Abstract
Efficient transcription of long polycistronic operons in bacteria frequently relies on accessory proteins but their molecular mechanisms remain obscure. RfaH is a cellular elongation factor that acts as a polarity suppressor by increasing RNA polymerase (RNAP) processivity. In this work, we provide evidence that RfaH acts by reducing transcriptional pausing at certain positions rather than by accelerating RNAP at all sites. We show that ‘fast’ RNAP variants are characterized by pause-free RNA chain elongation and are resistant to RfaH action. Similarly, the wild-type RNAP is insensitive to RfaH in the absence of pauses. In contrast, those enzymes that may be prone to falling into a paused state are hypersensitive to RfaH. RfaH inhibits pyrophosphorolysis of the nascent RNA and reduces the apparent Michaelis–Menten constant for nucleotides, suggesting that it stabilizes the post-translocated, active RNAP state. Given that the RfaH-binding site is located 75 Å away from the RNAP catalytic center, these results strongly indicate that RfaH acts allosterically. We argue that despite the apparent differences in the nucleic acid targets, the time of recruitment and the binding sites on RNAP, unrelated antiterminators (such as RfaH and λQ) utilize common strategies during both recruitment and anti-pausing modification of the transcription complex.
INTRODUCTION
RNAP is an obligatory processive enzyme that must complete synthesis of the entire RNA chain since the transcripts that are released prematurely cannot re-enter transcription cycle. In bacteria, even in the absence of the tightly condensed chromatin, RNAP still encounters many roadblocks that either stall it temporarily or trigger RNA release. DNA-bound proteins, DNA lesions and various nucleic acid signals that induce pausing, arrest and termination (1) can hinder RNAP progression along the template. Even at saturating substrate concentrations in vitro, RNAP is moving in leaps, with its fast movement along the template punctuated by pauses (2). Pausing plays numerous regulatory roles, is an obligatory step in termination pathways, and likely controls the overall rate of RNA chain elongation (3).
RNAP is capable of making very long RNA chains (30 000 nt long in bacteria) but its rate is rather modest compared to DNA replicases: in Escherichia coli, elongating RNAP (a complex of α2ββ′ω subunits) moves at 20–90 nt/s (4) whereas the replication fork advances 1000 nt/s (5). This relatively inefficient operation of RNAP does not represent the limit of its catalytic potential since ‘fast’ substitutions in the β and β′ subunits that significantly increase its overall rate in vitro have been described (6–10). An attractive explanation rests on an assumption that the relatively slow rate of transcription is necessary for efficient regulation of gene expression where it provides for timely recruitment of, and response to regulatory factors, attenuation control, as well as determines folding pathways of the nascent RNA. Moreover, in bacteria transcription and translation are coupled, imposing additional restrictions on the speed that RNAP can attain without placing the nascent RNA in danger of release by Rho, which terminates the untranslated messages (4). In other words, a catalytically perfect RNAP would leave little room for regulation and likely uncouple transcription and translation, while much slower RNAP would not be nimble enough to keep up with sustaining the RNA pool as it adapts to changing environmental and physiological conditions. Indeed, while different ‘fast’ and ‘slow’ viable alleles of RNAP have been isolated, they alter the apparent elongation rate in vitro by less than 3- to 5-fold in each direction (7,8,10–12), whereas mutations coding for much faster or slower enzyme variants are lethal (6,9,13–15).
As substitutions that constitutively change the overall rate of RNA chain elongation appear to have a negative impact on fitness and are being removed by natural selection, the stage is set for transient alteration of RNAP kinetic properties by regulatory proteins. A subset of such factors (known as antiterminators) reduces pausing and termination (in other words, confers a fast phenotype) thereby helping RNAP transcribe long operons. These proteins use different nucleic acid targets during recruitment: λN binds the nascent RNA structure, λQ is recruited to the double-stranded DNA near the promoter, RfaH is recruited to the single-stranded non-template (NT) DNA strand during elongation (16–19). The sites on RNAP to which these proteins bind are likely also distinct: we have recently concluded (20) that RfaH binds to the β′-subunit clamp helices (β′ CH), whereas the target sites for λ regulators are still unknown but are thought to be quite different (21,22). Yet all antiterminators share the ability to accelerate RNAP, suggesting that they induce similar changes in the transcription elongation complex (TEC).
To date, the changes that lead to the ‘antitermination’ modification of the RNAP have not been characterized in detail, and the molecular mechanism(s) by which elongation factors or substitutions in RNAP make the enzyme faster or slower is not known: they may control nucleotide addition at every template position by affecting the common rate-limiting step (which has not been elucidated for RNAP), or influence the TEC isomerization into off-pathway states at pause and termination sites (23). We have proposed that at a pause site RNAP isomerizes into a state in which nucleotide addition is slowed due to transient changes in the active site architecture (Figure 1), and from which different classes of pause and termination complexes arise (24). We further speculated that substitutions in RNAP may alter its propensity towards the isomerization into the slow state. In a fast RNAP, the productive alignment of the 3′ RNA end in the active site, and consequently nucleotide addition, is favored. In contrast, a slow RNAP is more likely to lose the 3′end from the active site and enter a paused state, escape from which can be delayed by two orders of magnitude. Antiterminators may act in the same regulatory pathway, switching RNAP into the fast state. Slow, pause-prone enzymes should then be hypersensitive to modification by antiterminators, whereas fast RNAPs should appear resistant to further acceleration.
Figure 1.
Unactivated intermediate mechanism. During rapid elongation (top horizontal arrows) RNAP active site (represented by two circles denoting the i and i−1 subsites) is optimized for facile catalysis: the 3′ OH of the nascent RNA is perfectly positioned to accommodate the incoming substrate NTP. At some sites, the active site undergoes a rearrangement to yield an unactivated intermediate in which the nucleotide addition is slowed (paused state). The precise nature of this rearrangement is unknown; it may include the 3′ end fraying (as shown), slight changes in the position of the catalytic residues or Mg2+ ion, etc. From the unactivated intermediate, RNAP can isomerize into more stable, long-lived paused states upon the hairpin formation (at class I sites) or backtracking (at class II sites), or into a termination complex. Substitutions in RNAP that confer ‘fast’ and ‘slow’ elongation phenotypes (such as βP560S,T563I and βQ513P, respectively) may affect isomerization into the unactivated intermediate by favoring different states of the RNAP active site (24).
To test this hypothesis, we have determined effects of the E. coli RfaH on RNA chain elongation by enzymes from an expanded panel of fast and slow RNAPs, including many previously uncharacterized kinetic variants. RfaH is recruited to the TEC at specific sites (called ops) and is required for expression of several long operons (25). Reports from several labs indicated that RfaH acts as an antiterminator both in vivo and in vitro (16), although the exact mechanism of its action remains elusive. RfaH structure and its binding site on the TEC are known (20). RfaH increases the overall elongation rate in vitro, reduces pausing at mechanistically distinct regulatory sites (known as class I and II, Figure 1), and facilitates bypass of some terminators (16,26). Unlike other well-studied antiterminators (18,27), RfaH does not require any accessory proteins (e.g. NusA or NusG), and its action is not dramatically affected by addition of the cellular extract. These features allow us to dissect RfaH effect on the TEC in a highly purified model system.
