Abstract
One way herpes simplex virus type-1 (HSV-1) spreads in vivo is by polykaryocytes formation. Here we demonstrate that polykaryocyte production during HSV-1 spread in cultured human corneal fibroblasts (CF) required heparan sulfate (HS) and more specifically 3-O sulfated HS (3-OS HS). The polykaryocyte formation heavily depended on the expression of HS on target (CF) cells but not on glycoprotein expressing effector cells. Furthermore, we provide the first visual evidence of 3-OS HS and HSV-1 gD colocalization during the membrane fusion process. Taken together our results provide novel insight into the significance of HS in polykaryocyte formation.
1. Introduction
Cell surface heparan sulfate (HS) glycosaminoglycans (GAG) contain alternating disaccharide units of uronic acid and N-acetylglucosamine. During biosynthesis, HS is modified by multiple enzymes including a last 3-O sulfation step that requires 3-O sulfotransferases (3-OSTs) [1, 2]. These modifications not only add molecular diversity to HS but also provide specific binding sites for a variety of proteins including entry receptors for many viruses [3-6].
In case of herpes simplex virus type-1 (HSV-1), it was first proposed that HS chains can serve as an initial attachment site for the viral envelope glycoproteins gB and gC [1, 7]. Subsequent insights into the role of HS revealed that modification of HS by 3-OSTs (except isoform-1) can result in generation of 3-O sulfated HS (3-OS HS) [8-12]. The 3-OS HS can act as a receptor for gD, a glycoprotein necessary for HSV-1 entry into cells [2]. The ability to bind gD is acquired when HS chains contain at least one of the following disaccharides, -IdoA2S-GlcNH23S6S- or -IdoA2S-GlcNH23S- [2]. Corneal fibroblasts (CF), a primary cell type cultured from the human cornea, are known to express 3-OS HS specific disaccharides [13, 14].
HSV-1 infection of the cornea, especially the inner layer or stroma from where the CFs were isolated, can result in significant vision loss and is considered a leading infectious cause of blindness [15]. The corneal herpetic lesions can contain polykaryocytes that are formed by membrane fusion of infected cells with neighboring uninfected cells induced by viral glycoproteins [16]. Although the polykaryocyte formation is considered an important means of viral spread and a clue for HSV-1 keratitis [16-20] its mechanism remains poorly understood for natural target cell types such as human CF. To increase our understanding of the mechanism, the current study focuses on the role of HS and uniquely modified HS chains in this phenomenon. While studies in the past have suggested no significant role of HS [19,21], we provide evidence to suggest that HS is required by primary cultures of human CF to form polykaryocytes. We also demonstrate the presence of 3-OS HS at the cell-to-cell contact sites during membrane fusion.
2. Materials and methods
2.1. Cell cultures
As previously described [22], cultures of human CF were derived from the stroma of corneal tissues obtained from the Illinois Eye Bank, Chicago, IL, using institution approved protocol and culture conditions in accordance with the Declaration of Helsinki). CF from the 3rd passage was used for the study. P.G. Spear (Northwestern University, Chicago) provided wild-type CHO-K1 cells and CHO-745 cells that are deficient in HS or GAG biosynthesis. Both wild-type CHO-K1 cells and CHO-745 cells were grown in Ham’s F12 (Invitrogen Corp, Carlsbad, CA) supplemented with 10% FBS.
2.2. Plasmids
The plasmids used in this study were pPEP98 (gB), pPEP99 (gD), pPEP100 (gH) and pPEP101 (gL) that express HSV-1 glycoproteins, plasmid pT7EMCLuc that express firefly luciferase gene under T7 promoter, plasmid pCAGT7 that expresses T7 RNA polymerase [19]. HSV-1 gD receptor plasmid pDS43 that expresses 3-OST-3B [2] and nectin-1 (pBG38) were also used. The control plasmid, pCDNA3.1, was purchased from Invitrogen (Carlsbad, CA, USA).
