Abstract
For most aminoacyl-tRNA synthetases (aaRS), their cognate tRNA is not obligatory to catalyze amino acid activation, with the exception of four class I (aaRS): arginyl-tRNA synthetase, glutamyl-tRNA synthetase, glutaminyl-tRNA synthetase and class I lysyl-tRNA synthetase. Furthermore, for arginyl-, glutamyl- and glutaminyl-tRNA synthetase, the integrated 3' end of the tRNA is necessary to activate the ATP-PPi exchange reaction. Tryptophanyl-tRNA synthetase is a class I aaRS that catalyzes tryptophan activation in the absence of its cognate tRNA. Here we describe mutations located at the appended β1–β2 hairpin and the AIDQ sequence of human tryptophanyl-tRNA synthetase that switch this enzyme to a tRNA-dependent mode in the tryptophan activation step. For some mutant enzymes, ATP-PPi exchange activity was completely lacking in the absence of tRNATrp, which could be partially rescued by adding tRNATrp, even if it had been oxidized by sodium periodate. Therefore, these mutant enzymes have strong similarity to arginyl-tRNA synthetase, glutaminyl-tRNA synthetase and glutamyl-tRNA synthetase in their mode of amino acid activation. The results suggest that an aaRS that does not normally require tRNA for amino acid activation can be switched to a tRNA-dependent mode.
INTRODUCTION
Aminoacyl-tRNA synthetases (aaRS) catalyze the esterification of an amino acid to the 3′-end of its cognate tRNA in a two-step reaction. In the first step, the amino acid is activated in the form of an enzyme-bound aminoacyl–adenylate intermediate, while the second step involves the transfer of the amino acid to the 3′-end of its cognate tRNA to produce an aminoacyl-tRNA (1). Based on two distinct ATP-binding cores, the 20 aaRSs are equally divided into two classes (2,3). In most cases, formation of the aminoacyl-adenylate does not require the presence of its cognate tRNA and can be performed by the isolated catalytic domain of some synthetases (4,5). However, for three class I synthetases, arginyl-, glutaminyl- and glutamyl-tRNA synthetase (ArgRS, GlnRS and GluRS, respectively), as well as the exceptional class I lysyl-tRNA synthetase (LysRS), tRNAs are absolutely required to accomplish the first step of the reaction.
The requirement of tRNA in aminoacyl-adenylate formation has been confirmed for ArgRS from different sources (6–11) and is likely a common feature of this enzyme. An intact 3′-terminal adenosine of its cognate tRNA is also required, because arginine activation cannot be induced by periodate-treated tRNA (7,9,10,12,13). Proper tRNA structure is also necessary for arginine activation (13). For GluRS and GlnRS, amino acids also cannot be activated without their cognate tRNA (14–17). The activator function also requires the integrity of the 3′ terminus of the tRNA, and chemical modification of this terminus abolishes this activity (14,16,18). Exceptionally, several bacteria and archaea have the class I-type LysRS instead of the class II-type LysRS (19,20). Class I LysRS is structurally related to GluRS (21), and does not catalyze lysyl-adenylate formation in the absence of tRNA (22). However, the biological significance of requiring tRNA to activate amino acids remains puzzling. The fact that ArgRS requires its cognate tRNA to activate arginine has led to the suggestion that the arginyl–adenylate intermediate does not exist in the arginylation reaction (23), although Kern and Lapointe (24,25) later demonstrated that GluRS charges tRNA with glutamate in two steps and that the glutamyl–adenylate intermediate exists, despite the fact that tRNA is needed for the first step.
Tryptophanyl-tRNA synthetase (TrpRS) is considered a class I aaRS, and together with all other synthetases except for ArgRS, GluRS, GlnRS and class I LysRS, can activate an amino acid substrate smoothly in the absence of cognate tRNA (1). We recently showed that the ATP-PPi exchange reaction of a mutated Bacillus subtilis TrpRS can be modulated by its inactivated tRNATrp (26).
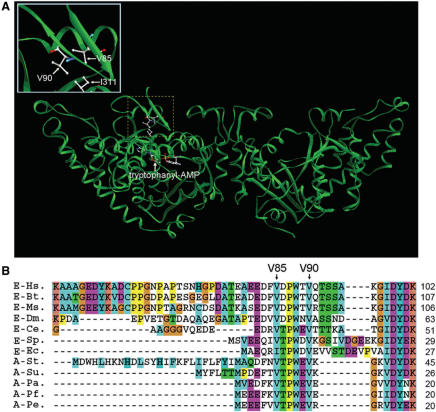
Eukaryotic TrpRS differs from prokaryotic TrpRS in containing an appended N-terminus. For human TrpRS, the N-terminal domain is composed of an appended peptide and an eukaryote-specific patch (E82–K154) containing a β1–β2 hairpin structure adjacent to the ATP-binding pocket and the KMSAS loop (27). Deletion of this hairpin severely reduces aminoacylation activity, suggesting that human TrpRS has a more complicated aminoacylation mechanism than B. subtilis TrpRS. At the β1–β2 hairpin of human TrpRS (1R6T from the Protein Data Bank), the backbone of V85 and V90 form a hydrogen bond. The side chains of the two valines protrude into the catalytic domain, which looks like a buckhorn (Figure 1A).
Figure 1.
Primary and tertiary structure of human TrpRS. (A) Crystal structure of full-length human TrpRS (40). The tryptophanyl–AMP intermediate is indicated by an arrowhead. The V85 and I311 interface between the β1–β2 hairpin and main body is enlarged in the boxed inset. (B) Primary sequence alignment of the region of human TrpRS containing the β1–β2 hairpin. The conserved V85 and V90 in TrpRSs are indicated by arrowheads.
In this study, we mutated V85 and V90 and unexpectedly found that the V85S mutant could no longer activate tryptophan even though its tryptophanylation activity still proceeded normally. Furthermore, we confirmed that the tryptophan activation of V85S can be partially rescued by the addition of bovine tRNATrp. We constructed further mutants at V85 to further investigate and found that the V85E mutation also caused a switch to a tRNA-dependent mode of amino acid activation, just like ArgRS, GluRS, GlnRS and class I LysRS. Structural analysis showed that V85 can interact with I311 through a hydrophobic interaction to affect tryptophan activation, which was confirmed by I311 mutations. The results indicate that an aaRS that does not normally require tRNA for amino acid activation can be switched to a tRNA-dependent mode.
