Abstract
The premature aging and cancer-prone disease Werner syndrome stems from loss of WRN protein function. WRN deficiency causes replication abnormalities, sensitivity to certain genotoxic agents, genomic instability and early replicative senescence in primary fibroblasts. As a RecQ helicase family member, WRN is a DNA-dependent ATPase and unwinding enzyme, but also possesses strand annealing and exonuclease activities. RecQ helicases are postulated to participate in pathways responding to replication blockage, pathways possibly initiated by fork regression. In this study, a series of model replication fork substrates were used to examine the fork regression capability of WRN. Our results demonstrate that WRN catalyzes fork regression and Holliday junction formation. This process is an ATP-dependent reaction that is particularly efficient on forks containing single-stranded gaps of at least 11–13 nt on the leading arm at the fork junction. Importantly, WRN exonuclease activity, by digesting the leading daughter strand, enhances regression of forks with smaller gaps on the leading arm, thus creating an optimal structure for regression. Our results suggest that the multiple activities of WRN cooperate to promote replication fork regression. These findings, along with the established cellular consequences of WRN deficiency, strongly support a role for WRN in regression of blocked replication forks.
INTRODUCTION
Werner syndrome (WS) is a rare, autosomal recessive disease characterized by early onset and increased frequency of many phenotypes normally associated with human aging including graying and loss of hair, wrinkling and ulceration of skin, cancer, atherosclerosis, cataracts, osteoporosis, diabetes and hypertension (1,2). Intriguingly, all of these phenotypes result from loss of function of a single gene product, WRN, belonging to the RecQ family of DNA helicases (3) that includes the prototype RecQ in E. coli, Sgs1 in S. cerevisiae, Rqh1 in S. pombe and four other family members in humans. Importantly, the highly cancer-prone Bloom syndrome (BS) is caused by mutations in human RecQ family member BLM (4), while Rothmund–Thomson (RTS), RAPADILINO and Baller–Gerold syndromes are caused by different mutations in family member RECQL4 (5–7). The RECQL4-related syndromes are collectively characterized by abnormalities in skeletal development and skin pigmentation (poikiloderma), but RTS also shows an elevated incidence of osteosarcoma. At the cellular level, loss of function of a RecQ family member generally results in increased spontaneous and damage-induced chromosomal aberrations, suggesting crucial functions for these proteins in maintaining large-scale genome stability. In agreement with this notion, WRN-deficient cells have higher frequencies of chromosomal deletions, insertions and translocations and are more sensitive to selected DNA damaging agents (including replication inhibitors, topoisomerase I inhibitors and interstrand crosslinking agents) than cells derived from normal individuals (8–13). Moreover, primary fibroblasts from individuals with WS rapidly undergo senescence in culture, apparently as a result of the inability to properly maintain their telomeres (14–16). Accumulation of senescent cells or the cumulative loss of cells by apoptosis is almost assuredly the cause of premature aging in WS; these mechanisms are also postulated to play a role in normal aging (17–19). Thus, the WS phenotypes may point to specific tissues in which these mechanisms may be at work during the development of aging phenotypes in normal individuals.
All RecQ helicases including WRN are highly homologous within defined amino acid sequence motifs that are also identifiable but less conserved in a larger group of enzymes from helicase superfamilies 1 and 2. These sequence motifs anchor a domain that, in RecQ helicases, uses the energy derived from ATP hydrolysis to unwind DNA with a 3′ to 5′ polarity defined by the strand upon which the enzyme translocates. WRN and other RecQ helicases unwind short duplexes and a variety of unusual DNA structures including forks, D-loops, G-quartets and triplexes (20). Both WRN and BLM also readily branch migrate Holliday junctions (21,22). Recently, a number of RecQ helicases, including the human WRN, BLM, RECQ1, RECQL4 and RECQ5β proteins, have been shown to facilitate annealing of complementary DNA strands (23–27). Under certain circumstances, the unwinding and annealing activities of WRN, BLM or RecQ5β can function coordinately to achieve strand exchange (23,28). Taken together, biochemical studies suggest that some RecQ helicases are structurally designed to act on three- or four-stranded replication or recombination intermediates. Importantly, WRN is the only human RecQ homolog to have a nuclease domain in its N-terminal region (29) that confers a 3′ to 5′ exonuclease activity that is particularly robust on complex DNA structures and thus similar to the specificity of its unwinding activity (30–32). Although each of these DNA-dependent activities has been independently examined in vitro, it remains unclear whether and how they might act together in a specific DNA metabolic pathway to help maintain genome stability.
Although multiple DNA repair systems are present in all cells, encounters between replication forks and persistent DNA damage cannot be completely avoided. Recent investigations indicate that cells have evolved important pathways to respond to and overcome replication fork blockage caused by lesions in the DNA template or other circumstances (33–37). It has been proposed that the initial step in dealing with a blocked replication fork involves its regression—a process by which the parental strands re-anneal and the daughter strands are paired to generate a Holliday junction or so-called ‘chicken foot’ structure (Figure 1A). Following Holliday junction formation, several alternative pathways might be employed for removing or circumventing the obstacle and restarting replication. With each pathway, eventual re-establishment of a functional replication fork is crucial for maintaining genomic stability and permitting cell survival. Because of the cellular phenotypes caused by RecQ deficiencies, they are often postulated to participate in pathways responding to fork blockage (38–40). More specifically, WRN-deficient cells show an extension of S phase and specific replication abnormalities including asymmetry in the normal bidirectional progression of replication forks, suggesting difficulty in overcoming obstacles to replication (41,42). They are also hypersensitive to compounds such as hydroxyurea, topoisomerase inhibitors and interstrand crosslinking agents that inhibit replication fork progression (8,11,43,44). Moreover, immunofluorescence studies in normal cells demonstrate that WRN is present in some replication foci and is actively recruited to these foci by treatment with certain genotoxic agents (21,42,45–48). If WRN or other RecQ helicases act in pathways that respond to replication fork blockage, loss of their function might cause sporadic replication fork collapse, leading to generation of double-strand breaks, elevated genomic instability and an increased likelihood of cell death.
Figure 1.
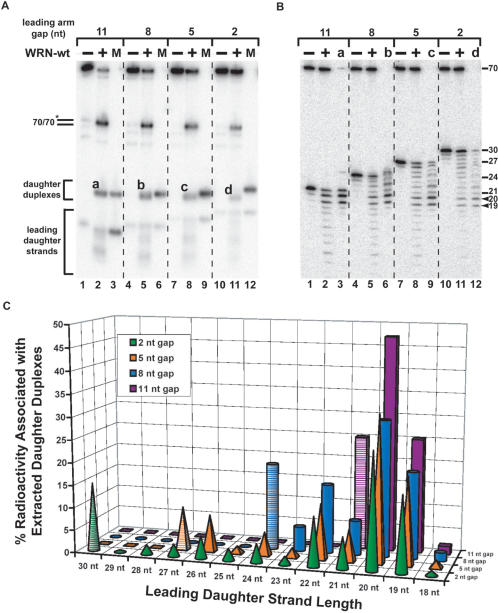
Short replication fork substrates and the influence of leading arm gap size on regression efficiency of WRN-E84A. (A) A series of model replication fork substrates with homologous parental–daughter arms of the indicated lengths was generated by a two-step annealing process (see ‘Materials and methods’ section). The parental strands (gray) were entirely complementary except for 5 nt (indicated in dark gray) precisely at the fork junctions to prevent spontaneous branch migration, while the daughter strands (black) are completely complementary except where they overlap this 5 nt region. For different short fork substrates, the length of the leading daughter strand ranged from 32 to 21 nt (denoted by dashed line) resulting in a leading arm gap of 0–11 nt at the fork junction. The putative WRN- or BLM-mediated conversion of these substrates through Holliday junction intermediates to parental and daughter duplex products is diagramed. (B) Reactions containing fork substrates (50 pM) with leading strand gaps of 0, 2, 5, 8 or 11 nt and WRN-E84A (25–150 pM) were incubated at 37°C for 5 min and analyzed by native PAGE and visualized by phosphorimaging. The migration of specific DNA markers is indicated at left, with brackets encompassing the positions of different daughter duplexes and leading daughter strands generated from different fork substrates. (C) Quantitation (presented as% conversion, in molar terms, from fork substrate) of WRN-E84A-concentration dependent formation of daughter duplex products from fork substrates with leading strand gaps from 0 to 11 nt. (D) Reactions containing WRN-E84A (200 pM) and fork substrates (50 pM) with leading strand gaps from 0 to 11 nt were incubated at 37°C for the indicated times and analyzed as in (B). Quantitation (as described earlier) of enzyme-dependent formation of daughter duplex and leading daughter strand (inset) products over time is graphed for each fork substrate. (E) Reactions containing WRN-E84A (200 pM) and fork substrate (50 pM) with an 11 nt leading arm gap (21lead fork) were incubated for the indicated times and analyzed as in (B). Lane 6 contains markers for the daughter duplex (21lead/30lag) and leading daughter strand (21lead). (F) Radioactivity associated with detectable DNA species in panel E (lanes 1–5) was quantitated and the percentage that each species contributed to the total radioactivity (100%) at each time point is plotted as a bar graph, with legend at right. The numbers between lanes correspond to the decreases in four-stranded fork substrate (top) and the sum of the increases in daughter and parental duplex species (bottom) from the previous to the subsequent time point. The near exact correspondence of these increases to the reductions in the four-stranded fork at each time point indicates that daughter and parental duplex are generated simultaneously and directly from the fork substrate.
At the minimum, the regression of blocked replication forks to form Holliday junctions would entail unwinding of both parental–daughter duplex arms and pairing of the nascent daughter strands with concomitant re-annealing of the parental strands. Accurate completion of this complex process would be facilitated by an enzyme that possesses both unwinding and strand annealing capability such as WRN, or perhaps another RecQ helicase. Some exonucleolytic processing of either the leading or lagging daughter strand may also be involved in the fork regression process. A preliminary report from our laboratory has shown that BLM and an exonuclease-deficient WRN mutant, WRN-E84A, can catalyze fork regression (49). In this study, a series of replication fork substrates have been used to determine the effect of leading arm structure on the fork regression capabilities of WRN and BLM. Our results show a pronounced effect of leading arm structure on the efficiency of regression mediated by WRN-E84A and BLM. Importantly, the 3′ to 5′ exonuclease activity of wild-type WRN enhances regression on a number of these structures through limited degradation of the leading daughter strand. Thus, our results indicate that the multiple enzymatic activities of WRN act together to mediate regression of replication forks. Furthermore, we demonstrate (on another model fork substrate) that WRN can mediate fork regression to form the Holliday junction or ‘chicken foot’ structure characteristic of a bona fide fork regression process. A function of WRN to specifically regress blocked forks during replication fork repair would be highly consistent with the specific genomic instability phenotypes associated with WS.
