Abstract
Using sensitive structure similarity searches, we identify a shared α+β fold, RAGNYA, principally involved in nucleic acid, nucleotide or peptide interactions in a diverse group of proteins. These include the Ribosomal proteins L3 and L1, ATP-grasp modules, the GYF domain, DNA-recombination proteins of the NinB family from caudate bacteriophages, the C-terminal DNA-interacting domain of the Y-family DNA polymerases, the uncharacterized enzyme AMMECR1, the siRNA silencing repressor of tombusviruses, tRNA Wybutosine biosynthesis enzyme Tyw3p, DNA/RNA ligases and related nucleotidyltransferases and the Enhancer of rudimentary proteins. This fold exhibits three distinct circularly permuted versions and is composed of an internal repeat of a unit with two-strands and a helix. We show that despite considerable structural diversity in the fold, its representatives show a common mode of nucleic acid or nucleotide interaction via the exposed face of the sheet. Using this information and sensitive profile-based sequence searches: (1) we predict the active site, and mode of substrate interaction of the Wybutosine biosynthesis enzyme, Tyw3p, and a potential catalytic role for AMMECR1. (2) We provide insights regarding the mode of nucleic acid interaction of the NinB proteins, and the evolution of the active site of classical ATP-grasp enzymes and DNA/RNA ligases. (3) We also present evidence for a bacterial origin of the GYF domain and propose how this version of the fold might have been utilized in peptide interactions in the context of nucleoprotein complexes.
INTRODUCTION
Several recent structural studies indicate that a number of protein folds have been repeatedly deployed as scaffolds for a biochemically diverse set of interactions with nucleic acids. Some notable examples of such folds are the RNA recognition motif (RRM)-like fold, double ψ-beta barrel (and the related EI barrel), β-grasp, S5-like fold, HhH (helix-hairpin-helix) and HTH (helix-turn-helix) [for further details see the SCOP database (1)]. These folds are not only found in proteins that passively interact with nucleic acids, but also form the catalytic domains of several key enzymes involved in nucleic acid metabolism, such as nucleic acid polymerases, pseudouridine synthases, topoisomerases, RNA phosphatases and nucleases (2–7). Detection of conserved folds and the characterization of common structural features shared by different representatives of a fold often illuminate several functional aspects of the proteins in which they are found (8–11). In particular, such studies are useful in interpreting nucleic acid–protein interactions, predicting the active sites of enzymes that operate on nucleic acids, and uncovering the evolutionary history of complex biochemical functions observed in extant organisms(12–16).
While these nucleic acid binding domains display folds spanning the entire structural spectrum, certain generic structural classes are frequently encountered amongst them. These include small β-barrel folds (e.g. double ψ-beta barrel and the related EI barrel), several two-layered α+β folds (e.g. RRM-like, β-grasp and S5-like fold) and simple helical bundles (e.g. HhH and HTH) [see SCOP database (1)]. Though, the conserved core of these nucleic acid binding domains are small compact structures, they might show several elaborations in the form of insertions and extensions that are associated with acquisition of diverse biochemical activities. Furthermore, some structures show signs of having been assembled from simpler structural units that usually need to form obligate dimers in order to attain stability. In particular, such a pathway has been invoked to explain origins of some β-barrel folds, like the 6-stranded double ψ-beta barrel (DPBB) and the EI-barrel folds, which are found in several ancient domains with major roles in nucleic acid binding and metabolism (9,10,17,18). The former domain, amongst other contexts, forms the catalytic domain of both DNA- and RNA-templated RNA polymerases, while the latter domain is found in translation elongation factors (10,19,20). Both of these domains have been derived from duplication of the same 3-stranded precursor, followed by dimerization. Despite being assembled from a common ancestral precursor, the two folds have very different dimerization patterns of the monomer units: in the DPBB the two units interlock to form the two characteristic ψ-loops, whereas in the EI barrel they are placed adjacently without any cross-over (19). The β-clip fold found in the SET methyltransferase domain, and sandwich-barrel hybrid motif fold also found in the RNA polymerases, are other comparable examples of assembly of barrel-like folds from simple 3-stranded elements (19,21,22).
The small size of many nucleic acid binding folds makes identification of their members, through entirely automatic methods, difficult. This difficulty is further compounded by the possibility of circular permutation, insertions and alternative structural arrangements seen in folds potentially evolving from accretion of simple structural elements. However, given the relatively small number of globular non-helical folds in the protein universe, these could be identified using a combination of transitive structural and topological similarity searches, and case-by-case analysis of individual folds. We were especially interested to explore if structural themes analogous to those observed in the small β-barrel folds might also be operational in small, ancient α+β folds, specifically those with functions related to nucleic acid metabolism. In particular, we sought to identify simple α+β folds with internal symmetries that might have been potentially assembled through duplication in the manner of the above-mentioned β-barrel folds. Using the above-stated multi-pronged approach, we present the discovery of a small two-layered α+β fold constructed from simple units with two strands and a helix, showing multiple topological variants emerging from different circular permutations. This fold appears to have been utilized in diverse biochemical contexts in key roles related to nucleic acid and nucleotide metabolism. Its characterization laid out in this article helps in understanding the substrate (nucleotide, nucleic acid or protein) interaction and evolution of various proteins containing this fold.
MATERIAL AND METHODS
The non-redundant (NR) database of protein sequences (National Center for Biotechnology Information, NIH, Bethesda, MD) was searched with the BLASTP program (23). Profile searches were conducted using the PSI-BLAST program (24) with either single sequences or multiple alignments as queries, with a profile inclusion expectation (e) value threshold of 0.01. Searches were iterated until convergence. For queries and searches containing computationally biased segments, the statistical correction option built into the BLAST program was used. Multiple alignments were constructed using the MUSCLE and/or T-COFFEE programs (25,26), followed by manual adjustment based on PSI-BLAST hsp results and information provided by solved three-dimensional structures. All large-scale sequence and structure analysis procedures were carried out with the TASS software package (V. Anantharaman, SB and LA, unpublished results), a successor to the SEALS package (27). Protein structures were visualized using the Swiss-PDB viewer (28) and cartoons were constructed with the PyMOL program (http://www.pymol.org). Protein secondary structure predictions were made with the JPRED program (29), using multiple alignments as queries. Phylogenetic analysis was carried out using a variety of methods including maximum-likelihood, neighbor-joining and minimum evolution (least squares) methods (30–32). Maximum-likelihood distance matrices were constructed using the TreePuzzle 5 program (33) and were used as input for the construction of neighbor-joining with the Weighbor program (30).
Structure similarity searches were conducted using the standalone version of the DALI program called DaliLite (34,35) with the query structures scanned against local current version of PDB that has all chains as separate entries. The structural hits for each query was collected and parsed for congruence of strand orientation with the template structure (L3-I, PDBID: 1JJ2, chain B, 80-190).This was further confirmed by visual examination of each structure. The interacting residues of various proteins of the fold with their interacting molecules have been deduced using custom-written PERL scripts. The scripts encode interacting distance cut-off values of 5.0 and 3.5 Å between appropriate atoms in 3D for deducing the hydrophobic and polar interactions, respectively. These inferred interactions were further examined manually using Swiss-PDB viewer for confirming the contacts between residues of the fold and atomic groups of interacting partners.
RESULTS AND DISCUSSION
Recognition of RAGNYA fold
In the search for novel domains of unknown provenance, we surveyed the folds in the SCOP database (1) for uncharacterized globular inserts. In the archaeo-eukaryotic ribosomal protein L3 from the ribosomal large subunit (50S) (36), which is classified as an EI barrel fold (10,17,19,37), we observed an insert (PDB: 1JJ2 chain B residues 80–190) folding into a distinct un-classified α+β domain. Examination of this domain showed that it formed a two-layered structure with a 4-stranded β-sheet, and two α-helices packing against one of the faces (Figure 1). The topology of the L3 insert (L3-I) domain indicated that it was comprised of a tandem repeat of two β–β–α units. When viewed from the exposed face of the sheet, the strands show a characteristic down-up-down-up polarity (Figure 1). Comparison of the topology with other two-layered α+β folds with β–β–α elements, such as (1) Ribonuclease PH fold, (2) DcoH fold and (3) homing endonuclease and glucose permease fold in the SCOP database showed that the L3-I was distinct from those folds (1). The topology of the L3-I domain also showed that the N- and C-termini were juxtaposed, potentially allowing circularly permuted versions of such a fold to exist. In order to identify other domains with an equivalent fold, we set up a search procedure incorporating multiple criteria: (1) structure similarity searches of a local, current version of the PDB database were initiated with the DaliLite program. These searches were conducted transitively to account for the possibility of extreme structural divergence acting on a fold of relatively small size. (2) Results of these searches were filtered such that the recovered modules completely mapped on the L3-I fold and did not overlap with any previously characterized globular fold. (3) The resulting hits were further constrained for equivalence of strand polarity with the query, and not just topology (to account for circularly permuted versions). (4) Iterative sequence similarity searches, using the PSI-BLAST program, were set up with each true positive recovered in the above structure similarity searches to identify sequence homologs and the phyletic patterns of the concerned domains.
