Abstract
Sterols such as cholesterol are a significant component of eukaryotic cellular membranes, and their unique physical properties influence a wide variety of membrane processes. It is known that the concentration of sterol within the membrane varies widely between organelles, and that the cell actively maintains this distribution through various transport processes. Vesicular pathways such as secretion or endocytosis may account for this traffic, but increasing evidence highlights the importance of nonvesicular routes as well. The structure of an oxysterol-binding protein homologue (OSH) in yeast (Osh4p/Kes1p) has recently been solved, identifying it as a sterol binding protein, and there is evidence consistent with the role of a cytoplasmic, nonvesicular sterol transporter. Yeast have seven such proteins, which appear to have distinct but overlapping functions with regard to maintaining intracellular sterol distribution and homeostasis. Control of sterol distribution can have far-reaching effects on membrane-related functions, and Osh proteins have been implicated in a variety of processes such as secretory vesicle budding from the Golgi and establishment of cell polarity. This review summarizes the current body of knowledge regarding this family and its potential functions, placing it in the context of known and hypothesized pathways of sterol transport in yeast.
Keywords: yeast, oxysterol binding protein, sterol, lipids, membrane transport, lipid binding proteins
1. Introduction
It is well established that sterols such as cholesterol are distributed heterogeneously throughout the eukaryotic cell [1, 2]. This phenomenon is most dramatically represented in the plasma membrane (PM), which contains 60−80% of cellular free cholesterol, about 35−45% of the lipid in the PM [1, 3]. This is a critical aspect of cellular homeostasis, since alterations in sterol concentrations within a membrane may dramatically alter the physical properties of a membrane (such as fluidity), affecting such diverse processes as signal transduction, membrane trafficking, or the function of integral proteins such as ion channels [4, 5]. Intracellular cholesterol storage disorders such as Niemann-Pick Disease Type C and Tangier disease [6, 7] highlight the importance of understanding the mechanisms of storage and transport. It is increasingly clear that maintaining this intracellular sterol distribution is dependent upon a tightly controlled system of synthesis, transport, and storage.
As sterols are ubiquitous components of cellular membranes, they are transported throughout the cell by means of vesicular trafficking mechanisms. This process would rapidly equilibrate the sterol concentration in cellular membranes if there were no lipid sorting during transport vesicle formation. Yet this is clearly not the case: it has been noted that there is a gradient of sterol concentration along the secretory pathway. From the endoplasmic reticulum, where sterols are synthesized, the concentration of cholesterol increases across the membranes of the Golgi, until the highest concentration of cholesterol is found in the plasma membrane [8-10]. This requires a mechanism to sort cholesterol into or out of transport vesicles, a process that may be driven by the concentration of cholesterol into lipid rafts [11]. One study has demonstrated that cholesterol and sphingomyelin are partially excluded from nascent COPI vesicles (retrograde Golgi-to-ER transport), implying an active mechanism to direct sterols forward through the secretory pathway [12].
There is also evidence that sterols can be moved between cellular compartments by nonvesicular mechanisms, which entail the movement of monomeric sterol between donor and acceptor membranes [13-15]. Sterols are capable of spontaneous diffusion into and out of membranes, but efficiency and perhaps directionality can be imparted to this process through the action of transport proteins. In mammalian systems, NPC2, sterol carrier protein-2/nonspecific lipid-transfer protein (SCP-2/nsLTP), and certain members of the START-domain family have been proposed as potential sterol carrier proteins [16, 17], with the promise of other as-yet unknown participants waiting to be discovered.
The budding yeast Saccharomyces cerevisiae has also been established as a model organism for the study of sterol transport (major known pathways of transport are depicted in Figure 1) [18, 19]. Though the species of sphingolipids differ significantly between yeast and mammals, the metabolic pathways of fatty acids and glycerophospholipids are largely conserved [20], and intracellular transport mechanisms may also be homologous. In the case of sterols, for instance, yeast maintain a similar ‘gradient’ of sterol distribution through the secretory pathway [21]. In the last decade, many laboratories have taken advantage of the well-studied genome of this organism and the panoply of genetic manipulation techniques in order to elucidate pathways of sterol transport and identify components homologous to mammalian systems. While yeast lack homologues of some mammalian lipid carriers, such as the SCP-2/nsLTP family [22], there are many other protein families and pathways that are conserved in yeast. Here we review the current state of knowledge regarding sterol transport in budding yeast, and the role of the OSH proteins (oxysterol binding protein homologues), a family of proteins that share a novel domain with the mammalian oxysterol binding protein (OSBP).