MATERIALS AND METHODS
Enzymes
Wild-type WRN (WRN-wt), WRN-E84A, and WRN-K577M were overexpressed in insect cells and purified essentially as described previously (50), except that 0.1% Nonidet P40 (NP40) was included in all liquid chromatography buffers. WRN-E84A contains a point mutation in the conserved nuclease domain that inactivates its 3′ to 5′ exonuclease activity (29); this mutant retains DNA unwinding and annealing activities (23,51). WRN-K577M contains a point mutation inactivating its unwinding activity but still retains exonuclease and annealing activities (23,31,52). Recombinant human BLM, purified after overexpression in yeast as described previously (53), was provided by Joanna Groden (Ohio State University). The E. coli Holliday junction resolvase RusA was purified as previously described (54), except that RusA overexpression in E. coli was at 25°C and the lysis step was performed in 1.5 M KCl. UvrD was provided by Steven Matson (University of North Carolina) while both PriA and Rep were from Ken Marians (Sloan-Kettering); these proteins were purified by previously described methods (55,56) Standards of known concentration were used to determine protein concentrations using the Bradford assay and/or SDS–PAGE. All proteins were stored at −80°C prior to use.
DNA substrate construction
Nucleotide sequences of gel-purified oligonucleotides (Midland Certified Reagent Company, Midland, TX) are specified in Table 1. The 3′ ends of the 70lag, 70lead and 30lag oligomers were modified with phosphate groups that block the 3′ to 5′ exonuclease activity of WRN-wt and WRN-K577M (57). For construction of short fork substrates with both the lagging parental and leading daughter strands labeled, the 70lag, 21lead, 24lead, 27lead, 30lead and 32lead oligomers were 5′ end-labeled with 32P-γ-ATP and T4 polynucleotide kinase, 3′-phosphatase free (Roche Molecular Biologicals, Indianapolis, IN) and unincorporated nucleotides were removed using standard procedures. In an initial annealing step to form parental-daughter partial duplexes, labeled 70lag was heated to 90°C and slow-cooled with excess unlabeled 30lag, while unlabeled 70lead was treated similarly in individual reactions with excess labeled 21lead, 24lead, 27lead, 30lead or 32lead. The resulting lagging and leading parental–daughter partial duplexes were then mixed together at 37°C for 18 h. The long fork substrate was prepared similarly, except it contained radiolabels on both the lagging daughter (82lag) and leading parental (122lead) strands. Three-stranded forks were also prepared from sequential high and low-temperature annealing reactions, but without one of the leading or lagging daughter strands. Double-stranded substrates were prepared from single-step annealing reactions. Single-stranded oligonucleotides used for markers and in annealing reactions were simply labeled and gel-purified. After separation by native 8% polyacrylamide gel electrophoresis (PAGE), all DNA substrates were excised, extracted into TEN buffer (10 mM Tris, pH 8.0, 1 mM EDTA and 10 mM NaCl), and stored at 4°C prior to use.
Table 1.
Oligonucleotides used to construct model replication forksa
| Short fork substrate series |
| 21lead |
| GCTATCGTACATGATATCCTC |
| 24lead |
| GCTATCGTACATGATATCCTCACA |
| 27lead |
| GCTATCGTACATGATATCCTCACACTC |
| 30lead |
| GCTATCGTACATGATATCCTCACACTCACT |
| 32lead |
| GCTATCGTACATGATATCCTCACACTCACTTA |
| 30lag |
| TCAGAGTGTGAGGATATCATGTACGATAGC |
| 70lead |
| CGTGACTTGATGTTAACCCTAACCCTAAGAATTCGGCTTAAGTGAGTGTGAGGATATCATGTACGATAGC |
| 70lag |
| GCTATCGTACATGATATCCTCACACTCTGAATAGCCGAATTCTTAGGGTTAGGGTTAACATCAAGTCACG |
| Long fork substrate |
| 72lead |
| GCAGCGTCGCTGCTAGCGTGCAGCGCTTGTACTTCAGCTGATAGACACGTGGCAATTGCCTACATGTAT-CCT |
| 82lag |
| TCAGAGTGTGAGGATACATGTAGGCAATTGCCACGTGTCTATCAGCTGAAGTTGTTCGCGACGTGCGAT-CGTCGCTGCGACG |
| 122lead |
| CGTGACTTGATGTTAACCCTAACCCTAAGATATCGCGTTAAGTGAGTGTGAGGATACATGTAGGCAATT-GCCACGTGTCTATCAGCTGAAGTACAAGCGCTGCACGCTAGCAGCGACGCTGC |
| 122lag |
| CGTCGCAGCGACGATCGCACGTCGCGAACAACTTCAGCTGATAGACACGTGGCAATTGCCTACATGTAT-CCTCACACTCTGAATACGCGATATCTTAGGGTTAGGGTTAACATCAAGTCACG |
aAll sequences are depicted in 5′ to 3′ orientation.
Enzymatic assays
All enzymatic assays were conducted in WRN reaction buffer (40 mM Tris–HCl, pH 7.0, 4 mM MgCl2, 0.1% NP40, 100 ug/ml bovine serum albumin and 5 mM dithiothreitol); fork regression, exonuclease and helicase assays also contained ATP (1 mM) unless otherwise indicated. For these assays, labeled fork (regression), partial duplex (helicase) or oligomeric (annealing) DNA substrate (50–200 pM) was pre-incubated for 5 min at 4°C with enzyme (WRN-E84A, WRN-K577M, WRN-wt, BLM, UvrD, Rep or PriA) at the concentrations in figure legends, then transferred to 37°C for the indicated times. In annealing reactions, complementary single-stranded oligomer (50 pM) was added just prior to incubation at 37°C. For potential detection of Holliday junctions during regression assays with long fork substrate, RusA (10–40 nM) was added 1 min into the 37°C incubation. Reactions (or aliquots thereof) were stopped by adding either one-sixth volume of helicase dyes (30% glycerol, 50 mM EDTA, 0.9% SDS, 0.25% bromphenol blue and 0.25% xylene cyanol) or an equal volume of formamide dye (95% formamide, 20 mM EDTA, 0.1% bromphenol blue and 0.1% xylene cyanol) for analysis by native 8% PAGE or denaturing 14% PAGE, respectively. Specific DNA species (daughter duplexes and RusA-generated products) identified on native PAGE were excised and extracted using a gel extraction kit (Qiagen) then re-analyzed by denaturing 14% PAGE. DNA products on native and denaturing gels were visualized and quantitated using a Storm 860 phosphoimager and ImageQuant software (GE Healthcare).
In fork regression assays, radioactivity associated with individual DNA species was measured. For specific kinetic experiments (Figure 1F), the amounts (as a percentage of total radioactivity) of each DNA species were determined and directly compared. To calculate enzyme-mediated generation of DNA products, the percentage of each product with respect to the total (molar) amount of original fork substrate in that reaction was quantitated following subtraction of background levels of respective DNA species in reactions without enzyme. For analysis of WRN exonuclease activity during regression reactions, the amounts of radioactivity associated with intact and digested products of the leading daughter strand were determined and the percentage of each length product with respect to the total radioactivity derived from the leading daughter strand in that lane was quantitated. This data for each product derived from the leading daughter strand from individual lanes is comparatively presented for daughter duplexes extracted from native PAGE (Figure 5C). Alternatively, the percentages of each product formed after 5 min of digestion are compared with the amount of the respective product in the undigested substrate (0 min time point), and the percent change over time for each product is plotted (Figure 4B).
Figure 5.
The exonuclease activity of WRN-wt performs limited digestion of the leading daughter strand of fork substrates to create an optimal structure for fork regression. Reactions containing fork substrates (50 pM) with leading strand gaps of 11, 8, 5 and 2 nt were incubated for 5 min at 37°C without (−) or with (+) WRN-wt (300 pM). (A) Aliquots of these reactions were analyzed by native PAGE along with markers (M) for undigested daughter duplexes and leading daughter strands specific for each fork substrate (lanes 3, 6, 9 and 12). Positions of migration of parental duplex (70lag/70lead), daughter duplex and leading daughter strand products are indicated at left. The letters (a–d) associated with daughter duplexes (generated from each fork substrate) identify the specific products that were excised and analyzed by denaturing PAGE. (B) In parallel, aliquots of the same reactions were subject to denaturing PAGE with the total DNA products from reactions containing WRN-wt (lanes 2, 5, 8 and 11) run alongside the corresponding daughter duplex products (a–d) individually extracted from native PAGE (lanes 3, 6, 9 and 12). The positions of migration and sizes (in nt) of both undigested leading daughter strands for each fork substrate and the primary 19 and 20 nt digestion products (denoted with arrowheads) are indicated at right. (C) The percentage of the total radioactivity associated with each band in the daughter duplex digestion profiles (B, lanes 3, 6, 9 and 12) was quantitated and then plotted with respect to the product (leading daughter strand) lengths derived from each original fork substrate [back to front, 11 nt gap (purple blocks), 8 nt gap (blue cylinders), 5 nt gap (orange pyramids) and 2 nt gap (green cones) substrates, respectively]. Striped shapes represent the amount of undigested leading daughter strand for each substrate, also signifying the maximum length possible for each corresponding substrate.
Figure 4.
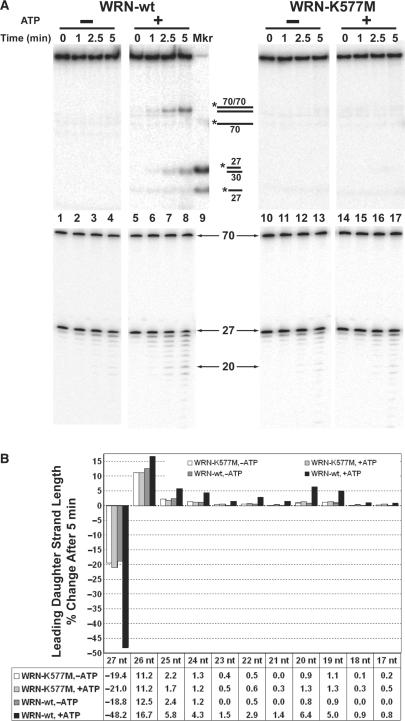
Effects of ATP on WRN regression and exonuclease activities on fork substrates. (A) Reactions containing 27lead fork substrate (50 pM) with a leading arm gap of 5 nt and either WRN-wt (150 pM, lanes 1–8) or WRN-K577M (150 pM, lanes 10–17) were incubated at 37°C in the presence or absence of ATP as indicated. Duplicate aliquots were removed from each reaction at 0, 1, 2.5 and 5 min for analysis in parallel by native (top panels) and denaturing PAGE (bottom panels). The native PAGE analysis also contained daughter duplex (27lead/30lag) and leading daughter strand (27lead) markers (lane 9) and the migration of relevant DNA structures (top) and various lengths of single-stranded species (bottom) are indicated between panels. (B) For the 0 and 5 min time points in A analyzed by denaturing PAGE, the amount of radioactivity in each band of ≤27 nt was quantitated and the percentage of each band with respect to total signal derived from the leading daughter strand was determined. Then, the relative (loss or gain) change in the percentage of each DNA species ranging between 17 and 27 nt from the 0 to the 5 min time point is presented in bar graph form to directly compare exonuclease digestion profiles under the following conditions: WRN-K577M minus ATP (white), WRN-K577M plus ATP (light gray), WRN-wt minus ATP (dark gray), WRN-wt plus ATP (black). The table at bottom shows these numerical values for the relative change in each DNA species over 5 min. Negative values associated specifically with the 27-mer reflect the reduction in the amount of intact leading daughter strand as a result of WRN exonuclease activity.