MATERIALS AND METHODS
Strains and plasmids
Bovine tRNATrp was expressed in Escherichia coli JM109 and human TrpRS and its mutants were expressed in E. coli BL21-CodonPlus (DE3)-RIL. BL21-CodonPlus (DE3)-RIL was purchased from Stratagene (La Jolla, CA, USA). The bovine tRNATrp gene was cloned into the pGEM-9Zf (–) plasmid obtained from Promega (Madison, WI, USA) and was a kind gift from Dr Xue Hong at the Hong Kong University of Science and Technology. The plasmid pTrc-hTrpRS containing the human trpS gene was also a gift from Dr Xue Hong. The human trpS gene expression vector pET24a (+) was purchased from Novagen.
Sequence homology analysis and structure comparison
TrpRS protein sequences from archaebacteria and eukaryotes were downloaded from the aaRS database (www.pozman.edu.pl/aars) (28). Abbreviations of the species are as follows: E-Hs, Homo sapiens; E-Bt, Bos taurus; E-Ms, Mus musculus; E-Dm, Drosophila melanogaster; E-Ce, Caenorhabditis elegans; E-Sp, Schizosaccharomyces pombe; E-Ec, Encephalitozoon cuniculi; A-St, Sulfolobus tokodaii; A-Su, Sulfolobus solfataricus; A-Pa, Pyrococcus abyssi; A-Pf, Pyrococcus furiosus; A-Pe, Pyrobaculum aerophilum. The first letter E in the abbreviation means eukaryote, and A means archaea. Multiple sequence alignments were carried out with Clustal X 1.83 software (ftp-igbmc.u-strasbg.fr/pub/ClustalX/). The pdb files of human TrpRSs (1O5T, 1R6T, 1ULH and 2DR2) were downloaded from the Protein Data Bank (http://www.rcsb.org/pdb/). Structure comparisons and figure production were carried out with Swiss-PdbViewer 3.7 software, which was downloaded from http://www.expasy.ch/spdbv/text/download.htm. The operation pathway of Swiss-PdbViewer 3.7 software for structure comparison was Fit\Magic Fit\CA (carbon alpha) only\.
Expression and purification of human TrpRS and bovine tRNATrp
Expression and purification of human TrpRS were carried out as described previously (29). Concentrations of purified proteins were determined with the Bradford reagent (30). Mutagenesis was carried out by PCR as described previously (31). Mutant human TrpRS genes were verified by DNA sequencing. Abbreviations of the mutations were as follows: V85A, valine 85 to alanine; V85E, valine 85 to glutamate; V85K, valine 85 to lysine; V85L, valine 85 to leucine; V85S, valine 85 to serine; V90A, valine 90 to alanine; V90S, valine 90 to serine; V85A/V90A, V85A/V90A double mutations; I311V, isoleucine 311 to valine; I311E, isoleucine 311 to glutamate. The bovine tRNATrp used for the enzyme activity assay was expressed and purified as described previously (32).
Periodate oxidation of tRNA
The oxidation of tRNA by sodium periodate was carried out as previously described (33,34) with some modifications. Bovine tRNATrp was treated with 10 mM sodium periodate in 1 ml of 150 mM sodium acetate (pH 5.3), incubated at 4°C in the dark for 1 h. Glucose was then added to a final concentration of 10 mM to remove excess sodium periodate. The tRNA was incubated at 37°C for 30 min and then precipitated with ethanol.
Enzymatic assays
The ATP-PPi exchange reaction was assayed at 30°C in a reaction mixture containing 100 mM Tris–HCl pH 7.8, 10 mM potassium fluoride, 10 mM magnesium chloride, 4 mM ATP, 0.02 μCi [γ-32P]ATP, 4 mM sodium pyrophosphate and 2 mM tryptophan in total volume of 20 μl. Reactions were initiated by the addition of 0.2 μM enzyme. At each time point, samples were quenched on ice. One microliter of the reaction mixture was spotted onto PEI cellulose TLC plates (purchased from Merck, Germany) and developed in 1 M KH2PO4 and 1 M urea to separate the ATP and PPi. The radioactivity was revealed and quantified using a PhosphorImager™ (Molecular Dynamics, Little Chalfont, Bucks, UK).
For mutants V85S, V85E, V85K, V85A/V90A and I311E, the tRNA-dependent ATP-PPi exchange reaction was assayed. Wild-type bovine tRNATrp and oxidized bovine tRNATrp were added to the PPi exchange reaction mixture to a final concentration of 60 μM. The other components for the reaction were identical to that described above.
The aminoacylation assay was also carried out at 30°C in a mixture containing 4 mM ATP, 0.8 mM DTT, 1 μCi l-[5-3H]tryptophan, 12 μM l-tryptophan, 8 mM MgCl2, 80 mM Tris–HCl, pH 7.5 and 12 μM purified bovine tRNATrp in a total volume of 50 μl. Reactions were initiated by the addition of 50 nM enzyme. At each time point, samples were quenched on ice and 20 μl aliquots were spotted onto filter paper disks, which were washed three times with ice-cold 5% trichloroacetic acid containing 0.05% tryptophan and with cold anhydrous ethanol, dried and transferred into vials for determination of radioactivity. For all kinetic assays, the concentration of tRNATrp varied from 0.2 to 12.8 μM. Each reaction was repeated at least four times under the same conditions. The kcat/Km for aminoacylation was calculated from Eadie–Hofstee plots. All data were fitted to the Michaelis–Menten equation.
RESULTS
The V85S mutation enables human TrpRS to switch to tRNA-dependent tryptophan activation
Based on a structural analysis of human TrpRS, we found that there were two adjacent valines in the middle of the β1 and β2 sheet, V85 and V90, whose side chains both approach the substrate-binding pocket (Figure 1A). The β1–β2 hairpin is essential to the aminoacylation activity of human TrpRS (27), and therefore these two valine residues may be important for enzyme activity. Based on sequence alignment, we found that V85 and V90 are conserved in eukaryotes and archaebacteria (Figure 1B). Therefore, we constructed mutations at these two positions, V85 and V90, to generate the V85A, V85S, V90A and V90S variants.