Figure 1.
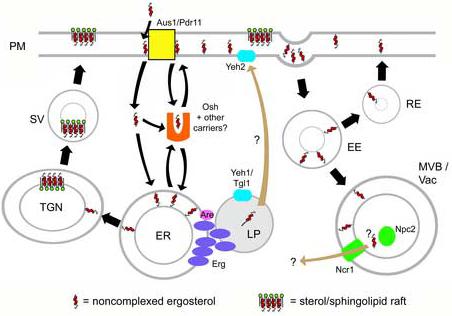
Known pathways of vesicular sterol transport, and hypothetical models of nonvesicular sterol transport. Straight arrows, vesicular routes; curved arrows, nonvesicular pathways (brown curved arrows represent pathways for which there is no direct evidence, or evidence only by homology to mammalian systems). (Vesicular routes may comprise diffusion through cytoplasm as well as translocation across membranes in close apposition.) Pathways are as follows. Ergosterol is synthesized by a suite of enzymes (Erg) located in the endoplasmic reticulum (ER) and lipid particle (LP). Sterols in the ER may either by converted to steryl esters by acyl-CoA:sterol acyltransferases (Are) for storage in the LP; transported to the cell surface by a sterol carrier protein (Osh); or sent to the Golgi (TGN), where it associates with sphingolipid to form ‘rafts’ or detergent-resistant membranes, and is transported to the plasma membrane in secretory vesicles (SV). Steryl esters in the LP can be mobilized to free ergosterol by steryl ester hydrolases (Yeh1/2, Tgl1) at the ER/LP and PM. Extracellular sterol, or sterol within the PM, may be internalized by transporters within the membrane (Aus1/Pdr11), whereupon it may be transported to the ER by carrier proteins. PM sterol can also be internalized by endocytosis; early endosomes (EE) may send sterol to the late endosome/multivesicular body (MVB) and vacuole (Vac), whereupon the yeast homologues of the human Niemann-Pick C genes (Ncr1, Npc2) might facilitate sterol movement to other organelles. Alternately, recycling endosomes (RE) may take sterol from the EE back to the surface.
2. An overview of sterol transport in budding yeast
2.1 Ergosterol biosynthesis and delivery to PM
Rather than cholesterol, yeast utilize ergosterol, which contains two extra double bonds and a methyl group. Though this confers slightly different physical properties, in the broad strokes ergosterol exhibits many of the same properties as cholesterol, such as its effect on the fluidic properties of the membrane and its tendency to associate with detergent-resistant membranes (DRMs) [23, 24]. In ergosterol-deficient yeast mutants, exogenous cholesterol may even substitute for ergosterol with no apparent ill effects.
The ergosterol biosynthetic pathway has been extensively characterized and previously reviewed [21]. It is carried out by a suite of enzymes identified as belonging to the ERG gene family, localized mostly in the endoplasmic reticulum (Figure 1). The Erg enzymes found later in the pathway exhibit somewhat low substrate specificity, acting on sterol precursors other than their normal physiological substrate. This can result in an array of non-ergosterol sterol products that differ primarily in their placement of double bonds and methyl groups [25]. Normally present at minimal levels in wild-type cells, they predominate in erg mutants, substituting for ergosterol [26] and sometimes severely altering the fluidic properties of the plasma membrane [27]. This can result in enhanced permeability to ions and lipophilic drugs [28, 29] and significant depletion of sterol-sphingolipid rafts, as evidenced by the heightened detergent accessibility of raft-associated proteins such as Gas1p or Hup1p [30, 31]. These defects ultimately lead to the disruption of a variety of ergosterol-dependent events such as endocytosis [32], protein sorting to the plasma membrane [33], and homotypic vacuole fusion [34].