RESULTS
Fork regression by exonuclease-deficient WRN and the influence of leading arm structure
Our earlier experiments (49) indicated that WRN and BLM could act on a model fork structure with homologous arms to generate both parental and daughter duplexes, consistent with the possibility that these enzymes might regress replication forks in vivo. We next wanted to determine whether and how the precise structure at the fork junction might influence these novel regression activities. To this end, a series of model four-stranded replication fork substrates was constructed from individual oligomers (for details, see ‘Materials and methods’ section and Table 1 for nucleotide sequences). These short fork substrates (Figure 1A) contained a 38 bp parental duplex region, a lagging parental–daughter arm of 30 bp plus a 2 nt single-stranded gap at the fork junction, and a leading parental–daughter arm with a parental strand region of 32 nt but, on individual fork substrates, the leading daughter strand varied in length from 32 to 21 nt resulting in single-stranded gaps of 0–11 nt at the fork junction. Individual short fork substrates are identified below by their leading daughter strand (32lead fork) and/or the size of the single-stranded gap on the leading arm. Importantly, lagging and leading parental–daughter arms of these substrates were entirely homologous (except for 5 non-complementary nt on each parental strand precisely at the fork junction included to prevent spontaneous branch migration), permitting pairing between daughter strands and re-annealing of parental strands to form Holliday junction intermediates and eventually produce both parental and daughter duplexes (Figure 1A). With the exception of the leading daughter strand, the other 3′ ends of these substrates were modified to block the 3′ to 5′ exonuclease activity of WRN. These short fork substrates were radiolabeled on the 5′ ends of both the lagging parental strand (70lag) and the leading daughter strand (21lead, 24lead, 27lead, 30lead or 32lead) to facilitate identification of multiple DNA products.
In assays on this series of replication fork substrates, we initially used an exonuclease-deficient protein, WRN-E84A, to avoid potential degradation of the leading daughter strand that might complicate interpretation of our results. To determine the influence of leading arm structure on regression activity, WRN-E84A was incubated with individual fork substrates with gaps of 0, 2, 5, 8 and 11 nt on the leading arm at the fork junction. Our fork substrates contain three duplex regions potentially subject to unwinding when treated with a DNA helicase such as WRN. Specifically, forward unwinding of the parental duplex region of the fork would yield two parental–daughter partial duplexes (PDs), while unwinding of either parental–daughter arm would yield a three-stranded fork and a displaced daughter strand. However, experiments performed with these substrates with low (sub-equimolar to a 3-fold molar excess) concentrations of WRN-E84A produced, within 5 min, primarily two species that co-migrated with markers for the parental (70 bp) and daughter duplexes (Figure 1B). In comparison, other DNA species (some present in low amounts in substrate preparations) including three-stranded forks, parental–daughter partial duplexes, and leading daughter strands were not produced in significant amounts by WRN-E84A. The parental duplex could be the result of a fork regression event but also might be generated from spontaneous annealing of the partially hybridized parental strands following unwinding of both parental–daughter duplex regions. However, the daughter duplex could only arise from the unwinding of both parental–daughter arms of the fork combined with annealing of the daughter strands and thus specifically reflects fork regression. Importantly, the generation of daughter duplex (as well as parental duplex) products was observed for all fork substrates but was dramatically increased using the fork substrate (21lead fork) with an 11 nt gap as compared to fork substrates with smaller gaps (Figure 1B). Higher concentrations of WRN-E84A mediated increased conversion to daughter duplexes for each substrate, but the relative efficiencies of regression were preserved between substrates. Quantitation of data obtained over a wider range of WRN-E84A concentration (Figure 1C) demonstrated clearly that daughter duplex formation preferentially occurred when the fork substrate contained an 11 nt gap. Using a fork with a smaller gap of 8 nt reduced daughter duplex formation precipitously, while further shortening of the gap lowered the efficiency of this reaction further. This data was corroborated by kinetic experiments performed using a fixed concentration of WRN-E84A on each substrate. In these assays (Figure 1D), the daughter duplexes formed were clearly detectable and increased linearly with time but were relatively modest for fork substrates with gaps of 0, 2, 5 and 8 nt. In contrast, daughter duplex formation from the substrate with an 11 nt gap was markedly higher at each time point (reaching about 60% conversion by 5 min) than for the substrates with smaller gaps. It is notable that generation of the leading daughter strand product is minimal over the same time frame for each substrate (Figure 1D, inset), again suggesting that the daughter duplex formation occurs through direct coordination between unwinding of both parental–daughter arms and pairing of the daughter strands. Taken together, our data indicates that daughter duplex formation (indicative of fork regression) by exonuclease-deficient WRN-E84A occurs much more readily on replication fork substrate with a larger (11 nt) gap on the leading strand than on forks with smaller gaps. The efficiency of regression by WRN-E84A drops considerably when the gap is shortened to 8 nt and decreases further on substrates with even smaller gaps. Our results on these substrates confirm our earlier observation (using a structurally different fork substrate) that WRN catalyzes a reaction reminiscent of fork regression (49). Moreover, they suggest that, although WRN-E84A has a certain amount of structural flexibility, the efficiency of this reaction is determined by structure of the leading arm at the fork junction.
Mechanistic considerations
Since daughter duplex formation catalyzed by WRN-E84A on 21lead fork substrate containing an 11 nt leading arm gap was so much more efficient than on other substrates, a more in-depth analysis of WRN-E84A action on this substrate was performed. The amount of each detectable DNA species from a kinetic experiment on this substrate (Figure 1E) was determined at each time point. Then, the contribution (expressed as percentage) of each DNA species to the total radioactivity was plotted over time (Figure 1F). Before the beginning of the reaction, the fork substrate contained 91.6% of the total radioactivity with no other individual species contributing more than 2.5%. After initiation of the WRN-E84A-mediated reaction, only the amounts of four-stranded fork, daughter duplex and parental duplex changed significantly; at any time point, not one of the other DNA species (lagging parental strand, parental-daughter partial duplexes, three-stranded fork or leading daughter strand) ever contributed more than 6.3% to the total radioactivity. Most notably, the amount of fork substrate decreased dramatically with time while the amounts of parental and daughter duplex increased (Figure 1E and F). Importantly, the combined increases in parental and daughter duplex products (positive values shown between bars, at bottom) almost exactly reflected the decreases (negative values between bars, at top) in fork substrate between individual time points (Figure 1F). Thus, it can be concluded that, during the course of this reaction, the radioactivity in the fork substrate (labeled on one parental and one daughter strand) was distributed in a concerted manner between the parental and daughter duplex products without significant generation of other intermediates. This strongly suggests that WRN-E84A, in a reaction mimicking fork regression, catalyzes direct and coordinated conversion of our short fork substrate with the 11 nt gap on the leading arm to parental and daughter duplexes. In all likelihood, this mechanism also applies to WRN-mediated (and possibly BLM-mediated) action on other, less favorable, fork substrates.
The most straightforward analysis of these results is that WRN-E84A produces daughter (and parental) duplex from fork substrates by coordinately unwinding the parental–daughter arms and annealing the leading and lagging daughter strands by an intramolecular strand exchange reaction. However, we wanted to determine whether daughter duplex formation might occur by strand exchange between independent DNA molecules. To this end, we examined the action of WRN-E84A on two different three-stranded forks, one containing only the lagging daughter strand and the other only the leading daughter strand. When WRN-E84A was incubated with only one of these three-stranded forks, no formation of daughter duplex was possible and, indeed, only other DNA species (leading daughter strand, parental duplex and parental–daughter duplexes) are produced (Supplemental Figure 1, lanes 1–6). As expected, production of daughter duplex (indicated by asterisk) is observed when WRN-E84A is incubated with four-stranded fork containing both leading and lagging daughter strands (Supplemental Figure 1, lanes 13–15). If WRN-E84A is incubated with both three-stranded forks simultaneously, a daughter duplex might conceivably occur via intermolecular strand exchange between the different forks. When this experiment was performed (with concentrations of both three-stranded fork substrates either half or equal to that of the four-stranded fork in the positive control), no daughter duplex was produced; instead, we observed only DNA species corresponding to those formed by unwinding of individual three-stranded forks (Supplemental Figure 1, lanes 7–12). This result indicates that WRN-mediated intermolecular strand exchange does not detectably occur between these three-stranded replication forks under the same conditions in which daughter duplex is produced from a four-stranded replication fork substrate. Thus, daughter duplex formation from a four-stranded fork by WRN-E84A likely occurs by an intramolecular strand exchange mechanism, a concept even more strongly supported by our experiments showing that WRN can regress our longer fork substrates to form Holliday junctions (Figure 6).
Figure 6.
