Abstract
Structural variation is an important cause of genetic variation. Whole genome analysis techniques can efficiently identify copy-number variable regions but there is a need for targeted methods, to verify and accurately size variable regions, and to diagnose large sample cohorts. We have developed a technique based on multiplex amplification of size-coded selectively circularized genomic fragments, which is robust, cheaper and more rapid than current multiplex targeted copy-number assays.
INTRODUCTION
Genetic variation occurs on multiple levels, from single nucleotide polymorphisms (SNPs) to larger events involving contiguous blocks of DNA sequence that vary in copy number between individuals. The structural diversity in the human genome is much higher than previously assumed, and attracts an increased interest within the genetics community. It is now becoming increasingly clear that submicroscopic variations are major contributors to genetic diversity and human disease (1,2).
The interest in copy-number variation (CNV) has led to the establishment of a number of analytical methods, using either global or targeted approaches. Microarray-based comparative genome hybridization (array-CGH) is a commonly used global approach to CNV detection (3,4), enabling genome-wide scans for detection of novel CNVs. CGH arrays are manufactured with different resolution and coverage, using different approaches to probe genomic samples, ranging from BAC clones to short oligonucleotides attached to the array surface (5). High-throughput SNP analysis can also be employed for CNV-detection, as revealed by long stretches of apparently homozygous loci or unusual heterozygous signal ratios (6,7). Although global array-based approaches can provide high resolution data on CNVs in individuals, there remains a need for simple, cost-efficient, accurate methods to validate and test candidate CNVs across larger populations.
One established targeted approach for CNV analysis is quantitative PCR (qPCR) (8). However, this technique requires setting up a large number of replicate reactions to score individual deletions and duplications, and is generally not suitable for multiplexing. Similarly, fluorescence in situ hybridization (FISH) is a labor-intensive technique which is not usually highly multiplexed, though it is well-established in diagnostics laboratories. Examples of multiplexed targeted copy-number analysis approaches are Quantitative multiplex PCR of short fluorescent fragments (QMPSF) (9), multiplex amplifiable probe hybridization (MAPH) (10) and multiplex ligation-dependent probe amplification (MLPA) methods (11–13). In MLPA, which has become perhaps the most commonly used one, up to 40 loci can be analyzed in parallel.
Here, we present an approach based on the selector technique (14), called multiplex ligation dependent genome amplification (MLGA). In contrast to MLPA, genomic DNA is amplified rather than probe molecules, and a single probe is required for each target instead of two. This leads to increasing reaction kinetics and decreasing probe amplification background. Furthermore, these shorter probes are easily manufactured by conventional oligonucleotide synthesis. These properties allow for cost-efficient design of custom MLGA assays with a short turnover time. This is demonstrated in an accompanying paper, where a candidate duplication was verified, sized, and diagnosed in a very cost-efficient approach (Salmon Hillbertz,N.H.C. et al., Nat. Genet. in press).
MATERIALS AND METHODS
Selector probe design
A set of 14 human target genes were chosen on five different chromosomes (Table 1). Sequences for each target were collected from the Ensembl database (www.ensembl.org, assembly NCBI 36, Oct 2005). These sequences were processed in the PieceMaker software (15) to generate a set of restriction fragments using a restriction enzyme of choice. A single fragment for each target sequence was then chosen in such a way that the fragments in each pool were between 100 and 400 nt in length with each fragment having a different length, with a minimum size difference of 6 nt.
Table 1.
List of target loci for the MLGA probe set
| Gene/Probe name | Chromosome | Position in chromosome | |
|---|---|---|---|
| Start (nt) | Length (nt) | ||
| AR | X | 66 684 790 | 102 |
| SRY | Y | 2 715 319 | 112 |
| MADH4 | 18 | 46 820 996 | 119 |
| SIM2 | 21 | 37 017 473 | 132 |
| L1CAM | X | 152 785 534 | 141 |
| SOD1 | 21 | 31 954 636 | 157 |
| TYMS | 18 | 649 034 | 164 |
| ABCC4 | 13 | 94 484 282 | 196 |
| SERPINB2 | 18 | 59 706 873 | 213 |
| BRCA2 | 13 | 31 792 206 | 236 |
| STCH | 21 | 14 669 412 | 252 |
| SRY | Y | 2 714 956 | 290 |
| RPS6KA3 | X | 20 079 076 | 339 |
| NFATC1 | 18 | 75 257 833 | 358 |
Chromosome position according to Ensembl assembly NCBI 36, October 2005.