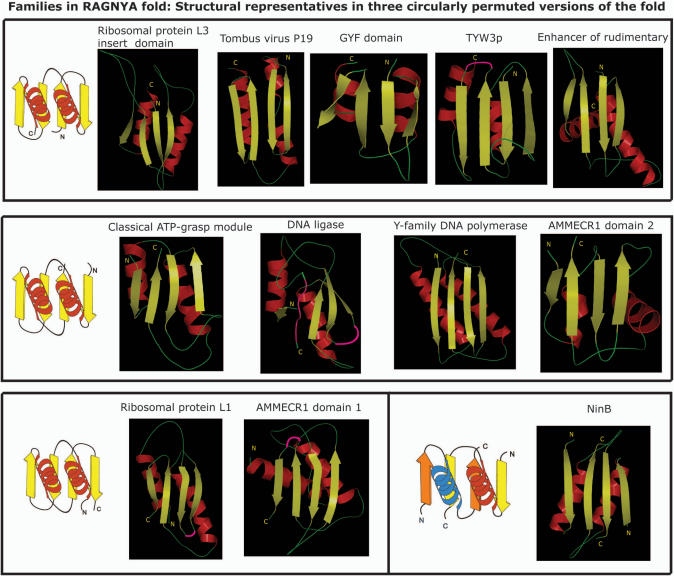
Figure 1.
Representative structures from various families of the RAGNYA fold are shown in the ‘open face’ view. These families encompass the three distinct circularly permuted versions of the fold, whose topology diagrams are shown beside the corresponding structures. The topology diagrams correspond to a 180° rotation about a vertical axis in the plane of paper with respect to the view of the actual structures. The two identical subunits of NinB are shown in different colors in the topology diagram. The figure were made using Pymol. The PDB IDs of the structures are respectively 1JJ2, 1RPU, 1WH2, 1TLJ, 2NML, 1WR2, 1V9P, 1JIH, 1VAJ, 1DWU, 1WSC and 1PC6 (from top to bottom).
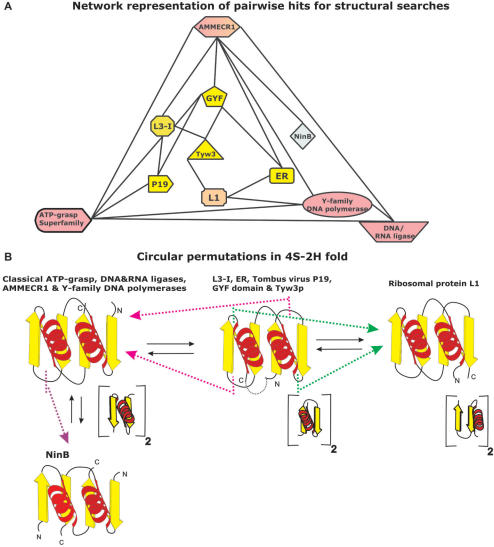
The results of the procedure were represented as a network and true positives form a completely connected graph (Figure 2A), which was not reproduced with a comparably high degree of inter-connectivity using other 4-stranded, two-layered folds as starting points. As a result we identified 11 different domains containing an equivalent fold, namely: (1) L3-I (PDB: 1JJ2 chain B residues 80–190); (2) siRNA silencing repressor of Tombusviruses (CIRV p19; PDB: 1RPU, chain A); (3) The GYF domain (PDB 1L2Z, chain A; 1WH2); (4) Enhancer of rudimentary proteins (ER; PDB: 1WWQ); (5) one domain of the tRNA Wybutosine biosynthesis enzyme Tyw3p, typified by the SSO0622 protein from Sulfolobus solfataricus (PDB: 1TLJ, chain A, residues 53–102 and 147–172); (6) the two related globular domains of AMMECR1 (PDB: 1WSC, chain A); (7a) the N-terminal domain of ATP-grasp enzyme superfamily (PDB: 1WR2, chain A, residues 37–116) (7b) the related domain from the DNA/RNA ligase-type nucleotidyltransferases (1V9P chain B, residues 2085–2119 and 2255–2293); (8) C-terminal DNA-interacting domain of DinB-like (Y-family) DNA polymerases (PDB: 1JX4, Chain A, residues 241–341); (9) The DNA-recombination proteins of the NinB family from λ and other related caudate bacteriophages (PDB: 1PC6 A and B); (10) the ribosomal protein L1 (1DWU, chain B) (Figure 1). Visual examination of the above structures, when positioned equivalently as shown in Figure 1, confirmed their structural congruence, strongly indicating the presence of a shared fold in these proteins. All versions are unified by the presence of a sheet in which the strands show a characteristic down-up-down-up polarity as was first noted in L3-I (Figure 1). We refer to the exposed surface of the sheet as the open face, and the one packed against the two helices as the obscured face (Figure 1). Accordingly, we termed this fold as RAGNYA after certain key proteins in which it was detected, encompassing its major structural variations (Ribosomal protein L1 and L3, ATP grasp modules, GYF domain, NinB, Y-family DNA polymerases, AMMECR1).
Figure 2.
(A) A network representation of structural relationships revealed by the transitive search procedure. The nodes, represented as different shapes in the network, correspond to protein structures while the edges denote the recovery of a hit in the structure similarity search. L3-I, P19, Tyw3 and ER, respectively denote insert domain in Ribosomal protein L3, siRNA silencing repressor of tombusviruses, tRNA Wybutosine biosynthesis enzyme and enhancer of rudimentary. L1 refers to Ribosomal protein L1. The nodes have been colored according to contained circularly permuted version of RAGNYA fold. (B) The three distinct circularly permutated variants and the split version (NinB) of the RAGNYA fold. The existence of circular permutations between domains with topology like L3-I (shown in the middle) and the topologies seen in domains like the classical ATP-grasp module (shown on the left) and Ribosomal protein L1 (shown on the right) are illustrated using pink and green lines and arrows respectively. Also shown below each of the topology diagram are the underlying repeating units of each version of the fold.
Structural diversity of the RAGNYA fold
Four distinct structural variations were found in the above-mentioned 11 domains with the RAGNYA fold. Not surprisingly, three of the four major variants of the fold are related by circular permutations that result from a connection of the juxtaposed N- and C-termini of the original configuration (i.e. L3-I), and corresponding generation of new termini elsewhere in the fold (Figure 2B). The first variant, typified by the original configuration observed in the L3-I domain, is additionally represented by the globular domain of the Enhancer of rudimentary proteins (ER) (38,39), tombusvirus p19 proteins (40,41), GYF domains (42,43) and tRNA Wybutosine biosynthesis enzyme Tyw3p (44,45). The second variant is characterized by a circular permutation resulting in the connection of the N- and C-termini of the first version, and concomitant generation of new N- and C-termini either just N-terminal to strand-2 or just C-terminal to strand-3 of the first version (Figure 2B). This variant is represented by the C-terminal domain of the DinB-like (Y-family) DNA polymerases (46,47), the N-terminal domain of classical ATP-grasp module (48,49) and in the AMMECR1 proteins (50,51). The third major version has a single representative in the form of the ribosomal protein L1 (52). It is typified by a circular permutation that connects the N- and C-termini of the original L3-I-like configuration while generating new termini just C-terminal to either strand-2 or to strand-4 in the original topology (Figure 2B). The fourth variant represents a ‘broken-up’ version of the fold, in that it is comprised of a dimer of two identical subunits. Each monomer contributes a unit of two strands and one helix for the assembly of a complete fold (Figures 1 and 2B). This version is currently only represented by the NinB proteins of lambdoid bacteriophages (53,54). The configuration of the monomeric subunit of these proteins is a simple β–α–β unit, which is effectively equivalent to the internal repeats seen in the second variant of the fold (Figure 2B).