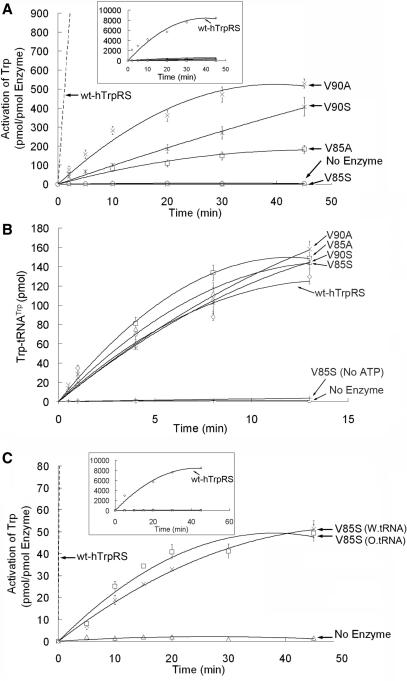
In the ATP-PPi exchange reaction, the V85A, V90A and V90S mutants showed decreased, but visible tryptophan activation activity, with V85A having the lowest enzyme activity (Figure 2A). Furthermore, when V85 was mutated to serine, no ATP-PPi exchange activity was detected, even at a very high enzyme concentration (5.1 µM) (Figure 2A). Unexpectedly, the aminoacylation activity of the V85S mutant remained comparable to the other three mutant enzymes (Figure 2B) and could not catalyze the tryptophanylation reaction in the absence of ATP (Figure 2B). This suggested that the aminoacylation reaction of the V85S mutant proceeds normally and that the mutation does not change the recognition of ATP. These results implied that the V85S mutant might be able to activate tryptophan only in the presence of tRNA. To investigate whether the V85S mutant assumes a tRNA-dependent ATP-PPi exchange activity, wild-type or sodium periodate-treated bovine tRNATrp was added to the ATP-PPi exchange reaction mixture. Unexpectedly, both the wild type and the sodium periodate-treated bovine tRNATrp were able to partially rescue the ATP-PPi exchange activity of the V85S mutant (Figure 2C). Sodium pyrophosphate can greatly inhibit the tryptophanylation reaction. After adding the wild-type tRNAs to the ATP-PPi exchange reaction, the tryptophanyl-tRNATrp could hardly be detected (data not shown). Thus, the tRNA-independent ATP-PPi exchange reaction for human TrpRS is switched to a tRNA-dependent mode by the single V85S mutation. However, in contrast to ArgRS, GlnRS and GluRS (7,9,10,12–14,16,18), the integrity of the 3′ terminus of the tRNA was not necessary for the ATP-PPi exchange activity of the V85S mutant (Figure 2C). Therefore, it is reasonable to conclude that human TrpRS is switched to a ‘nonproductive’ form by V85S mutation but can be induced to a ‘productive’ form by tRNA binding. Comparing Figure 2A B, we conclude that V85 mutations have a greater impact on tryptophan activation than the tryptophanylation reaction.
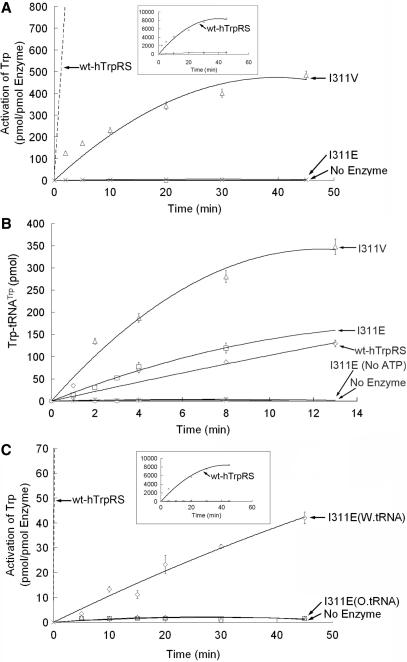
Figure 2.
Enzymatic activities of the V85A, V85S, V90A and V90S mutants. (A) ATP-PPi exchange activity of the four mutants and wild-type human TrpRS. To display the whole curve of wt-hTrpRS, we enlarged the scale of the Y-axis in the boxed inset. (B) Aminoacylation activity of the four mutants and wild-type human TrpRS. The enzyme concentrations are 5 nM for wild-type human TrpRS (wt-hTrpRS), 20 nM for V85A and V90A and 50 nM for V85S and V90S. (C) PPi exchange activity of the V85S mutant in the presence of wild-type or oxidized bovine tRNATrp. The wt-hTrpRS curve is shown in the boxed inset.
Additional mutants of valine 85
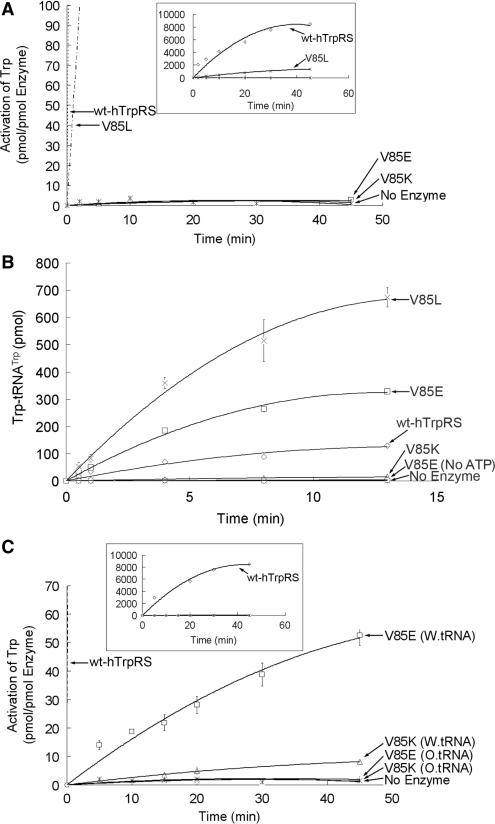
To further investigate the role of the V85 residue in the ATP-PPi exchange reaction, we constructed three more mutations at this position including V85L, V85E and V85K. Relative to the V85E and V85K mutations, the V85L mutant did not show a large decrease in tryptophan activation (Figure 3A). However, the V85E and V85K mutants lacked the ATP-PPi exchange activity of human TrpRS under normal conditions (Figure 3A), just like the V85S mutant (Figure 2A). In the tryptophanylation reaction, the V85L mutant was able to acylate bovine tRNATrp with very high efficiency (Figure 3B). Similar to the V85S mutant, the V85E mutant has low but noticeable activity (Figure 3B). In contrast, the V85K mutant had barely detectable aminoacylation activity (Figure 3B). In the presence of oxidized bovine tRNATrp, the V85E TrpRS was unable to catalyze the ATP-PPi exchange reaction, but the wild-type bovine tRNATrp could still serve as an activator (Figure 3C), which suggests that the hydroxyl groups of the 3′ adenosine of tRNA can also activate human TrpRS. For the V85K mutant, wild-type tRNA could slightly rescue its PPi exchange activity while tRNA treated with sodium periodate could not (Figure 3C).
Figure 3.
Enzymatic activities of the V85E, V85K and V85L mutants. (A) ATP-PPi exchange activity of the three mutations and wild-type human TrpRS. The boxed inset displayed the whole curve of wt-hTrpRS and the V85L mutant. (B) Aminoacylation activity of the three mutations and wild-type human TrpRS. The enzyme concentrations are 5 nM for wt-hTrpRS and V85L and 200 nM for V85E and V85K. (C) PPi exchange activity of the V85E and V85K mutants in the presence of wild-type or oxidized bovine tRNATrp. The enlarged scale for wt-hTrpRS and the 85L mutant are shown in the boxed inset.