Selector probes serving as templates for the circularization of each chosen target fragment (Table 1) were designed using the ProbeMaker software (16). Each selector consists of two synthetic oligonucleotides; a target-specific selector probe (70–74 nt), and a universal vector oligonucleotide (34 nt). Oligonucleotides were synthesized by DNA Technology A/S, Denmark (Table 2). The central part of each selector probe is complementary to the vector oligonucleotide so that hybridization between the two generates the recognition sequence for the Hind III restriction enzyme and a universal primer pair site for parallel PCR amplification. The ends of the selector probes (18–20 nt each) have sequences complementary to the ends of the restriction fragments targeted for selection.
Table 2.
List of oligonucleotides (selectors, vector and primers) used in the MLGA protocol
| Oligonucleotide | Sequence (5′ to 3′) |
|---|---|
| AR | GAAATCCTACCCTCCTCTTTACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGTCTTGTAAGTCAAACATTAA |
| SRY_2 | AGCCGAAAAATGGCCATTACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGGCGATCAGAGGCGCAAGA |
| MADH4 | TTAAACAGGCTGAATACTGGACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGTGCTATTAATTGTAAGCTGT |
| SIM2 | GCTGGAACATCCTCCTAAAAACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGCTCCAGAGGCGGTGGCTC |
| L1CAM | AACCAACTCCTCTTCTGCACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGGGGACATGAGGCCATGAC |
| SOD1 | TAGAGCGCTGAAGCCGGAACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGTAGAACAGAGGCCAGCAA |
| TYMS | TCTAAGCAGAAAGGTGGGTACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGCCGCACTCGCTTGTGGTA |
| ABCC4 | GGGTTTTCCCCTCATTCTTACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGTGCTGTTGAGGTACATACAG |
| SERPINB2 | TTGGCACAGGGAAGGAAGACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGCAGGTATACCTGTTGTGAAT |
| BRCA2 | ACATATTCTTCCTCATGTTGACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGACAAAGGGAGGTGATCTAAG |
| STCH | TCATGGTGATGGTGAAGAAAACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGAGTTGAAGAGGTTTGGGC |
| SRY_1 | ACTTACAGCCCTCACTTTCACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGAGGCGAAGATGCTGCCGA |
| RPS6KA3 | TTACTATCAGCCTCACATTTACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGACCCCAGGTTGCTTACAT |
| NFATC1 | CCTGGGGAATTCAGGGGCACGATAACGGTAGAAAGCTTTGCTAACGGTCGAGGGTATTTTCAAAGCCACTTG |
| Vector | CTCGACCGTTAGCAAAGCTTTCTACCGTTATCGT |
| Fwd. primer | AGCTTTGCTAACGGTCGAG |
| Rev. primer | AGCTTTCTACCGTTATCGT |
DNA samples
Six genomic DNA samples were extracted from blood (Flexigene, Qiagen), collected with the appropriate permissions from individuals diagnosed with Down syndrome, and admitted to the Department of Clinical Genetics, Uppsala University. DNA samples were also extracted from the aneuploid cell cultures NA04626, NA01416 and NA06061 (Coriell Cell Repositories) with 3, 4 and 5 X-chromosomes. Pooled samples of male and female DNA from Promega (cat# G147A 20745001, cat# G152 20215001) were used as references.