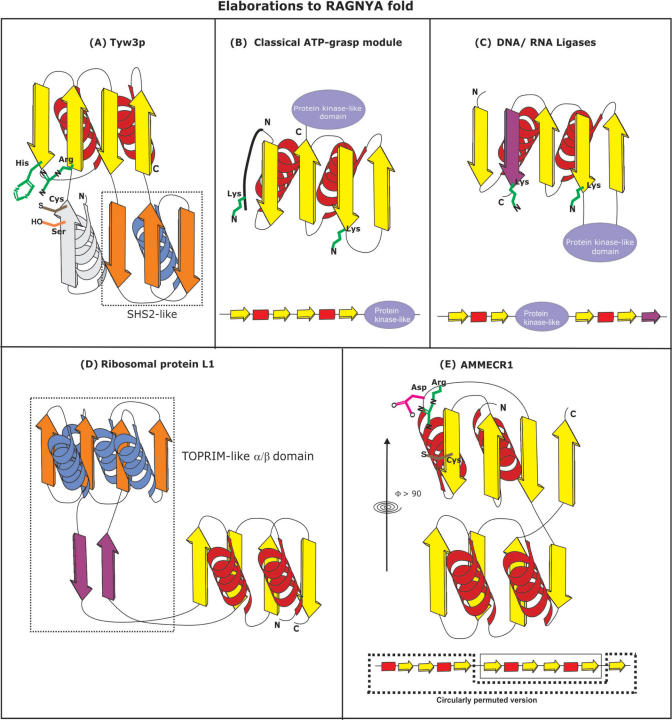
Some representatives of these basic variants show additional elaborations in the form of domain insertions and extensions, as well as further duplications and permutations. Insertions of other domains into the fold are seen in versions found in Tyw3p and the ribosomal protein L1 (Figure 3A). While the insertion is in an equivalent position in both of these versions, the inserted domains themselves are unrelated, implying that they occurred independently. In Tyw3p, the insert is an α+β globular domain that assumes a topology similar to the SHS2 domain (55) (Figure 3A). The core RAGNYA domain and the insert domain together with an N-terminal extension form a pseudo-symmetric structure with a large C-shaped cleft which contains the catalytic residues required for Wybutosine synthesis in its center (see later for details). In the case of the ribosomal protein L1, the insert is a catalytically inactive version of the TOPRIM domain, which assumes a 4-stranded form of the Rossmannoid fold (56,57). This domain is held away from to the core RAGNYA domain by means of an extended linker and forms an independent surface for interaction with other proteins in the ribosomal subunit (Figure 3D). AMMECR1 is a two domain protein that appears to have arisen from duplication of the entire RAGNYA domain. However, it additionally displays a higher order circular permutation that has resulted in the first strand of the first RAGNYA domain being permuted to the extreme C-terminus of the protein (Figure 3E). This permutation results in the first RAGNYA domain effectively acquiring a topology comparable to the form seen in ribosomal protein L1. The sheets of the duplicated RAGNYA domains face each other at an angle greater than 90° resulting in a deep cleft that superficially resembles the situation in Tyw3p.
Figure 3.
Topology diagrams illustrating various elaborations to the RAGNYA fold seen in the following domains: (A) Tyw3p, which has a SHS2-like domain, shown within the dotted box, inserted between the last two strands (third and fourth strands) of the RAGNYA fold. (B) Classical ATP-grasp module has a protein-kinase-like domain fused to the C-termini of core RAGNYA fold. They possess two conserved lysines, shown in green, one at the C-termini of first strand and the other at the N-terminal extension to the RAGNYA fold. (C) DNA and RNA ligases, possessing close structural congruence to the classical ATP-grasp, have a protein-kinase-like domain inserted between second and third strands of the RAGNYA fold unlike the classical ATP-grasp module. However, they too possess the two conserved lysines, shown in green, at the C-termini of the second and the extension strands, shown in purple color. (D) Ribosomal protein L1 has a TOPRIM-like domain, shown within the dotted box, inserted between first and second strands of the RAGNYA fold. (E) AMMECR1 has two domains with the RAGNYA fold that are interlocked with each other. The exposed cysteine which is potentially involved in likely catalytic role of the protein is shown in brown. The two domains are rotated with respect to each other by more than 90° about a vertical axis indicated in the figure. The sequence of secondary structures shown below reveals that the two domains are related by circular permutations.
The classical ATP-grasp modules have an N-terminal RAGNYA fused to a C-terminal domain related to protein kinases and PIPK C-terminal domains (58,59) (Figure 3B). RNA/DNA ligases and the closely related capping enzymes have a peculiar version of the ATP-grasp module, wherein the two internal β–α–β repeats of the RAGNYA domain flank the kinase-like domain of the ATP-grasp module respectively at the N- and C-termini (Figure 3C). This configuration could have arisen through: (1) a circular permutation of the classical ATP-grasp module resulting in N-terminal β–α–β unit of the original module being displaced to the C-terminus. (2) Alternatively, the kinase-like domain might have been secondarily inserted into the RAGNYA domain between the two β–α–β units. However, in both the classical ATP-grasp module and the nucleic acid ligases, the critical phosphate-binding lysine and base interacting residue are found in the second strand of the N-terminal β–α–β unit (Figure 3B and C) (60). If there was indeed a circular permutation in the ligases, then the β–α–β unit containing these residues would have been at the C-terminus. Hence, the presence of the equivalent lysine in the N-terminal β–α–β unit in both versions argues for the kinase-like domain being inserted into the middle of the RAGNYA fold in the nucleic acid ligases (Figure 3C). Interestingly, this version of the RAGNYA domain is distorted due to a C-terminal extension which assumes an extended configuration and is incorporated as an additional stranded inserted in the middle of 4-stranded sheet in the core RAGNYA fold (Figures 1 and 3C).
A common nucleic acid/nucleotide interaction mode is utilized by most members of the RAGNYA fold
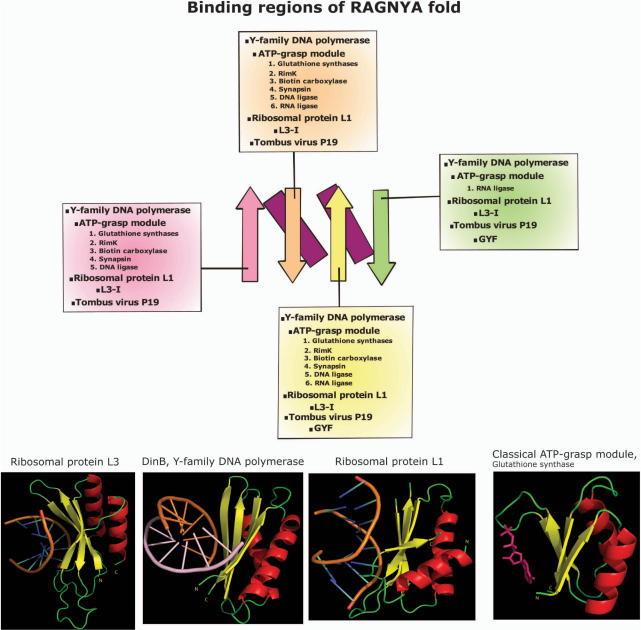
Of the eleven distinct domains with the RAGNYA fold, seven have been shown to directly interact with either RNA or DNA. The L3-I, Tombusvirus p19 and ribosomal protein L1 interact with double-stranded (ds) regions of rRNA or siRNA-mRNA duplexes, tRNA Wybutosine biosynthesis enzyme Tyw3p with tRNA, the family Y DNA polymerase C-terminal domains and phage NinB proteins interact with DNA and the RNA/DNA ligases interact with both nucleotides and either RNA or DNA. In classical ATP-grasp modules, as well as nucleic acid ligases, the RAGNYA fold interacts with ATP. Other members of the fold, namely the enhancer of rudimentary proteins and the GYF domain have been shown to be parts of nucleoprotein complexes involved in pre-mRNA splicing and transcription or DNA replication, respectively, but there is no evidence for their direct interaction with nucleic acids (61,62). Structures with bound substrates are available in the case of the L3-I, Tombusvirus p19, ribosomal protein L1, Y-family DNA polymerase C-terminal and the two versions of the ATP-grasp module (Figure 4). Examination of these structures reveals a common mode of substrate interaction for the RAGNYA domains, with the open face of the sheet being primarily involved in contacting the nucleic acid or nucleotide (Figure 4). In the case of bound nucleic acids both hydrophobic and polar contacts are made with the backbone and the bases. For example, the RAGNYA domain of the Y-family DNA-polymerase appears to be involved in the binding of damaged DNA close to the site of abasic lesions (46,63–65). In this family the conserved basic residues from the N-terminus of strand-1 and end of strand-4 contact phosphates of the dsDNA's backbone, whereas a conserved arginine from the N-terminus of strand 4 participates in binding bases close to the site of abasic lesion.