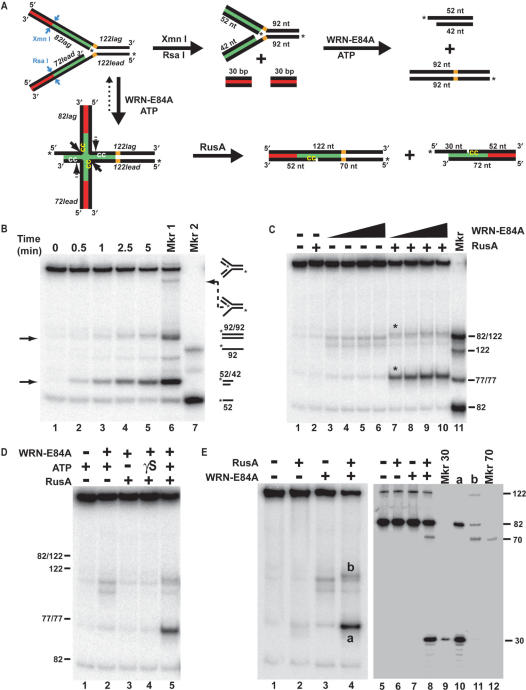
WRN regresses long fork substrate to Holliday junction structures cleaved by RusA. (A) A model replication fork substrate (top left) was constructed using four oligomers (identified in italics) in a two-step annealing process (see ‘Materials and methods’ section). Lagging and leading arms were homologous (green) proximal and heterologous (red) distal to the fork junction; unique restriction sites for Xmn I and Rsa I specified blunt end cleavage at the boundary between homologous and heterologous regions on the lagging and leading arms, respectively (blue arrows). Parental strands contained non-complementarity (5 nt, in orange) precisely at the fork junction to prevent spontaneous branch migration. Digestion with Xmn I and Rsa I generated a replication fork with shorter, homologous arms that, upon fork regression (by WRN-E84A) would yield daughter and parental duplex products (top pathway). In contrast, potential WRN-E84A-mediated regression of the unrestricted fork would yield a Holliday junction structure (bottom left) in which branch migration would be limited by the heterologous regions. Since both the labeled and unlabeled strands within the homologous region of each arm contain consensus 5′-CC-3′ sequences for RusA cleavage (sequences indicated in yellow and white while cleavage sites denoted by solid and dotted arrows, respectively), putative Holliday junctions may be resolved by RusA to yield two nicked duplexes (bottom right). Although resolution by RusA may occur on the labeled or unlabeled strands of the Holliday junction, only products generated by cleavage of the labeled strands are depicted. (B) Rsa I- and Xmn I-restricted long fork substrate (50 pM) was incubated at 37°C for the indicated times with WRN-E84A (100 pM). DNA products were analyzed by native PAGE with phosphorimaging, along with heat-denatured fork preparations either slow-cooled to produce various annealing products (Mkr 1) or rapidly cooled to maintain oligomers in single-stranded form (Mkr 2). Migration of relevant DNA structures is denoted at right. (C) Reactions containing unrestricted long fork substrate (50 pM) without or with WRN-E84A (100, 200, 400 or 600 pM) were initiated at 37°C, supplemented with RusA (10 nM) after 1 min as indicated, and stopped after 5 min total. DNA products were analyzed as in B, along with a marker (lane 11) containing labeled 82lag/122lag, 122lead, 82lag and a 77 bp duplex. (D) Reactions containing unrestricted long fork (50 pM) and WRN-E84A (200 pM) and RusA (10 nM) where indicated were performed without (−) or with (+) ATP (1 mM) or ATPγS (1 mM, denoted γS) and analyzed as in C, with the position of specific markers at right. (E) Reactions containing unrestricted long fork substrate (200 pM), WRN-E84A (800 pM) and ATP were incubated at 37°C for 15 min total with RusA (40 nM) added at 1 min where indicated. Aliquots were analyzed in parallel by native (left panel, lanes 1–4) and denaturing PAGE (right panel, lanes 5–8). For native PAGE, the bands denoted a and b represent DNA species that were excised, extracted and analyzed subsequently by denaturing PAGE (right panel, lanes 10 and 11, respectively). Also run on this gel were labeled 30-mer (lane 9) and 70-mer (lane 12) as markers for RusA cleavage at its specific sites on the labeled 82lag and 122lead strands, respectively, of putative Holliday junction structures formed from unrestricted long fork substrate.
Although our kinetic experiments showed little or no production of free leading daughter strands at the near equimolar WRN-E84A concentrations that mediated daughter duplex formation, we wanted to more thoroughly examine whether daughter duplex formation was a concerted process or the result of possibly independent unwinding and annealing steps. Thus, annealing reactions were performed both with complementary daughter and parental oligomers. Although WRN-mediated annealing of 80-mers can be achieved in reactions with or without ATP (51), these reactions were performed without ATP to minimize potential unwinding of duplex products. In a concentration-dependent manner, WRN-E84A annealed the parental oligomers to generate the 70 bp parental duplex (Supplemental Figure 2, lanes 27–31), confirming its previously reported annealing capability (23). However, at WRN-E84A concentrations equal to and significantly higher than needed for fork regression, enzyme-mediated annealing of lagging daughter oligomer (30lag) to any of the leading daughter oligomers was not detected (Supplemental Figure 2, lanes 1–25). These experiments and others (A. Machwe, unpublished results) indicate that WRN does not facilitate annealing of free oligomers when both are relatively short (<30 nt). A similar effect of oligomer length on BLM-mediated annealing has been previously reported (24). More importantly, the inability of WRN-E84A (and BLM) to anneal these oligomers when free in solution demonstrates that formation of daughter duplex from our fork substrates does not occur by independent enzyme-mediated unwinding and annealing steps. Instead, daughter strands appear to be paired while still associated with the fork, a process likely facilitated by some structural property inherent in WRN and BLM. These results indicate that daughter duplex formation results from an intimate linkage between unwinding of the parental–daughter arms and pairing of the daughter strands. Together with our analysis of the production of daughter and parental duplexes from 21lead fork (Figure 1F), WRN-mediated conversion of our fork substrates to daughter (and parental) duplexes reflects a bona fide fork regression process.
Figure 2.
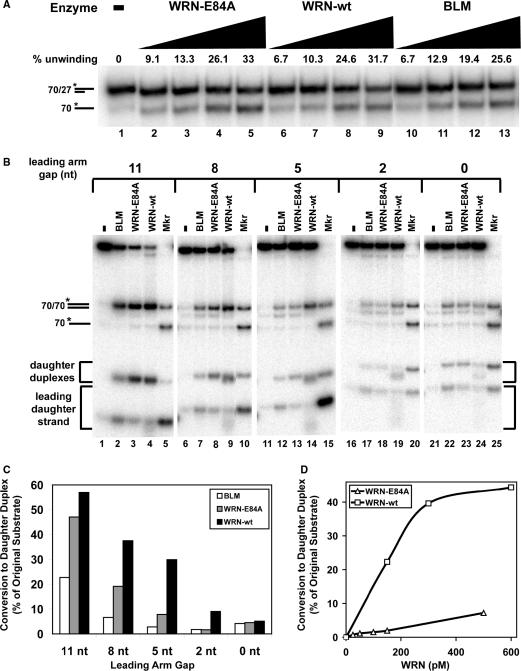
Wild-type WRN (WRN-wt) catalyzes more efficient regression of fork substrates with shorter leading arm gaps than exonuclease-deficient WRN-E84A or BLM. (A) Unwinding activities of WRN-E84A (60, 120, 360, 480 pM), WRN-wt (60, 120, 360, 480 pM), or BLM (60, 120, 180, 240 pM) on partial duplex substrate (*70lag/27lag) over 5 min at 37°C were compared. Duplex and single-stranded DNA products were separated by non-denaturing PAGE and quantitated after phophorimaging, with amounts of enzyme-mediated unwinding presented above each lane. (B) Reactions containing fork substrates (50 pM) with leading strand gaps of 11 nt (lanes 1–4), 8 nt (lanes 6–9), 5 nt (lanes 11–14), 2 nt (lanes 16–19) or 0 nt (lanes 21–24) without or with BLM (160 pM), WRN-E84A (120 pM) or WRN-wt (120 pM) as indicated were incubated at 37°C for 5 min. DNA products were analyzed by native PAGE and visualized by phosphorimaging. Also subject to PAGE in parallel were marker sample mixtures (asterisks below denoting the radiolabeled strands) each containing parental duplex (*70lag/70lead) and single-stranded 70-mer (*70lag) markers but distinguished by substrate-specific daughter duplex and leading daughter strand markers as follows: *21lead/30lag and *21lead (lane 5), *24lead/30lag and *24lead (lane 10), *27lead/30lag and *27lead (lane 15), *30lead/30lag and *30lead (lane 20), and *32lead/30lag and *32lead (lane 25). (C) For the reactions analyzed in B containing various fork substrates and BLM, WRN-E84A or WRN-wt, the percentage of daughter duplex formation relative to the original (molar) amount of intact fork substrate is quantitated as described in ‘Materials and methods’ section. This data is plotted in bar graph form showing the relative efficiencies of BLM (white), WRN-E84A (gray) and WRN-wt (black) in forming daughter duplex from replication fork substrates with leading strand gaps of 11, 8, 5, 2 and 0 nt. (D) Regression reactions containing fork substrate (50 pM) with a leading strand gap of 5 nt and either WRN-wt (150, 300 or 600 pM) or WRN-E84A (25, 50, 100, 150 or 500 pM) were incubated 5 min at 37°C and analyzed as described in B. The molar amount of daughter duplex (with respect to the amount of fork substrate) generated at each WRN-wt (squares) or WRN-E84A (triangles) concentration is plotted.
With regard to protein specificity, it is worthwhile to determine whether this fork regression function is limited to WRN (and BLM) or perhaps common to other enzymes with helicase activity. Since the UvrD, Rep and PriA proteins have been implicated in the response to replication fork blockage in E. coli (56,58), we examined the action of these helicases on our replication fork substrates. When increasing concentrations of these proteins were incubated with the 21lead, 27lead and 32lead fork substrates containing leading arm gaps of 11, 5 and 0 nt, respectively, no significant production of daughter duplex (reflective of fork regression) was observed (Supplemental Figure 3), in contrast to our results with WRN-E84A. Both UvrD and Rep produced almost exclusively parental duplex and free leading daughter strand. This pattern suggests that UvrD and Rep displace both daughter strands, allowing the already linked parental strands to completely re-anneal but (unlike WRN) without pairing of the displaced daughter strands. The behavior of PriA varied depending on fork structure. On forks with 11 and 5 nt leading arm gaps, PriA not only produced parental duplexes and free leading daughter strand but also generated a significant amount of parental–daughter partial duplexes, the latter due to forward unwinding of the fork. However, on 32lead fork without a leading arm gap, PriA generates predominantly a three-stranded fork (without concomitant release of labeled leading daughter strand). This indicates that PriA preferentially unwinds the lagging daughter strand on forks without a leading arm gap, in agreement with previous biochemical analyses (56). Thus, fork regression is not a common property of all helicases, but limited to a subset that includes WRN and BLM. Notably, E. coli RecG can also regress our model replication fork substrates (data not shown), as would be expected from previous studies (59).
Figure 3.
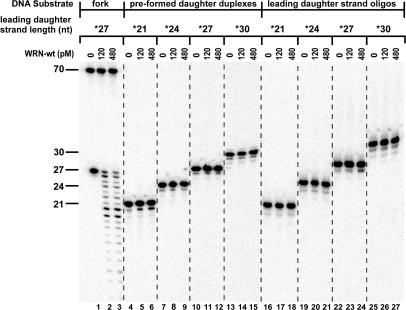
Comparison of WRN exonuclease activity on substrates and products of fork regression reactions. WRN-wt (0, 120 or 480 pM) was incubated at 37°C for 5 min with 50 pM of fork substrate containing a leading arm gap of 5 nt [*27lead/70lead-*70lag/30lag (lanes 1–3)], pre-formed daughter duplex substrates [*21lead/30lag (lanes 4–6)], *24lead/30lag (lanes 7–9), *27lead/30lag (lanes 10–12) and *30lead/30lag (lanes 13–15)] or isolated leading daughter strand oligomers [*21lead (lanes 16–18), *24lead (lanes 19–21), *27lead (lanes 22–24) and *30lead (lanes 25–27)], with the asterisks above indicating the radiolabeled strands. The resulting DNA species were analyzed by denaturing PAGE and phosphorimaging, with the lengths (in nt) and positions of migration of the labeled, undigested oligomers within various substrates indicated at left. Importantly, WRN exonuclease activity is only detectable on the leading daughter strand in the context of the intact fork substrate.