MLGA
Restriction digestion was performed for 30 min at 37° C using 8 U of restriction enzyme Mnl I (Fermentas) and 200 ng genomic DNA in 5 µl of NEB4 buffer (New England Biolabs) supplemented with 0.1 µg/µl BSA. The restriction enzyme was inactivated during 5 min at 95° C. Much less DNA can be used, however, and as little as 12.5 ng genomic DNA has been used in successful assays (Salmon Hillbertz,N.H.C. et al., Nat. Genet. in press).
Circularization of restriction fragments was performed by adding 10 µl of a solution containing 33 nM vector oligonucleotide, 0.15 nM of each selector, 14.5 mM MgCl2, 1.2 mM NAD, 0.3 U/µl Ampligase (Epicentre) and 0.75 × PCR buffer (Invitrogen) to the restriction digested DNA. The reactions were incubated with the following temperature profile: 95° C for 5 min, followed by three cycles of 75° C 5 min, 65° C 5 min, 60° C 5 min, 55° C 5 min and 50° C 10 min. To enrich for circularized DNA, 15 µl of a solution containing 7.5 U exonuclease I (New England Biolabs), 0.13 M Tris–HCl (pH 9.0), 3.4 mM MgCl2 and 0.02 µg/µl BSA was added. The reaction was incubated at 37° C for 30 min, followed by 70° C for 10 min to inactivate the enzyme.
Amplification of selected targets was performed by adding 6 µl of the reaction (∼40 ng DNA) to 19 µl PCR-mix, containing 0.9 × PCR buffer (Invitrogen), 0.66 mM MgCl2, 0.33 mM dNTP, 0.13 µM each of forward and reverse primer, 5 U Hind III (Fermentas) and 0.5 U Platinum Taq DNA polymerase (Invitrogen). Hind III was added in the PCR-mixture to create a linear template for the PCR amplification, decreasing the risk of amplifying multiple laps of the circular DNA template. Temperature cycling was performed as follows: 37° C for 30 min, 95° C for 5 min followed by 30 cycles of 95° C 15 s, 55° C 30 s and 72° C for 60 s followed by 72° C for 10 min.
PCR products were analyzed using an Agilent Bioanalyzer 2100™ instrument and quantified using the Agilent 2100 expert software, version B.02.02.SI238.
RESULTS
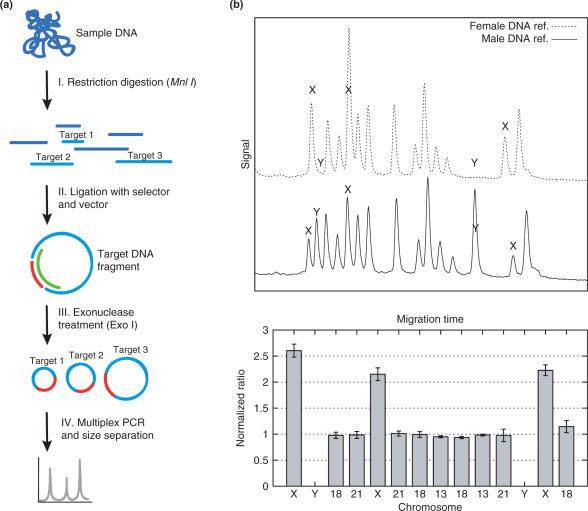
The MLGA technique employs a number of enzymatic processing steps (Figure 1a). In brief, sample DNA is first restriction digested to generate genomic fragments with defined ends. DNA is then denatured and mixed with a pool of selector probes and a thermostable DNA ligase to allow hybridization and circularization of the corresponding target fragments. The sample is treated with exonuclease I to enrich for circularized DNA, and finally, the circularized fragments are PCR amplified using a universal primer pair. The selector probes are designed such that the amplified fragments are of different size, allowing for rapid and simple detection and quantification using electrophoretic separation. In order to evaluate the approach on a model system for CNV, selector probes targeting 14 different loci on human chromosomes X, Y, 13, 18 and 21, were designed to analyze male and female genomic DNA (Figure 1b). To obtain relative quantification, each peak area was normalized by dividing with the sum of areas of all peaks originating from autosomal targets. The ratios of each individual probe area were then compared between the male and female DNA samples. The results show only marginal differences in autosomal peak ratios between the male and female samples, and the expected lack of Y-chromosome and duplication of X-chromosome signals in the female sample compared with the male sample (Figure 1b).