Our results indicate that RfaH acts primarily to reduce pausing: it fails to further accelerate the already fast mutants, as well as the wild-type RNAP transcribing under pause-free conditions, but is particularly effective with the enzymes that are prone to pausing because of amino acid changes or substrate deprivation. In contrast, RfaH cannot correct those slow phenotypes, which are due to the defects in the elementary catalytic steps rather than to the off-pathway events like pausing. We also show that the enzymes traditionally regarded as fast are better characterized as pause-resistant. We discuss our results within the framework of the current structural analysis of bacterial TECs.
MATERIALS AND METHODS
Plasmids used for protein purification
pIA238 encodes the wild-type rfaH ORF in pET28 (Novagen); the protein carries a His6 tag followed by a thrombin cleavage site (16). pIA777 is a derivative of pET36b(+) (Novagen) that encodes RfaH N-domain–TEV–C-domain–[His6]. Plasmid pVS10 (20) encodes the wild-type E. coli core RNAP. Other plasmids used to purify RNAP variants were: pIA542, β′Y795A; pIA627, β′T786V; pIA433, βH625Y; pIA434, βQ513P; pVS48, β′F773V; pIA645, βD675A; pVS14, β′Δ[943-1130]; pIA639, β′S793F; pVS50, β′L672D,V673D; pIA622, βS1105A; pIA648, βM1107A; pIA780, β′Δ [282-292]Gly4. Sequences of all plasmid constructs were verified at the OSU PMGF center and will be made available upon request.
Proteins and reagents
All general reagents were obtained from Sigma and Fisher; NTPs and [α32P]-NTPs, from GE; PCR reagents, restriction and modification enzymes, from NEB. Oligonucleotides were obtained from Integrated DNA Technologies. DNA purification kits were from Qiagen and Zymo Research. The full-length RfaH, RfaH N-domain, and RNAPs were purified as described in (20). βP560S,T563I RNAP was purified as described in (24).
Transcription elongation assays
For pause assay with RNAP variants and full-length RfaH, linear DNA template generated by PCR amplification (30 nM), holo RNAP (40 nM), ApU (100 μM), and starting NTP subsets (1 μM CTP, 5 μM ATP and GTP, 10 μCi [α32P]-CTP, 3000 Ci/mmol) were mixed in 100 μl of 20 mM Tris–acetate, 20 mM Na-acetate, 2 mM Mg acetate, 5% glycerol, 1 mM DTT, 0.1 mM EDTA, pH 7.9. Reactions were incubated for 15 min at 37°C. RfaH was added to 50 nM where indicated (for 3 min at 37°C) and transcription was restarted by addition of nucleotides (10 μM GTP, 150 μM ATP, CTP and UTP) and rifapentin to 100 μg/ml at 37°C. Samples were removed at 10, 20, 30, 40, 60, 90, 120, 180, 240, 360, 480, 600, 720 s and after a final 5-min incubation with 200 μM GTP, quenched by addition of an equal volume of STOP buffer (10 M urea, 20 mM EDTA, 45 mM Tris–borate; pH 8.3), and loaded on 8% denaturing urea/acrylamide (19:1) gels in 0.5× TBE. The gels were dried and analyzed using Storm 820 and ImageQuant (GE). Pause half-life (the time during which half of the complexes re-enter the elongation pathway) was determined by non-linear regression analysis. Termination assays were performed as described in (26). Assays with the N-domain of RfaH were performed in 20 mM Tris–HCl, 14 mM MgCl2, 20 mM NaCl, 5% glycerol, 1 mM DTT, 0.1 mM EDTA as described in (20).
Pyrophosphorolysis
Linear pIA226 DNA template generated by PCR amplification (100 nM), holo RNAP (120 nM), ApU (100 μM), N-domain (120 nM) (or storage buffer) and starting NTPs (1 μM GTP, 5 μM ATP and UTP, 10 μCi [α32P]-GTP, 3000 Ci/mmol) were mixed in 25 μl of buffer TGC2 (20 mM Tris–Cl, 20 mM NaCl, 2 mM MgCl2, 5% glycerol, 1 mM DTT, 0.1 mM EDTA, pH 7.9), and incubated for 15 min at 37°C. Halted A26 complexes were purified by gel filtration through AutSeq50 spin columns (GE) equilibrated in TGC2, and diluted 2-fold. Reactions were initiated by the addition of 1/10 vol of 250 μM PPi in TGC2, samples were removed at times shown in Figure 6, and quenched.
Figure 6.
RfaH effects on pyrophosphorolysis. (A) TEC may interconvert between states in which the 3′ end of the nascent RNA occupies different positions in the RNAP active site. (Left) In the optimal configuration, the 3′ OH is in the i site; the i+1 site is ready to accept the incoming NTP, and the complex is resistant to PPi. (Middle) In PPi -sensitive complexes, the 3′ end occupies the i+1 site, hindering the NTP binding. (Right) The 3′ end of the RNA may become frayed thereby inhibiting catalysis in both forward and reverse directions; this rearrangement may accompany isomerization into a short-lived, elemental pauses and long-lived hairpin-stabilized pauses. (B) Halted A26 transcription complexes formed in the absence or in the presence of the N-domain of RfaH were incubated with 25 μM PPi at 37°C for the times indicated. The reactions were stopped by the addition of urea (to 5 M) and EDTA (to 30 mM) and analyzed on a 12% denaturing gel. (C) The fraction of the 26-mer full-length RNA was quantified from the gels shown in panel B.
RESULTS
RfaH effects on RNAPs depend on their elongation properties
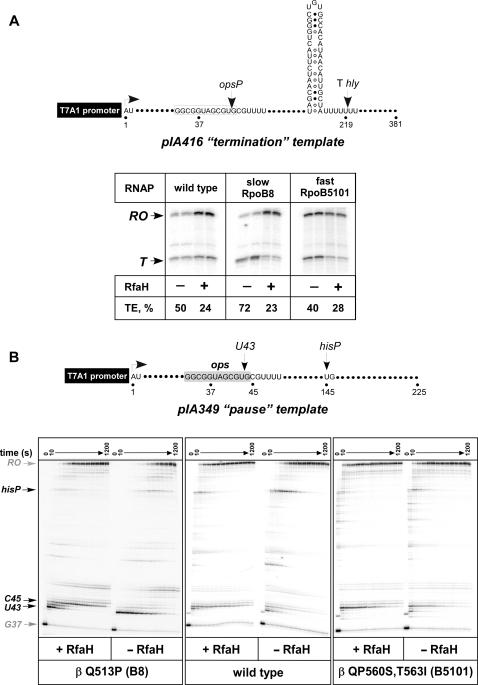
We first tested RfaH effect on two model enzymes, the slow RpoB8 (βQ513P) and the fast RpoB5101 (βP560S,T563I), whose elongation, pausing and termination properties are well known (12,24): B8 pauses and terminates more efficiently and elongates RNA less rapidly than the wild-type RNAP, whereas B5101 displays the opposite phenotypes. We first employed the standard single-round termination assay on Thly, a typical intrinsic terminator from the hemolysin operon that responds to RfaH in vivo (28) and in vitro (26), to test if these enzymes differ in their response to RfaH. We used the pIA416 linear transcription template on which the Thly was positioned downstream from a strong T7A1 promoter and a canonical ops element (Figure 2A). On this template, radiolabeled transcription complexes can be halted at position G37 when transcription is initiated in the absence of UTP, with ApU dinucleotide, ATP, GTP and α–[32P]CTP. The halted G37 complexes can then be chased upon addition of all four NTP substrates and RfaH. We found that RfaH (at 50 nM) decreased termination by the wild-type RNAP by ∼2-fold, but was much more effective with the slow RNAP (∼3-fold effect), and less effective (∼1.4-fold effect) with the fast variant (Figure 2A).
