S gene of A. gambiae was assembled from the sequence of 31 cDNA sequences obtained from the AnoEST database. Sequence analysis indicated the presence of the full 29 bp I-PpoI recognition site. The assembled gene sequence was used to design a set of primers (rDfwd and rDrev, Figure 4a) that amplify a 2 kb fragment of the A. gambiae 28S rDNA gene containing the I-PpoI site (Figure 4a). The PCR product from A. gambiae strains KWA and G3 was digested with I-PpoI to confirm the presence of the site in both strains (data not shown).
Figure 4.
I-PpoI expression and cleavage of genomic rDNA repeats in Sua 4.0 cell lines. (a) Schematic map of A. gambiae rDNA clusters and the location of the I-PpoI site within the 28S rDNA. ITS, internally transcribed spacer; Hc, HincII (HindII). (b) Southern blot analysis of A. gambiae Sua 4.0 cells transfected with pEGFP-Ppo or pEGFP-Ppo H98A. Genomic DNA was digested with ClaI and a 2 kb fragment of rDNA amplified with primers rDfwd and rDrev was used as probe. DNA in lanes marked by a (+) was digested with I-PpoI in vitro. (c) Primer extension analysis. Genomic DNA extracted from cells transfected with pEGFP-Ppo or pEGFP-Ppo H98A was digested with HincII and extended with primer rPrex. DNA in lanes marked by a (+) was digested with I-PpoI in vitro.
I-PpoI cleaves chromosomal rDNA repeats in vivo
To determine whether I-PpoI can bind and cleave chromosomal rDNA repeats in vivo, A. gambiae Sua 4.0 cells were transfected with pEGFP-Ppo or pEGFP-Ppo H98A plasmids that express a functional and an inactive version of I-PpoI respectively. Genomic DNA was extracted 24 and 48 h post-transfection and digested with ClaI or ClaI and I-PpoI (Figure 4a). ClaI is predicted to cut 1.5 kb up and 1.4 kb downstream of the I-PpoI site. Southern blot analysis using the 2 kb 28S rDNA PCR product as a probe indicated that chromosomal 28S rDNA was cut efficiently by I-PpoI but not I-PpoI H98A (Figure 4b). The probe hybridized to several fragments, the shortest of which corresponds to the expected 2.9 kb ClaI fragment. All but one of these fragments were cleaved by I-PpoI (Figure 4b). This seems to indicate some heterogeneity in the 28S genes and we obtained a similar result with another enzyme combination (data not shown). Presumably this is a result of retrotransposon insertions downstream of the I-PpoI site (43). The appearance of resistant bands, retrieved when using I-SceI in Anopheles cells (Figure 1b, lanes 7 and 10), was not observed when analysing genomic rDNA cleaved with I-PpoI in vitro (Figure 4b, lanes 7–10). To confirm the result obtained by Southern blotting, we performed a primer extension analysis using primer rPrex, which binds 21 bases downstream of the I-PpoI cleavage site (Figure 4a). Figure 4c shows that I-PpoI expression causes premature stops of the primer extension at a position corresponding to the I-PpoI site. These data together indicate that I-PpoI is able to efficiently cut mosquito rDNA in transfected cells.