Figure 1.
(a) Multiplex ligation-dependent genome amplification (MLGA), reaction scheme. (I) Genomic DNA is digested by restriction enzyme to generate targets with defined ends. (II) Each MLGA probe consists of two oligonucleotides, one selector oligo of 70–74 nt (green) and one general vector oligo of 34 nt (red). MLGA probe together with DNA-ligase forms circular DNA of target molecules after denaturation and hybridization. (III) To reduce background signal in the assay, undesirable, linear DNA is degraded by exonuclease I (Exo I). (IV) Multiplex PCR is facilitated by using universal primers that hybridize to a sequence in the vector. PCR products are analyzed using the Agilent Bioanalyzer 2100™ electrophoresis system. (b) Data from an MLGA set of 14 probes targeting loci on human chromosomes 13, 18, 21, X and Y. The upper graph shows the resulting elution diagrams from analyses of male and female DNA pools.
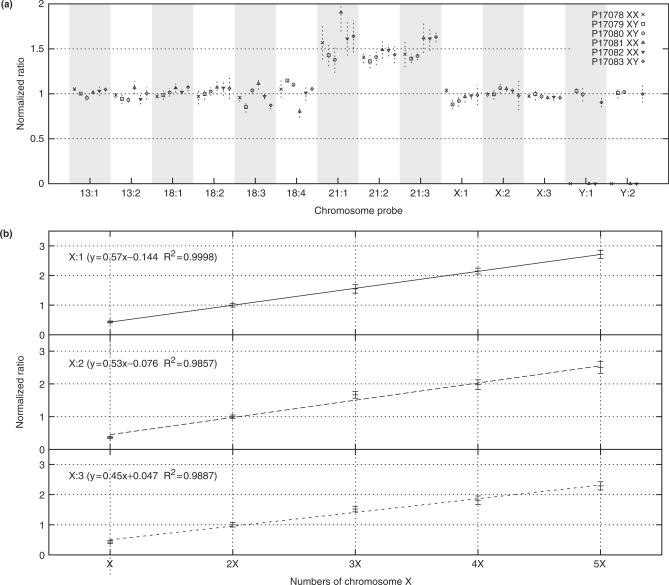
DNA samples prepared from six different Down syndrome patients were analyzed using the same probe set to confirm that duplications of autosomal chromosomes can be detected. All chromosome 21 probes show a ratio around 1.5 indicating a trisomy, as expected (Figure 2a). In order to test the linearity and sensitivity of copy-number measurements, a series of cell lines carrying 1, 2, 3, 4 and 5 copies of the X-chromosome were analyzed. Normalization was performed by dividing each peak area with the sum of the autosomal peak areas (Figure 2b). The increase in signal is linear with a slope of 0.5 units per additional X-chromosome, implying that the MLGA method is accurate and sensitive enough to quantify a broad range of copy-number changes.
Figure 2.
(a) DNA samples prepared from six different Down syndrome patients, three males and three females, were analyzed with a set of 14 selector probes distributed over chromosomes 13, 18, 21, X and Y. Data was normalized by dividing each peak area with the sum of the peak areas of all non chromosome 21 probes. On the x-axis, probes are ordered according to chromosomal position. Ratios between patient and reference DNA sample values are shown on the y-axis, using sex-matched reference samples. (b) Graph shows the response of X targeting probes to an increasing number of chromosomes. On the x-axis samples with 1–5 copy of chromosome X are shown, where samples for 1X is male, 2X is female and 3–5X are aneuploid cell cultures from Coriell Cell Repositories. To illustrate the results, each normalized value was divided by the 2X diploid sample value.