2.3. Cell fusion assays and estimation of polykaryocytes
Cell fusion assays were performed as described previously [8, 11, 17-20]. Briefly, the CHO- K1 cells and/or CHO-745 cells designated “effector” cells were co-transfected using Lipofectamine (Gibco/BRL) with plasmids gB, gD, gH and gL, along with the plasmid pT7EMCLuc (0.5 μg DNA of each plasmid). The cultured CF cells or “target” cells grown up to 50-60% confluency were co-transfected with pCAGT7 that expresses T7 RNA polymerase [19]. The effector cells expressing pT7EMCLuc and pcDNA3.1 and the target cells expressing 3-OST-3 with T7 RNA polymerase were considered as controls. A group of multinucleated cells (10 or more nuclei) were scored positive for polykaryocytes formation.
2.4. Cell surface immunofluorescence assays
Both cultured CF and HSV-1 glycoprotein-expressing CHO-K1 cells were co-cultured in chamber polystyrene vessels (BD Falcon) at around 30% confluency. Cells were then incubated with primary antibodies at 4°C for 45 min, washed 10 times with cold PBS, and fixed with acetone for 10 min at -20°C. The primary antibodies used were rabbit polyclonal (R7) to HSV-1 gD (kindly provided by R. Eisenberg and G. Cohen, University of Pennsylvania) diluted 1: 20,000 in PBS/10% normal goat serum (NGS) and the anti 3-OS HS mouse monoclonal antibody [23] diluted 1: 100 in PBS/10% NGS with 0.5 M NaCl. After fixation, cells were dried at room temperature for 5 min, blocked in PBS/10% calf serum, and incubated with secondary antibody: anti-mouse polyclonal FITC conjugated (1: 500 dilution) or anti-rabbit polyclonal with H&L Texas Red-conjugated (1: 5000 dilution) for 45 min at 37°C. The cells were again washed in PBS before mounting in Vectashield mounting medium (Vector Laboratories, Inc.Burlingame, CA). Leica confocal microscope SP2 was used for sequential scanning of the samples.
3. Results and Discussion
The ability of CF to use its endogenous gD receptor to form polykaryocytes with HSV-1 glycoprotein expressing CHO-K1 cells was examined. These cells express cell surface HS but lack functional gD receptors, including 3-OS HS [2]. As a result, they are resistant to both HSV-1 entry and virus induced membrane fusion [8]. Polykaryocyte formation was examined by mixing “target” CF with HSV-1 glycoproteins (gB, gD, gH, gL)-expressing “effector” cells [17-20]. The polykaryocytes formed by CF was compared with those seen with 3-OST-3-expressing and nectin-1-expressing cells. Two set of positive control cells were selected because CF expresses 3-OST-3 but not nectin-1, which is a relatively common receptor for HSV-1 [13, 14, 24]. As a negative control, mock effector cells transfected with empty vector (pCDNA3.1) were used. As shown in Fig. 1A (panels a, b, c), polykaryocytes with sizes similar to those in nectin-1 or 3-OST-3-expressing CHO-K1 cells were detected in CF. During the same time interval, little, if any, polykaryocyte formation was detected in control (pCDNA3.1) cells (Fig. 1A, panels d,e,f). It was also evident (Fig. 1B; solid bars) that the average number of polykaryocytes (with 10 or more nuclei) formed in one well of a 6-well cell culture dish was very similar in all three types of cells examined. These results demonstrate that the four glycoproteins (gB, gD, and gH-gL) are sufficient for comparable level of polykaryocyte formation to occur in CF against nectin-1 or 3-OST-3-expressing cells.
Fig. 1.