I-PpoI but not I-SceI expression causes cytotoxicity in A. gambiae cell lines
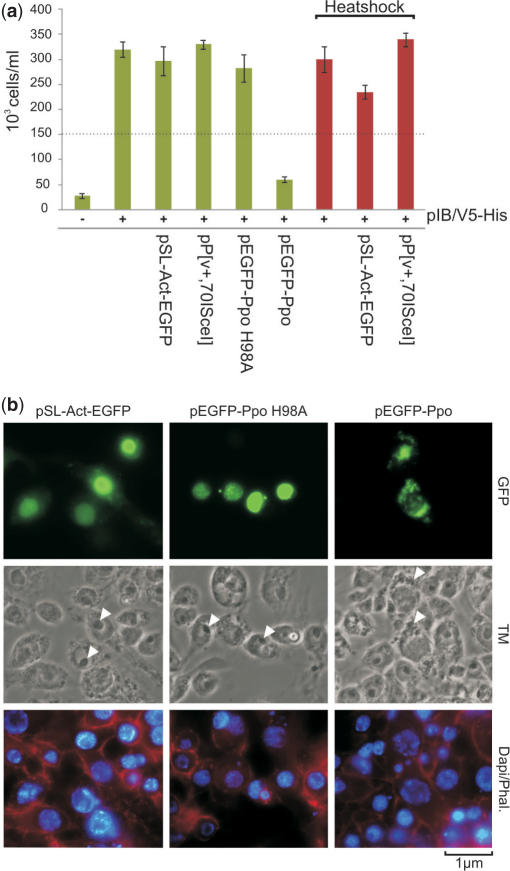
In several mosquito species including A. gambiae, the 28S rDNA genes are clustered as tandem repeats on the X chromosome. The use of I-PpoI offers therefore the possibility to selectively disrupt the X chromosome. While I-PpoI cleaves the essential 28S rDNA genes, I-SceI is not predicted to have a target site in the A. gambiae genome. To study the effect of expressing these two HEGs on the viability of mosquito cells, we transfected Sua 4.0 cells with pP[v+,70I-SceI], pEGFP-Ppo, pEGFP-Ppo H98A or pSL-Act-EGFP, together with the plasmid pIB/V5-His, which confers resistance to the translation inhibitor blasticidin S. Cell proliferation was assessed as a measure of growth in medium containing 50 μg/ml blasticidine, which will kill all cells that do contain the pIB/V5-His plasmid. The results of these experiments are shown in Figure 5a. While neither I-SceI nor mutant I-PpoI H98A expression interferes with cell proliferation, I-PpoI expression leads to growth arrest. Expression of the I-PpoI-GFP fusion protein seemed to induce nuclear fragmentation (Figure 1d) as observed by fluorescent microscopy. It appears that the nucleoli of cells transfected with pEGFP-Ppo (Figure 5b) are disintegrating. This can be explained by the fact that ribosomal DNA repeats which are cut by I-PpoI form the nucleolus organizer regions of the cell.
Figure 5.
Cell proliferation analysis and morphology of I-PpoI and I-SceI expressing cells. (a) I-PpoI expression arrests proliferation of Sua 4.0 cells. Cells were co-transfected with HEG expression vectors and a GFP control in combination with pIB/V5-His which confers resistance to blasticidin and counted after 5 days of growth in medium supplemented with blasticidin S. Red bars indicate cells that were heat-shocked to induce expression of I-SceI. The dotted line indicates the 150 000 cells seeded for the experiment. (b) Sua 4.0 cells were transfected with pEGFP-Ppo, pEGFP-Ppo H98A and control pSL-Act-GFP. Forty-eight hour post-transfection, cells were fixed and stained with DAPI as well as phalloidin. White arrows indicate nucleoli of cells. TM, transmission.
Whereas natural HEGs have evolved under constant selection pressure towards reduced toxicity and are selected to cut only at their native homing sites, engineered HEGs might be less specific. Experiments with designed zinc finger nucleases have shown that expression of these enzymes is often accompanied by general toxicity (44–46). Our assay confirms that I-SceI, although highly active, is not toxic when expressed in cells. I-PpoI on the other hand cleaves the essential rDNA genes and induces proliferation arrest, as has been described for human cells (42). The HEG activity, GFP reporter and cell proliferation assays described can be applied to the initial assessments for the suitability of any engineered HEG candidates designed against Anopheles genes.
DISCUSSION
Our results demonstrate that two different HEGs, I-SceI and I-PpoI, when expressed in A. gambiae cells and embryos, retain their ability to recognize and cleave their cognate target sequences. Analysis of I-SceI and I-PpoI function using an interplasmid activity assay in cultured mosquito cells showed that, in the absence of a template for homologous recombination, recovered target sites contained deletions ranging from 1 to 80 bp created by HEG-induced cleavage and subsequent NHEJ repair. Sequences recovered from embryos injected with the I-SceI gene contained both insertions (1–66 bp) and deletions (2–62 bp). It is not yet known whether the observed bias towards deletions in cells reflects a feature of Anopheles biology or whether it is a consequence of our plasmid-based selection system. All targeted sites analysed were resistant to subsequent I-SceI and I-PpoI cleavage in vitro as nucleotide deletions or insertions in the recognition site significantly impair endonuclease activity (47).