A role for WRN exonuclease activity in fork regression
Our data above indicate that fork substrates containing larger single-stranded gaps on the leading arm are superior structures for regression by exonuclease-deficient WRN-E84A. Since an increase in gap size within these forked structures could be accomplished by degradation from the 3′ end of the leading daughter strand, we hypothesized that the 3′ to 5′ exonuclease activity of WRN-wt might also participate in the fork regression process. To initially test the merit of this theory, we compared the action of exonuclease-proficient WRN-wt with that of WRN-E84A on our fork substrates with different gap sizes on the leading arm. Wild-type BLM, which does not contain exonuclease activity, was also examined to directly compare its action with that of both WRN proteins. First, we measured unwinding by WRN-wt, WRN-E84A, and BLM on a 27 bp partial duplex substrate (70lag/27lag, with both 3′ ends modified to block WRN exonuclease) containing a 3′ overhang of 43 nt (Figure 2A) to determine the amounts of each enzyme needed for comparable activity within the linear range of unwinding. Notably, the same molar concentrations of WRN-E84A and WRN-wt catalyzed nearly equal levels of unwinding; BLM-mediated unwinding was also of similar strength on this 3′ overhang substrate. Then, the action of these enzymes (using concentrations with the same relative unwinding strength) was directly compared on our fork substrates containing leading strand gaps of 11, 8, 5, 2 and 0 nt. These experiments confirm that each of our replication fork substrates can be acted upon by both WRN-E84A and WRN-wt to primarily yield parental duplex and daughter duplex products as well as minor amounts of leading daughter strand (Figure 2B). As judged by formation of daughter duplex, the regression activities of WRN-wt and WRN-E84A on fork substrate with an 11 nt gap are comparable. However, as the gap size on the leading arm decreases, daughter duplex formation becomes much more efficient with WRN-wt than with WRN-E84A (Figure 2C). This difference is particularly notable on the 27lead fork with a 5 nt leading arm gap (Figure 2B, lanes 13 versus 14, and C). A titration of both proteins on this fork substrate also demonstrated that WRN-wt is consistently much more efficient than WRN-E84A in forming the daughter duplex reflective of the fork regression process (Figure 2D). BLM is less efficient at fork regression than WRN-wt on all forks except the substrate without a gap on the leading arm (for which regression activity is very weak for each protein), as evidenced by consistently lower daughter duplex formation and higher production of free leading daughter strand (Figure 2B and C). In assessing the action of WRN-wt on these fork substrates, two other differences from WRN-E84A (and BLM) were observed. In reactions containing WRN-wt, there was a faint smear extending down from the leading daughter strand band, reflecting minor generation of shorter leading daughter strands. More importantly, for the fork substrates with gaps of less than 11 nt, daughter duplexes produced by WRN-wt migrated slightly faster than those formed by WRN-E84A or BLM, indicating that one or both strands that compose this DNA species had been altered by WRN-wt. As the other strands in the fork substrates are blocked at their 3′ ends, these effects appear to be due to the 3′ to 5′ exonuclease activity inherent in WRN-wt specifically on the leading daughter strand during the course of the reaction. Importantly, this data could imply that the exonuclease activity of WRN-wt may digest the leading daughter strand prior to or during formation of daughter duplexes, particularly on replication fork substrates with gaps on the leading arm shorter than 11 nt.
The above analysis of WRN-wt action on our replication fork substrates suggests that its 3′ to 5′ exonuclease activity clearly participates in production of faster-migrating daughter duplexes. Since the 3′ ends of other strands in our fork substrates are blocked, WRN-mediated degradation is likely targeted specifically to the leading daughter strand. However, the timing of this exonucleolytic digestion with respect to other enzymatic events (i.e. regression) was unclear. One way to approach this question was to determine whether (and how) WRN exonuclease activity might digest possible reaction products that contain the leading daughter strand. Thus, the action of WRN-wt on both isolated lead daughter strand oligomers (21lead, 24lead, 27lead and 30lead) and daughter duplex substrates (21lead/30lag, 24lead/30lag, 27lead/30lag and 30lead/30lag) as well as our replication fork substrate (27lead fork) with a 5 nt gap was examined by denaturing PAGE. The 27lead fork substrate was utilized here and subsequently due to the obvious benefit that the exonuclease activity of WRN-wt provides for the regression efficiency on this substrate (Figure 2B–D). In this experiment (as well as later ones), the labeled lagging parental strand (70 nt) of the fork substrate that is modified at its 3′ end is not detectably digested by WRN exonuclease (Figure 3, lanes 2–3). Importantly, at enzyme concentrations comparable to those used in regression assays above, the exonuclease activity of WRN-wt could not detectably digest either pre-formed daughter duplexes (Figure 3, lanes 4–15) or isolated leading daughter strand oligomers (Figure 3, lanes 16–27). In dramatic contrast, the leading daughter strand was extensively digested in the context of the intact 27lead fork substrate (Figure 3, lanes 1–3). This result on single-stranded oligomers is in agreement with earlier studies showing that oligomers of <30 nt are very poor substrates for WRN exonuclease (60). These experiments on isolated daughter duplexes and daughter strand oligomers are highly relevant to the timing of WRN exonuclease activity in our reactions containing short fork substrates. Specifically, once daughter duplexes are formed or leading daughter strands are displaced, they are not subject to WRN exonuclease activity. Therefore, digestion of the leading daughter strand of the fork substrate by WRN-wt must have occurred before or concomitant with (but not after) the formation of daughter duplex.
To further investigate a possible relationship between digestion of the leading daughter strand and regression, the effect of ATP on WRN exonuclease activity was examined. Reactions with 27lead fork substrate and either WRN-wt or the ATPase- and helicase-deficient WRN-K577M mutant were analyzed in parallel for regression and exonuclease activity in the presence or absence of ATP. For these experiments, concentrations of WRN-wt and WRN-K577M that yielded approximately equal levels of exonuclease activity on the leading daughter strand within this fork substrate in the absence of ATP were used. When DNA products of kinetic assays over 5 min with and without ATP were analyzed by native PAGE, formation of daughter (and parental) duplexes occurs readily with WRN-wt in the presence of ATP but not detectably in its absence; moreover, WRN-K577M does not catalyze formation of daughter duplex regardless of the presence or absence of ATP (Figure 4A, top panels). WRN-wt and WRN-E84A also do not catalyze formation of daughter duplex in the presence of the weakly hydrolyzable analog ATPγS (data not shown). These results demonstrate that formation of daughter duplex indicative of fork regression requires the ATPase and helicase activities of WRN. When these same reactions were analyzed by denaturing PAGE, some degradation of the leading daughter strand by WRN-wt and WRN-K577M is observed whether or not ATP is present (Figure 4A, bottom panels). However, digestion of the leading daughter strand is much more pronounced with WRN-wt in the presence of ATP in comparison to each of the other conditions, as indicated both by the direct digestion patterns (Figure 4A, bottom panels) and by the quantitation of relative changes in leading daughter strand length ranging from 27 nt (undigested) to 17 nt after 5 min (Figure 4B). While the other conditions have remarkably similar digestion profiles, reactions containing WRN-wt in the presence of ATP show, by 5 min, a much higher level of exonucleolytic activity on the leading daughter strand by two criteria. They have 1) a much more dramatic (approximately 2.5-fold greater) loss of signal associated with undigested leading daughter strand (27 nt) and 2) a concomitantly increased generation of shorter leading daughter strand products with lengths between 26 and 19 nt (Figure 4B, compare black bars to others). Also notable in this digestion pattern are elevated amounts of 19- and 20-mers compared to several longer leading daughter strand products (see next paragraph for further clarification). Since only reactions including WRN-wt with ATP showed both formation of daughter duplex and enhanced exonuclease activity on the leading daughter strand, these results suggest that optimal WRN exonuclease activity may be connected to partial unwinding of the leading daughter strand and perhaps regression of the fork substrate. These experiments not only confirm that regression activity specifically requires the ATPase and helicase activities of WRN, but also indicate that the exonuclease activity of WRN on the leading daughter strand of replication fork substrates is positively correlated to its ability to perform regression.
Finally, we tested how the 3′ to 5′ exonuclease activity of WRN processed the leading daughter strands of fork substrates with different leading arm gaps in relation to the specific formation of daughter duplexes. For these experiments, fork substrates with leading arm gaps of 2–11 nt were incubated with WRN-wt and total DNA products were analyzed in parallel by native and denaturing PAGE. In addition, daughter duplex products from native PAGE were excised, extracted, and then analyzed by denaturing PAGE alongside the total DNA products from the regression reaction. As before, native PAGE (Figure 5A) showed that WRN-wt generated daughter and parental duplexes along with some short single-stranded products from these substrates. Significantly, the migration of daughter duplexes generated from fork substrates with gap sizes of 2–8 nt was slightly faster than their respective (undigested) daughter duplex markers (Figure 5A, compare lanes 5, 8 and 11 with lanes 6, 9 and 12, respectively). Analysis of total DNA products by denaturing PAGE shows that WRN-wt degrades the labeled leading daughter strand of each substrate, although the number of nucleotides removed decreases as the gap size increases (Figure 5B, lanes 2, 5, 8, 11 and 14). Conspicuously, for each fork substrate, there is substantial degradation until the leading daughter strand reaches 19–20 nt, but digestion beyond that point decreases precipitously. In this analysis of total products, the leading daughter strand could be associated with daughter duplex, displaced leading daughter strand and/or four-stranded fork, but not to any significant extent with other DNA structures. Parallel analysis of the extracted daughter duplexes (denoted a-d in Figure 5A and B) shows that the leading daughter strand component of these duplexes is strictly degraded in a range extending down to 19 nt (Figure 5B, lanes 3, 6, 9, 12 and 15). Thus, the lengths of leading daughter strand specifically associated with the daughter duplex product reflects a defined subset of the WRN exonuclease activity occurring during the entire reaction. Keeping in mind that WRN exonuclease does not act on the daughter duplex once formed (Figure 3), this result indicates that the exonuclease activity of WRN-wt digests the leading daughter strand of each fork substrate, decreasing its length and thereby increasing the leading arm gap size to as much as 13 nt, prior to or concomitant with unwinding and pairing of daughter strands to yield the (faster-migrating) daughter duplexes. To scrutinize this further, the relative amounts of radioactivity associated with each band present in lanes corresponding to the extracted daughter duplexes produced from each fork substrate were determined and directly compared (Figure 5C). This analysis shows that, for each substrate, a minor portion of the daughter duplexes contained undegraded leading daughter strands—i.e. 15.1, 9.5, 19.1 and 25% (Figure 5C, see striped shapes) for the daughter duplexes from the fork substrates with 2, 5, 8 and 11 nt gaps, respectively. Thus, digestion of the leading daughter strand (prior to or concomitant with daughter duplex formation) occurs during the vast majority of fork regression events catalyzed by WRN-wt. The most favored length of the leading daughter strand within the daughter duplex product was, by far, 20 nt followed closely by 19 nt, regardless of the leading strand gap size in the original fork substrate. Although the length of the leading daughter strand in these daughter duplexes is certainly not uniform, there is a definite preference for a leading daughter strand length of 19 or 20 nt corresponding to a gap size of 13 or 12 nt on the original fork substrate. When considered in combination with our comparison of WRN-E84A and WRN-wt on fork substrates with different leading arm gaps (Figure 1), these findings convincingly demonstrate that the exonuclease activity inherent in wild type WRN enhances the regression of forks with small (2–8 nt) leading strand gaps, apparently by generating a better structure for the regression process. Furthermore, our data thus far indicate that the favored structure for regression by the coordinated helicase and annealing activities of WRN is a fork with a leading strand gap of at least 11–13 nt.