Thus, we conclude that mutation of V85 to a hydrophilic amino acid such as serine, lysine and glutamate abolishes the normal ATP-PPi exchange activity of human TrpRS (Figures 2A and 3A). On the other hand, replacing V85 with a neutral amino acid such as alanine, or a hydrophobic amino acid such as leucine, did not produce the same result (Figures 2A and 3A). Moreover, the more hydrophobic the side chains at the V85 position, the higher the enzyme activity.
Interaction between the β1–β2 hairpin and tRNA
The V85S single mutation switches human TrpRS from a tRNA-independent to a tRNA-dependent mode of tryptophan activation (Figure 2). For the V85S mutant, the tRNA molecule, not the hydroxyl group at the 3′ end, serves as an activator, which is different from ArgRS, GluRS and GlnRS (Figure 2C). The V85E mutant was activated to catalyze the ATP-PPi exchange reaction only in the presence of wild-type bovine tRNATrp (Figure 3C). This suggests that the 2′ or 3′ hydroxyl group at the 3′ adenosine of tRNA also can act as an activator, similar to ArgRS, GluRS and GlnRS. The V85K mutant had barely detectable enzyme activity.
Based on the recent co-crystal structure of human TrpRS and bovine tRNA, we concluded that V85 should be close to a tRNA acceptor stem when tRNA is bound to human TrpRS (27,35). Thus, the phosphate groups of tRNAs should be easily affected by the V85 mutation. To verify our hypothesis, we calculated the Km of tRNA for the V85S, V85E and V85K mutants. For the V85S mutant, the Km value was 2.14 μM, similar to 1.30 μM for wild-type human TrpRS (Table 1). The Km value was 8.50 μM for V85E and 0.294 μM for V85K (Table 1). Therefore, it is reasonable to conclude that the V85S mutant that leaves the side chain uncharged has little effect on tRNA binding, so not only wild type but also oxidized tRNA is able to properly bind to the enzyme to prompt the ATP-PPi exchange reaction. Because the side chain of glutamate is negatively charged, the V85E mutation would push out the negatively charged phosphate groups of tRNAs, weakening tRNA binding. The V85K mutant, with a positively charged side chain, can attract the negatively charged phosphate groups of tRNAs. As a result, tRNA may be bound to human TrpRS at an incorrect position when V85 was mutated to a lysine. Therefore, the fact that the V85K mutant has the lowest acceptor activity is reasonable.
Table 1.
The aminoacylation kinetics constant for wt-hTrpRS, V85A, V85S, V90A, V90S, V85E, V85K, V85L, and V85A/V90A mutants. All Km are for tRNA
| Enzymes | Km (μM) | kcat (s−1) | kcat/Km (μM−1 s−1) | Relative activity |
|---|---|---|---|---|
| wt-hTrpRS | 1.30 ± 0.16 | 1.37 ± 0.14 | 1.05 | 1.00a |
| V85A | 1.41 ± 0.15 | 0.476 ± 0.026 | 0.338 | 0.322 |
| V85S | 2.14 ± 0.54 | (2.77 ± 0.77) × 10−2 | 1.29 × 10−2 | 1.23 × 10−2 |
| V90A | 18.4 ± 2.2 | 1.39 ± 0.070 | 7.55 × 10−2 | 7.19 × 10−2 |
| V90S | 5.20 ± 0.90 | (6.25 ± 0.63) × 10−2 | 1.20 × 10−2 | 1.14 × 10−2 |
| V85E | 8.50 ± 1.4 | (1.97 ± 0.37) × 10−2 | 2.32 × 10−3 | 2.21 × 10−3 |
| V85K | 0.294 ± 0.047 | (9.44 ± 2.4) × 10−4 | 3.21 × 10−3 | 3.06 × 10−3 |
| V85L | 24.8 ± 5.6 | 5.11 ± 0.99 | 0.206 | 0.196 |
| V85A/V90A | 2.23 ± 0.24 | (4.71 ± 0.49) × 10−2 | 2.11 × 10−2 | 2.01 × 10−2 |
aThe activity of wild-type human TrpRS was set to 1.00.
We next calculated the Km for tRNA and the kcat values of all the above mutations (Table 1). These varied dramatically at the β1–β2 hairpin, with Km ranging from 0.294 to 24.8 μM and kcat ranging from 9.44 × 10−4 S−1 to 5.11 S−1. The Km values suggest that V85 and V90 can interact with tRNA when tRNA binds to human TrpRS. Thus, it is logical that charge variations of the side chains at V85 and V90 have the greatest impact on tRNA acceptor activity.
V85 interacts with I311 of the AIDQ sequence via by hydrophobic side chains
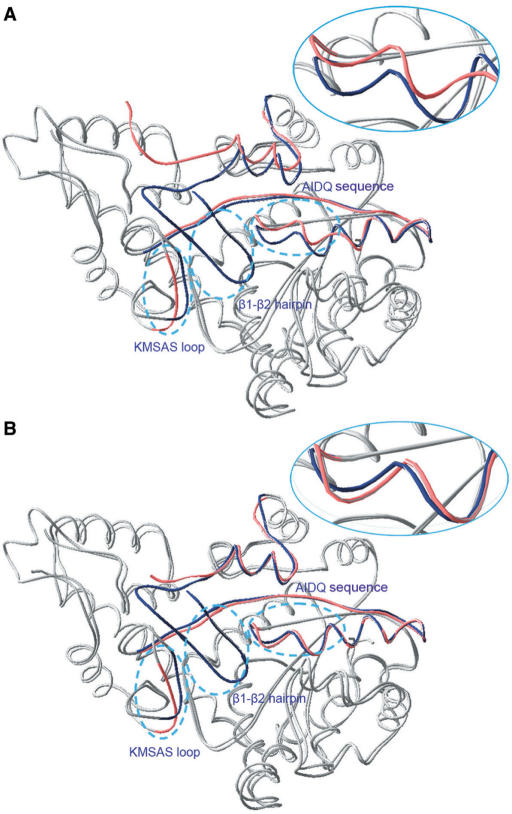
Based on the crystal structure of the full-length human TrpRS (1R6T), we found that V85 is far away from the substrate-binding pocket (Figure 1A) and therefore could not directly affect tryptophan activation. However, the side chain of V85 is only 4.41 Å away from the side chain of I311 (Figure 1A). I311 is a residue in the conserved AIDQ sequence. A310 and D312 interact with the ribose of ATP through hydrogen bonds to stabilize ATP binding (35). Combining all findings, we concluded that V85 may affect I311 through hydrophobic interactions between their side chains to affect tryptophan activation. Furthermore, we compared the crystal structure of unliganded human mini-TrpRS (1ULH), which contains the β1–β2 hairpin structure at one monomer, with the unliganded crystal structure of T2-TrpRS (1O5T) in which the β1–β2 hairpin structure is hydrolyzed. Both the mini-TrpRS and T2-TrpRS are truncated forms of human TrpRS at the N-terminus with mini-TrpRS encoding residues 48–471 and T2-TrpRS encoding residues 94–471. We found that the AIDQ sequence of mini-TrpRS assumes a compact form (Figure 4A), while the AIDQ sequence of T2-TrpRS adopts a loose form (Figure 4A). When bovine tRNATrp binds to T2-TrpRS (2DR2), as expected, its AIDQ sequence switches to the compact form (Figure 4B). Therefore, it is reasonable to conclude that, at least partially, the interaction of V85 with I311 can induce the AIDQ sequence to an active compact form. If V85 is mutated to a hydrophilic amino acid that disrupts the hydrophobic interaction between V85 and I311 (such as serine, glutamate and lysine), the conformational change in the AIDQ sequence might not be induced correctly. Alternatively, because the 3′ end of tRNA and the AIDQ sequence interact (35), tRNA can substitute for the function of the β1–β2 hairpin.