Figure 2.
RfaH has different effects on the fast and slow RNAPs. (A) Single-round termination assay on pIA416 template encoding the RfaH-responsive Thly terminator with the wild-type, slow (βQ513P) and fast (βP560S,T563I) RNAPs. Top. The relevant template features, start site (+1), transcript end (run-off, RO), the ops pause site (opsP, U43) and Thly terminator, are indicated in the schematic shown on top. The termination efficiency (TE) was determined as a ratio of terminated RNA (T) to the total RNA, the sum of the terminated and run-off products. A representative assay is shown on the right. (B) Single-round pause assay on a linear pIA349 template (top). Halted radiolabeled G37 TECs were pre-incubated with RfaH or storage buffer for 5 min at 37°C, and then challenged with rifapentin at 100 μg/ml and NTPs (10 μM GTP, 150 μM ATP, CTP, UTP). Aliquots were withdrawn at times ranging from 10 to 720 s and after a high GTP chase and analyzed on 8% denaturing gels.
The termination assay lends support to our hypothesis but does not allow us to distinguish whether RfaH fails to bind to the βP560S,T563I TEC or is unable to trigger the post-recruitment RNAP modification. We then used the pIA349 template (16) that allows to monitor both RfaH recruitment to the ops site and its post-recruitment effect at the his pause site (Figure 2B); this template is identical to pIA416 except for the sequence downstream of the ops element. The ops element induces RNAP pausing at position 43 with nearly 100% efficiency in vitro; when present, RfaH reduces pausing at U43 but dramatically delays RNAP at a site located 2 nt downstream (C45). This characteristic delay at the C45 position can be used to ascertain RfaH recruitment to the NT DNA. When halted G37 complexes were formed with wild-type RNAP and chased in the presence of RfaH, the half-life of the hisP was reduced ∼2.7-fold (Figures 2B and 3A). This effect was independent of the source and method used for RNAP purification: the same results were obtained with the chromosomally-encoded RNAP purified from MRE600 cells by the standard procedure (16) or expressed from the multi-cistronic vector and purified using chitin-binding domain-intein tag on β′(6) or hexahistidine tag on β′ (20) during the first, affinity purification step. When the wild-type RNAP was replaced by fast or slow variants, RfaH failed to reduce pausing at the hisP site by βP560S,T563I, and was particularly effective with the βQ513P enzyme (Figures 2B and 3A). Addition of RfaH produced the delay at C45 by all three enzymes, suggesting that it was recruited to the TEC in a similar fashion.
Figure 3.
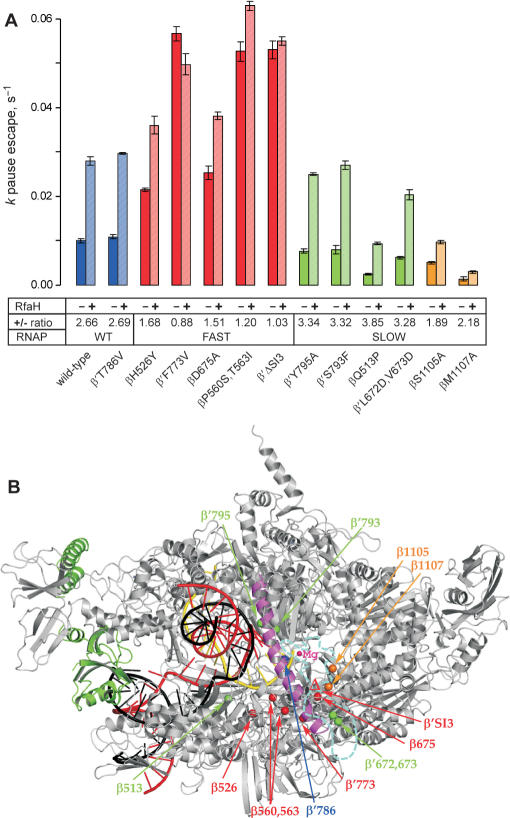
The summary of the RfaH effects on pausing at the hisP site by the RNAP variants. (A). The data from experiments similar to those shown in Figures 2B were analyzed as described in (20); for each enzyme, the assay was repeated at least twice. The kp for escape from the pause was measured in the presence and in the absence of RfaH; the ratio is presented below each variant. The pause efficiency cannot be determined reliably, particularly for the slow enzymes which display significant asynchrony during elongation. The substitutions that are fast and resistant to RfaH are colored in red, those that displays the wild-type elongation rate and response to RfaH are colored in blue, the substitutions that are slow and hypersensitive to RfaH—in green. Finally, the β S1105 and β M1107 residues, whose substitutions to Ala both reduce the elongation rate and the response to RfaH, are shown in orange. (B). A model of RfaH bound to the TEC. The Thermus thermophilus RNAP (46) is shown in gray with the bridge helix highlighted in magenta, the template DNA—in red, the NT DNA—in black, the nascent RNA—in yellow. Position of the RNAP active site is marked by the high-affinity catalytic Mg2+ ion (small magenta sphere). RfaH (green) is bound to the β′ CH. The Cα atoms of the RNAP substitutions tested in this work are shown as spheres and colored according to their response to RfaH as in panel A. The disordered β′ trigger loop is depicted as a cyan dashed line, with the β′SI3 domain (missing in T. thermophilus) inserted in the middle; the βΔ′SI3 deletion variant used here lacks E. coli β′ residues 943–1130 (6). The distance between the RfaH contact site on the β′ CH and the nearest residue shown (β Q513) is 53 Å.