Recently, it has been shown that HEGs can be engineered to confer new sequence specificities (15–18), thus offering the possibility of using these highly specific nucleases for gene disruption and gene therapy (48,49). Our findings indicate that engineered HEGs could be used to selectively disrupt mosquito genes either in the germ line or in a tissue-specific manner depending on the spatial and temporal expression pattern of the driving promoter.
It has been suggested that the ‘selfish’ genetic behaviour of HEGs could be exploited to rapidly spread a genetic modification affecting mosquito vectorial capacity from a few individuals to an entire population (11,19). This process would require both DNA cleavage and repair via homologous recombination at the targeted site using the HEG-carrying allele as template. To determine the feasibility of such an approach, we determined whether repair of HEG-mediated cleavage occurred by homologous recombination when appropriate template sequences are present. A functional study carried out in mosquito cells showed, that in the presence of I-SceI, a plasmid carrying a transcriptionally silent and incomplete eGFP coding sequence could function as template to repair (as inferred by the gain of fluorescence) another non-functional eGFP sequence disrupted by an I-SceI cleavage site. The frequency of homologous repair events in mosquito cells is comparable to that previously described in mammalian cell lines (34). To extend this observation, we adapted the SacRB-based interplasmid assay to score homologous recombination events. We used for this purpose a relatively large conversion cassette containing a Tet resistance transcription unit (1.5 kb), in an attempt to simulate the size of a complete HEG element. While no recombinant clones were observed in the absence of the HEG, the precise transfer of the Tet cassette between two plasmids was found in 2 out of 98 events. We also observed nine recombination events showing larger insertions encompassing regions of the pSL plasmid flanking the Tet cassette. We speculate that these events arose by imprecise extensions during homologous recombination. Results from these experiments suggest an overall frequency of homologous repair in embryos of ∼10%. Our Tet selection system and GFP reporter system only allow us to recover events that have resulted in the complete insertions of the Tet/GFP sequences. The actual rate of homologous repair might be higher as the occurrence of putative recombination events that resulted in the transfer of smaller fragments of Tet or GFP can be expected but were selected against. Also, in none of our experiments were the repair donor fragments flanked by HEG half sites, a fact that might reduce the rate of homologous repair. In our experiments, DNA DSBs are induced in vivo by the activity of HEGs. Efficient homologous repair in A. gambiae Ag55 cells has also been demonstrated by the transfection of linear recombination substrates (50).
Together, these findings indicate in two highly predictive experimental systems that HEG would retain their gene-driving activity in mosquitoes. Furthermore, HEG-mediated cleavage of a target gene in embryos followed by gene conversion using a repair template provided in trans could represent an alternative approach to traditional transposon or integrase-mediated germ line mosquito transformation.
We have shown that I-PpoI specifically recognizes and cleaves a vital target sequence in A. gambiae cells inducing cell proliferation arrest and presumably cell death. The I-PpoI recognition site is located in the 28S rDNA gene within domain V of the 28S rRNA, a region that forms the peptidyl transferase centre of the ribosome. This sequence is among the most conserved sequences in the entire eukaryotic kingdom. In some mosquito species, including at least two members of the A. gambiae complex (51–53), the rDNA repeats are exclusively located in the centromeric region of the X chromosome. This arrangement raises the possibility of using I-PpoI to selectively attack the X chromosome: if specifically expressed during male meiosis, I-PpoI could distort the sex ratio towards males by selectively incapacitating X-carrying spermatozoa (19). Natural driving Y chromosomes in Aedes aegypti and Culex pipiens have been described and can produce extreme sex ratios of more than 90% males (54). Although the molecular details of how these distorters act are unknown, cytological evidence suggests that they are associated with breaks in the X chromosome during male meiosis I (55,56). These observations are relevant for the development of malaria vector control measures. A male sex distorter combining the specificity of I-PpoI together with its potential genetic drive activity if placed on the Y chromosome could knock down a wild-type Anopheline population in a few tens of generations (57).
ACKNOWLEDGEMENTS
We are indebted to Raymond Monnat for his generosity in providing the I-PpoI expression vectors as well as pDR-GFP and for helpful comments. We would also like to thank Kalle Magnusson, Sofia Pinto, Michael Povelones and Rita Tewari for technical help. F.C. is currently sponsored by an MRC Career Development Award. Funded by a Grant from the Foundation for the National Institutes of Health through the Grand Challenges in Global Health initiative. Funding to pay the Open Access publication charges for this article was provided by the Foundation for the National Institutes of Health through the Grand Challenges in Global Health initiative.