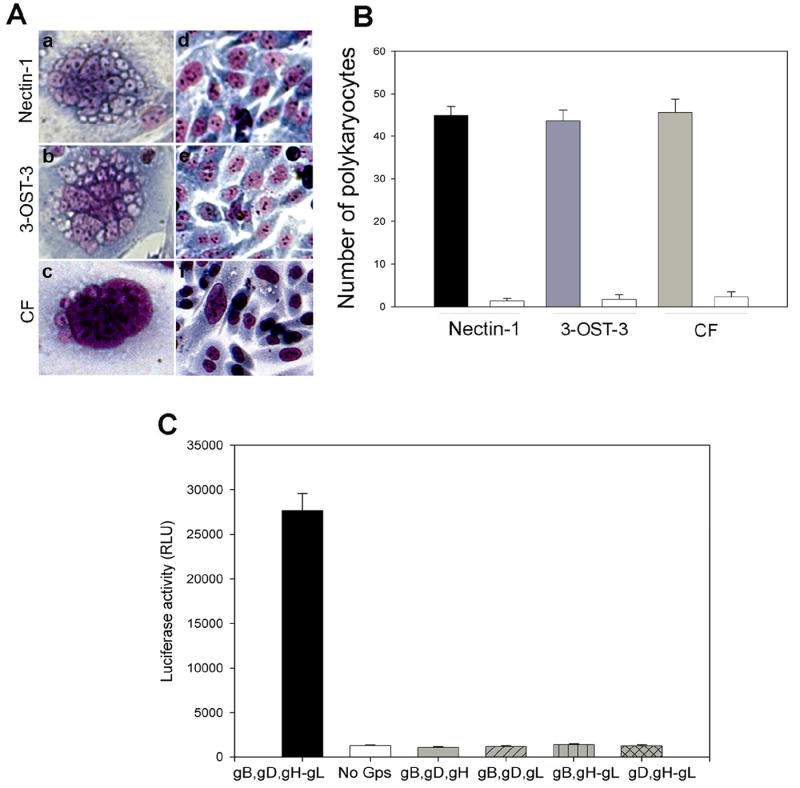
(A) Microscopic visualization of HSV-1 glycoproteins induced polykaryocyte formation in CF. Wild-type CHO- K1 effector cells transfected with expression constructs for HSV-1 glycoproteins (gB, gD, and gH-gL) were co-cultured with target cells expressing nectin-1 (panel a), 3-OST-3 (panel b) or CF (panel c). The corresponding control effector cells were transfected with an empty vector pCDNA3.1 and co-cultured with nectin-1 (panel d), 3-OST-3 (panel e) or CF (panel f). After 24 h, the cells were fixed with 2% formaldehyde and 0.2% glutaraldehyde at room temperature for 30 min and stained with Giemsa stain (Fluka) for 20 min. Shown are photographs of representative cells.
(B) Quantitative analysis of polykaryocyte formation. Effector cells were transfected with gB, gD, and gH-gL (dark bars) or pCDNA3.1 (empty bars) and co-cultured in 24-well dishes with target cells as indicated. The polykaryocytes were stained (as described above) and counted. Cells containing 10 or more nuclei were considered a polykaryocyte.
(C) Expression of gB, gD and gH-gL is required. The effector CHO-K1 cells were transfected with expression plasmids for glycoproteins indicated and mixed with CF. Membrane fusion due to polykaryocyte formation was detected by monitoring Luciferase activity. Relative luciferase units (RLUs) determined using a Sirius luminometer (Berthold detection systems).