We extended this analysis to include a panel of enzymes that were obtained during previous studies of resistance to antibiotics (13,14,29,30), analysis of species-specific differences in response to regulatory signals (6) and the mechanism of substrate selection (31); these RNAPs displayed a range of elongation rates in vitro. All these RNAPs were purified through several chromatographic steps, and were free from accessory proteins. We excluded from this analysis a number of enzymes with gross defects in transcription, such as the very slow β′N458D (31) or β′R1106A (32). We used the rate of RNAP escape from the hisP site (in the same assay system as in Figure 2B) to evaluate the post-recruitment effects of RfaH; RfaH also reduces the his pause efficiency (16) but this parameter is difficult to measure accurately, particularly with the slow RNAPs that pause at the upstream sites more prominently, and thus arrive at the hisP site asynchronously. We found that RfaH acted similarly on the wild-type enzyme and a variant with the ‘wild-type’ overall elongation rate (β′T786V) while failing to accelerate the fast enzymes (βH526Y/RpoB2, βD675A, β′F773V, β′ΔSI3, and βP560S,T563I) to the same extent (the defects varied, though). The slow enzymes fell into two categories: four variants (β′S793F, β′Y795A, βQ513P, and β′L672D,V673D) displayed augmented response, whereas two others (βS1105A and βM1107A)—reduced response to RfaH (Figure 3A). The summary of our findings is presented in Figure 3B, where the positions of these RNAP substitutions are shown in the context of the model of RfaH bound to the TEC.
RfaH is still recruited to fast RNAPs
The observation that RfaH cannot further accelerate those RNAPs that are already fast is consistent with the proposed switch. However, there remain two trivial explanations: (i) the RfaH-binding site is altered by substitutions or (ii) RfaH does not become recruited to the fast elongation complexes because these TECs move through the ops signal too rapidly.
Several observations argue against the first possibility. First, substitutions that confer fast phenotypes are located in different regions, far apart from each other, and in the regions that are not accessible from the enzyme surface in the context of the TEC (Figure 3B). Second, RfaH-binding site is located on the tip of the β′ CH (20), 50 Å away from the closest fast substitution. Third, RfaH still delays the fast enzymes immediately downstream from the ops site (Figure 2B). Finally, RfaH affects the kinetic parameters of the fast enzymes (see subsequently). In contrast, RNAPs with substitutions or deletions in the β′CH possess ‘wild-type’ elongation properties but are totally defective in both the recruitment of RfaH at the ops site and the post-recruitment response to RfaH at the downstream sites (20).
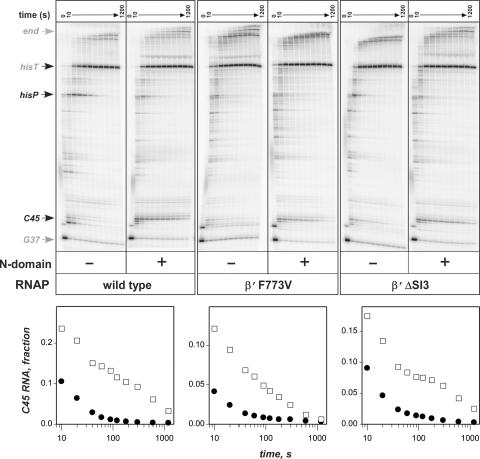
To exclude the second possibility, we utilized the isolated N-terminal domain of RfaH in place of the full-length protein. The N-domain contains all the DNA- and RNAP-binding determinants and retains all the elongation enhancement properties of RfaH (20). However, it no longer requires the ops site for binding to the TEC because its RNAP-binding site is always exposed (see Discussion section)—thus, the N-domain binds to (presumably) all TECs formed by the wild-type RNAP (in particular, the halted G37 in Figure 2B; data not shown) and has ample opportunities to become recruited to the G37 TECs formed by the fast enzymes, provided that the RfaH-binding site remains intact. We found that the N-domain accelerated the wild-type RNAP but not the two fastest mutants, β′F773V and β′ΔSI3 (Figure 4); however, the N-domain was clearly recruited to the altered TEC since it delayed all three RNAPs at the C45 site similarly. We conclude that RfaH binds to the fast RNAPs but fails to accelerate them further.
Figure 4.
The isolated N-domain of RfaH fails to accelerate the fast RNAPs. The assay was performed on pIA349 template (Figure 2B).
RfaH does not accelerate RNAP in the absence of pausing
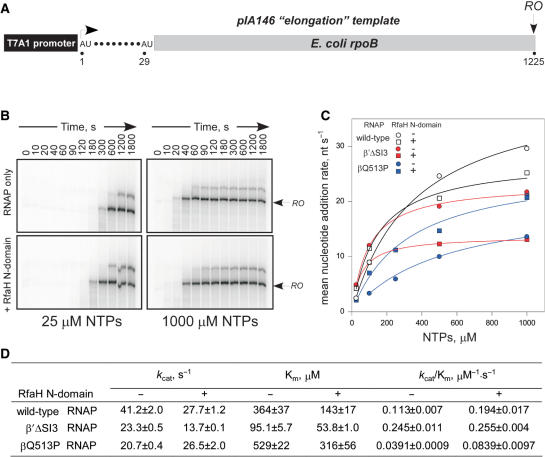
If RfaH works as an anti-pausing factor, its failure to accelerate the fast enzymes could be due to their inability to pause. Indeed, all the templates that we have tested for RfaH effects on elongation thus far contained pause sites (opsP and hisP), at which the wild-type RNAP pauses with high efficiency, but the fast enzymes do not. A simple prediction of this mechanism would be that if the wild-type RNAP transcribed a template devoid of any strong pauses, particularly at saturating concentrations of substrate NTPs, it would also become resistant to RfaH action because pausing would no longer determine the overall elongation rate. Since the ops site is in itself a pause, we utilized an ops-less template pIA146 (Figure 5A) that encodes the rpoB gene fragment and the ops-independent N-domain. While it is impossible to find a truly pause-less natural DNA template, the rpoB gene is the best model system known: it is devoid of strong regulatory pauses and has been used extensively in studies of RNA chain elongation (2,33). We monitored the overall elongation rate by accumulation of 1225 nt run-off RNA (Figure 5B). At 1 mM NTPs, the N-domain failed to accelerate wild-type RNAP, and the mean rate was actually reduced from 30 to 25 nt/s (Figure 5C). However, wild-type RNAP was modestly (1.7-fold) accelerated by the N-domain at subsaturating (25 μM) NTPs. The net effect of the RfaH N-domain on wild-type RNAP (Figure 5D) was the reduction in Km and, more importantly, the 1.7-fold increase in the kcat/Km ratio that approximates the second order rate constant for substrate binding. The increase in the kcat/Km ratio was more pronounced (2.1-fold) with the slow βQ513P RNAP, which was accelerated by the RfaH N-domain even at 1 mM NTPs; apparently this enzyme was still limited by substrate availability under such conditions. Interestingly, the fastest RNAPs in our collection, β′F773V and β′Δ SI3, were actually slower than the wild type at 1 mM NTPs, and were not accelerated by N-domain at all NTPs concentrations tested (from 25 μM to 1 mM; Figure 5D and data not shown). We conclude that the fast phenotype of these enzymes is due to their inability to pause rather than the higher intrinsic elongation rate. Consequently, it appears that RfaH acts by reducing pausing rather than by increasing the elongation rate, as it fails to act under the conditions that already suppress pausing.