Figure 4.
Binding modes of the RAGNYA fold. The four strands of the RAGNYA fold are colored differently and shown in the ‘open face’ view. The topological connectivities which are distinct between the circularly permuted topologies are not shown. The boxes connected to the strands list the domain families that use the strand to bind various ligands. All known structures of RAGNYA domains suggest that the RAGNYA domains bind ligands mainly from the ‘open face’ with the exception of the GYF domain which binds ligands in a ‘side-on’ mode, perhaps in addition to the face on mode. The representative structures from three distinct permuted versions of the RAGNYA fold along with their duplex nucleic acid ligands and a structure of the classical ATP-grasp module with its nucleotide ligand are shown to illustrate the similarity of the binding modes. The PDBIDs of these structures are respectively 1JJ2, 1JX4, 1MZP and 1GSA.
NinB proteins function in a similar capacity to the bacterial RecFOR complex, downstream of the λ-exonuclease in the early stages of recombination of λ-like phages. They have been shown to bind strongly to ssDNA and weakly to dsDNA (53,54). The use of different gapped substrates with ssDNA and dsDNA segments have suggested that the DNA binds across a surface cleft, whose base is formed by the open face of the RAGNYA fold in NinB. The clear preference for ssDNA as against dsDNA is atypical, given that most other nucleic acid binding members of the RAGNYA fold interact with dsDNA or dsRNA. Examination of the crystal structure of NinB revealed that the predominantly α-helical C-terminal domain of NinB obscures a part of the open face of the RAGNYA fold that is available for interaction in the other representatives of the fold. Hence, the version of the fold in NinB appears to have sufficient space only to accommodate ssDNA, thereby explaining its preferential binding properties. A comparable mode of substrate interaction (with nucleic acids, nucleotides and peptides), using the open face of the sheet, has also been observed in other structurally distinct two-layered folds with comparably sized β-sheets, such as the RRM-like, β-grasp and the S5 folds (6,66,67). Furthermore, the preservation of a common mode of substrate binding in the RAGNYA fold, irrespective of circular permutation or constitution from separate 2-strand-1-helix elements (NinB), strongly suggests that this mode of interaction is the preferred binding mode preserved throughout the fold. These observations on the binding mode also help in predicting the mode of interaction of Tyw3p with its tRNA substrate. Tyw3p is an enzyme required for the in situ synthesis of the modified base 2-methylthio-N6-isopentenyladenosine or wybutosine (yW) in the anticodon loop of phenylalanine tRNA (44). Based on the precedence of the other RNA–protein interactions seen in this fold, we propose that the Tyw3p would probably bind the dsRNA of the anticodon stem and present the anticodon loop to the catalytic residues. The insert domain seen in the Tyw3p is additionally likely to cooperate with the RAGNYA fold by forming a ‘roof ’ over the bound anticodon stem.
Adaptation of the common substrate-binding platform of the RAGNYA domain for diverse enzymatic roles
Despite the common mode of substrate interaction used by most members of the RAGNYA fold, it has often been utilized for very distinct biochemical functions. A careful analysis of the structures and the underlying sequence conservation pattern revealed the different adaptations that emerged in functionally distinct versions of the fold. In Tyw3p, the nucleic acid ligases/capping enzymes, the ATP-grasp enzymes and AMMECR1 the fold has been adapted to perform enzymatic functions in very distinct ways. Tyw3p catalyzes the fourth step of the six-step synthesis of yW, in which an AdoMet donor provides a methyl group for the methylation of the available nitrogen in the central ring of the tricyclic yW precursor (44). Superposition of the conservation pattern of the Tyw3p proteins on to the structure of the Sulfolobus ortholog of Tyw3p shows that the three blocks of nearly universally conserved residues are spatially closely clustered (see Supplementary information). The first of these is an asparate derived from the N-terminal extension, the second is a motif of the form [ST]xSCxGR that lies in the junction between the SHS2-like insert and the RAGNYA fold, and the third is a conserved histidine from the end of the strand-2 of the RAGNYA fold (see Figure 3A and Supplementary information). The polar nature of these conserved residues and their spatial clustering strongly indicate that they constitute the active site of Tyw3p (Figure 3A). Furthermore, this predicted active site lies close to ‘one edge’ of the protein (Figure 3A), suggesting that this location allows interaction with the target guanine 37 in the anticodon loop, when the anticodon stem is bound along the open face of the RAGNYA domain as proposed earlier. The presence of the single absolutely conserved cysteine suggests that the methyl transfer reaction catalyzed by this enzyme is probably very different from that catalyzed by the Rossmann fold methyltransferases, like Trm5p, which catalyzes the first step of yW synthesis (44). It is likely that the cysteine actually receives the methyl group from the AdoMet cofactor and then relays it to the target Nitrogen atom on the yW precursor. Thus, emergence of a set of residues in the core RAGNYA fold, as well as in the associated insert and N-terminal extension, which were located on the ‘edge’ of the structure, appears to have given rise to a highly specific RNA modifying enzyme on the ancestral platform provided by the core RNA-binding domain.
Interestingly, in addition to the superficial similarity to Tyw3p in its C-shaped structure with a deep cleft, the AMMECR1 protein also shows a comparable set of nearly absolutely conserved residues in the form of two motifs. The first of these is a RGChG (where ‘h’ is any hydrophobic residue) signature in the middle of strand-2 of the first RAGNYA domain, and a DxRa signature (where ‘a’ is any aromatic residue) at the beginning of the helix-2 of the same domain (see Supplementary information). These conserved residues form a spatially close group, when mapped on to the structures of the AMMECR1, indicating that they are likely to constitute the active site of the AMMECR1 proteins (Figure 3E). In particular, the thiol group of the absolutely conserved cysteine of the AMMECR1 protein projects into the central cleft and is potentially available for a catalytic reaction (see Figure 3E and Supplementary information). The second RAGNYA fold forms the floor of the cleft and does not appear to contribute any obvious catalytic residues. Hence, it appears likely that the duplicated RAGNYA folds have specialized, with the second one probably being involved mainly in substrate contact, while the first provides the catalytic residues. Thus, a similar shape and potential set of catalytic residues appear to have convergently emerged in the Tyw3p and AMMECR1 proteins. While the reaction catalyzed by AMMECR1 remains uncharacterized, examination of contextual information derived from phyletic profiles, gene neighborhood, domain fusion and protein interaction network analysis hint certain definitive possibilities. AMMECR1 is highly conserved in archaea and eukaryotes and sporadically found in certain bacterial lineages (see later for details). The strong archaeo-eukaryotic phyletic pattern is indicative of a role in core cellular functions, including RNA metabolism. In the yeast protein–protein interaction network it appears to belong to complexes including RNA transport and processing proteins such as Nup114p, Soh1p, Yra1p and Jsn1p (68). In prokaryotes it shows a persistent gene-neighborhood association with a conserved radical SAM enzyme related to MiaB and a ring-opening dioxygenase. It also shows gene fusions with the gene for latter enzyme (see Supplementary information). This observation implies that AMMECR1 catalyzes a reaction in the same pathway as the radical SAM enzyme and the dioxygenase, perhaps transferring an organic radical on the conserved cysteine. Notably, Tyw1p, an enzyme prior to Tyw3p in the yW biosynthetic pathway is also a radical SAM enzyme involved in production of one of the rings of yW (44). Thus, the combined contextual information points to the possibility that AMMECR1 might catalyze an as yet uncharacterized RNA base modification, like Tyw3p.