Next, we decided to determine which glycoproteins were essential for fusion of primary cultures of CF with the effector cells. We transfected effector CHO-K1 cells with various combinations of HSV-1 glycoproteins and performed a luciferase reporter (Promega, Madison, WI) based cell-to-cell fusion assay [19]. As shown in Fig. 1C, it was clear that fusion of cultured CF required expression of all four glycoproteins, gB, gD, gH, and gL in effector cells [17-20]. In the absence of any of the four glycoproteins luciferase signals were identical to the background (no glycoproteins). Next, we sought to determine the role of HS in polykaryocyte formation. We hypothesized that since cultured CFs express 3-OS HS as the gD receptor [13], HS in general will be required for polykaryocyte formation. In order to test this hypothesis we examined the effect of HS degrading enzyme, heparinase-I. Along with CF, nectin-1 and 3-OST-3-expressing CHO-K1 cells were used as controls. The experiment was performed by dividing each type of the target cell population (CF, nectin-1 or 3-OST-3-expressing CHO-K1) into two equal pools. Heparinase-I (1.5 U ml-1, Sigma) was added to one pool while the other pool was compensated with identical volume of phosphate-buffered saline (PBS) [2]. Both treated and untreated pools were then mixed with equal amounts of effector cells and co-cultivated. As shown in Fig. 2A, heparinase treatment drastically impaired the ability of CF and 3-OST-expresssing CHO-K1 to from giant multinucleated cells. By contrast, heparinase-I treatment had virtually no effect on the fusion of nectin-1 expressing CHO-K1 cells, which is in line with the fact that nectin-1 mediated cell fusion does not depend on HS expression [21]. Likewise, the number of polykaryocytes formed was significantly lowered upon heparinase treatment of 3-OST-3 expressing CHO-K1 cells and CF but not when nectin-1 cells were treated. This indicates that CF and as expected, CHO-K1 3-OST-3 cells, exhibit heavy dependence for polykaryocyte formation on HS expression, implicating 3-OS HS as the mediator of polykaryocyte formation by CF. In subsequent experiments we investigated whether the effector, target or both types of cells require HS expression. To determine this we made use of a mutant CHO cell line (CHO-745) that is defective in HS or GAG biosynthesis [2]. The CHO-745 cells transiently expressing the four glycoproteins (gB, gD, gH, gL) were co-cultured with either CF or wild type CHO-K1 cells that express nectin-1 or 3-OST-3. The cells were either treated with heparinase or un-treated. Since the effector cells used here naturally lack HS, it was reasoned that if HS expression was required for the effector cells, we should observe a significant decrease in the size and the number of polykarocytes formed in the untreated cells. On the other hand, if HS expression was needed only for target cells that use 3-OS HS as the gD receptor then a treatment with heparinase should reduce polykaryocyte formation. As seen in Fig. 2C-2D untreated cells in each case formed polykaryocytes that were similar in size and numbers and thus, ruled out any significant need of HS expression for effector cells. A significant difference was seen in the case of treated cells. While nectin-1-expressing cells were largely unaffected by the treatment, both CF and 3-OST-3-expressing cells exhibited a marked reduction in the size and the number of polykaryocytes formed upon heparinase treatment. This data, therefore, suggests that HS expression on the target cell is important, which further affirms the possibility that 3-OS HS is the mediator required for making contacts for fusion with gD on the effector cells. Finally, to determine the expression of 3-OS HS on CF and to confirm that it is co-localized with HSV-1 gD expressed on effector cells we performed immunocytochemistry. The target cells (CF) were co-cultured with effector cells and fixed. A primary monoclonal antibody (HS4C3) that specifically recognizes 3-OS HS [23] was used in combination with an anti-gD polyclonal antibody (R7). The primary antibodies were detected by corresponding secondary antibodies conjugated with FITC (for 3-OS HS detection) or Texas Red (for gD detection). As shown in Fig. 3B, 3-OS HS was detected on CF (green) and gD (red) on the effector cells (Fig. 3C). It was also evident (Fig. 3D) that gD and 3-OS HS colocalized (yellow) at the cell-to-cell contact sites for fusion. The insets in figure 3 are shown at a higher magnification in the lower panels to highlight the gD and 3-OS HS colocalization (Fig 3E-G). Overall, the colocalization suggests a possible interaction between 3-OS HS and gD, which may lead to the membrane fusion.
Fig. 2.