Figure 5.
RfaH does not accelerate transcription in the absence of pausing. (A) The pIA146 template. (B) Halted radiolabeled A29 TECs formed with the wild-type (WT) RNAP were extended with NTPs in the absence (upper panels) or in the presence (lower panels) of the RfaH N-domain, aliquots were withdrawn at indicated times and analyzed on 4% denaturing gels. Only the gel region near the full-length transcript (RO) is shown. (C) The mean elongation rate of the WT, βQ513P and β′ΔSI3 RNAPs as a function of [NTPs] in the absence (circles) and the presence (squares) of the RfaH N-domain. The mean rate was estimated by fitting the RO product accumulation (data from panel B and analogous experiments) to exponential (with an offset) cumulative distribution function using Scientist 3.0 software (Micromath). (D) Michaelis–Menten parameters kcat, Km and kcat/Km ratio estimated from data in C.
RfaH inhibits pyrophosphorolysis
Molecular motions that underlie the formation of the paused state are not known in detail, but this isomerization likely occurs from the pre-translocated state and involves some rearrangements of the active site (see Discussion section). The increase in NTP concentration is known to reduce pausing by shifting the TEC into the post-translocated state. RfaH increases kcat/Km ratio, thus effectively mimicking the increase in the NTP concentration, and may therefore also act to stabilize the post-translocated state either directly or indirectly, by increasing the TEC′s affinity for substrate NTPs during elongation. To distinguish between these possibilities, we examined the effect of RfaH on pyrophosphorolysis, a reversal of the nucleotide addition reaction that occurs in pre-translocated TEC (Figure 6A) in the absence of NTPs.
We used halted A26 complexes that exist predominantly in pre-translocated or even backtracked state and are sensitive to PPi—or Gre-mediated transcript cleavage, respectively (13). We formed halted radiolabeled A26 complexes in the absence or in the presence of the RfaH N-domain, and then challenged these complexes with low concentrations of PPi (25 μM) for 10–180 s at 37°C (Figure 6A); as reported previously, A26 complexes were highly sensitive to cleavage, generating 25- and 24-mer products within 10 s.
We found that complexes formed with the wild-type RNAP were stabilized against PPi-induced cleavage by the N-domain (Figure 6C). This effect was dependent on specific RfaH–RNAP interaction because RNA cleavage in the complexes formed with the β′ ΔCH enzyme, which lacks the RfaH contact site and is unable to respond to either full-length RfaH or N-domain (20), was not altered in the presence of the N-domain. The apparent protection against pyrophosphorolysis is consistent with the hypothesis that RfaH stabilizes the post-translocated state of the TEC. These results are inconsistent with the direct effect of RfaH on NTP binding: if RfaH acted primarily to increase the substrate affinity, it would facilitate forward and reverse reactions similarly at limiting substrate concentrations (such as used here) because RNAP contacts to the β and γ phosphates (pyrophosphate moiety) constitute the major fraction of the NTP interactions in the substrate-bound TEC (34). In summary, our data argue that RfaH increases NTP binding indirectly by favoring the particular (post-translocated) TEC conformation that serves as a target for NTP, thereby facilitating the catalysis in the forward (nucleotide addition) direction.
DISCUSSION
In this work, we demonstrate that the response of the E. coli RNAP to bacterial antiterminator RfaH is modulated by the kinetic properties of the enzyme itself: enzyme variants with the same or slower elongation rate than the wild-type RNAP are accelerated by RfaH, whereas the fast enzymes are resistant to its action. The widely spread positions of the residues altered in RfaH-resistant RNAPs (Figure 3B) and the fact that the RfaH-binding target, the β′CH, is located 50 and 75 Å away from the closest substitution and the RNAP active site, respectively, argue against a direct effect of fast substitutions on RfaH binding and in support of an allosteric mechanism of control. Below we offer structural insights into the origins of the anti-pausing behavior of RfaH. Our analysis also allows us to draw many parallels to other antitermination systems—we compare the RfaH mechanism of action to that of λQ to conclude that in spite of the apparent differences in the nucleic acid targets, the time of recruitment, and the targets on RNAP, the two proteins share many details of the molecular mechanism during both recruitment and post-recruitment modification of the TEC.
RfaH and fast substitutions increase TEC processivity exclusively by reducing pausing
Our data demonstrate that the elongation enhancer RfaH as well as fast and majority of slow substitutions modulate the RNAP rate by altering its propensity to isomerize into an off-pathway paused state. Specifically, we show that β′F773V and β′ΔSI3 RNAPs, the two fastest variants among the ones we studied, display pause-free RNA chain elongation, yet have the same or lower turnover numbers than does the wild-type enzyme (Figure 5 and data not shown). Similarly, RfaH fails to accelerate transcription by the wild-type RNAP on the template devoid of pauses at saturating NTPs or further enhance elongation by pause-resistant RNAPs even in the presence of regulatory pauses. These results are consistent with RfaH and fast substitutions acting upon the same, ‘pausing’ step.
On the other hand, the slow enzymes fall into two groups. The first group is comprised from the catalytically competent but pause-prone variants bearing allosterically acting substitutions (e.g. βQ513P); consistent with its exclusive effect on pausing, RfaH is able to rescue these variants. In contrast, two enzymes with substitutions near the active site (βS1105A, βM1107A) are resistant to the RfaH-induced acceleration. These residues are located near the three basic side chains (βR678, βR1106 and β′R731) that hold the substrate phosphates in the active site (34)—thus substitutions of βS1105 and βM1107 may trigger the displacement of the Arg side chains, consequently altering the substrate positioning in the active site and slowing catalysis. We surmise that RfaH is unable to correct defects in the active site caused by βS1105A and βM1107A substitutions which confer diminished catalytic competence.
RfaH may function by facilitating RNAP translocation
RfaH (or another factor/fast RNAP) may reduce pausing by (i) stabilizing the pre-translocated state from isomerization into a paused state or (ii) favoring the TEC transition into the post-translocated state, in which the active site is programmed for facile nucleotide addition. While at least some fast RNAPs (e.g. βP560S,T563I) are thought to utilize the first mechanism (35) (Figure 1), RfaH likely acts by shifting equilibrium towards the post-translocated state. Indeed, the RfaH-induced increase in kcat/Km ratio, and thus presumably the apparent rate of substrate binding, is a predictable consequence of the increase in the fraction of post-translocated TEC. Moreover, we show that RfaH inhibits pyrophosphorolysis of the nascent RNA and thus likely facilitates translocation directly and independently on the substrate NTP (Figure 6). Interestingly, the RfaH paralog NusG is also thought to reduce transcriptional pausing by affecting the TEC translocation; specifically, by preventing RNAP backtracking (24,36,37).