Figure 4.
Structural comparisons of human T2-TrpRS and mini-TrpRS. (A) Comparison of unliganded T2-TrpRS (1O5T) and mini-TrpRS (1ULH). (B) Comparison of T2-TrpRS bound to bovine tRNATrp (2DR2) and mini-TrpRS (1ULH). For clarity, the tRNA molecule of 2DR2 is not displayed, and only part of the N-terminus, the AIDQ sequence, and the KMSAS loop are colored dark blue for mini-TrpRS and pink for T2-TrpRS. In both Figure 4A and B, the β1–β2 hairpin, the AIDQ sequence and the KMSAS loop are indicated with sky blue dashed ellipses. The conformational change of the AIDQ sequence is enlarged in the circled ellipse inset.
To confirm our hypothesis, we mutated I311 to valine and glutamate and determined their enzyme activity. As we had expected, when the isoleucine was mutated to acidic glutamate, the normal ATP-PPi exchange activity of human TrpRS was abolished (Figure 5A), but the mutant enzyme could still catalyze the tryptophanylation reaction with decreased efficiency (Figure 5B). Wild-type bovine tRNATrp, not the oxidized form, was able to promote the ATP-PPi exchange reaction (Figure 5C), just like the V85E mutation. Compared with wild-type human TrpRS, the I311V mutation had little effect on both the ATP-PPi exchange reaction and tryptophanylation reaction relative to the large effect of the I311E mutant on both reactions (Figure 5A and B). Therefore, the above inference should be true. Relative to the V85E mutant, the I311E mutant has even lower aminoacylation activity. The I311E mutant might interfere with the interaction between the A76 ribose of bovine tRNATrp and AIDQ sequence (35).
Figure 5.
Enzymatic activities of the I311E and I311V mutants. (A) ATP-PPi exchange activity of the two mutants and wild-type human TrpRS. The boxed inset displayed the whole curve of wt-hTrpRS. (B) Aminoacylation activity of the two mutants and wild-type human TrpRS. The enzyme concentrations are 5 nM for wild-type human TrpRS, 50 nM for the I311V mutant and 200 nM for the I311E mutant. (C) PPi exchange activity of the I311E mutant in the presence of wild-type or oxidized bovine tRNATrp and wild-type human TrpRS. The enlarged scale for wt-hTrpRS is shown in the boxed inset.
The V90 mutation enhances the hydrophobic interaction between V85 and I311
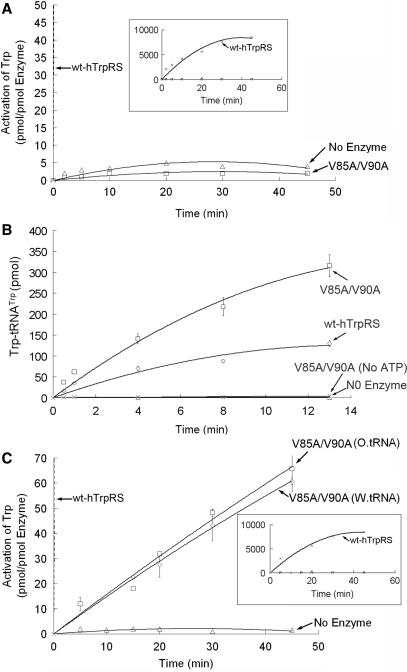
We have found that the V90 mutations had little effect on the ATP-PPi exchange reaction. Mutants of human TrpRS at position 90 only showed reduced ATP-PPi exchange activity. Based on structural analysis of the full-length human TrpRS (1R6T), we found that the side chain of V90 is farther away from the side chain of I311 than that of V85. Thus, V90 should only act as a subsidiary residue to enhance the hydrophobic interaction between V85 and I311. Therefore, we constructed a double mutant of human TrpRS, V85A/V90A. As expected, the behavior of V85A/V90A was similar to the V85S mutant (Figure 6). We also calculated the Km and kcat for the V85A/V90A double mutant, which were 2.23 μM and 0.0471 S−1, respectively (Table 1).
Figure 6.
Enzymatic activities of the V85A/V90A double mutants. (A) ATP-PPi exchange activity of this mutant and the wild-type human TrpRS. The enlarged scale is shown in the boxed inset for wt-hTrpRS. (B) Aminoacylation activity of the V85A/V90A double mutant and wild-type human TrpRS. The enzyme concentrations are 5 nM for wild-type human TrpRS and 200 nM for the V85A/V90A mutant. (C) PPi exchange activity of the V85A/V90A double mutant in the presence of wild-type or oxidized bovine tRNATrp and wild-type human TrpRS. The wt-hTrpRS curve is shown in the boxed inset.
There are no Schiff bases formed between oxidized bovine tRNATrp and human TrpRS mutants
Previous studies have shown that tRNA oxidized by sodium periodate can form Schiff bases with the ε-NH2 of lysine residues close to the binding site (33,36), irreversibly inhibiting enzyme activity. To clarify whether the oxidized bovine tRNATrp can form Schiff bases with human TrpRS, we pre-incubated the V85E, V85K, V85S, V85A/V90A and I311E mutants with oxidized tRNA for 10 min, 20 min, 30 min and 40 min, and then performed the aminoacylation assay (see Materials and Methods section for aminoacylation conditions). After 10 min, reactions were stopped and enzyme activity was determined. The results showed that oxidized tRNA did not inhibit the activity of the enzymes (Supplementary Figure S1). Therefore, Schiff bases do not form between oxidized tRNA and TrpRS.