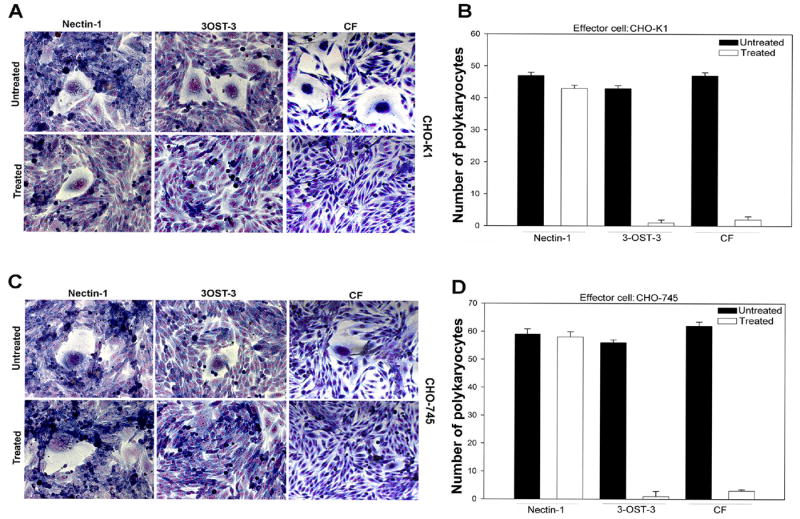
(A-B) Enzymatic removal of HS blocks polykaryocyte formation. (A). Target cells (as indicated) were either treated with heparinase I or left untreated prior to co-cultivation with effector CHO-K1 cells expressing gB, gD, gH-gL. (B). The number of polykaryocytes formed in target cells treated with heparinase or untreated was quantitated. Both effectors and target cells were mixed in 1:1 ratio and cultured for additional 16 h in a 24 well dishes. Cells with 10 or more nuclei were counted.
(C-D) HS is required by target but not the effector cells. HS negative CHO-745 cells were used as effector cells with the target cells as indicated. Heparinase treated or untreated cells were visualized for polykaryocyte formation (C) and quantified as described above (D).
Fig. 3.
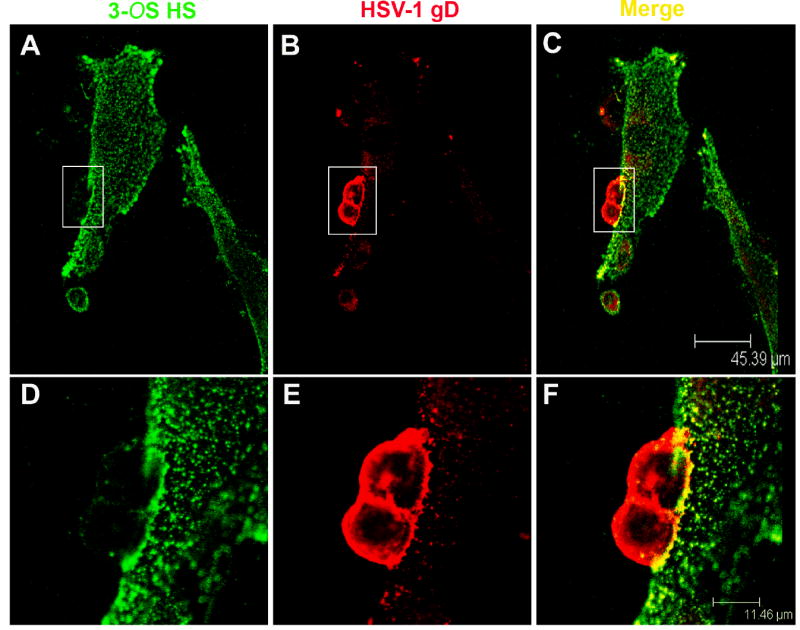
(A-F). Sequential confocal analyses demonstrating co-localization of HSV-1 gD with 3-OS HS. The fusion assay and the staining were performed as described in the text. 3-OS HS was detected with an anti-3-OS HS monoclonal antibody, HS4C3 [23] and a FITC conjugated secondary anti-body (green), gD with a rabbit polyclonal antibody (R7) and a Texas Red conjugated secondary antibody. The merging of green and red channels demonstrates the co-localization (yellow).