WRN regresses replication forks to form Holliday junction intermediates
The experiments above demonstrate that WRN (and BLM) can readily produce a daughter duplex from model replication forks with homologous parental–daughter arms of limited length. However, in vivo the parental-daughter arms are essentially continuous, extending back to the origin of replication. Thus, during the process of fork regression, the daughter strands would remain associated with the parental strands resulting in a Holliday junction or ‘chicken foot’ structure. Such a structure is believed to be the key intermediate in pathways that cope with blocked forks and restart replication (33–37). To determine whether WRN could generate Holliday junction intermediates, a longer, more complex replication fork substrate was designed. Precisely at the fork junction, this substrate (Figure 6A, top left) contained 5 nt of non-complementarity on each arm (again to prevent spontaneous branch migration) as well as the optimal size gap (12 nt) on the leading arm for regression by WRN that obviated the need for its exonuclease activity. Most importantly, both parental–daughter arms of this substrate were significantly longer and contained homologous sequences proximal to the fork junction (denoted in green) but non-homologous sequences (30 bp on each arm) at the distal ends (denoted in red). Theoretically, this design would allow initiation of fork regression with branch migration and daughter strand pairing up to the point of heterology, at which further regression might be inhibited. Nevertheless, if regression was initiated, the fork should be converted, at least transiently, to a Holliday junction with limited mobility (Figure 6A, lower pathway). The high efficiency of WRN-mediated regression of fork substrates with similar-sized leading arm gaps (Figures 1 and 2) made us hopeful that we could detect transient formation of such Holliday junction structures by using known resolvases. Thus, consensus sequences (5′-↓CC-3′) for cleavage by RusA, a Holliday junction-specific resolvase, were included on each strand within the homologous but not the heterologous regions of the parental–daughter arms. Notably, 5′-CC-3′ sequences were also present in the parental duplex region, serving as an internal control for the Holliday junction specificity of RusA. Unique Rsa I and Xmn I sites were included precisely at the junctions between homologous and heterologous regions on the leading and lagging arms, respectively. For this substrate, radioactive labels were placed at the 5′ ends of the lagging daughter strand (82lag) and the leading parental strand (122lead). Notably for this series of experiments, the specific activity of the former was approximately 18 times that of the latter, influencing the relative intensities of individual DNA products. Moreover, this particular placement of radiolabels determines the nature of DNA products that can be observed following treatment of both intact fork substrate with WRN-E84A plus RusA (Figure 6A, bottom right) and restricted fork with only WRN-E84A (Figure 6A, top right).
We initially wanted to determine whether this longer fork substrate could be acted on by WRN to yield a daughter duplex product in a manner identical to the substrate above. To this end, this substrate was restricted with Rsa I and Xmn I (Figure 6A, top center) to release the heterologous (and unlabeled) regions of both arms; the resulting fork with entirely homologous leading and lagging arms was purified. When this substrate was treated with WRN-E84A, two new DNA products (Figure 6B, lanes 2–5, indicated by arrows) appeared with migration consistent with that of the potential daughter duplex (a 42 bp plus 10 nt 5′ overhang) and the parental duplex (92 bp); note that the daughter duplex appears more intense due to the higher specific activity of the lagging daughter strand. As above, production of both species required ATP hydrolysis and occurred at extremely low WRN concentrations without detectable amounts of single-stranded products (data not shown). These experiments confirm that this restricted fork could be regressed by WRN-E84A in the same manner as the shorter fork substrate (21lead fork) that contained a similar structure at the fork junction. These findings also suggest that the length of the homologous arms in our substrates does not significantly affect WRN's regression function, as the homologous arms of the restricted fork are approximately twice as long as those in our short fork substrates.
Next, the unrestricted long fork substrate was treated with WRN-E84A in the presence or absence of RusA. Over the same range of concentration used above, WRN-E84A alone did not generate a product resulting from pairing of the daughter strands (Figure 6C, lanes 3–6), in contrast to our results on forks with completely homologous arms. This suggests that the heterology on the distal ends of each arm inhibits WRN-mediated generation of ‘daughter duplex’ in a manner that cannot be overcome by unwinding activity at these limiting concentrations of WRN-E84A. Thus, if fork regression intermediates are formed by WRN-E84A from this substrate, they revert (by enzyme-mediated or spontaneous reversal of branch migration) back to the fork structure during the reaction and/or electrophoresis. Small amounts of parental–daughter duplexes were detected, consistent with weak forward unwinding of this fork. In order to detect whether Holliday junction intermediates are transiently generated, RusA was added one min following the start of a 37°C incubation of substrate with WRN-E84A. Addition of RusA to WRN-containing reactions yielded new DNA products (indicated by asterisks) when analyzed by native PAGE (Figure 6C, lanes 7–10). The more prominent product migrated a little slower than a 77 bp marker, while the other migrated just slightly above a 122/82 partial duplex marker, consistent with RusA resolution of a Holliday junction formed from this fork substrate—i.e. a 72 bp duplex with a 10 nt 5′ overhang (more prominent because it contains the labeled strand with higher specific activity) and a 122 bp duplex, each putatively containing a nick (see Figure 6A, lower right). In an important control, RusA without WRN-E84A does not detectably act on the fork substrate (Figure 6C, lane 2). These results are consistent with the previously characterized specificity of RusA for Holliday junctions and also indicate that WRN-E84A is generating the Holliday junction structure for RusA cleavage. Importantly, RusA cleavage required not only WRN-E84A but also ATP hydrolysis, as cleavage was not detected in the absence of ATP or in the presence of the poorly hydrolyzable analog ATPγS (Figure 6D), demonstrating that WRN ATPase and helicase activities were necessary to convert the fork substrate to a Holliday junction intermediate.
Similar reactions were analyzed to determine whether these new DNA products indeed corresponded with RusA-specific cleavage events on Holliday junctions generated by WRN-dependent fork regression. If a Holliday junction were generated by fork regression, positioning of RusA consensus (5′-↓CC-3′) sequences in the labeled lagging daughter (82lag) and leading parental (122lead) strands within the homologous regions on the arms of this substrate would cause RusA to cut 30 and 70 nt from the labeled 5′ ends of the respective strands (see Figure 6A, lower right). When reactions containing WRN-E84A and/or RusA were analyzed by denaturing PAGE, neither WRN-E84A alone nor RusA alone detectably produced any change to the labeled (122lead and 82lag) strands of the fork substrate (Figure 6E, lanes 6 and 7). In contrast, reactions that contained WRN-E84A followed by RusA showed new bands that co-migrated with 30 and 70 nt markers (Figure 6E, lane 8). To further clarify the nature of the events catalyzed by WRN-E84A plus RusA, the new DNA products detected by native PAGE (Figure 6E, lane 4, denoted a and b) were individually excised, extracted, and subjected to denaturing PAGE. This analysis indicated that the lower product (a) contained both cleaved and uncleaved 82-mer (Figure 6E, lane 10). The upper product (b) contained both cleaved and uncleaved 122-mer with also some 82-mer (Figure 6E, lane 11), the latter apparently derived from lagging parental–daughter duplex that nearly co-migrates by native PAGE with this RusA cleavage product. Regarding this outcome, it is important to reiterate that CC sequences were also present on the unlabeled (122lag and 72lead) strands within the homologous regions. Thus, we conclude that after WRN-E84A mediates fork regression, RusA can cleave either both labeled or both unlabeled strands of the resulting Holliday junction (see Figure 6A, bottom left, solid and dashed arrows, respectively). While each set of cleavages generates different locations of nicks, analysis by native PAGE shows only two new products, a 72 bp duplex with 10 nt 3′ overhang plus a 122 bp duplex, regardless of whether the nicks are present in the labeled or unlabeled strands. When these products are extracted and analyzed by denaturing PAGE, concerted RusA cleavage of the labeled strands yields shorter single-stranded products (30- and 70-mers) while concerted cleavage of the unlabeled (undetectable) strands leaves the labeled strands intact (82- and 122-mers). These results clearly demonstrate that WRN-E84A regresses this fork substrate to generate a Holliday junction or ‘chicken foot’ structure that is cleaved by RusA at its specific recognition sequences.
DISCUSSION
Replication fork blockage is such a common event that cells have evolved specialized pathways to handle these situations. Regression of a blocked replication fork is theorized to be the initial step in dealing with these serious challenges to completion of DNA replication, genome stability and cell survival (33–37). Fork regression would involve pairing of nascent daughter strands and re-annealing of parental strands to form a Holliday junction or ‘chicken foot’ intermediate. Following fork regression, obstacles to replication might be addressed by alternative pathways including (1) repair of a blocking lesion and reverse branch migration to regenerate a forked structure, (2) a strand-switching replication step in which the lagging daughter strand serves as template followed again by reverse branch migration, bypassing the blocking lesion and re-establishing the fork or (3) resolution of the Holliday junction to generate a double-strand break that could initiate recombinational pathways to restore a functional replication fork. A preliminary report from our lab established that the human RecQ helicases WRN and BLM have the ability to regress a specific replication fork substrate (49). In this study, we have examined the ability of WRN to act on several model replication fork substrates, including a series of short fork substrates containing structural differences at the fork junction and another longer substrate that allowed Holliday junction formation and detection during a potential regression reaction. Importantly, fork regression by WRN on our long fork substrate directly forms a Holliday junction that is detected using the RusA resolvase (Figure 6). Fork regression requires the ATPase and helicase activities of WRN, as it does not occur in reactions (1) lacking ATP or including the poorly hydrolyzable analog ATP-γ-S or (2) using the WRN-K577M mutant that lacks both ATPase and unwinding activity. Most notably, our experiments with shorter fork substrates indicate a specific role for the 3′ to 5′ exonuclease activity of WRN during fork regression—i.e. controlled digestion of the leading daughter strand to generate a more favorable structure for regression and thereby increase the efficiency of this process. These findings suggest a novel role for the exonuclease activity of WRN during fork regression that operates in coordination with its unwinding and annealing activities. Thus, all of the DNA-dependent activities of WRN may cooperate to promote replication fork regression.