DISCUSSION
ArgRS, GluRS, GlnRS and Class I LysRS all share the common feature that is the absolute requirement of tRNA in the amino acid activation step. For these aaRSs, the tRNA is postulated to serve as the enzyme activator in the first step and as the substrate in the second step of aminoacylation. Except for these four enzymes, the other class I aaRSs do not require the presence of tRNA to activate amino acid, such as TrpRS. Here, we combined mutations to illustrate that human TrpRS can be switched to a tRNA-dependent mode to activate tryptophan by single or double mutation(s) in the β1–β2 hairpin and AIDQ sequence. These mutant enzymes were all tested at a high concentration (>2.5 μM) and were unable to activate tryptophan in the absence of tRNA. This finding was unexpected. These mutations abolished the PPi exchange activity of TrpRS, which can be partially rescued by the addition of tRNA. For ArgRS, GluRS and GlnRS, comparisons of the tRNA-free and tRNA-bound enzyme crystals have revealed tRNA-induced enzyme conformational changes (37–39), which should be responsible for amino acid activation. The situation for human TrpRS should be the same and was confirmed by structural comparison (Figure 4). On the other hand, for wild-type human TrpRS, V85 should perform the same function, at least partially, as tRNA in the tryptophan activation reaction (Figure 4).
Conformational changes in human TrpRS induced by V85 would presumably require specific molecular interactions. We have identified an interface between the eukaryotic-specific patch and the catalytic center, which might be responsible, at least in part, for molecular interactions that would facilitate tryptophan activation. This site is defined in human TrpRS by the eukaryotic-specific patch that contains V85 at the β1–β2 hairpin and I311 at the conserved AIDQ sequence which directly interacts with ATP, based on the crystal structure of full-length human TrpRS (40). The side chains of V85 and I311 are as little as 4.41 Å apart. A comparison of the X-ray crystal structures of T2-TrpRS and mini-TrpRS (both not bound by substrates) illustrates that the β1–β2 hairpin is responsible for the ‘productive’ form of human TrpRS (Figure 4). We concluded that, at least partially, the compact form of the AIDQ sequence of human TrpRS is ensured by hydrophobic interactions between the side chains of V85 and I311, because the side chain of V90 is much farther away from the side chain of I311 than that of V85. V90 should be a minor residue to interact with I311 by hydrophobic interaction, which was confirmed by the V85A/V90A double mutant. A310 and D312 are known to be able to form hydrogen bonds with the ribose of ATP to stabilize ATP binding. Therefore, if V85 is mutated to a hydrophilic amino acid, the hydrophobic interaction between V85 and I311 will be disrupted and the AIDQ sequence might assume a loose form. As a result, the hydrogen bond interaction between ATP and the AIDQ sequence will be disrupted and ATP may not be able to bind TrpRS stably. Thus, mutated human TrpRS cannot catalyze tryptophan activation until tRNA is also bound.
For V85E and I311E, the oxidized tRNA cannot rescue their PPi exchange activity, but wild-type tRNA still can (Figures 3C and 5C). After treatment with sodium periodate, the 2′ and 3′ hydroxyl groups of tRNA are oxidized to aldehyde groups. This should not result in any large changes in the hydrogen bonds between the A76 of tRNA and residues D312 and Q313 of the AIDQ sequence (35), otherwise the tRNA oxidized by sodium periodate would not be able to bind to human TrpRS properly and rescue the PPi exchange activity of the V85S mutant and the V85A/V90A double mutants. These results imply that the 2′ or 3′ hydroxyl group also can serve as an activator, just like ArgRS, GluRS and GlnRS. As enzyme activity proceeds, the 2′ or 3′ hydroxyl group of A76 will consequentially interact with the Trp–adenylate intermediate to accept activated tryptophan. The interaction between tRNA and Trp-adenylate may stabilize ATP binding allowing the human TrpRS to activate tryptophan, even though V85 or I311 was mutated to glutamate. Furthermore, the sequence alignments show that V85 and V90 are conserved in eukaryotes and archaebacteria, including the proline and tryptophan in the β1–β2 hairpin (Figure 1B). Therefore, the β1–β2 hairpin structure of human TrpRS should be conserved in eukaryotes and archaebacteria and its function should be the same as that in human TrpRS.
Wild-type human TrpRS or mini-TrpRS does not require its cognate tRNA to activate tryptophan (41). However, we can switch its tRNA-independent PPi exchange activity to a tRNA-dependent mode by introducing mutations within the eukaryote-specific patch (E82–K154), which is not found in prokaryotic enzymes. It raises the question of whether the tRNA-independent PPi exchange activity of human TrpRS is an ancestral feature or has been more recently acquired with the acquisition of the eukaryote-specific patch. Our results cannot give a definite answer, although it seems that the partition between tRNA-independent and tRNA-dependent PPi exchange activity within a single aaRS can shift dramatically with relative evolutionary ease. Based on sequence alignment, we recently found a TrpRS from Pyrococcus horikoshii, a hyper-thermophilic archaebacterium. This TrpRS has eukaryotic features, but the eukaryotic-specific patch is completely absent in this enzyme. This archeabacterial TrpRS could provide further evidence to define the evolutionary connection between TrpRS and ArgRS-like aaRSs.
For ArgRS, GluRS, GlnRS and class I LysRS, the biological significance of requiring tRNA as a cofactor to activate amino acid is unclear. However, a previous study showed that tRNA binding can help these four class I aaRSs specially recognize their cognate amino acid. The co-crystal structure of Thermus thermophilus GluRS and tRNAGlu with or without glutamate demonstrated that the amino acid-binding site of T. thermophilus GluRS is incomplete and the presence of the cognate tRNAGlu facilitates glutamate binding by the enzyme (39). In the absence of tRNA, T. thermophilus GluRS binds not only l-glutamate but also noncognate amino acids such as d-glutamate, l-aspartate and l-glutamine (42). The binding of the noncognate amino acids is eliminated by tRNAGlu binding to the enzyme. Similarly, the co-crystal structure of E. coli GlnRS indicated that the tRNA itself is involved in the Gln-AMP-binding site (38). For ArgRS, the Bacillus stearothermophilus and Neurospora crassa ArgRSs can bind the cognate arginine only in the presence of tRNAArg (7,10). A similar phenomenon was also reported for class I LysRS (22). The recently discovered dual specificity enzyme, prolyl-tRNA synthetase (ProRS), can acylate not only proline but also cysteine, although it can only recognize cysteine in the presence of Trna (43,44). The existence of aaRSs that can catalyze the synthesis of more than one aminoacyl-tRNA is assumed to be an important step in the evolution of these enzymes (45). tRNA-dependent amino acid recognition may be a relic of synthetase evolution. However, a recent paper by Carter et al. (46) showed that the minimal TrpRS catalytic domain from B. stearothermophilus can still catalyze the tryptophan activation step without tRNA and also mischarge tRNA. Therefore, the biological significance of tRNA-dependent amino acid recognition remains puzzling. Otherwise, is the tRNA-dependent amino acid recognition inherent to class I aaRSs? Maybe we can get the answers in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work is supported by the National Natural Science Foundation (No. 30370325) and 973 program (No. 2005CB724602). Funding to pay the Open Access publication charges for this article was provided by No.2005CB724602 (973 program).