After synthesis in the ER, ergosterol is delivered to the plasma membrane by a route that was, until recently, largely uncharacterized. Research in mammalian cells had found newly synthesized cholesterol in a novel, low-density membrane fraction but not in the soluble fraction [35, 36]. Transfer of newly synthesized sterol to the PM was ATP dependent but brefeldin A (BFA)-insensitive, suggesting either a nonvesicular pathway, or a vesicular transport mechanism that bypasses the BFA-sensitive route through the Golgi [37, 38]. In budding yeast, it was found that temperature-sensitive mutants defective in Sec18p-dependent secretory vesicle trafficking were still capable of rapid (t1/2=10 min) transport of newly synthesized ergosterol from the ER to the plasma membrane [39]. These findings led to a proposed model wherein only non-raft associated, chemically active sterol is available for nonvesicular transfer between the PM and ER and that both organelles have similar amounts of chemically active sterol. By sequestering chemically active sterol, raft formation in the PM would therefore drive the net accumulation of sterol in this compartment. The efficiency of sterol transfer between the ER and PM would seem to rule out passive diffusion and suggest it could be mediated by soluble carrier proteins or proteins at sites of apposition of the ER and PM. However, certain other temperature-sensitive sec mutants are capable of partially blocking ergosterol delivery to the PM, concomitant with a partial block in protein secretion [40]. A possible Sec18p-independent secretory pathways [41] may therefore account for some portion of cholesterol traffic, though direct confirmation of this has yet to be obtained. Taken together, the picture of ER-to-PM sterol transport is intriguing but still incomplete, and the relative importance of vesicular versus nonvesicular mechanisms remains to be determined.
Additionally, sterol intermediates may be subjected to some form of transport during the biosynthetic pathway, especially to and from lipid particles. The lipid particle (LP, also known as the lipid droplet) is an unusual organelle consisting of a neutral-lipid core (steryl esters and TAG in roughly equal amounts) surrounded by a phospholipid monolayer, with some attendant membrane-anchored and peripheral proteins [42]. Whereas most of the enzymes along the ergosterol biosynthetic pathway are ER-localized, Erg6p and Erg7p are found almost exclusively in the lipid particle [43-45], and Erg1p and Erg27p can be found in both of these organelles [46, 47]. How sterol intermediates move between these organelles is unknown, but a study of erg11Δerg3Δ double mutants found that lanosterol, the product of Erg7p and substrate for Erg11p accumulated not at the site of its synthesis (LP) or delivery (ER) but is equally distributed among all organelles. The authors speculated that pathways of inter-organellar transport exist, are independent of the synthetic enzymes, and are specific for ergosterol or for certain intermediates [48]. Another possibility is that if rafts are important for intracellular sorting of sterol, then missorting in these mutants may occur because sterol intermediates could have differing affinities for rafts relative to ergosterol. The reasons for the distribution of Erg enzymes between the ER and LP remain obscure, but if mechanisms of sterol transport exist between these organelles, then control of sterol transporters, or of ER-LP contact, may offer an additional level of regulation of ergosterol biosynthesis.
2.2 Exogenous sterol uptake and transport
Under aerobic conditions, yeast are fully autotrophic for ergosterol and do not take up exogenous sterols from the medium. However, sterol biosynthesis requires molecular oxygen at several steps. Thus, sterol uptake is enabled under anaerobic conditions, or in mutants unable to synthesize the heme prosthetic group (a required component of several Erg proteins including Erg11p, Erg 3p and Erg5p) [49]. In these cases, exogenous sterol must be supplied in order to enable growth. Uptake may also be induced in aerobic conditions in two ways. The first is a hypermorphic mutation in the UPC2 gene (upc2−1), which encodes a transcription factor regulating the expression of numerous genes, including some of the ERG family [50-52]. The second is in the constitutive overexpression of the transcription factor SUT1, itself normally expressed under anaerobic conditions [53]. Sterol is efficiently taken up in both of these mutants, though it is not required for survival.
Transcriptional analyses of upc2−1 and SUT1 + strains identified three upregulated genes that could potentially act as sterol transporters: AUS1, PDR11, and DAN1 [52, 54]. In a upc2−1 background, it was found that deletions of aus1, pdr11, and dan1 severely reduced sterol uptake [52]. Aus1p and Pdr11p belong to the ATP-binding cassette (ABC) family of transporters and localize primarily to the plasma membrane, making them ideal candidates for a direct role in sterol movement [55]. In mammals, a number of ABC transporters are responsible for the efflux of cholesterol or other sterols from the cell [56]. Though Aus1p and Pdr11p are required for sterol uptake rather than efflux, they may work by a similar mechanism. The function of Dan1p, a putative cell wall mannoprotein, is less clear. In a wild type background, constitutive expression of either AUS1 or DAN1 alone failed to promote uptake, but coexpression of both allowed influx [54]. Dan1p may therefore work in tandem with the ABC transporters, perhaps by allowing sterol penetration of the cell wall or by “presenting” sterols to the PM-localized transporters.