The NT strand and the β pincer as the targets for RfaH action
We envision at least two mechanisms by which RfaH may facilitate translocation. First, RfaH directly cross-links to the NT strand (16) and may act as a ratchet to favor the forward translocation by binding to the upstream portion of the NT DNA on the surface of the TEC; such an effect does not necessarily require a change in the active site conformation. To evaluate this possibility, we need an experimental model for the RfaH/NT/RNAP interactions within the TEC. However, none of the TEC structures currently available contains the NT DNA strand, and the only bacterial species for which the high-resolution structures exist (Thermus) lack RfaH and differ from E. coli in many RNAP surface features, which are likely essential for RfaH action.
Second, RfaH may allosterically control the positioning of RNAP elements that affect its translocation along the DNA. These elements are yet unknown but several models that emphasized the key role of the conformational transitions (bending/straightening) of the bridge helix (BH) in translocation were proposed (36,38). However, our structural analysis argued against the BH flexibility and instead revealed two mobile structural elements, the β′-subunit trigger loop (TL) and the β-subunit pincer, that change their positions in the bacterial TECs (34,39).
In the current view of the nucleotide addition cycle (Figure 7), the TEC translocation between the pre- and post-states is driven by thermal motions and is biased toward the post-translocated state by binding of the incoming NTP substrate in the pre-insertion site. The TL, which is unstructured in the substrate-free post-translocated TEC, refolds into two α-helices (trigger helices, TH) which align with the BH to form the three-helical bundle in the presence of substrate; this dramatic structural transition effectively ‘closes’ the active site and likely represents the rate-limiting step for catalysis (34). After catalysis and PPi release, the reverse transitions occur and the active site opens. The TH may unfold upon PPi release or during translocation. We favor the latter pathway, in which unfolding occurs in concert with the β pincer closure, because this scenario is simpler, does not contradict any data that we are aware of, and readily explains the formation of the paused state, which occurs from the pre-translocated state and is accompanied by the TL folding into an unknown conformation (TL*; Figure 7) likely stabilized by the frayed 3′ RNA nt (35). Accordingly, both structuring (TL→TH) and unstructuring (TL*→TL) of the TL may, theoretically, provide an anti-pausing effect.
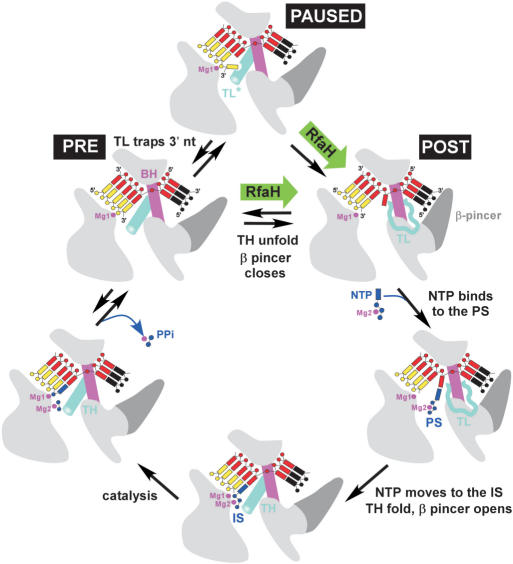
Figure 7.
The nucleotide addition cycle (34). The TEC elements shown are as follows: RNAP (light gray), BH (magenta), RNA (yellow), NT DNA (black), template DNA (red), TL (cyan) and β pincer (dark gray). Substrate NTP (blue) in complex with Mg2 binds to the post-translocated TEC in the pre-insertion site (PS) and then moves to the active, insertion site (IS); this transition is accompanied by the active site closure through folding of the TH and opening of the β pincer. At a pause site, the TL apparently folds into an alternative state (TL*), trapping the frayed 3′ nt (35). RfaH may facilitate the β pincer closure, thereby reducing pausing (paused→post) or favoring the pre→post transition during the elongation pathway.
Factors that control the TL→TH transition would be expected to affect both the nucleotide binding (and NTP-driven translocation) and the formation of the off-pathway intermediates. Indeed, deletion of a large β′ domain inserted into the middle of TL in the E. coli RNAP (in the β′ΔSI3 enzyme) confers catalytic defects [Figure 5 and (34)] and a pause-resistant, fast phenotype (Figure 3). Studies in the yeast system also highlighted the regulatory importance of the TL and suggested that regulatory proteins may exert their effects through altering the TL position (40). However, RfaH binds on the opposite side of the TEC in the model (Figure 3B) and thus cannot control the TL directly. Allosterically mediated unfolding of the TL (that would destabilize both the pre- and paused states) by RfaH cannot be ruled out off hand, although we cannot envision a plausible structural mechanism for this action.
The TEC transition between the open and the closed, catalytically competent state is also accompanied by rearrangements of several bulky β domains that trigger the displacement of the β-pincer and subsequently to opening (by ∼4.5 Å) of the downstream DNA channel (34,39) (Figure 7). We speculate that the β pincer may be the ultimate target of the RfaH-induced allosteric signal: RfaH may facilitate the β pincer closure, and thus favor the TEC transition to the post-translocated state. Consistently, RfaH interacts with the RNAP domain (β459–505) (20) that belongs to the group of the interconnected β domains undergoing systematic structural rearrangement in the TEC. Interestingly, this β domain is directly connections to the rifamycin-binding site, wherein a hypothetical allosteric signal propagated to the active site originates (29) and in which slow (βQ513P) and fast (βH526Y) RNAP substitutions are located—these substitutions can therefore also affect the β pincer movements. Finally, the stabilization of the β pincer in a closed conformation may explain the inhibitory effect of RfaH on the pause-free elongation rate (Figure 5) that is likely limited by the concerted TH-folding/β pincer opening (Figure 7). Overall, our results suggest that RfaH reduces pausing by disfavoring the pause-forming TEC state. Distinguishing whether RfaH acts as a ratchet through its contacts to the NT DNA strand, as an allosteric switch which controls domain rearrangements that accompany transitions between closed and open RNAP states, or as a combination thereof is not trivial, and awaits not only the atomic resolution structure of the RfaH-bound TEC but also a better understanding of the physicochemical properties of the elemental nucleotide addition cycle of the RNAP.
Fundamental congruity of antitermination mechanisms
Our analysis revealed many unexpected similarities between RfaH and λQ, from the mechanism of recruitment to the downstream RNAP modification (Figure 8). Below we compare RfaH and Q mechanisms step by step, highlighting their similarities and differences.
Figure 8.
Comparison of the RfaH and λQ mechanisms. (A). Mechanism of recruitment. (Left) RNAP pauses downstream from the λ pR′ promoter start site (shown by a bent arrow); the pause depends on the σ70 interaction with a –10-like sequence element positioned 12 nt downstream from the promoter –10 hexamer. Q (shown as a dimer) binds to the QBE DNA site located between the –35 and the –10 promoter elements and to the region 4 of the σ70 subunit (47) in the promoter-proximal paused complex. Domain rearrangements in σ (likely triggered by the nascent RNA extrusion) accompany the paused complex formation: σ4 is repositioned relative to σ2, and apparently interacts with the –35-like element adjacent to the pause-inducing sequence (18). Destabilization of the σ4–β flap interactions unmasks the Q target on core RNAP (22). (Right) RNAP pauses at a promoter-distal ops site and the N-domain of RfaH binds specifically to the NT DNA, triggering domain dissociation that in turn unmasks the site on the N-domain that binds to the β′ CH (20). (B). Post-recruitment TEC modification. (Left) Q (likely together with NusA) remains bound to RNAP throughout transcription of the late λ operon making RNAP resistant to pause and all termination signals. (Right) RfaH also remains bound to RNAP through contacts between the N-domain and the β′ CH (and non-specific interactions with the NT DNA); RNAP becomes resistant to pause and some termination signals. The C-domain of RfaH may mediate binding to translation and secretion factors.