Conflict of interest statement. None declared.
REFERENCES
- 1.WHO. Geneva, 2005.
- 2.Collins FH, Paskewitz SM. Malaria: current and future prospects for control. Annu. Rev. Entomol. 1995;40:195–219. doi: 10.1146/annurev.en.40.010195.001211. [DOI] [PubMed] [Google Scholar]
- 3.Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- 4.Catteruccia F, Nolan T, Loukeris TG, Blass C, Savakis C, Kafatos FC, Crisanti A. Stable germline transformation of the malaria mosquito Anopheles stephensi. Nature. 2000;405:959–962. doi: 10.1038/35016096. [DOI] [PubMed] [Google Scholar]
- 5.Grossman GL, Rafferty CS, Clayton JR, Stevens TK, Mukabayire O, Benedict MQ. Germline transformation of the malaria vector, Anopheles gambiae, with the piggyBac transposable element. Insect Mol. Biol. 2001;10:597–604. doi: 10.1046/j.0962-1075.2001.00299.x. [DOI] [PubMed] [Google Scholar]
- 6.Abraham EG, Donnelly-Doman M, Fujioka H, Ghosh A, Moreira L, Jacobs-Lorena M. Driving midgut-specific expression and secretion of a foreign protein in transgenic mosquitoes with AgAper1 regulatory elements. Insect Mol. Biol. 2005;14:271–279. doi: 10.1111/j.1365-2583.2004.00557.x. [DOI] [PubMed] [Google Scholar]
- 7.Ito J, Ghosh A, Moreira LA, Wimmer EA, Jacobs-Lorena M. Transgenic anopheline mosquitoes impaired in transmission of a malaria parasite. Nature. 2002;417:452–455. doi: 10.1038/417452a. [DOI] [PubMed] [Google Scholar]
- 8.Moreira LA, Ito J, Ghosh A, Devenport M, Zieler H, Abraham EG, Crisanti A, Nolan T, Catteruccia F, et al. Bee venom phospholipase inhibits malaria parasite development in transgenic mosquitoes. J. Biol. Chem. 2002;277:40839–40843. doi: 10.1074/jbc.M206647200. [DOI] [PubMed] [Google Scholar]
- 9.Catteruccia F, Benton JP, Crisanti A. An Anopheles transgenic sexing strain for vector control. Nat. Biotechnol. 2005;23:1414–1417. doi: 10.1038/nbt1152. [DOI] [PubMed] [Google Scholar]
- 10.James AA. Gene drive systems in mosquitoes: rules of the road. Trends Parasitol. 2005;21:64–67. doi: 10.1016/j.pt.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 11.Sinkins SP, Gould F. Gene drive systems for insect disease vectors. Nat. Rev. Genet. 2006;7:427–435. doi: 10.1038/nrg1870. [DOI] [PubMed] [Google Scholar]
- 12.Burt A, Trivers R, editors. Genes In Conflict. Belknap Harvard; 2006. [Google Scholar]
- 13.Stoddard BL. Homing endonuclease structure and function. Q. Rev. Biophys. 2005;38:49–95. doi: 10.1017/S0033583505004063. [DOI] [PubMed] [Google Scholar]
- 14.Chevalier BS, Stoddard BL. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 2001;29:3757–3774. doi: 10.1093/nar/29.18.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volna P, Jarjour J, Baxter S, Roffler SR, Monnat J., Jr, Stoddard BL, Scharenberg AM. Flow cytometric analysis of DNA binding and cleavage by cell surface-displayed homing endonucleases. Nucleic Acids Res. 2007;35:2748–2758. doi: 10.1093/nar/gkm182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashworth J, Havranek JJ, Duarte CM, Sussman D, Monnat R.J., Jr, Stoddard BL, Baker D. Computational redesign of endonuclease DNA binding and cleavage specificity. Nature. 2006;441:656–659. doi: 10.1038/nature04818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosen LE, Morrison HA, Masri S, Brown MJ, Springstubb B, Sussman D, Stoddard BL, Seligman LM. Homing endonuclease I-CreI derivatives with novel DNA target specificities. Nucleic Acids Res. 2006;34:4791–4800. doi: 10.1093/nar/gkl645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith J, Grizot S, Arnould S, Duclert A, Epinat JC, Chames P, Prieto J, Redondo P, Blanco FJ, et al. A combinatorial approach to create artificial homing endonucleases cleaving chosen sequences. Nucleic Acids Res. 2006;34:e149. doi: 10.1093/nar/gkl720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 2003;270:921–928. doi: 10.1098/rspb.2002.