The exact mechanism by which the ABC transporters facilitate sterol movement is unknown, but there have been recent clues that the transporters facilitate movement of sterols from the plasma membrane to the ER, where they are esterified. Energy-depleted upc2−1 cells were still able to accumulate small amounts exogenous cholesterol in the plasma membrane [55]. When the cells were once again permitted to grow, Aus1p and Pdr11p were found to catalyze efficient sterol transfer from the PM to ER (as gauged by subsequent esterification in the ER). Sec mutants failed to inhibit this process, and sterols with a lower affinity for detergent-resistant membrane fractions were transported more efficiently [55]. This strongly implies a non-vesicular pathway of PM-to-ER sterol movement that selectively affects non-raft-associated sterols. This in itself does not necessarily imply soluble sterol carrier proteins; the extensive contacts between PM and ER membranes in yeast leave open the possibility that Aus1p and Pdr11p can directly insert sterols into closely apposed ER membranes. They may also facilitate the diffusion of sterols from the periplasmic space (between the PM and cell wall) into the PM. It is also possible that the yeast ABC transporters induce perturbations in the shape, structure, and/or phase behavior of the local lipidic environment, permitting noncomplexed sterol to diffuse more freely from the cytosolic leaflet of the PM. Further structural analyses of these proteins are needed to determine their exact mechanism of action.
One recent study opens the door to the possibility that sterol uptake is connected somehow to mitochondrial function. Schneiter and colleagues [57] conducted a broad screen for mutants deficient in sterol uptake. Interestingly, a number of genes responsible for mitochondrial structure and function were found to result in uptake deficiency when mutated. One such candidate, UGO1, codes for an outer-membrane protein that participates in the mitochondrial fusion apparatus [58], and might catalyze other fusion events that permit sterol movement between the fused membranes. However, it is more difficult to see the connection between sterol movement and the other candidates, which include several subunits and chaperones of F1F0 ATP synthase; aconitase; and a pair of less-characterized genes, CAT5 and MGM101, which may participate in ubiquinone biosynthesis and mtDNA oxidative damage repair, respectively. Though the connections are not immediately apparent, future work may provide novel insights into the relationship between mitochondria and sterol uptake.
2.3. Endocytosis and post-endocytic sterol efflux
In mammalian cells, endocytosed PM cholesterol is sorted in endosomes and rapidly returned to the PM [13, 15]. Though a similar process must occur in yeast, little is known about how it is mediated. In yeast, evidence strongly suggests that the accumulation of non-ergosterol intermediates and aberrant sorting of sterols can affect endocytosis at the internalization step [30, 32] as well as at post-internalization steps [30, 59]. These effects could be due to hypothetical endocytic regulators having high specificity for ergosterol but not the sterol intermediates. It is also possible that the accumulation of sterol intermediates results in more generalized defects in the physical properties of the membrane such as changes in fluidity or the ability to curve, resulting in unfavorable conditions for vesicular budding and fusion [60].