DISCUSSION
The MLGA is a multiplex targeted approach for copy-number analysis, which seems well-suited for CNV measurement and validation. MLGA has several advantages over the commonly used multiplexed targeted copy-number assay MLPA. First, MLGA probes are easier and cheaper to manufacture, as only one probe is required per locus and they are similar in size and relatively short (70–74 nt). Extensive purification is not required since the probes do not need functional ends in contrast to MLPA probes, and also no modification of the 5′ end. Second, a uni-molecular circularization reaction is inherently more rapid and efficient than a bi-molecular ligation reaction (17). Moreover, probe amplification methods, such as MLPA, suffer from probe-dependent and target-independent amplification artifacts (17). Therefore, very low concentrations of probes are used in the MLPA, resulting in a requirement for long hybridization times to saturate the target sequences.
For assays that could be applied in the diagnostic setting, time is a critical factor. With MLGA, the total assay time, including electrophoresis, is 5 h, relative to the ∼24 h assay time for the MLPA. Another important aspect for custom loci is the turn-over time in assay design, particularly when sizing duplications/deletions, which typically requires an iterative process to map the chromosomal break points. An MLGA assay can be set up in ∼5 days, including oligonucleotide design and synthesis, and two rounds of experimental optimization and verification of the assay. Finally, the MLGA assay can potentially create longer PCR products than MLPA, since the length of the product is defined by the genomic DNA sequence rather than by the length of synthetic probes. This flexibility in PCR product length may allow for higher levels of multiplexing.
In a separate study, we applied the technique for verification and sizing of a CNV (Salmon Hillbertz,N.H.C. et al., Nat. Genet. in press). We investigated a suspected duplication involving an SNP identified during the course of genome-wide SNP analyses of different dog breeds. The distance between the closest flanking SNPs was 930 kb, so we initially designed MLGA probes with 100 kb spacing over 2 Mb, including a fragment containing the SNP. Two probes, including the fragment with the SNP, responded with a 2-fold increase in homozygous dogs compared with control fragments, and with 1.5-fold increase in heterozygous dogs, thus verifying that the region indeed was duplicated. We then designed a new set of probes with 10 kb spacing, flanking the copy-number positive fragments, to try to more precisely define the size of the duplication. Using a final set of probes, the duplicated region could be defined sufficiently well to design a PCR primer pair that amplified across the duplication break point. The PCR fragment was sequenced and the size of the duplication was determined to be 133.4 kb. Finally, a diagnostic MLGA assay was compiled to screen 72 dogs. The phenotype, experimental details and implication of the duplication are described in Salmon Hillbertz,N.H.C. et al., (Nat. Genet. in press)
One potential disadvantage compared to the MLPA approach, is that a larger proportion of the sequence in the final PCR product is defined by genomic DNA sequences. This can potentially introduce a bias in amplification rates due to the diversity in sequence. This though can be addressed by applying stringent criteria during the in silico design process of each probe set. From a number of applied MLGA projects, we have learnt that 75–80% of designed selector probes reproducibly select a fragment of the intended size. There may be several reasons for this non-complete assay conversion rate. We have previously shown that high GC content (>60%) in the selected fragment decreases the probability of a probe being successful (14). The GC content can affect both the circularization and PCR amplification yield, possibly due to secondary structures interfering with probe and primer hybridization and/or extension. Such secondary structures may also be present in DNA fragments with a lower GC-content, escaping our GC-content design threshold.
We expect to be able to develop better design criteria to improve success as more assays are developed and can be evaluated. Since selected fragments are in the order of 100 bp and CNVs are often in the order of several kilobases, positional constraints on design are quite low. There are, on average, four selectable restriction fragments to choose among per kilobase of genomic DNA sequence, since we are using restriction enzymes with 4 bp recognition sequences. It is possible to further increase freedom of design by introducing a site-specific cleavage of the target strand, making the design only depending on a restriction recognition site at the 3′ end of the target fragment (14). The first-trial success rate among different DNA samples is about 90–95% (data not shown). Data from failing subjects can be rescued by collecting new DNA samples.