RAGNYA domain in both versions of the ATP-grasp module has a set of common interactions with the nucleotide substrate. These primarily include a phosphate contact using a conserved lysine and a base interaction via hydrophobic or polar contacts mediated by an equivalently positioned residue, both from strand-2 (Figure 3B and C). However, they also possess unique additional phosphate contacts. In the case of the classical ATP-grasp module this contact comes from a conserved basic residue in the N-terminal helical extension to the core RAGNYA fold, while in nucleic acid ligases it is from the C-terminal extension, which is inserted as a strand into the sheet of the RAGNYA domain (Figure 3C). Furthermore, many members of the classical ATP-grasp version of the RAGNYA domain (e.g. glutathione synthetase) have a glycine-rich loop between the two β–α–β units of the fold, which provides additional contacts with the phosphates, in a manner reminiscent of the glycine rich loops seen in several other NTP-binding domains (49). In both cases, the RAGNYA domain is stacked against the kinase-like fold with the bound nucleotide in between them. This suggests that the two domains might have originally functioned as stand-alone partners; with the RAGNYA domain supplying the chief nucleotide contacts and the kinase-like domain providing other key catalytic residues. This is consistent with the observation that the kinase-like domain has been independently linked either to the C-terminus or inserted between the two β–α–β units of the RAGNYA in two versions of the ATP-grasp module respectively. This is also in agreement with observations reported in previous studies, which suggest that the kinase-like domain has similarly partnered with other globular domains in classical eukaryote-type protein kinases and PIPKs (58,59). It appears likely that the ancestral version of the ATP-grasp RAGNYA domain had a single conserved lysine for phosphate contact and a base-contacting position on the strand-2 (Figure 3B and C), which were further augmented by the additional innovations for phosphate contact as described earlier.
This adaptation of the RAGNYA domain for enzymatic functions again parallels the similar deployment of the RRM-like fold as a scaffold for the catalytic activities of numerous enzymes functioning in nucleotide and nucleic acid metabolism (6). Prominent examples of such RRM-like fold enzymatic domains include pseudouridine synthases, nucleic acid polymerases and nucleotide diphosphate kinases. In all these domains the exposed face of the sheet plays some role in substrate-binding, just as in the RAGNYA domains (69–71).
Adaptation of the RAGNYA fold for protein–protein interactions
While most characterized members of the RAGNYA fold interact with nucleotides or nucleic acids, the version in the GYF domain is currently known to interact mainly with peptides (42,43). The only structurally characterized interaction, namely that with proline-rich peptides, occurs in a ‘side-on’ fashion (Figure 4), via the edge of the domain, involving several conserved hydrophobic residues that are part of the domain's hydrophobic core (43). This interaction appears to be important for the function of several eukaryotic GYF domains that exhibit preferential binding for different types of proline-rich peptides (72,73). Our discovery of bacterial GYF domains (see later for details) provides several additional leads regarding the ancestral-binding properties and functions of this domain: (1) In both eukaryotic and bacterial versions the aromatic and hydrophobic residues of the core are strongly conserved suggesting that interaction with proline-rich peptides via these residues is a conserved feature. (2) Several bacterial proteins with GYF domains also contain flanking proline-rich stretches suggesting an interaction between these and the GYF domains. (3) The bacterial versions are often fused to transmembrane (TM) domains or occur in conserved gene-neighborhoods encoding adjacent TM domain proteins (Figure 5 and see Supplementary information). Many bacterial GYF domains are also found fused in the same polypeptide with tetratricopeptide repeat domains suggesting a role in protein–protein interactions (Figure 5). These observations taken together suggest that the role in protein–protein interactions via a ‘side-on’ contact with proline is likely to be an ancestral specialization of both bacterial and eukaryotic GYF domains.
Figure 5.
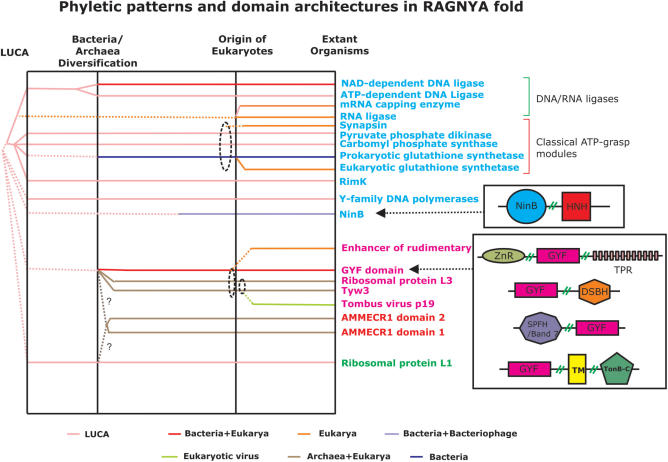
Phyletic patterns and prominent architectures of the RAGNYA fold domain families are shown. The domain families belonging to distinct permuted versions of the fold are colored differently. The superkingdom-based coloring scheme for the phyletic distribution is indicated below. The dotted lines from a common point denote a hypothesized common origin between families, while the solid lines indicate the presence of evidence to support a common origin. Dotted ellipses encircle a set of families from which a family of more limited phyletic distribution is likely to have diversified. The ‘?’ indicates that the evolutionary origin of the family is unclear. While shown as originating from a common precursor, it should be noted that individual complete versions of the RAGNYA fold might have been independently re-assembled from precursor two strand-one helix units. The prominent architectures of the GYF domain and NinB families are shown. In these architectures abbreviations used for the domain families are: ZnR – zinc ribbon, SPFH – Band 7/SPFH, TPR – tetratricopeptide repeat, DSBH – double-stranded beta helix (cupin-like), TM – transmembrane region, TonB-C – C-terminal domain of TonB and HNH – HNH endonuclease. Representative GIs corresponding to the architectures are the following: NinB+HNH: 116333759, ZnR+GYF+TPR: 121536395, GYF+DSBH: 77747736, SPFH/Band 7+GYF: 32472385 and GYF+TM+TonB-C: 108761942.
However, recent studies have suggested that the GYF domain also interacts with U5-15k protein in the U5 ribonucleoprotein complex independent of proline-rich sequences (62). Furthermore, the GYF domains contain a conserved position (almost always tryptophan in the bacterial versions) in the strand-3 whose side chain is exposed on the open face, which could potentially mediate contact with a substrate through hydrophobic interactions (see Supplementary information). These observations suggest that the GYF domain might contain an uncharacterized interaction mode involving the open face, as seen in other RAGNYA domains. The most conserved versions of the GYF domain in eukaryotes are found associated with the U5 RNP complex. In bacteria, conserved gene-neighborhoods and gene fusions suggest a functional association with a potential nucleic acid binding protein with Zinc-ribbons similar to TFIIB and PriN’ (see Figure 5 and Supplementary information). This might suggest that the GYF domain was originally derived from ancestral nucleic acid binding RAGNYA domain proteins, with a function shift for peptide interactions in nucleoprotein complexes. Subsequently, it appears to have been utilized more widely in peptide-binding contexts in other nucleoprotein complexes.
The enigmatic, highly conserved ER protein has been identified in several independent protein-interaction screens to associate with the RNA polymerase complex, and as the DNA-polymerase associated protein PDIP46 (61). Thus, like the GYF domain, it might represent another case of secondary adaptation of the RAGNYA domain for protein–protein interaction. Genetic studies have implicated the ER protein in regulation of pyrimidine biosynthesis, cell-cycle progression and transcriptional regulation (38,74,75), but its exact role is yet to be uncovered. Examination of the structure and conservation pattern of the ER proteins from diverse eukaryotes suggests that the open face of the RAGNYA domain contains several patches of polar residues that could be potentially critical for its interactions with other proteins (see Supplementary information).