Research into the human disorder Niemann-Pick Disease Type C has resulted in the identification of novel sterol-binding proteins conserved from yeast to mammals. The disease state is characterized by the accumulation of cholesterol in aberrant late endosomal/lysosomal structures [61]. This disease is caused by mutations in either of two genes, NPC1 and NPC2 [6]. The gene products are found in the endosomal pathway and act in concert to promote efflux of LDL-derived cholesterol out of the lysosome to the PM and other membrane pools [62]. NPC1 is a transmembrane protein with a sterol-sensing domain oriented within the lumen of the late endosome, while NPC2 is a soluble cholesterol-binding protein located primarily within the late endosome and lysosome. It has recently been found that the yeast homologues of these proteins, Ncr1p and Npc2p, respectively, are functionally conserved from yeast to mammals. Cultured mammalian cells with mutations in NPC1 or NPC2 accumulated cholesterol in lysosomal structures, but expression of the respective yeast homologue reverted this phenotype [63-65]. Human NPC2 is capable of transporting cholesterol to acceptor membranes in vitro, and this activity is enhanced in acidic environments and when acceptor membranes contained significant concentrations of the lysosome/late endosome-specific lipid lysobisphosphatidic acid [66]. Since cholesterol efflux in vivo is tied to the presence of critical cholesterol-binding residues [67], the collective evidence suggests that NPC2 acts as a soluble cholesterol carrier in mammalian lysosomes/late endosomes, though direct in vivo evidence has not yet been presented. It is likely that yeast Npc2p, which shares several of the putative sterol-binding residues, has a similar capacity to bind and transport sterol in vitro, though this has not yet been directly demonstrated.
In mammals, lysosomal cholesterol efflux may be mediated by an association between NPC1 and certain cholesterol-binding START domain proteins [68-71]. Yeast, however, have no identifiable START domains in any open reading frame. It may be that other, similar proteins stand in for START proteins in the Ncr1p/Npc2p system or other lipid-transport pathways. One of the most well characterized START proteins is the ceramide transporter CERT. In addition to the C-terminal START domain, CERT also possesses a pleckstrin homology (PH) domain that directs targeting to the Golgi via binding to PI(4)P, and a motif (FFAT, or “two phenylalanines in an acidic tract”) that allows binding to VAP (VAMP-associated protein), an ER transmembrane protein. This dual targeting allows nonvesicular transfer of ceramide from the ER to Golgi, as determined by the work of Hanada and colleagues [72]. The mammalian oxysterol binding protein (OSBP) is similar to CERT in that it also possesses a PH domain, FFAT motif, and a novel C-terminal lipid-binding domain (Figure 2). Budding yeast have seven OSBP homologues (termed “OSH”) that share this latter domain, making them potential candidates for sterol carrier proteins [73, 74].
Figure 2.

Domain structure, subcellular localization (in yeast) and known lipid affinites of human oxysterol binding protein (OSBP) and the yeast OSBP homologues (Osh1p-Osh7p). The relative positioning of the ORDs and other domains is based on the ORD sequence alignments from the Supplementary Material in [78]. Black, oxysterol binding protein-related domain (ORD). Purple, pleckstrin homology (PH) domain. Green, ‘two phenylalanines in an acid tract’ (FFAT) motif. Red, ankyrin repeats. Yellow, Golgi dynamics (GOLD) domain.
3. Structure of oxysterol binding protein homologues (OSH)
OSBP was initially purified and characterized in mammalian cells due to its ability to bind oxysterols, which can serve as potent regulators of cholesterol metabolism [75]. There are at least twelve OSBP-related proteins (ORPs) in mammals. All contain an OSBP-related domain (ORD) that, in OSBP, mediates oxysterol binding. However, the specific lipid ligands of most ORDs have not been determined. While the functions of many ORPs are unknown, collectively they are thought to affect numerous processes involved in lipid distribution and metabolism, as well as vesicular trafficking [76][see also this issue]. The ability to bind lipids and the general similarity in domain organization to the ceramide transfer protein CERT, suggest that some ORPs could similarly act as soluble lipid transporters. Evidence suggests that at least one mammalian ORP acts as a lipid sensor, modulating the activity of two ERK phosphatases in its cholesterol-bound state [77].
Budding yeast have seven ORPs (Figure 2). This group contains three paralogous pairs, which likely arose from an ancestral whole-genome duplication event [77]: Osh1p (Swh1p) and Osh2p; Osh4p (Kes1p) and Osh5p (Hes1p); and Osh6p and Osh7p. Each pair shares a higher degree of similarity than with the other Osh proteins. Four Osh proteins, Osh4 to Osh7, consist of only an ORD, while the other three are structured similarly to OSBP, possessing additional targeting domains (Figure 2).