Enzymatic reactions containing WRN-E84A or WRN-wt showed a concentration- and time-dependent conversion of our short fork substrates to both parental and daughter duplex products (see Figures 1 and 5A). Daughter duplex formation is specific for a regression event as it requires both unwinding and pairing of the physically unlinked daughter strands (Figure 1A). Although generation of free leading daughter strand product is detectable in reactions containing relatively high WRN concentrations, several lines of evidence demonstrate that daughter duplex formation occurs through an intimate linkage between unwinding of parental–daughter arms and pairing of daughter strands. First, WRN does not anneal the free daughter oligomers (due to their short length) that comprise daughter duplexes (Supplemental Figure 2), indicating that annealing does not independently follow unwinding of the parental-daughter arms. Second, at limiting concentrations of WRN-E84A (Figure 1), there is almost exclusive production of daughter and parental duplexes without significant generation of three-stranded forks or free leading daughter strands. Furthermore, free leading daughter strands are not significantly produced at any time during our kinetic experiments (Figures 1D–F and 4A), strongly suggesting that daughter duplexes are formed from the fork substrates without release of daughter strands. Most convincingly, a quantitative analysis of WRN-E84A-mediated regression of 21lead fork substrate over time demonstrates that the parental and daughter duplexes are produced directly and essentially simultaneously from intact four-stranded fork (Figure 1F). Lastly, pairing of daughter strands during regression is a process mediated by WRN (or BLM) but not all unwinding enzymes, as other helicases such as UvrD and Rep unwind both daughter strands of our short fork substrates but do not produce daughter duplexes. These results indicate that daughter duplex formation does not simply occur spontaneously during daughter strand unwinding. Taken together, our results suggest that, during regression, the partially unwound daughter strands are juxtaposed by WRN in a manner that promotes their pairing to form a Holliday junction that, for our short fork substrates, is subsequently converted to daughter and parental duplexes. This is further supported by the WRN-dependent conversion of our long fork substrate to Holliday junctions (Figure 6) in which the daughter strands are paired while still associated with the parental strands. It seems very likely that WRN-mediated fork regression occurs through coordination between its unwinding function and its previously reported strand annealing activity (23).
The results with our short fork substrates indicated that the leading arm structure had a major influence on the efficiency of fork regression. Specifically, daughter duplex formation by exonuclease-deficient WRN-E84A was greatly enhanced when the single-stranded gap on the leading arm increased to 11 nt from ≤8 nt (Figures 1 and 2). Although WRN-wt was similarly effective as WRN-E84A in daughter duplex formation from substrate with an 11 nt gap, it was much more efficient on fork substrates with shorter leading strand gaps (Figure 2), suggesting that WRN's 3′ to 5′ exonuclease activity might be digesting the leading daughter strand to increase the gap size. In addition, daughter duplexes formed by WRN-wt migrated slightly faster by native PAGE than those formed by WRN-E84A, in agreement with putative 3′ to 5′ processing of the leading daughter strand. Analysis by denaturing PAGE confirmed that WRN-wt was specifically degrading the leading daughter strand during these regression reactions (Figures 3–5). However, the exonuclease activity of WRN-wt could not detectably digest daughter duplexes (or displaced leading daughter strands) once formed (Figure 3), indicating that digestion of the leading daughter strand was occurring prior to or concomitant with formation of daughter duplex. Further analysis indicated that, during regression, exonucleolytic digestion of the leading daughter strand by WRN-wt was essentially limited to a defined range, regardless of the original length of this strand in the fork substrate. The observed preference for digestion to a length of 20 or 19 nt corresponds to generation of a 12 or 13 nt single-stranded gap on the leading arm, respectively (Figure 5B and C). Taken together, these results indicate that WRN exonuclease activity promotes regression on substrates with gaps shorter than 11 nt by digesting the leading daughter strand to increase the leading arm gap size. The exonuclease-deficient WRN-E84A protein is unable to alter the gap size and thus acts efficiently only on our short fork substrate with an 11 nt gap. By this reasoning, the optimum structure for regression by WRN, without assistance from its exonuclease function, is a fork with a leading arm gap of at least 11–13 nt. This conclusion is further supported by the highly efficient regression and Holliday junction formation by WRN-E84A on the longer fork substrate containing a leading arm gap of 12 nt (Figure 6B and C). These experiments suggest WRN exonuclease activity may participate in processing the leading daughter strand during regression of blocked replication forks, but its degree of involvement may depend on the precise structure at the fork junction and perhaps the spatial relationship between leading and lagging daughter strands.
The efficiency of WRN-mediated regression on forks with ≥11 nt leading arm gaps suggests that WRN may preferentially act on forks in which leading strand synthesis is blocked while lagging strand synthesis continues, leaving a single-stranded gap on the leading arm. During fork regression, WRN exonuclease activity may further digest the leading daughter strand, a processing step that may simply generate the optimum structure for enzyme-mediated pairing of the daughter strands. However, there may be other advantages to this arrangement. On this series of substrates, WRN-mediated regression only occurs efficiently when the leading daughter is at least 10–12 nt shorter than the lagging daughter strand. If this structural relationship was maintained (or perhaps further exaggerated) in vivo, fork regression would always yield a Holliday junction containing a single free end with a 5′ overhang. Since formation of Rad51-mediated filaments occurs on 3′ overhangs (61), this structure might inhibit recombination, perhaps in favor of alternate, less error-prone pathways such as (1) repair of the blocking lesion and reverse branch migration to re-establish a viable replication fork or (2) strand switching synthesis and reverse branch migration with concomitant bypass of the obstacle (on the parental leading strand) that originally impeded fork progression. Recent studies in E. coli suggest that lesion repair might be the first alternative attempted following fork blockage caused by DNA damage (62). It is tempting to speculate that enzymatic regression processes have evolved to at least initially favor less error-prone pathways such as repair or strand switching. Cells from WS patients have a higher rate of spontaneous RAD51 foci formation than normal cells, supporting the idea that recombination might be utilized more often when WRN is non-functional (43). Importantly, the notion that WRN regresses replication forks to specifically generate intermediates that suppress instead of promote recombination would be consistent with the hyperrecombination phenotypes of cells that have lost WRN function.
Fork structures with heterologous arms have previously been shown to be excellent substrates for WRN helicase (63). Here, we demonstrate that WRN specifically regresses forks with homologous arms. With regard to its action on fork structures, WRN binds in the vicinity of the fork junction as judged by DNase I footprinting (Machwe et al., unpublished results). It is quite relevant that WRN-wt and WRN-E84A regression activities are highly efficient when the structure of the fork substrate is favorable. As judged by daughter duplex formation, regression of the fork substrate with an 11 nt gap on the leading arm is readily detectable after 5 min using sub- and near-equimolar levels of enzyme compared to substrate (Figure 1B, lanes 19–23). Furthermore, at low concentrations, WRN is more effective at catalyzing regression of this fork substrate than unwinding a 27 bp partial duplex (Figure 2). WRN also efficiently regresses our longer fork substrate containing a 12 nt gap at near-equimolar concentrations (Figure 6B and C). Although these experiments cannot determine the stoichiometry of WRN with respect to the DNA substrate, the requirement for unwinding of both parental-daughter arms during regression might imply at least a dimeric structure. Irregardless, the efficiency by which WRN catalyzes this multi-faceted regression reaction suggests that it is particularly suited to this task. It is noteworthy that BLM, another human RecQ helicase that possesses unwinding and strand pairing activities and DNA substrate specificity similar to WRN, also performs fork regression (49,64). Like WRN, BLM produces Holliday junctions from our long unrestricted fork substrate that are recognized and cleaved by RusA (data not shown). However, BLM appears to be consistently less efficient in fork regression of each of our short fork substrates than WRN-wt (Figure 2A and B). Although RecG can also regress model forks as previously reported (59), other helicases (including UvrD, Rep and PriA implicated in resolution of fork blockage in E. coli) tested thus far could not perform this function, suggesting that fork regression capability appears relatively limited to a small group of helicases that includes WRN and BLM. Thus, some RecQ helicases are structurally designed to catalyze fork regression by combining their helicase and strand pairing activities. Notably, two other human RecQ family members, RecQ1 and RecQ5β, have also been shown to possess both DNA unwinding and annealing activities (25,26). It may be relevant in a physiological context that BLM and other human RecQ helicases do not possess exonuclease activity and alone cannot modify the structure at the fork junction in the way that WRN-wt can. We speculate that WRN is preferentially involved in regression of blocked forks, while BLM may be more likely to participate (in combination with topoisomerase IIIα and BLAP75) in other DNA transactions, such as double Holliday junction resolution to prevent crossing over during homologous recombination (65).