In summary, our study demonstrates for the first time the significance of HS in general and 3-OS HS in particular in polykaryocyte formation. Also for the first time, using a specific monoclonal antibody against 3-OS HS [23] we demonstrate its co-localization with gD on cell-to-cell contact sites. In addition, our results revise previous indications that HS has no significant role in this process [19,21]. Obviously, HS may not be necessary for polykaryocyte formation by all cell types, but it clearly can prove important for certain natural target cell types such as CF, where the virus can use it for entry [13-14] and cell-to-cell spread by polykaryocyte formation. The significance of our work is amplified by the fact that many viruses of different families have also been found to interact with HS [6]. These include the Picornaviridae (foot-and-mouth disease virus), Togaviridae (Sindbis virus and dengue virus), Parvoviridae (adeno-associated virus 2), Poxviridae (vaccinia virus), Retroviridae (human immunodeficiency virus) and Herpesviridae (virtually all human herpesviruses) [1, 6]. Our study, therefore, opens the door for future studies analyzing the significance of HS and more specifically, 3-OS HS, in other virus-induced membrane fusion mechanisms as well.
Acknowledgments
This investigation was supported by NIH RO1 grants Al057860 (DS), EY03890 (BY) and P30 EY01792 (core grant) and a RPB career award (DS). VT was supported by American Heart Association Postdoctoral Fellowship (AHA0525768Z) and a grant award from Illinois Society for Prevention to Blindness (ISPB).
Abbreviations
- HS
heparan sulfate
- 3-OST
3-O- sulfotransferase
- 3-OS HS
3-O sulfated heparan sulfate
- CF
corneal fibroblasts
- HSV-1
herpes simplex virus type-1
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Shukla D, Spear PG. Herpesviruses and heparan sulfate: an intimate relationship in aid of viral entry. J Clin Invest. 2001;108:503–510. doi: 10.1172/JCI13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shukla D, Liu J, Blaiklock P, Shworak NW, Bai X, Esko JD, Cohen GH, Eisenberg RJ, Rosenberg RD, Spear PG. A novel role for 3-O-sulfated heparan sulfate in herpes simplex virus 1 entry. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl U, Kusche-Gullberg M, Kjellen L. Regulated diversity of heparan sulfate. J Biol Chem. 1998;273:24979–24982. doi: 10.1074/jbc.273.39.24979. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg RD, Shworak NW, Liu J, Schwartz JJ, Zhang L. Heparan sulfate proteoglycans of the cardiovascular system. Specific structures emerge but how is synthesis regulated? J Clin Invest. 1997;99:2062–2070. doi: 10.1172/JCI119377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esko D, Lindahl U. Molecular diversity of heparan sulfate. J Clin Invest. 2001;108:169–173. doi: 10.1172/JCI13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Throp SC. Cell surface heparan sulfate and its roles in assisting viral 311 infections. Med Res Rev. 2002;22:1–25. doi: 10.1002/med.1026. [DOI] [PubMed] [Google Scholar]
- 7.WuDunn D, Spear PG. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J Virol. 1989;63:52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiwari V, Clement C, Duncan MB, Chen J, Liu J, Shukla D. A role for 3-O-sulfated heparan in cell fusion induced by herpes simplex virus type 1. J Gen Virol. 2004;85:805–809. doi: 10.1099/vir.0.19641-0. [DOI] [PubMed] [Google Scholar]
- 9.Xia G, Chen J, Tiwari V, Ju W, Li JP, Malmstrom A, Shukla D, Liu J. Heparan sulfate 3-O-sulfotransferase isoform 5 generates both an antithrombin-binding site and an entry receptor for herpes simplex virus, type 1. J Biol Chem. 2002;277:37912–37919. doi: 10.1074/jbc.M204209200. [DOI] [PubMed] [Google Scholar]