The structure of one of these small Osh proteins, Osh4p (also known as Kes1p), was recently solved both in the absence of ligand and complexed with cholesterol, ergosterol, and a number of oxysterols [78]. The ORD is essentially a beta-barrel with a hydrophobic interior that can accommodate a single sterol molecule. The barrel is blocked at one end by a bundle of α-helices, while the other is occluded by a “lid” region (residues 1−29) containing an amphipathic α-helix connected by a flexible linker (Figure 3). Sterols and oxysterols bind in the interior of the barrel at nanomolar affinity, oriented with the 3β–hydroxyl group of the sterols at the bottom of the hydrophobic binding tunnel. The sterol side chain makes contact with the inside of the lid, possibly stabilizing the closed conformation and creating an entirely sealed environment for the sterol.
Figure 3.

Proposed mechanism of sterol transport by Osh4p. Unliganded Osh4p possesses a relatively flexible lid domain, and binds to membranes most likely through highly conserved charged residues at the surface (yellow). This binding may be stabilized by the presence of PI(4,5)P2 in the membrane (green headgroup). Sterol enters the interior of the ORD, inducing conformational changes. Upon dissociation from the membrane, hydrophobic residues in the lid domain (black) contact the hydrocarbon tail of sterol, stabilizing Osh4p in a ‘closed’ conformation. Sterol delivery to the acceptor membrane is likely a mirror of the extraction from the donor membrane.
This structure provides insights into how Osh4p and other ORPs might interact with membranes. A number of highly conserved hydrophilic residues are near the entrance of the ligand-biding tunnel. Mutation of these residues revealed that they are required for Osh4p function in vivo and, as discussed below, for the ability of Osh4p to extract and transfer sterols between membranes [78, 79]. These residues are likely required for the interaction of Osh4p with the charged surface of membranes. Overall, the structure of Osh4p suggests that ORPs bind membranes and facilitate the movement of sterol (and perhaps other lipids) into or out of its hydrophobic binding tunnel. Thus, ORPs like the Osh proteins could function as lipid transfer proteins and lipid sensors.
4. Sterol transport by Osh proteins
4.1 Osh4p mutational analyses and in vitro transport assays
Evidence that Osh4p and other Osh proteins transfer sterols between cellular membranes in vivo comes from studies on the uptake and trafficking of exogenous sterols in yeast. Exogenous sterols enter the PM and can move from there to the ER by a pathway that does not require any of number of the SEC genes needed for vesicular transport [55]. Cells depleted of all seven Osh proteins showed a dramatic decline in PM-to-ER transport, as well as in the delivery of newly synthesized ergosterol from the ER to the PM [79]. The transport of newly synthesized ergosterol from the ER to the PM also slows dramatically in these mutants [80]. Cells lacking all the Osh proteins except Osh4p transferred exogenous cholesterol to the ER only slightly faster than strains missing all of these proteins, suggesting that Osh proteins other than Osh4p are also required. Analysis of mutants missing just one of the Osh proteins suggested that Osh3p and Osh5p probably also play significant roles in PM to ER sterol transfer. Consistent with a role for Osh proteins in intracellular sterol trafficking, it has been shown that sterol distribution is severely altered in mutants missing all of these proteins [74, 81].
The finding that Osh4p can move sterols between liposomes in vitro supports a direct role for the Osh protein in PM to ER sterol transfer [79]. Osh5p, which shares the greatest degree of similarity with Osh4p, can also bind and transport sterol at a level comparable to that of Osh4p (Schulz and Prinz, unpublished observation). Since OSH5 expression is upregulated in upc2−1 strains [52], Osh5p is a promising candidate for an Aus1p/Pdr11p-associated transporter under conditions that enable sterol uptake. It remains possible that other proteins also transfer sterols between the PM and ER, however. Sterols are still moved between the ER and PM in cells lacking all the Osh proteins, albeit at a significantly slower rate than in wild-type cells [79, 80]. This transfer might reflect diffusion of sterols through the aqueous phase unassisted by any protein, or the action of other undiscovered sterol transfer proteins. Furthermore, some Osh proteins may not transfer sterols at all; Osh6p and Osh7p display little to no capacity to transport or extract cholesterol from membranes in vitro (Schulz and Prinz, unpublished observation).