ACKNOWLEDGEMENTS
We would like to thank Brian Peter of Agilent Laboratories for DNA samples, preliminary data and advice on the article. This work was supported by the Wallenberg Foundation, the Olle Engkvist Byggmästare Foundation, the Swedish Research Council and the EU FP-6 Integrated Project MolTools. Funding to pay the Open Access publication charges for this article was provided by the Swedish Research Council.
Conflict of interest statement. F.D and M.N. have licensed the commercial rights to the technology to Olink AB (Uppsala, Sweden), a company in which M.N. also hold stock. M.I., J.S., A-C.T., M-L.B. declare no conflict of interest.
REFERENCES
- 1.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 2.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solinas-Toldo S, Lampel S, Stilgenbauer S, Nickolenko J, Benner A, Dohner H, Cremer T, Lichter P. Matrix-based comparative genomic hybridization: biochips to screen for genomic imbalances. Gen. Chrom. Cancer. 1997;20:399–407. [PubMed] [Google Scholar]
- 4.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat. Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 5.Barrett MT, Scheffer A, Ben-Dor A, Sampas N, Lipson D, Kincaid R, Tsang P, Curry B, Baird K, et al. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc. Natl Acad. Sci. USA. 2004;101:17765–17770. doi: 10.1073/pnas.0407979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Moorhead M, Karlin-Neumann G, Falkowski M, Chen C, Siddiqui F, Davis RW, Willis TD, Faham M. Allele quantification using molecular inversion probes (MIP) Nucleic Acids Res. 2005;33:e183. doi: 10.1093/nar/gni177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, Wei W, Zhang J, Liu G, Bignell GR, Stratton MR, Futreal PA, Wooster R, Jones KW, et al. Whole genome DNA copy number changes identified by high density oligonucleotide arrays. Hum. Genom. 2004;1:287–299. doi: 10.1186/1479-7364-1-4-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 9.Casilli F, Di Rocco ZC, Gad S, Tournier I, Stoppa-Lyonnet D, Frebourg T, Tosi M. Rapid detection of novel BRCA1 rearrangements in high-risk breast-ovarian cancer families using multiplex PCR of short fluorescent fragments. Hum. Mutat. 2002;20:218–226. doi: 10.1002/humu.10108. [DOI] [PubMed] [Google Scholar]
- 10.Armour JA, Sismani C, Patsalis PC, Cross G. Measurement of locus copy number by hybridisation with amplifiable probes. Nucleic Acids Res. 2000;28:605–609. doi: 10.1093/nar/28.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stern RF, Roberts RG, Mann K, Yau SC, Berg J, Ogilvie CM. Multiplex ligation-dependent probe amplification using a completely synthetic probe set. Biotechniques. 2004;37:399–405. doi: 10.2144/04373ST04. [DOI] [PubMed] [Google Scholar]
- 13.White SJ, Vink GR, Kriek M, Wuyts W, Schouten J, Bakker B, Breuning MH, den Dunnen JT. Two-color multiplex ligation-dependent probe amplification: detecting genomic rearrangements in hereditary multiple exostoses. Hum Mutat. 2004;24:86–92. doi: 10.1002/humu.20054. [DOI] [PubMed] [Google Scholar]
- 14.Dahl F, Gullberg M, Stenberg J, Landegren U, Nilsson M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stenberg J, Dahl F, Landegren U, Nilsson M. PieceMaker: selection of DNA fragments for selector-guided multiplex amplification. Nucleic Acids Res. 2005;33:e72. doi: 10.1093/nar/gni071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stenberg J, Nilsson M, Landegren U. ProbeMaker: an extensible framework for design of sets of oligonucleotide probes. BMC Bioinform. 2005;6:229. doi: 10.1186/1471-2105-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardenbol P, Baner J, Jain M, Nilsson M, Namsaraev EA, Karlin-Neumann GA, Fakhrai-Rad H, Ronaghi M, Willis TD, et al. Multiplexed genotyping with sequence-tagged molecular inversion probes. Nat. Biotechnol. 2003;21:673–678. doi: 10.1038/nbt821. [DOI] [PubMed] [Google Scholar]