Phyletic patterns and early evolutionary history of RAGNYA fold proteins
Though the sequence similarity between most individual superfamilies of domains containing the RAGNYA fold has largely eroded, they retain a striking congruence of their structural elements and their polarities. More importantly, the different circularly permutated versions of the fold, as well as the forms modified by inserts, bind their substrates in a common mode. These features support the RAGNYA fold being a monophyletic assemblage of domains. However, given its simplicity and symmetry it cannot be ruled out that the complete fold could have been possibly re-assembled independently in certain instances by the duplication of the basic two-strand-one helix unit. This possibility is supported by the NinB structure that is composed of a non-covalent dimer of identical two-strand-one helix units. Furthermore, the strong symmetry of the two β–α–β units of the Y-family DNA polymerase C-terminal domain might indicate such an independent assembly of this version of the fold through reduplication of an ancestral β–α–β unit.
To understand better the early history of the fold, we compared the phyletic patterns of all domains containing it (Figure 5). Most groups appear to have deep evolutionary histories—the ribosomal protein L1, three families of the classical ATP-grasp module, and at least one family of nucleic acid ligase are conserved in most groups of organisms, and include representatives from all the three superkingdoms of Life (bacteria, archaea and eukaryotes). This suggests that they trace back to the last universal common ancestor (LUCA) of all extant life forms. Of the nucleic acid ligases, the archaeo-eukaryotic clade universally conserves an ATP-dependent DNA ligase, whereas the bacterial clade universally contains a NAD-dependent form. This suggests that they possibly diverged from each other in early evolution from a precursor present in LUCA. Three families of the classical ATP-grasp domain that might potentially trace back to LUCA are the carbamoyl phosphate synthetase, pyruvate phosphate dikinase and the ribosomal protein S6 α-l glutamate ligase (RimK). These observations imply that by the time of LUCA not only had the ATP-grasp module diversified into its classical and nucleic acid ligase-like versions, but the classical version had itself further radiated to occupy very distinct functional niches related to amino acid and nucleotide metabolism, protein modification and pyruvate metabolism. Thus, the cooperation between the RAGNYA domain and the kinase-like domain, and the distinct associations between these two domains (fusion or domain insertions) had all taken place prior to LUCA. Additionally, presence of a distinct dsRNA-binding version in the form of the L1 protein implies that the differentiation between the RNA-binding and nucleotide-binding versions had also occurred in this period. A possible corollary of this long pre-LUCA history of the RAGNYA domain is that it first emerged in the RNA world itself, in the form of a generic nucleic acid/nucleotide-binding domain. It subsequently appears to have further differentiated into specialized nucleotide-binding versions as in the ATP-grasp domain and RNA-binding versions.
The situation in the L3-I domain is more complicated—the ribosomal protein L3 itself is present throughout the three superkingdoms of life, but both structural comparisons and sequence similarity searches detect the L3-I domain only in the archaeo-eukaryotic orthologs. An examination of the bacterial L3 ortholog reveals that an insert is present in the equivalent region, which appears to be poorly structured in comparison to the L3-I domain. Nevertheless, at least two extended regions and one helical segment can be identified in this insert suggesting that it could have emerged through the loss or degeneration of part of the original L3-I domain. Thus, the L3-I module was potentially present in the ancestral L3 protein, and emerged as a part of the radiation of RAGNYA fold domains prior to LUCA. New RNA-binding roles appear to have been acquired later in evolution as suggested by the Tyw3p protein (in the archaeo-eukaryotic lineage) and perhaps in the AMMECR1 protein. The tombusvirus siRNA repressor appears to be a late virus-specific innovation perhaps acquired from the structurally closely related L3-I domain of the eukaryotic host. The distribution of the Y-family DNA polymerases with the RAGNYA domain in all the three superkingdoms might imply their presence in LUCA. However, their sporadic distribution in archaea, along with the lack of a clear signal for vertical evolutionary relationship between the versions from the three superkingdoms raises the possibility of a later origin and dispersal through lateral gene transfers, especially amongst the prokaryotes. The DNA-binding NinB versions of the RAGNYA domains are distributed only in lambdoid siphoviruses, a few podoviruses or their prophage remnants in bacterial genomes. In many lambdoid siphoviruses of low GC Gram-positive bacteria we observed fusions between their NinB ortholog and a nuclease domain of the EndoVII (HNH) fold (see Supplementary information) (76,77). This implies that NinB might collaborate in different phages with unrelated families of nucleases (e.g. lambda exonuclease and HNH) in genome recombination. The potential secondary reassembly of the version of the RAGNYA fold in the Y-family DNA polymerases and the presence of an equivalent stand-alone monomeric unit in NinB suggest that these DNA-binding versions might share a more recent ancestral monomeric precursor.
Until now the peptide-binding version of the RAGNYA fold, the GYF domain was found only in eukaryotes, including the basal most eukaryotic lineages. However, using a sequence profile constructed from eukaryotic representatives of the GYF domain we were able to detect bacterial homologs with significant e-values (e.g. RB6375, gi: 32474220 from Rhodopirellula was recovered with e = 10−3 in iteration 3). Conversely, reciprocal searches initiated with bacterial proteins (e.g. gi: 84704887, PB2503_12664 from Parvularcula bermudensis) recovered eukaryotic GYF domains with significant e-values (e < 10−3) within six iterations. As a result of these searches we identified numerous bacterial GYF domain proteins from diverse bacterial lineages including planctomycetes-chlamydia, bacteroidetes, proteobacteria, firmicutes, actinobacteria and cyanobacteria (see Supplementary information). This wide distribution in bacteria is also accompanied by considerable domain architectural diversity greater than that observed in eukaryotes (Figure 5). However, no GYF domains were found in archaea. Given the widespread presence in bacteria, especially α-proteobacterial lineages that spawned the eukaryotic mitochondrion (78), it is likely that the GYF domain first arose in bacteria, and was transferred to the ancestral eukaryote perhaps during the mitochondrial endosymbiosis. Interestingly, its recruitment to roles in spliceosomal complexes, like the U5 snRNP, and in endosomal-trafficking proteins like RME8 suggest that the acquisition of the GYF domain from the bacteria might have played an important role in the emergence of quintessentially eukaryotic systems (62,79).
GENERAL CONCLUSIONS
We present the identification of a new fold with nucleic acid, nucleotide or peptide-binding properties, shared by 11 distinct functionally diverse protein domains. The RAGNYA fold is characterized by the presence of an internal symmetry constituted by two topologically identical units, each with two strands and one helix. This structural peculiarity of the fold has resulted in it displaying at least three different circular permuted versions, and one case of re-assembly from two monomeric units derived from different polypeptides. In spite of this, the fold retains a distinctive exposed β-sheet with a unique strand polarity, and most members of the fold bind nucleic acids or nucleotides via this face. The RAGNYA fold appears to have been utilized as a scaffold on multiple occasions in the generation of novel enzymatic activities, as exemplified by the ATP-grasp enzymes, the nucleic acid ligases, the Tyw3p AdoMet-dependent tRNA modifying enzyme and the experimentally uncharacterized AMMECR1 enzyme. Based on our analysis of this fold we present functional predictions that help in explaining the reaction mechanisms and substrate-binding of the Tyw3p enzyme, and the possible function of the AMMECR1 protein. We also provide evidence that the two versions of the ATP-grasp module have been assembled independently either via a C-terminal fusion to a kinase-like domain or insertion of the kinase-like domain into the RAGNYA domain. Analysis of the RAGNYA domain also helped us to identify the common denominator in the nucleic acid-binding mode of the Y-family polymerase C-terminal DNA binding domain, phage NinB proteins, the siRNA repressor of Tombusviruses and ribosomal proteins L1 and L3. We also identify for the first time the bacterial GYF domains, which might lead to better understanding of the different modes of interaction of this version of the RAGNYA fold with peptide substrates.
We hope that the results presented here open up new avenues for the experimental investigation of this diverse group of proteins unified by a subtle, yet functionally significant structural feature.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
S.B. and L.A. acknowledge the Intramural research program of National Institutes of Health, USA for funding their research. We thank Lakshminarayan Iyer, Maxwell Burroughs and S. Geetha for carefully reading through the manuscript and providing useful suggestions.