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hahn MW, Nuzhdin SV. The fixation of malaria refractoriness in mosquitoes. Curr. Biol. 2004;14:R264–R265. doi: 10.1016/j.cub.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 21.Klinakis AG, Loukeris TG, Pavlopoulos A, Savakis C. Mobility assays confirm the broad host-range activity of the Minos transposable element and validate new transformation tools. Insect Mol. Biol. 2000;9:269–275. doi: 10.1046/j.1365-2583.2000.00183.x. [DOI] [PubMed] [Google Scholar]
- 22.Catteruccia F, Nolan T, Blass C, Muller HM, Crisanti A, Kafatos FC, Loukeris TG. Toward Anopheles transformation: minos element activity in anopheline cells and embryos. Proc. Natl Acad. Sci. USA. 2000;97:2157–2162. doi: 10.1073/pnas.040568397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muscarella DE, Vogt VM. A mobile group I intron in the nuclear rDNA of Physarum polycephalum. Cell. 1989;56:443–454. doi: 10.1016/0092-8674(89)90247-x. [DOI] [PubMed] [Google Scholar]
- 24.Muscarella DE, Ellison EL, Ruoff BM, Vogt VM. Characterization of I-Ppo, an intron-encoded endonuclease that mediates homing of a group I intron in the ribosomal DNA of Physarum polycephalum. Mol. Cell. Biol. 1990;10:3386–3396. doi: 10.1128/mcb.10.7.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowery L, Hung L, Knoche K, Bandziulis R. Properties of I-PpoI: a rare-cutting intron-encoded endonuclease. Promega Notes. 1992;38:8–12. [Google Scholar]
- 26.Jacquier A, Dujon B. An intron-encoded protein is active in a gene conversion process that spreads an intron into a mitochondrial gene. Cell. 1985;41:383–394. doi: 10.1016/s0092-8674(85)80011-8. [DOI] [PubMed] [Google Scholar]
- 27.Macreadie IG, Scott RM, Zinn AR, Butow RA. Transposition of an intron in yeast mitochondria requires a protein encoded by that intron. Cell. 1985;41:395–402. doi: 10.1016/s0092-8674(85)80012-x. [DOI] [PubMed] [Google Scholar]
- 28.Monteilhet C, Perrin A, Thierry A, Colleaux L, Dujon B. Purification and characterization of the in vitro activity of I-Sce I, a novel and highly specific endonuclease encoded by a group I intron. Nucleic Acids Res. 1990;18:1407–1413. doi: 10.1093/nar/18.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bellaiche Y, Mogila V, Perrimon N. I-SceI endonuclease, a new tool for studying DNA double-strand break repair mechanisms in Drosophila. Genetics. 1999;152:1037–1044. doi: 10.1093/genetics/152.3.1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 31.Rong YS, Golic KG. A targeted gene knockout in Drosophila. Genetics. 2001;157:1307–1312. doi: 10.1093/genetics/157.3.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyopadhyay P, Olivera BM, Brodsky M, Rubin GM, et al. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Horn C, Wimmer EA. A versatile vector set for animal transgenesis. Dev. Genes Evol. 2000;210:630–637. doi: 10.1007/s004270000110. [DOI] [PubMed] [Google Scholar]
- 34.Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MR4. Malaria Research and Reference Reagent Resource Center. Vol. 2007. [DOI] [PubMed]
- 36.Raghavan SC, Swanson PC, Ma Y, Lieber MR. Double-strand break formation by the RAG complex at the bcl-2 major breakpoint region and at other non-B DNA structures in vitro. Mol. Cell. Biol. 2005;25:5904–5919. doi: 10.1128/MCB.25.14.5904-5919.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gay P, Le Coq D, Steinmetz M, Ferrari E, Hoch JA. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J. Bacteriol. 1983;153:1424–1431. doi: 10.1128/jb.153.3.1424-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado CI. Positive selection procedure for entrapment of insertion sequence elements in gram-negative bacteria. J. Bacteriol. 1985;164:918–921. doi: 10.1128/jb.164.2.918-921.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borchert TV, Nagarajan V. Effect of signal sequence alterations on export of levansucrase in Bacillus subtilis. J. Bacteriol. 1991;173:276–282. doi: 10.1128/jb.173.1.276-282.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meng G, Futterer K. Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat. Struct. Biol. 2003;10:935–941. doi: 10.1038/nsb974. [DOI] [PubMed] [Google Scholar]