Conflict of interest statement. None declared.
REFERENCES
- 1.Ibba M, Söll D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 2000;69:617–650. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- 2.Eriani G, Delarue M, Poch O, Gangloff J, Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990;347:203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- 3.Cusack S. Eleven down and nine to go. Nat. Struct. Biol. 1995;2:824–831. doi: 10.1038/nsb1095-824. [DOI] [PubMed] [Google Scholar]
- 4.Zhang CM, Hou YM. Domain-domain communication for tRNA aminoacylation: the importance of covalent connectivity. Biochemistry. 2005;44:7240–7249. doi: 10.1021/bi050285y. [DOI] [PubMed] [Google Scholar]
- 5.Buechter DD, Schimmel P. Dissection of a class II tRNA synthetase: determinants for minihelix recognition are tightly associated with domain for amino acid activation. Biochemistry. 1993;32:5267–5272. doi: 10.1021/bi00070a039. [DOI] [PubMed] [Google Scholar]
- 6.Mitra K, Mehler AH. The role of transfer ribonucleic acid in the pyrophsphate exchange reaction of arginine-transfer ribonucleic acid synthetase. J. Biol. Chem. 1966;241:5161–5162. [PubMed] [Google Scholar]
- 7.Parfait R, Grosjean H. Arginyl-transfer ribonucleic-acid synthetase from Bacillus stearothermophilus. Purification, properties and mechanism of action. Eur. J. Biochem. 1972;30:242–249. doi: 10.1111/j.1432-1033.1972.tb02092.x. [DOI] [PubMed] [Google Scholar]
- 8.Char S, Gopinathan KP. Arginyl-tRNA synthetase from Mycobacterium smegmatis SN2: purification and kinetic mechanism. J. Biochem. (Tokyo) 1986;100:349–357. doi: 10.1093/oxfordjournals.jbchem.a121721. [DOI] [PubMed] [Google Scholar]
- 9.Gangloff J, Schutz A, Dirheimer G. Arginyl-tRNA synthetase from baker's yeast. Purification and some properties. Eur. J. Biochem. 1976;65:177–182. doi: 10.1111/j.1432-1033.1976.tb10403.x. [DOI] [PubMed] [Google Scholar]
- 10.Nazario M, Evans JA. Physical and kinetic studies of arginyl transfer ribonucleic acid ligase of Neurospora. A sequential ordered mechanism. J. Biol. Chem. 1974;249:4934–4936. [PubMed] [Google Scholar]
- 11.Lazard M, Agou F, Kerjan P, Mirande M. The tRNA-dependent activation of arginine by arginyl-tRNA synthetase requires inter-domain communication. J. Mol. Biol. 2000;302:991–1004. doi: 10.1006/jmbi.2000.4102. [DOI] [PubMed] [Google Scholar]
- 12.Mehler AH, Mitra SK. The activation of arginyl transfer ribonucleic acid synthetase by transfer ribonucleic acid. J. Biol. Chem. 1967;242:5495–5499. [PubMed] [Google Scholar]
- 13.Guigou L, Mirande M. Determinants in tRNA for activation of arginyl-tRNA synthetase: evidence that tRNA flexibility is required for the induced-fit mechanism. Biochemistry. 2005;44:16540–16548. doi: 10.1021/bi051575h. [DOI] [PubMed] [Google Scholar]
- 14.Ravel JM, Wang SF, Heinemeyer C, Shive W. Glutamyl and glutaminyl ribonucleic acid synthetases of Escherichia coli W. Separation, properties, and stimulation of adenosine triphosphate-pyrophosphate exchange by acceptor ribonucleic acid. J. Biol. Chem. 1965;240:432–438. [PubMed] [Google Scholar]
- 15.Deutscher MP. Rat liver glutamyl ribonucleic acid synthetase. II. Further properties and anomalous pyrophosphate exchange. J. Biol. Chem. 1967;242:1132–1139. [PubMed] [Google Scholar]
- 16.Lee LW, Ravel JM, Shive W. A general involvement of acceptor ribonucleic acid in the initial activation step of glutamic acid and glutamine. Arch. Biochem. Biophys. 1967;121:614–618. doi: 10.1016/0003-9861(67)90045-8. [DOI] [PubMed] [Google Scholar]
- 17.Lapointe J, Söll D. Glutamyl transfer ribonucleic acid synthetase of Escherichia coli. I. Purification and properties. J. Biol. Chem. 1972;247:4966–4974. [PubMed] [Google Scholar]
- 18.Kern D, Lapointe J. Glutamyl transfer ribonucleic acid synthetase of Escherichia coli. Study of the interactions with its substrates. Biochemistry. 1979;18:5809–5818. doi: 10.1021/bi00593a010. [DOI] [PubMed] [Google Scholar]
- 19.Ibba M, Bono JL, Rosa PA, Söll D. Archaeal-type lysyl-tRNA synthetase in the Lyme disease spirochete Borrelia burgdorferi. Proc. Natl Acad. Sci. USA. 1997;94:14383–14388. doi: 10.1073/pnas.94.26.14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ibba M, Morgan S, Curnow AW, Pridmore DR, Vothknecht UC, Gardner W, Lin W, Woese CR, Söll D. A euryarchaeal lysyl-tRNA synthetase: resemblance to class I synthetases. Science. 1997;278:1119–1122. doi: 10.1126/science.278.5340.1119. [DOI] [PubMed] [Google Scholar]
- 21.Terada T, Nureki O, Ishitani R, Ambrogelly A, Ibba M, Söll D, Yokoyama S. Functional convergence of two lysyl-tRNA synthetases with unrelated topologies. Nat. Struct. Biol. 2002;9:257–262. doi: 10.1038/nsb777. [DOI] [PubMed] [Google Scholar]
- 22.Ibba M, Losey HC, Kawarabayasi Y, Kikuchi H, Bunjun S, Söll D. Substrate recognition by class I lysyl-tRNA synthetases: a molecular basis for gene displacement. Proc. Natl Acad. Sci. USA. 1999;96:418–423. doi: 10.1073/pnas.96.2.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Craine J, Peterkofsky A. Evidence that arginyl-adenylate is not an intermediate in the orginyl-trna synthetase reaction. Arch. Biochem. Biophys. 1975;168:343–350. doi: 10.1016/0003-9861(75)90262-3. [DOI] [PubMed] [Google Scholar]
- 24.Kern D, Lapointe J. The catalytic mechanism of glutamyl-tRNA synthetase of Escherichia coli. Evidence for a two-step aminoacylation pathway, and study of the reactivity of the intermediate complex. Eur. J. Biochem. 1980;106:137–150. [PubMed] [Google Scholar]