Funding to pay the Open Access publication charges for this article was provided by the National Institutes of Health, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 2.Doherty AJ, Serpell LC, Ponting CP. The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA. Nucleic Acids Res. 1996;24:2488–2497. doi: 10.1093/nar/24.13.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol. Rev. 2005;29:231–262. doi: 10.1016/j.femsre.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Grishin NV. Two tricks in one bundle: helix-turn-helix gains enzymatic activity. Nucleic Acids Res. 2000;28:2229–2233. doi: 10.1093/nar/28.11.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murzin AG. A ribosomal protein module in EF-G and DNA gyrase. Nat. Struct. Biol. 1995;2:25–26. doi: 10.1038/nsb0195-25. [DOI] [PubMed] [Google Scholar]
- 6.Anantharaman V, Aravind L, Koonin EV. Emergence of diverse biochemical activities in evolutionarily conserved structural scaffolds of proteins. Curr. Opin. Chem. Biol. 2003;7:12–20. doi: 10.1016/s1367-5931(02)00018-2. [DOI] [PubMed] [Google Scholar]
- 7.Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bobay BG, Andreeva A, Mueller GA, Cavanagh J, Murzin AG. Revised structure of the AbrB N-terminal domain unifies a diverse superfamily of putative DNA-binding proteins. FEBS Lett. 2005;579:5669–5674. doi: 10.1016/j.febslet.2005.09.045. [DOI] [PubMed] [Google Scholar]
- 9.Coles M, Hulko M, Djuranovic S, Truffault V, Koretke K, Martin J, Lupas AN. Common evolutionary origin of swapped-hairpin and double-psi beta barrels. Structure. 2006;14:1489–1498. doi: 10.1016/j.str.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Castillo RM, Mizuguchi K, Dhanaraj V, Albert A, Blundell TL, Murzin AG. A six-stranded double-psi beta barrel is shared by several protein superfamilies. Structure. 1999;7:227–236. doi: 10.1016/s0969-2126(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 11.Anantharaman V, Aravind L. The PRC-barrel: a widespread, conserved domain shared by photosynthetic reaction center subunits and proteins of RNA metabolism. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-research0061. RESEARCH0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anantharaman V, Koonin EV, Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kinch LN, Ginalski K, Rychlewski L, Grishin NV. Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res. 2005;33:3598–3605. doi: 10.1093/nar/gki676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bycroft M, Hubbard TJ, Proctor M, Freund SM, Murzin AG. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 15.Liu Z, Macias MJ, Bottomley MJ, Stier G, Linge JP, Nilges M, Bork P, Sattler M. The three-dimensional structure of the HRDC domain and implications for the Werner and Bloom syndrome proteins. Structure. 1999;7:1557–1566. doi: 10.1016/s0969-2126(00)88346-x. [DOI] [PubMed] [Google Scholar]
- 16.Aravind L, Anantharaman V, Koonin EV. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins. 2002;48:1–14. doi: 10.1002/prot.10064. [DOI] [PubMed] [Google Scholar]
- 17.Mizuguchi K, Dhanaraj V, Blundell TL, Murzin AG. N-ethylmaleimide-sensitive fusion protein (NSF) and CDC48 confirmed as members of the double-psi beta-barrel aspartate decarboxylase/formate dehydrogenase family. Structure. 1999;7:R215–R216. doi: 10.1016/s0969-2126(99)80169-5. [DOI] [PubMed] [Google Scholar]
- 18.Coles M, Djuranovic S, Soding J, Frickey T, Koretke K, Truffault V, Martin J, Lupas AN. AbrB-like transcription factors assume a swapped hairpin fold that is evolutionarily related to double-psi beta barrels. Structure. 2005;13:919–928. doi: 10.1016/j.str.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 19.Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 2003;3:1. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nissen P, Kjeldgaard M, Thirup S, Polekhina G, Reshetnikova L, Clark BF, Nyborg J. Crystal structure of the ternary complex of Phe-tRNAPhe, EF-Tu, and a GTP analog. Science. 1995;270:1464–1472. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 21.Iyer LM, Aravind L. The emergence of catalytic and structural diversity within the beta-clip fold. Proteins. 2004;55:977–991. doi: 10.1002/prot.20076. [DOI] [PubMed] [Google Scholar]
- 22.Iyer LM, Koonin EV, Aravind L. Evolution of bacterial RNA polymerase: implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene. 2004;335:73–88. doi: 10.1016/j.gene.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 27.Walker DR, Koonin EV. SEALS: a system for easy analysis of lots of sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1997;5:333–339. [PubMed] [Google Scholar]
- 28.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 29.Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ. JPred: a consensus secondary structure prediction server. Bioinformatics. 1998;14:892–893. doi: 10.1093/bioinformatics/14.10.892. [DOI] [PubMed] [Google Scholar]
- 30.Felsenstein J. Inferring phylogenies from protein sequences by parsimony, distance, and likelihood methods. Methods Enzymol. 1996;266:418–427. doi: 10.1016/s0076-6879(96)66026-1. [DOI] [PubMed] [Google Scholar]
- 31.Hasegawa M, Kishino H, Saitou N. On the maximum likelihood method in molecular phylogenetics. J. Mol. Evol. 1991;32:443–445. doi: 10.1007/BF02101285. [DOI] [PubMed] [Google Scholar]
- 32.Adachi J, Hasegawa M. MOLPHY: Programs for Molecular Phylogenetics, I. —PROTML: Maximum Likelihood Inference of Protein Phylogeny. Tokyo: Institute of Statistical Mathematics; 1992. [Google Scholar]
- 33.Schmidt HA, Strimmer K, Vingron M, von Haeseler A. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 34.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 35.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–567. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 36.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: a new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coles M, Diercks T, Liermann J, Groger A, Rockel B, Baumeister W, Koretke KK, Lupas A, Peters J, et al. The solution structure of VAT-N reveals a ‘missing link' in the evolution of complex enzymes from a simple betaalphabetabeta element. Curr. Biol. 1999;9:1158–1168. doi: 10.1016/S0960-9822(00)80017-2. [DOI] [PubMed] [Google Scholar]
- 38.Wojcik E, Murphy AM, Fares H, Dang-Vu K, Tsubota SI. Enhancer of rudimentaryp1, e(r)p1, a highly conserved enhancer of the rudimentary gene. Genetics. 1994;138:1163–1170. doi: 10.1093/genetics/138.4.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wan C, Tempel W, Liu ZJ, Wang BC, Rose RB. Structure of the conserved transcriptional repressor enhancer of rudimentary homolog. Biochemistry. 2005;44:5017–5023. doi: 10.1021/bi047785w. [DOI] [PubMed] [Google Scholar]
- 40.Vargason JM, Szittya G, Burgyan J, Tanaka Hall TM. Size selective recognition of siRNA by an RNA silencing suppressor. Cell. 2003;115:799–811. doi: 10.1016/s0092-8674(03)00984-x. [DOI] [PubMed] [Google Scholar]
- 41.Ye K, Malinina L, Patel DJ. Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature. 2003;426:874–878. doi: 10.1038/nature02213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Freund C, Dotsch V, Nishizawa K, Reinherz EL, Wagner G. The GYF domain is a novel structural fold that is involved in lymphoid signaling through proline-rich sequences. Nat. Struct. Biol. 1999;6:656–660. doi: 10.1038/10712. [DOI] [PubMed] [Google Scholar]