- 10.Xu D, Tiwari V, Xia G, Clement C, Shukla D, Liu J. Characterization of heparan sulphate 3-O-sulphotransferase isoform 6 and its role in assisting the entry of herpes simplex virus type 1. Biochem J. 2005;385:451–459. doi: 10.1042/BJ20040908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiwari V, O’Donnell CD, Oh MJ, Valyi-Nagy T, Shukla D. A role for 3-O-sulfotransferase isoform-4 in assisting HSV-1 entry and spread. Biochem Biophys Res Commun. 2005;338:930–937. doi: 10.1016/j.bbrc.2005.10.056. [DOI] [PubMed] [Google Scholar]
- 12.O’Donnell C, Tiwari V, Jin-Oh M, Shukla D. A Role for 3-O-sulfotransferase isoform-2 in assisting HSV-1 entry and spread. Virology. 2006;346:452–459. doi: 10.1016/j.virol.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Tiwari V, Clement C, Xu D, Valyi-Nagy T, Yue BYJT, Liu J, Shukla D. Role for 3-O-sulfated heparan sulfate as a receptor for herpes simplex virus type-1 entry into primary human corneal fibroblasts. J Virol. 2006;80:8970–8980. doi: 10.1128/JVI.00296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clement C, Tiwari V, Scanlon PM, Vali-Nagy T, Yue BYJT, Shukla D. A novel for phagocytosis-like uptake in herpes simplex virus entry. J Cell Biol. 2006;174:1009–1021. doi: 10.1083/jcb.200509155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liesegang TJ. Herpes simplex virus epidemiology and ocular importance. Cornea. 2001;20:1–13. doi: 10.1097/00003226-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 16.Farhatullah S, Kaza S, Athmananthan S, Garg P, Reddy SB, Sharma S. Diagnosis of herpes simplex virus-1 keratitis using giemsa stain, immunofluorescence assay, and polymerase chain reaction assay on corneal scrapings. Br J Ophthalmol. 2004;88:142–144. doi: 10.1136/bjo.88.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muggeridge MI. Characterization of cell-cell fusion mediated by herpes simplex virus 2 glycoproteins gB, gD, gH and gL in transfected cells. J Gen Virol. 2000;81:2017–2027. doi: 10.1099/0022-1317-81-8-2017. [DOI] [PubMed] [Google Scholar]
- 18.Browne H, Bruun B, Minson T. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J Gen Virol. 2001;82:1419–1422. doi: 10.1099/0022-1317-82-6-1419. [DOI] [PubMed] [Google Scholar]
- 19.Pertel P, Fridberg A, Parish M, Spear PG. Cell fusion induced by herpes simplex virus glycoproteins gB, gD, and gH-gL requires a gD receptor but not necessarily heparan sulfate. Virology. 2001;279:313–324. doi: 10.1006/viro.2000.0713. [DOI] [PubMed] [Google Scholar]
- 20.Turner A, Bruun B, Minson T, Brwone H. Glycoprotein gB, gD, and gHgL of herpes simplex virus type 1 are necessary and sufficient to mediate membrane fusion in a cos cell transfection system. J Virol. 1998;72:873–875. doi: 10.1128/jvi.72.1.873-875.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Terry-Allison T, Montgomery RI, Warner MS, Geraghty RJ, Spear PG. Contribution of gD receptors and glycosaminoglycans sulfation to cell fusion mediated by herpes simplex virus 1. Virus Res. 2001;74:39–45. doi: 10.1016/s0168-1702(00)00244-6. [DOI] [PubMed] [Google Scholar]
- 22.Yue BYJT, Baum JL. Studies of corneas in vivo and in vitro. Vision Res. 1981;21:41–43. doi: 10.1016/0042-6989(81)90134-6. [DOI] [PubMed] [Google Scholar]
- 23.Ten Dam GB, Kurup S, van de Westerlo EM, Versteeg EM, Lindahl U, Spillmann DD, van Kuppevelt TH. 3-O-sulfated oligosaccharide structures are recognized by anti-heparan sulfate antibody HS4C3. J Biol Chem. 2006;281:4654–4662. doi: 10.1074/jbc.M506357200. [DOI] [PubMed] [Google Scholar]
- 24.Valyi-Nagy T, Sheth V, Clement C, Tiwari V, Scanlan P, Kavouras JH, Leach L, Guzman-Hartman G, Dermody TS, Shukla D. Herpes simplex virus entry receptor nectin-1 is widely expressed in the murine eye. Curr Eye Res. 2004;29:303–309. doi: 10.1080/02713680490516756. [DOI] [PubMed] [Google Scholar]