- 41.Chu G. Double strand break repair. J. Biol. Chem. 1997;272:24097–24100. doi: 10.1074/jbc.272.39.24097. [DOI] [PubMed] [Google Scholar]
- 42.Monnat R.J., Jr, Hackmann AF, Cantrell MA. Generation of highly site-specific DNA double-strand breaks in human cells by the homing endonucleases I-PpoI and I-CreI. Biochem. Biophys. Res. Commun. 1999;255:88–93. doi: 10.1006/bbrc.1999.0152. [DOI] [PubMed] [Google Scholar]
- 43.Besansky NJ, Paskewitz SM, Hamm DM, Collins FH. Distinct families of site-specific retrotransposons occupy identical positions in the rRNA genes of Anopheles gambiae. Mol. Cell. Biol. 1992;12:5102–5110. doi: 10.1128/mcb.12.11.5102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bibikova M, Golic M, Golic KG, Carroll D. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics. 2002;161:1169–1175. doi: 10.1093/genetics/161.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- 46.Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- 47.Argast GM, Stephens KM, Emond MJ, Monnat R.J., Jr I-PpoI and I-CreI homing site sequence degeneracy determined by random mutagenesis and sequential in vitro enrichment. J. Mol. Biol. 1998;280:345–353. doi: 10.1006/jmbi.1998.1886. [DOI] [PubMed] [Google Scholar]
- 48.Wickelgren I. Molecular biology. Spinning junk into gold. Science. 2003;300:1646–1649. doi: 10.1126/science.300.5626.1646. [DOI] [PubMed] [Google Scholar]
- 49.Uil TG, Haisma HJ, Rots MG. Therapeutic modulation of endogenous gene function by agents with designed DNA-sequence specificities. Nucleic Acids Res. 2003;31:6064–6078. doi: 10.1093/nar/gkg815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eggleston P, Zhao Y. A sensitive and rapid assay for homologous recombination in mosquito cells: impact of vector topology and implications for gene targeting. BMC Genet. 2001;2:21. doi: 10.1186/1471-2156-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Collins FH, Paskewitz SM, Finnerty V. Ribosomal RNA genes of the Anopheles gambiae species complex. Adv. Dis. Vector Res. 1989;6:1–28. [Google Scholar]
- 52.Collins FH, Mendez MA, Rasmussen MO, Mehaffey PC, Besansky NJ, Finnerty V. A ribosomal RNA gene probe differentiates member species of the Anopheles gambiae complex. Am. J. Trop. Med. Hyg. 1987;37:37–41. doi: 10.4269/ajtmh.1987.37.37. [DOI] [PubMed] [Google Scholar]
- 53.Paskewitz SM, Collins FH. Use of the polymerase chain reaction to identify mosquito species of the Anopheles gambiae complex. Med. Vet. Entomol. 1990;4:367–373. doi: 10.1111/j.1365-2915.1990.tb00453.x. [DOI] [PubMed] [Google Scholar]
- 54.Wood RJ, Newton ME. Sex-ratio distortion caused by meiotic drive in mosquitoes. Am. Nat. 1991;137:379–391. [Google Scholar]
- 55.Newton ME, Wood RJ, Southern DI. A cytogenetic analysis of meiotic drive in the mosquito, Aedes aegypti (L.) Genetica. 1976;46:297–318. [Google Scholar]
- 56.Sweeny TL, Barr AR. Sex ratio distortion caused by meiotic drive in a mosquito, Culex pipiens L. Genetics. 1978;88:427–446. doi: 10.1093/genetics/88.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang Y, Magori K, Lloyd AL, Gould F. Introducing desirable transgenes into insect populations using y-linked meiotic drive-a theoretical assessment. Evol. Int. J. Org. Evol. 2007;61:717–726. doi: 10.1111/j.1558-5646.2007.00075.x. [DOI] [PubMed] [Google Scholar]