Some Osh proteins may therefore transfer lipids other than sterols. In vitro, Osh4p can move phosphatidylserine (PS) and PI(4,5)P2 between liposomes [79]. This transfer likely differs from sterol transport by Osh4p. The charged headgroups of PS and PI(4,5)P2 probably cannot be accommodated by Osh4p with a closed lid-domain. Consistent with this, while Osh4p undergoes a conformational change after sterol binding [78], a similar change was not seen after binding of PS or PI(4,5)P2 (Raychaudhuri and Prinz, unpublished observations). Thus, Osh4p likely transfers these lipids in an open conformation. In addition, there is currently no evidence that Osh proteins move these lipids in vivo. The synthesis of phosphatidylethoanolamine (PE) from PS requires the nonvesicular transfer of PS from the ER to either a mitochondria or the Golgi complex [82] (see review in this issue). However, this transport was not affected in cells lacking all of the Osh proteins, suggesting that they probably play no role in this process [79]. Whether Osh4p or other Osh proteins transfer PIPs among cellular compartments remains to be determined.
4.2 PIPs and the regulation of sterol transfer by Osh proteins.
One intriguing aspect of sterol transfer by Osh4p is that it is specifically enhanced by PI(4,5)P2; no other PIPs had any effect on the efficiency of transport [79]. This is also true of Osh5p (Schulz and Prinz, unpublished observations). In vivo, depletion of PI(4)P and PI(4,5)P2 levels (via PI 4-kinase and PI(4)P 5-kinase mutations) dramatically slows PM-to-ER sterol transport, suggesting that PIPs regulate sterol transfer by Osh proteins in cells [79]. Since PI(4,5)P2 is highly enriched at the PM [83], sterol transfer by Osh4p and Osh5p (and perhaps other Osh proteins) may serve to regulate the sterol content of the PM, or promote uptake and esterification of exogenous sterol that diffuses into the PM.
The mechanism of PIP stimulation of sterol transfer by Osh proteins remains an open question. Surprisingly, PIPs may bind Osh4p at a site that is distant from the opening of the sterol-binding tunnel and distinct from the residues proposed to interact with the membrane [84]. How PIP-binding at this site could stimulate sterol extraction and transfer is not clear. PIPs stimulate transfer between membranes both when they are on donor and acceptor liposomes [79]. It is conceivable that PIPs stabilize binding to the membrane surface, increasing residence time of Osh4p and thereby increasing the probability of sterol entry into (or exit from) the binding pocket. This remains to be directly demonstrated, however.
4.3 Are Osh proteins really sterol sensors?
It is difficult to rule out that Osh proteins are not actually lipid sensors that indirectly affect non-vesicular sterol movement between the ER and PM, as has been suggested [13, 80]. These authors point out there is no evidence that any mammalian ORPs are lipid transfer proteins but substantial evidence that at least one, OSBP, functions as lipid sensor [77, 85]. Moreover, even in cells missing all the Osh proteins, sterols are still transferred between the ER and PM, albeit 5 to 10 times more slowly than in wild-type cells [79, 80]. Thus other, as yet unidentified proteins, could actually transfer sterols between these compartments. In addition, there is ample precedent for proteins that can transfer lipids between membrane in vitro but almost certainly do not in cells. Perhaps the best example is provided by Sec14p, an essential yeast phosphatidylinositol/phosphatidylcholine transfer protein (PITP) that is required for protein trafficking from the Golgi complex [86]. This protein is part of a large family of proteins, many of which can transfer phospholipids between liposomes in vitro. Nonetheless, it is widely accepted that Sec14p functions as a lipid sensor and not a lipid transfer protein in vivo. The case of Osh4p and the other Osh proteins, however, differs from this example in one important respect. It is not known whether non-vesicular transfer of PI or PC between cellular compartments is altered in cells with conditional defects in SEC14. In contrast, there is strong evidence that non-vesicular sterol transfer significantly slows in cells lacking Osh proteins. Thus, if Osh proteins do not directly transfer sterols between membranes in cells, they must regulate the proteins that do. Until such proteins are identified, the simplest interpretation of the findings summarized in sections 4.1 and 4.2 is that some Osh proteins directly transfer sterol in cells. It remains possible that some Osh proteins also function as lipid sensors.