Recruitment to the TEC
RfaH is recruited to the elongating RNAP at ops sequences, which are positioned 100 nt or more downstream from a promoter. The ops element induces RNAP pausing, and likely not only makes specific interactions with RfaH but also induces a conformational change in RNAP that is required for RfaH recruitment (20). In the context of the ops-paused TEC, the most conserved ops nts are exposed on the TEC surface in the NT DNA (16). Specific interactions between RfaH and the ops DNA delay RNAP downstream from the ops site (C45 in Figure 2), apparently by inducing backtracking (G.A.B., unpublished data), and facilitate dissociation of RfaH domains to unmask the RNAP-binding site located at the domain interface (20).
Q is recruited to RNAP at the late λ promoter. Q binds to the double-stranded DNA element (QBE) that overlaps the promoter; however, productive Q recruitment requires the σ initiation factor (18), which interacts with the NT DNA immediately downstream from the start site to induce a promoter-proximal pause. σ/DNA interaction is thought to induce a specific, Q-receptive state of the TEC, in which partial release of σ-promoter and σ-core contacts exposes a target for Q on the core enzyme (22). Specific contacts between σ and the –10-like element in the NT DNA also induce RNAP backtracking and thus delay its escape from the promoter-proximal pause (41).
Thus, in both cases the specific (but totally different) NT DNA sequence mediates recruitment of antiterminator to RNAP; RfaH recognizes the ops sequence directly, whereas Q relies on the σ factor. Both sets of specific contacts must be broken for RNAP escape from the recruitment site—transient retention of these contacts induces a characteristic pause. Interactions with the DNA induce conformational changes in the protein (RfaH or holoenzyme) that expose the binding site for its partner, core RNAP or Q, respectively. Finally, the same RNAP region, the β′ CH, mediates recruitment, either directly, by binding to RfaH (20), or indirectly, though binding to σ necessary for Q recruitment (42).
Post-recruitment modification
Following the escape from their respective recruitment sites, RfaH and Q likely lose all specific interactions with the nucleic acids but remain bound to the TEC, essentially turning into the accessory subunits. The interaction surfaces are likely distinct: Q may bind to the β flap domain near the nascent RNA exit channel, whereas RfaH remains bound to the β′CH. Both proteins, however, apparently induce allosteric signals that suppresses RNAP pausing. These signals may converge: the genetic screen for Q-resistant mutants (43) identified substitutions near the rifampicin-binding site and near the active site, the two regions where the substitutions conferring resistance to RfaH are located; substitutions of βD675 render RNAP defective in response to both Q and RfaH. Similarly to RfaH, the resistance to Q unlikely results from the loss of Q binding—most of the substituted residues are located in the interior of the TEC and far apart. This remarkable convergence in regulatory mechanisms between Q and RfaH may extend even further, as slow RNAP variant, βQ513P, was shown to be hypersensitive to Q-dependent acceleration, while the fast one, βH526Y, turned out to be resistant, with both enzymes retaining ability to bind Q (44).
Elongation enhancement versus TEC stabilization
There is one principal difference between the action of Q and RfaH: while both proteins increase the overall elongation rate, Q (and λ N) additionally stabilizes the TEC, thereby preventing RNA release at terminators. RfaH acts more as an anti-pausing factor, and is apparently specific for those signals that may be particularly sensitive to changes in the elongation rate. This difference likely stems from the distinct regulatory roles of these proteins: unlike the phage regulators, which ultimately cause cell death, RfaH modification has to be transient and must be turned off at the end of the operon. Thus, while RfaH apparently only reduces pausing (that serves as a prelude to termination) and does not actively stabilize the complex against dissociation, other antiterminators apparently employ additional mechanisms to inhibit RNA release; the anti-pausing effect alone is insufficient to explain the dramatic effect of Q (19) and N (45) on termination. The simplest explanation is that the other contacts within the TEC, such as that to the nascent RNA (45) and/or the β flap (22), may directly stabilize the RNA against the action of terminator hairpins or Rho protein.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported in part by the NIH grants GM67153 and AI064819 (to I.A.) and GM74252 (to D.G.V.). Funding to pay the Open Access publication charges for this article was provided by NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Mooney RA, Artsimovitch I, Landick R. Information processing by RNA polymerase: recognition of regulatory signals during RNA chain elongation. J. Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neuman KC, Abbondanzieri EA, Landick R, Gelles J, Block SM. Ubiquitous transcriptional pausing is independent of RNA polymerase backtracking. Cell. 2003;115:437–447. doi: 10.1016/s0092-8674(03)00845-6. [DOI] [PubMed] [Google Scholar]
- 3.Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem. Soc. Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- 4.Richardson J, Greenblatt J. In: Escherichia coli and Salmonella : Cellular and Molecular Biology, Vol. 1. Neidhardt F, Curtiss R. III, Ingraham J, Lin E, Low K, Magasanik B, Reznikoff W, Riley M, Schaechter M, et al., editors. Washington, DC: ASM Press; 1996. pp. 822–848. [Google Scholar]
- 5.Chandler M, Bird RE, Caro L. The replication time of the Escherichia coli K12 chromosome as a function of cell doubling time. J. Mol. Biol. 1975;94:127–132. doi: 10.1016/0022-2836(75)90410-6. [DOI] [PubMed] [Google Scholar]
- 6.Artsimovitch I, Svetlov V, Murakami KS, Landick R. Co-overexpression of Escherichia coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J. Biol. Chem. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- 7.Fisher RF, Yanofsky C. Mutations of the beta subunit of RNA polymerase alter both transcription pausing and transcription termination in the trp operon leader region in vitro. J. Biol. Chem. 1983;258:8146–8150. [PubMed] [Google Scholar]
- 8.Landick R, Stewart J, Lee DN. Amino acid changes in conserved regions of the beta-subunit of Escherichia coli RNA polymerase alter transcription pausing and termination. Genes Dev. 1990;4:1623–1636. doi: 10.1101/gad.4.9.1623. [DOI] [PubMed] [Google Scholar]
- 9.Tavormina PL, Landick R, Gross CA. Isolation, purification, and in vitro characterization of recessive-lethal-mutant RNA polymerases from Escherichia coli. J. Bacteriol. 1996;178:5263–5271. doi: 10.1128/jb.178.17.5263-5271.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weilbaecher R, Hebron C, Feng G, Landick R. Termination-altering amino acid substitutions in the beta′ subunit of Escherichia coli RNA polymerase identify regions involved in RNA chain elongation. Genes Dev. 1994;8:2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- 11.Heisler LM, Suzuki H, Landick R, Gross CA. Four contiguous amino acids define the target for streptolydigin resistance in the beta subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 1993;268:25369–25375. [PubMed] [Google Scholar]