- 43.Freund C, Kuhne R, Yang H, Park S, Reinherz EL, Wagner G. Dynamic interaction of CD2 with the GYF and the SH3 domain of compartmentalized effector molecules. EMBO J. 2002;21:5985–5995. doi: 10.1093/emboj/cdf602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noma A, Kirino Y, Ikeuchi Y, Suzuki T. Biosynthesis of wybutosine, a hyper-modified nucleoside in eukaryotic phenylalanine tRNA. EMBO J. 2006;25:2142–2154. doi: 10.1038/sj.emboj.7601105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noma A, Suzuki T. Ribonucleome analysis identified enzyme genes responsible for wybutosine synthesis. Nucleic Acids Symp. Ser. (Oxf.) 2006:65–66. doi: 10.1093/nass/nrl032. [DOI] [PubMed] [Google Scholar]
- 46.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 47.Ohmori H, Friedberg EC, Fuchs RP, Goodman MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, et al. The Y-family of DNA polymerases. Mol. Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 48.Hara T, Kato H, Katsube Y, Oda J. A pseudo-michaelis quaternary complex in the reverse reaction of a ligase: structure of Escherichia coli B glutathione synthetase complexed with ADP, glutathione, and sulfate at 2.0 A resolution. Biochemistry. 1996;35:11967–11974. doi: 10.1021/bi9605245. [DOI] [PubMed] [Google Scholar]
- 49.Galperin MY, Koonin EV. A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 1997;6:2639–2643. doi: 10.1002/pro.5560061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vitelli F, Meloni I, Fineschi S, Favara F, Tiziana Storlazzi C, Rocchi M, Renieri A. Identification and characterization of mouse orthologs of the AMMECR1 and FACL4 genes deleted in AMME syndrome: orthology of Xq22.3 and MmuXF1-F3. Cytogenet. Cell Genet. 2000;88:259–263. doi: 10.1159/000015533. [DOI] [PubMed] [Google Scholar]
- 51.Tajika Y, Sakai N, Tamura T, Yao M, Watanabe N, Tanaka I. Crystal structure of PH0010 from Pyrococcus horikoshii, which is highly homologous to human AMMECR 1C-terminal region. Proteins. 2005;58:501–503. doi: 10.1002/prot.20315. [DOI] [PubMed] [Google Scholar]
- 52.Nikonov S, Nevskaya N, Eliseikina I, Fomenkova N, Nikulin A, Ossina N, Garber M, Jonsson BH, Briand C, et al. Crystal structure of the RNA binding ribosomal protein L1 from Thermus thermophilus. EMBO J. 1996;15:1350–1359. [PMC free article] [PubMed] [Google Scholar]
- 53.Tarkowski TA, Mooney D, Thomason LC, Stahl FW. Gene products encoded in the ninR region of phage lambda participate in Red-mediated recombination. Genes Cells. 2002;7:351–363. doi: 10.1046/j.1365-2443.2002.00531.x. [DOI] [PubMed] [Google Scholar]
- 54.Maxwell KL, Reed P, Zhang RG, Beasley S, Walmsley AR, Curtis FA, Joachimiak A, Edwards AM, Sharples GJ. Functional similarities between phage lambda Orf and Escherichia coli RecFOR in initiation of genetic exchange. Proc. Natl Acad. Sci. U.S.A. 2005;102:11260–11265. doi: 10.1073/pnas.0503399102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Anantharaman V, Aravind L. The SHS2 module is a common structural theme in functionally diverse protein groups, like Rpb7p, FtsA, GyrI, and MTH1598/TM1083 superfamilies. Proteins. 2004;56:795–807. doi: 10.1002/prot.20140. [DOI] [PubMed] [Google Scholar]
- 56.Lima CD, Wang JC, Mondragon A. Three-dimensional structure of the 67K N-terminal fragment of E. coli DNA topoisomerase I. Nature. 1994;367:138–146. doi: 10.1038/367138a0. [DOI] [PubMed] [Google Scholar]
- 57.Aravind L, Leipe DD, Koonin EV. Toprim—a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–4213. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grishin NV. Phosphatidylinositol phosphate kinase: a link between protein kinase and glutathione synthase folds. J. Mol. Biol. 1999;291:239–247. doi: 10.1006/jmbi.1999.2973. [DOI] [PubMed] [Google Scholar]
- 59.Denessiouk KA, Lehtonen JV, Korpela T, Johnson MS. Two “unrelated” families of ATP-dependent enzymes share extensive structural similarities about their cofactor binding sites. Protein Sci. 1998;7:1136–1146. doi: 10.1002/pro.5560070507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aravind L, Koonin EV. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 61.Smyk A, Szuminska M, Uniewicz KA, Graves LM, Kozlowski P. Human enhancer of rudimentary is a molecular partner of PDIP46/SKAR, a protein interacting with DNA polymerase delta and S6K1 and regulating cell growth. FEBS J. 2006;273:4728–4741. doi: 10.1111/j.1742-4658.2006.05477.x. [DOI] [PubMed] [Google Scholar]
- 62.Laggerbauer B, Liu S, Makarov E, Vornlocher HP, Makarova O, Ingelfinger D, Achsel T, Luhrmann R. The human U5 snRNP 52K protein (CD2BP2) interacts with U5-102K (hPrp6), a U4/U6.U5 tri-snRNP bridging protein, but dissociates upon tri-snRNP formation. RNA. 2005;11:598–608. doi: 10.1261/rna.2300805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK. Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol. Cell. 2001;8:417–426. doi: 10.1016/s1097-2765(01)00306-9. [DOI] [PubMed] [Google Scholar]
- 64.Zhou BL, Pata JD, Steitz TA. Crystal structure of a DinB lesion bypass DNA polymerase catalytic fragment reveals a classic polymerase catalytic domain. Mol. Cell. 2001;8:427–437. doi: 10.1016/s1097-2765(01)00310-0. [DOI] [PubMed] [Google Scholar]
- 65.Silvian LF, Toth EA, Pham P, Goodman MF, Ellenberger T. Crystal structure of a DinB family error-prone DNA polymerase from Sulfolobus solfataricus. Nat. Struct. Biol. 2001;8:984–989. doi: 10.1038/nsb1101-984. [DOI] [PubMed] [Google Scholar]
- 66.Jessen TH, Oubridge C, Teo CH, Pritchard C, Nagai K. Identification of molecular contacts between the U1 A small nuclear ribonucleoprotein and U1 RNA. EMBO J. 1991;10:3447–3456. doi: 10.1002/j.1460-2075.1991.tb04909.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ramakrishnan V, White SW. The structure of ribosomal protein S5 reveals sites of interaction with 16S rRNA. Nature. 1992;358:768–771. doi: 10.1038/358768a0. [DOI] [PubMed] [Google Scholar]
- 68.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoang C, Ferre-D'Amare AR. Cocrystal structure of a tRNA Psi55 pseudouridine synthase: nucleotide flipping by an RNA-modifying enzyme. Cell. 2001;107:929–939. doi: 10.1016/s0092-8674(01)00618-3. [DOI] [PubMed] [Google Scholar]
- 70.Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- 71.Dumas C, Lascu I, Morera S, Glaser P, Fourme R, Wallet V, Lacombe ML, Veron M, Janin J. X-ray structure of nucleoside diphosphate kinase. EMBO J. 1992;11:3203–3208. doi: 10.1002/j.1460-2075.1992.tb05397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kofler MM, Freund C. The GYF domain. FEBS J. 2006;273:245–256. doi: 10.1111/j.1742-4658.2005.05078.x. [DOI] [PubMed] [Google Scholar]
- 73.Kofler M, Motzny K, Freund C. GYF domain proteomics reveals interaction sites in known and novel target proteins. Mol. Cell. Proteomics. 2005;4:1797–1811. doi: 10.1074/mcp.M500129-MCP200. [DOI] [PubMed] [Google Scholar]
- 74.Gelsthorpe M, Pulumati M, McCallum C, Dang-Vu K, Tsubota SI. The putative cell cycle gene, enhancer of rudimentary, encodes a highly conserved protein found in plants and animals. Gene. 1997;186:189–195. doi: 10.1016/s0378-1119(96)00701-9. [DOI] [PubMed] [Google Scholar]
- 75.Pogge von Strandmann E, Senkel S, Ryffel GU. ERH (enhancer of rudimentary homologue), a conserved factor identical between frog and human, is a transcriptional repressor. Biol. Chem. 2001;382:1379–1385. doi: 10.1515/BC.2001.170. [DOI] [PubMed] [Google Scholar]
- 76.Rafferty JB, Bolt EL, Muranova TA, Sedelnikova SE, Leonard P, Pasquo A, Baker PJ, Rice DW, Sharples GJ, et al. The structure of Escherichia coli RusA endonuclease reveals a new Holliday junction DNA binding fold. Structure. 2003;11:1557–1567. doi: 10.1016/j.str.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 77.Aravind L, Makarova KS, Koonin EV. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 79.Nielsen TK, Liu S, Luhrmann R, Ficner R. Structural basis for the bifunctionality of the U5 snRNP 52K Protein (CD2BP2) J. Mol. Biol. 2007;369:902–908. doi: 10.1016/j.jmb.2007.03.077. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.