- 25.Kern D, Lapointe J. The catalytic mechanism of the glutamyl-tRNA synthetase from Escherichia coli. Detection of an intermediate complex in which glutamate is activated. J. Biol. Chem. 1980;255:1956–1961. [PubMed] [Google Scholar]
- 26.Jia J, Chen XL, Guo LT, Yu YD, Ding JP, Jin YX. Residues Lys-149 and Glu-153 switch the aminoacylation of tRNATrp in Bacillus subtilis. J. Biol. Chem. 2004;279:41960–41965. doi: 10.1074/jbc.M401937200. [DOI] [PubMed] [Google Scholar]
- 27.Yang XL, Otero FJ, Ewalt KL, Liu J, Swairjo MA, Kohrer C, RajBhandary UL, Skene RJ, McRee DE, et al. Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis. EMBO J. 2006;25:2919–2929. doi: 10.1038/sj.emboj.7601154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Szymanski M, Deniziak MA, Barciszewski J. Aminoacyl-tRNA synthetases database. Nucleic Acids Res. 2001;29:288–290. doi: 10.1093/nar/29.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu F, Jia J, Jin Y, Wang DT. High-level expression and single-step purification of human tryptophanyl-tRNA synthetase. Protein Expr. Purif. 2001;23:296–300. doi: 10.1006/prep.2001.1500. [DOI] [PubMed] [Google Scholar]
- 30.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 31.Jia J, Xu F, Chen X, Chen L, Jin Y, Wang DT. Two essential regions for tRNA recognition in Bacillus subtilis tryptophanyl-tRNA synthetase. Biochem. J. 2002;365:749–756. doi: 10.1042/BJ20020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xue H, Shen W, Giege R, Wong JT. Identity elements of tRNATrp. Identification and evolutionary conservation. J. Biol. Chem. 1993;268:9316–9322. [PubMed] [Google Scholar]
- 33.Araya A, Hevia E, Litvak S. Study of the interactions between avian myeloblastosis virus reverse transcriptase and primer tRNA. Affinity labeling and inactivation of the enzyme by periodate-treated tRNATrp. Nucleic Acids Res. 1980;8:4009–4020. doi: 10.1093/nar/8.17.4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bullerwell CE, Gray MW. In vitro characterization of a tRNA editing activity in the mitochondria of Spizellomyces punctatus, a Chytridiomycete fungus. J. Biol. Chem. 2005;280:2463–2470. doi: 10.1074/jbc.M411273200. [DOI] [PubMed] [Google Scholar]
- 35.Shen N, Guo L, Yang B, Jin Y, Ding J. Structure of human tryptophanyl-tRNA synthetase in complex with tRNATrp reveals the molecular basis of tRNA recognition and specificity. Nucleic Acids Res. 2006;34:3246–3258. doi: 10.1093/nar/gkl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerlo E, Charlier J. Irreversible inactivation of arginyl-tRNA ligase by periodate-oxidized tRNA. FEBS Lett. 1979;99:25–28. doi: 10.1016/0014-5793(79)80240-9. [DOI] [PubMed] [Google Scholar]
- 37.Delagoutte B, Moras D, Cavarelli J. tRNA aminoacylation by arginyl-tRNA synthetase: induced conformations during substrates binding. EMBO J. 2000;19:5599–5610. doi: 10.1093/emboj/19.21.5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rath VL, Silvian LF, Beijer B, Sproat BS, Steitz TA. How glutaminyl-tRNA synthetase selects glutamine. Structure. 1998;6:439–449. doi: 10.1016/s0969-2126(98)00046-x. [DOI] [PubMed] [Google Scholar]
- 39.Sekine S, Shichiri M, Bernier S, Chenevert R, Lapointe J, Yokoyama S. Structural bases of transfer RNA-dependent amino acid recognition and activation by glutamyl-tRNA synthetase. Structure. 2006;14:1791–1799. doi: 10.1016/j.str.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Yang XL, Otero FJ, Skene RJ, McRee DE, Schimmel P, Ribas de Pouplana L. Crystal structures that suggest late development of genetic code components for differentiating aromatic side chains. Proc. Natl Acad. Sci. USA. 2003;100:15376–15380. doi: 10.1073/pnas.2136794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wakasugi K, Slike BM, Hood J, Otani A, Ewalt KL, Friedlander M, Cheresh DA, Schimmel P. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl Acad. Sci. USA. 2002;99:173–177. doi: 10.1073/pnas.012602099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hara-Yokoyama M, Yokoyama S, Miyazawa T. Conformation change of tRNAGlu in the complex with glutamyl-tRNA synthetase is required for the specific binding of L-glutamate. Biochemistry. 1986;25:7031–7036. doi: 10.1021/bi00370a041. [DOI] [PubMed] [Google Scholar]
- 43.Bunjun S, Stathopoulos C, Graham D, Min B, Kitabatake M, Wang AL, Wang CC, Vivares CP, Weiss LM, et al. A dual-specificity aminoacyl-tRNA synthetase in the deep-rooted eukaryote Giardia lamblia. Proc. Natl Acad. Sci. USA. 2000;97:12997–13002. doi: 10.1073/pnas.230444397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahel I, Stathopoulos C, Ambrogelly A, Sauerwald A, Toogood H, Hartsch T, Söll D. Cysteine activation is an inherent in vitro property of prolyl-tRNA synthetases. J. Biol. Chem. 2002;277:34743–34748. doi: 10.1074/jbc.M206928200. [DOI] [PubMed] [Google Scholar]
- 45.Nagel GM, Doolittle RF. Phylogenetic analysis of the aminoacyl-tRNA synthetases. J. Mol. Evol. 1995;40:487–498. doi: 10.1007/BF00166617. [DOI] [PubMed] [Google Scholar]
- 46.Pham Y, Li L, Kim A, Erdogan O, Weinreb V, Butterfoss GL, Kuhlman B, Carter CWJr. A minimal TrpRS catalytic domain supports sense/antisense ancestry of class I and II aminoacyl-tRNA synthetases. Mol. Cell. 2007;25:851–862. doi: 10.1016/j.molcel.2007.02.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.