RecQ helicases have previously been postulated to play roles in resolution of replication fork blockage (38,39,66). The finding that both WRN and BLM can readily regress model replication forks greatly strengthens this hypothesis. However, formation of Holliday junctions resulting from replication fork regression in vivo is somewhat speculative and further proof is needed of specific genome maintenance pathways utilizing fork regression and under what circumstances they are implemented. It is also noteworthy that WRN has been hypothesized to participate in several other pathways that preserve genome stability (such as telomere maintenance). Despite these caveats, potential involvement of WRN in replication fork regression is consistent with previous findings regarding WRN and certain properties of WRN-deficient cells. WRN-deficient cells are slower than their normal counterparts in completing S phase and show asymmetric replication fork progression, consistent with an inability to properly resolve fork blockage (41,42). They are also hypersensitive to agents that severely inhibit DNA replication including (1) hydroxyurea, which depletes deoxynucleotide pools, (2) topoisomerase inhibitors that induce strand breaks and DNA-protein crosslinks and (3) interstrand crosslinking agents (such as mitomycin C and cisplatin) that prevent unwinding of the parental strands (8,11,43,44). Furthermore, in normal cells, WRN migrates rapidly to sites of DNA synthesis following treatment with these and other DNA damaging agents (21,45–47), findings that suggest that WRN is recruited to sites where replication is blocked. Our results suggest the reason for this relocalization—i.e., WRN is brought to blocked replication forks to catalyze their regression as part of a pathway that maintains genome stability. It is possible that, although they lack exonuclease activity, other human RecQ helicases such as BLM can partially compensate for loss of WRN function in fork regression. However, if such redundancy exists, it is likely imperfect and there may be situations in which WRN-deficient cells are still compromised in dealing with blocked replication forks. As a result, cells lacking WRN are hyperrecombinant and accumulate chromosomal abnormalities that are almost assuredly responsible for the increased cancer incidence of WS patients. Alternatively, in response to these DNA metabolic problems some cell types may trigger apoptosis or permanent cell cycle exit (senescence). With time, the cumulative loss of either cells by apoptosis or reduction in proliferative capacity within a tissue may cause the premature aging phenotypes of WS. Although more research is needed to confirm these hypotheses, a specific function in fork regression is highly consistent with existing knowledge regarding WRN and WS.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
This work was supported by NCI Grant R01 CA113371-01 to D.K.O., while R.G.L. and E.B. were supported by the UK Medical Research Council. The authors would like to thank Joanna Groden, Ken Marians, Steven Matson, Vilhelm Bohr, Judy Campisi and M.D. Gray for providing important reagents and Deanna Edwards for critical reading of the manuscript. Funding to pay the Open Access publication charges for this article was provided by the Graduate Center for Toxicology, University of Kentucky College of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Goto M. Hierarchical deterioration of body systems in Werner's syndrome: implications for normal ageing. Mech. Ageing Dev. 1997;98:239–254. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- 2.Martin GM, Oshima J. Lessons from human progeroid syndromes. Nature. 2000;408:263–266. doi: 10.1038/35041705. [DOI] [PubMed] [Google Scholar]
- 3.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 4.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 5.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 6.Siitonen HA, Kopra O, Kaariainen H, Haravuori H, Winter RM, Saamanen AM, Peltonen L, Kestila M. Molecular defect of RAPADILINO syndrome expands the phenotype spectrum of RECQL diseases. Hum. Mol. Genet. 2003;12:2837–2844. doi: 10.1093/hmg/ddg306. [DOI] [PubMed] [Google Scholar]
- 7.Van Maldergem L, Siitonen HA, Jalkh N, Chouery E, De Roy M, Delague V, Muenke M, Jabs EW, Cai J, et al. Revisiting the craniosynostosis-radial ray hypoplasia association: Baller-Gerold syndrome caused by mutations in the RECQL4 gene. J. Med. Genet. 2006;43:148–152. doi: 10.1136/jmg.2005.031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lebel M, Leder P. A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc. Natl Acad. Sci. U.S.A. 1998;95:13097–13102. doi: 10.1073/pnas.95.22.13097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogburn CE, Oshima J, Poot M, Chen R, Hunt KE, Gollahon KA, Rabinovitch PS, Martin GM. An apoptosis-inducing genotoxin differentiates heterozygotic carriers for Werner helicase mutations from wild-type and homozygous mutants. Hum. Genet. 1997;101:121–125. doi: 10.1007/s004390050599. [DOI] [PubMed] [Google Scholar]
- 10.Poot M, Gollahon KA, Rabinovitch PS. Werner syndrome lymphoblastoid cells are sensitive to camptothecin-induced apoptosis in S-phase. Hum. Genet. 1999;104:10–14. doi: 10.1007/s004390050903. [DOI] [PubMed] [Google Scholar]
- 11.Poot M, Yom JS, Whang SH, Kato JT, Gollahon KA, Rabinovitch PS. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15:1224–1226. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- 12.Fukuchi K, Martin GM, Monnat RJ., Jr Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc. Natl Acad. Sci. U.S.A. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gebhart E, Bauer R, Raub U, Schinzel M, Ruprecht KW, Jonas JB. Spontaneous and induced chromosomal instability in Werner syndrome. Hum. Genet. 1988;80:135–139. doi: 10.1007/BF00702855. [DOI] [PubMed] [Google Scholar]
- 14.Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat. Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 15.Wyllie FS, Jones CJ, Skinner JW, Haughton MF, Wallis C, Wynford-Thomas D, Faragher RG, Kipling D. Telomerase prevents the accelerated cell ageing of Werner syndrome fibroblasts. Nat. Genet. 2000;24:16–17. doi: 10.1038/71630. [DOI] [PubMed] [Google Scholar]
- 16.Du X, Shen J, Kugan N, Furth EE, Lombard DB, Cheung C, Pak S, Luo G, Pignolo RJ, et al. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol. Cell. Biol. 2004;24:8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campisi J. Replicative senescence: an old lives’ tale? Cell. 1996;84:497–500. doi: 10.1016/s0092-8674(00)81023-5. [DOI] [PubMed] [Google Scholar]
- 18.Faragher RG, Kipling D. How might replicative senescence contribute to human ageing? Bioessays. 1998;20:985–991. doi: 10.1002/(SICI)1521-1878(199812)20:12<985::AID-BIES4>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 19.Shay JW, Wright WE. Aging. When do telomeres matter? Science. 2001;291:839–840. doi: 10.1126/science.1058546. [DOI] [PubMed] [Google Scholar]
- 20.Opresko PL, Cheng WH, Bohr VA. Junction of RecQ helicase biochemistry and human disease. J. Biol. Chem. 2004;279:18099–18102. doi: 10.1074/jbc.R300034200. [DOI] [PubMed] [Google Scholar]
- 21.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom's syndrome gene product promotes branch migration of Holliday junctions. Proc. Natl Acad. Sci. U.S.A. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 24.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, Andreoli L, Muzzolini L, Vindigni A, Brosh RM., Jr. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 27.Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst.) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5{beta} helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3′→5′ exonuclease. Nat. Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3′→5′ exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machwe A, Xiao L, Theodore S, Orren DK. DNase I footprinting and enhanced exonuclease function of the bipartite Werner syndrome protein (WRN) bound to partially melted duplex DNA. J. Biol. Chem. 2002;277:4492–4504. doi: 10.1074/jbc.M108880200. [DOI] [PubMed] [Google Scholar]
- 32.Orren DK, Theodore S, Machwe A. The Werner syndrome helicase/exonuclease (WRN) disrupts and degrades D-loops in vitro. Biochemistry. 2002;41:13483–13488. doi: 10.1021/bi0266986. [DOI] [PubMed] [Google Scholar]
- 33.Cox MM. The nonmutagenic repair of broken replication forks via recombination. Mutat. Res. 2002;510:107–120. doi: 10.1016/s0027-5107(02)00256-7. [DOI] [PubMed] [Google Scholar]
- 34.Haber JE. DNA recombination: the replication connection. Trends Biochem. Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 35.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 36.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell. Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 37.Michel B. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- 38.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen JC, Loeb LA. The Werner syndrome gene: the molecular basis of RecQ helicase-deficiency diseases. Trends Genet. 2000;16:213–220. doi: 10.1016/s0168-9525(99)01970-8. [DOI] [PubMed] [Google Scholar]
- 40.Orren DK. Werner syndrome: molecular insights into the relationships between defective DNA metabolism, genomic instability, cancer and aging. Front. Biosci. 2006;11:2657–2671. doi: 10.2741/1999. [DOI] [PubMed] [Google Scholar]
- 41.Poot M, Hoehn H, Runger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp. Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Lopez AM, Jackson DA, Iborra F, Cox LS. Asymmetry of DNA replication fork progression in Werner's syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- 43.Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat R.J.,, Jr Homologous recombination resolution defect in werner syndrome. Mol. Cell. Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakamoto S, Nishikawa K, Heo SJ, Goto M, Furuichi Y, Shimamoto A. Werner helicase relocates into nuclear foci in response to DNA damaging agents and co-localizes with RPA and Rad51. Genes Cells. 2001;6:421–430. doi: 10.1046/j.1365-2443.2001.00433.x. [DOI] [PubMed] [Google Scholar]
- 46.Karmakar P, Bohr VA. Cellular dynamics and modulation of WRN protein is DNA damage specific. Mech. Ageing Dev. 2005;126:1146–1158. doi: 10.1016/j.mad.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 47.Pichierri P, Rosselli F, Franchitto A. Werner's syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene. 2003;22:1491–1500. doi: 10.1038/sj.onc.1206169. [DOI] [PubMed] [Google Scholar]
- 48.Rodriguez-Lopez AM, Jackson DA, Nehlin JO, Iborra F, Warren AV, Cox LS. Characterisation of the interaction between WRN, the helicase/exonuclease defective in progeroid Werner's syndrome, and an essential replication factor, PCNA. Mech. Ageing Dev. 2003;124:167–174. doi: 10.1016/s0047-6374(02)00131-8. [DOI] [PubMed] [Google Scholar]
- 49.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 50.Orren DK, Brosh R.M, Jr, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Machwe A, Lozada EM, Xiao L, Orren DK. Competition between the DNA unwinding and strand pairing activities of the Werner and Bloom syndrome proteins. BMC Mol. Biol. 2006;7:1. doi: 10.1186/1471-2199-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA. The Werner syndrome protein is a DNA helicase. Nat. Genet. 1997;17:100–103. doi: 10.1038/ng0997-100. [DOI] [PubMed] [Google Scholar]
- 53.Karow JK, Chakraverty RK, Hickson ID. The Bloom's syndrome gene product is a 3′–5′ DNA helicase. J. Biol. Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 54.Chan SN, Harris L, Bolt EL, Whitby MC, Lloyd RG. Sequence specificity and biochemical characterization of the RusA Holliday junction resolvase of Escherichia coli. J. Biol. Chem. 1997;272:14873–14882. doi: 10.1074/jbc.272.23.14873. [DOI] [PubMed] [Google Scholar]
- 55.Mechanic LE, Hall MC, Matson SW. Escherichia coli DNA helicase II is active as a monomer. J. Biol. Chem. 1999;274:12488–12498. doi: 10.1074/jbc.274.18.12488. [DOI] [PubMed] [Google Scholar]
- 56.Heller RC, Marians KJ. Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. J. Biol. Chem. 2005;280:34143–34151. doi: 10.1074/jbc.M507224200. [DOI] [PubMed] [Google Scholar]
- 57.Harrigan JA, Fan J, Momand J, Perrino FW, Bohr VA, Wilson DM., III WRN exonuclease activity is blocked by DNA termini harboring 3′ obstructive groups. Mech. Ageing Dev. 2007;128:259–266. doi: 10.1016/j.mad.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Flores MJ, Bidnenko V, Michel B. The DNA repair helicase UvrD is essential for replication fork reversal in replication mutants. EMBO Rep. 2004;5:983–988. doi: 10.1038/sj.embor.7400262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McGlynn P, Lloyd RG. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc. Natl Acad. Sci. U.S.A. 2001;98:8227–8234. doi: 10.1073/pnas.111008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Machwe A, Xiao L, Orren DK. Length-dependent degradation of single-stranded 3′ ends by the Werner syndrome protein (WRN): implications for spatial orientation and coordinated 3′ to 5′ movement of its ATPase/helicase and exonuclease domains. BMC Mol. Biol. 2006;7:6. doi: 10.1186/1471-2199-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Thacker J. The RAD51 gene family, genetic instability and cancer. Cancer Lett. 2005;219:125–135. doi: 10.1016/j.canlet.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 62.Courcelle CT, Chow KH, Casey A, Courcelle J. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc. Natl Acad. Sci. U.S.A. 2006;103:9154–9159. doi: 10.1073/pnas.0600785103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brosh RM, Jr., Waheed J, Sommers JA. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 2002;277:23236–23245. doi: 10.1074/jbc.M111446200. [DOI] [PubMed] [Google Scholar]
- 64.Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 65.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 66.Courcelle J, Hanawalt PC. Participation of recombination proteins in rescue of arrested replication forks in UV-irradiated Escherichia coli need not involve recombination. Proc. Natl Acad. Sci. U.S.A. 2001;98:8196–8202. doi: 10.1073/pnas.121008898. [DOI] [PMC free article] [PubMed] [Google Scholar]