- 12.McDowell JC, Roberts JW, Jin DJ, Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- 13.Artsimovitch I, Chu C, Lynch AS, Landick R. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science. 2003;302:650–654. doi: 10.1126/science.1087526. [DOI] [PubMed] [Google Scholar]
- 14.Vassylyev DG, Svetlov V, Vassylyeva MN, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Artsimovitch I. Structural basis for transcription inhibition by tagetitoxin. Nat. Struct. Mol. Biol. 2005;12:1086–1093. doi: 10.1038/nsmb1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Severinov K, Markov D, Severinova E, Nikiforov V, Landick R, Darst SA, Goldfarb A. Streptolydigin-resistant mutants in an evolutionarily conserved region of the beta′ subunit of Escherichia coli RNA polymerase. J. Biol. Chem. 1995;270:23926–23929. doi: 10.1074/jbc.270.41.23926. [DOI] [PubMed] [Google Scholar]
- 16.Artsimovitch I, Landick R. RfaH stimulates chain elongation by bacterial transcription complexes after recruitment by the exposed nontemplate DNA strand. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 17.Rees W, Weitzel S, Yager T, Das A, von Hippel P. Bacteriophage lambda N protein alone can induce transcription antitermination in vitro. Proc. Natl Acad. Sci. USA. 1996;93:342–346. doi: 10.1073/pnas.93.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts JW, Yarnell W, Bartlett E, Guo J, Marr M, Ko DC, Sun H, Roberts CW. Antitermination by bacteriophage lambda Q protein. Cold Spring Harb. Symp. Quant. Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- 19.Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- 20.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovitch I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol. Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheeran A, Babu Suganthan R, Swapna G, Bandey I, Achary MS, Nagarajaram HA, Sen R. Escherichia coli RNA polymerase mutations located near the upstream edge of an RNA:DNA hybrid and the beginning of the RNA-exit channel are defective for transcription antitermination by the N protein from lambdoid phage H-19B. J. Mol. Biol. 2005;352:28–43. doi: 10.1016/j.jmb.2005.06.052. [DOI] [PubMed] [Google Scholar]
- 22.Nickels BE, Roberts CW, Roberts JW, Hochschild A. RNA-mediated destabilization of the sigma(70) region 4/beta flap interaction facilitates engagement of RNA polymerase by the Q antiterminator. Mol. Cell. 2006;24:457–468. doi: 10.1016/j.molcel.2006.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greive SJ, von Hippel PH. Thinking quantitatively about transcriptional regulation. Nat. Rev. Mol. Cell. Biol. 2005;6:221–232. doi: 10.1038/nrm1588. [DOI] [PubMed] [Google Scholar]
- 24.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl Acad. Sci. USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey MJ, Hughes C, Koronakis V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 1997;26:845–851. doi: 10.1046/j.1365-2958.1997.6432014.x. [DOI] [PubMed] [Google Scholar]
- 26.Carter HD, Svetlov V, Artsimovitch I. Highly divergent RfaH orthologs from pathogenic proteobacteria can substitute for Escherichia coli RfaH both in vivo and in vitro. J. Bacteriol. 2004;186:2829–2840. doi: 10.1128/JB.186.9.2829-2840.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason SW, Li J, Greenblatt J. Host factor requirements for processive antitermination of transcription and suppression of pausing by the N protein of bacteriophage lambda. J. Biol. Chem. 1992;267:19418–19426. [PubMed] [Google Scholar]
- 28.Koronakis V, Cross M, Hughes C. Expression of the E.coli hemolysin secretion gene hlyB involves transcript anti-termination within the hly operon. Nucleic Acids Res. 1988;16:4789–4800. doi: 10.1093/nar/16.11.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Artsimovitch I, Vassylyeva MN, Svetlov D, Svetlov V, Perederina A, Igarashi N, Matsugaki N, Wakatsuki S, Tahirov TH, et al. Allosteric modulation of the RNA polymerase catalytic reaction is an essential component of transcription control by rifamycins. Cell. 2005;122:351–363. doi: 10.1016/j.cell.2005.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Temiakov D, Zenkin N, Vassylyeva MN, Perederina A, Tahirov TH, Kashkina E, Savkina M, Zorov S, Nikiforov V, et al. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol. Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 31.Svetlov V, Vassylyev DG, Artsimovitch I. Discrimination against deoxyribonucleotide substrates by bacterial RNA polymerase. J. Biol. Chem. 2004;279:38087–38090. doi: 10.1074/jbc.C400316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, Goldfarb A. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tolic-Norrelykke SF, Engh AM, Landick R, Gelles J. Diversity in the rates of transcript elongation by single RNA polymerase molecules. J. Biol. Chem. 2004;279:3292–3299. doi: 10.1074/jbc.M310290200. [DOI] [PubMed] [Google Scholar]
- 34.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 35.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol. Cell. 2007 doi: 10.1016/j.molcel.2007.06.008. in press. [DOI] [PubMed] [Google Scholar]
- 36.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 37.Pasman Z, von Hippel PH. Regulation of rho-dependent transcription termination by NusG is specific to the Escherichia coli elongation complex. Biochemistry. 2000;39:5573–5585. doi: 10.1021/bi992658z. [DOI] [PubMed] [Google Scholar]
- 38.Epshtein V, Mustaev A, Markovtsov V, Bereshchenko O, Nikiforov V, Goldfarb A. Swing-gate model of nucleotide entry into the RNA polymerase active center. Mol. Cell. 2002;10:623–634. doi: 10.1016/s1097-2765(02)00640-8. [DOI] [PubMed] [Google Scholar]
- 39.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 40.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marr MT, Roberts JW. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol. Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 42.Ko DC, Marr MT, Guo J, Roberts JW. A surface of Escherichia coli sigma 70 required for promoter function and antitermination by phage lambda Q protein. Genes Dev. 1998;12:3276–3285. doi: 10.1101/gad.12.20.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santangelo TJ, Mooney RA, Landick R, Roberts JW. RNA polymerase mutations that impair conversion to a termination-resistant complex by Q antiterminator proteins. Genes Dev. 2003;17:1281–1292. doi: 10.1101/gad.1082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDowell JC. Ph.D. thesis. Ithaca: Cornell University; 1994. Relation of transcription termination and antitermination to the kinetics of transcription elongation. [Google Scholar]
- 45.Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell. 2001;107:437–449. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 46.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 47.Nickels BE, Roberts CW, Sun HI, Roberts JW, Hochschild A. The σ70 subunit of RNA polymerase is contacted by the λQ antiterminator during early elongation. Mol. Cell. 2002;10:611–622. doi: 10.1016/s1097-2765(02)00648-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.