5. OSH family as regulators of ergosterol homeostasis
The OSH family appears to share a general, mutually overlapping role in lipid homeostasis, particularly sterol distribution. An exhaustive mutational analysis showed that while individual OSH deletions were viable, deletion of all seven proved lethal [74]. Interestingly, strains remained viable as long as any one of the seven OSH genes was normally expressed (with the exception of OSH1, which requires overexpression). Some OSH disruptions showed minor perturbations in intracellular ergosterol levels, and these differences become more marked with further cumulative OSH deletions [73, 74]. Each single mutant displayed a slightly different profile of gene expression and resistance/susceptibility to toxins such as nystatin, lovastatin, and high salt concentrations. Depleting one cell line of all seven OSH proteins (by disrupting six and placing the seventh under an inducible promoter) during mid-log growth drastically elevated the total levels of ergosterol, zymosterol, and 22-dihydroergosterol [74], although this observation was not replicated in later studies using a line with six disrupted OSH genes and a temperature-sensitive mutant of the seventh (oshΔosh4ts) [81]. However, the consequent alterations in intracellular sterol distribution resulted in several serious membrane-related defects, such as fragmented vacuoles, accumulation of lipid particles and other vesicular structures, and inhibition of the internalization step of endocytosis [74, 81]. The collected evidence strongly points toward a model whereby each OSH protein possesses a unique function, but all share in the broader, essential function of maintaining a proper homeostatic distribution of sterol throughout the cell. It is possible that some OSH proteins are essential under certain specific conditions: expression of OSH3, for example, is inducible by alpha-factor mating pheromone, and appears to regulate nuclear fusion during mating [87]. It also appears to play an important regulatory role in filamentous/pseudohyphal growth in S. cerevisiae and C. albicans under nitrogen-starved conditions [87], though in both of these processes its specific mechanism of action remains unclear.
Many lines of evidence also suggest a reciprocal relationship between sterol and sphingolipid metabolism in yeast [19, 88, 89]. This is perhaps not surprising, given that these lipids may be present together in lipid rafts/DRMs, working in concert in some critical membrane processes. Some evidence exists that the OSBP-related proteins may represent one nexus of regulation between the pathways controlling sterol and sphingolipid activity. One recent study in mammalian cells has demonstrated that OSBP recruits CERT to the Golgi in a sterol-dependent manner, promoting ceramide transport and sphingomyelin synthesis [85]. In yeast, disruption of OSH2 drastically increases levels of certain sphingolipids and an OSH3 deletion grants resistance to an inhibitor of sphingolipid biosynthesis [90, 91]. Deletion of OSH4/KES1 conferred resistance to phytosphingosines in mutants unable to break down these signaling molecules (Δlcb3/Δdpl1), a phenotype similar to several ERG mutants [92]. This effect appeared to be mediated through decreased phosphorylation and mislocalization of the sphingoid long-chain base kinase Lcb4p, normally found at the plasma membrane. The authors hypothesize that the concentration of sterol in the plasma membrane (reduced in these mutants[74]) modulates this sphingolipid-derived signaling pathway. In this way, the OSH proteins may act as sterol sensors, responding to the overall levels and distribution of sterol, or as direct controllers of sterol distribution that in turn modulates other sensors.
6. Targeting signals and subcellular localization
As the members of the OSH family appear to share one essential function, so too does their distribution within the cell overlap to an extent. Using GFP-tagged constructs, it has been noted that OSH proteins are largely distributed throughout the cytoplasm, with a few exceptions, including Osh1p (discussed in greater detail below) as well as Osh2p, which concentrates at the plasma membrane, particularly at the bud and neck of S-phase cells [93]. Osh4p/Kes1p co-localizes with Golgi markers as well as being found throughout the cytoplasm [84]. Osh6p and Osh7p are primarily cytoplasmic, but with some patched distribution at the cell periphery; these patches seem to be absent in stationary phase cells [94] (Schulz and Prinz, unpublished observations).
Structurally, the yeast Osh proteins are broadly grouped into two main classes (Figure 2). Osh4p through Osh7p primarily consist of the conserved ORD. The ‘full length’ Osh proteins, Osh1p-Osh3p, correspond more closely to mammalian OSBP in that they consist of the ORD domain at the N-terminus, as well as other N-terminal targeting sequences. Osh1p-Osh3p have a PIP-binding PH domain which, in Osh1p, is specific for PI(4,5)P2 and strongly directs targeting to the Golgi [95]. Despite lacking a separate PH domain, Osh4p through Osh7p are all capable of binding PIPs via the ORD [79, 94], and it is this affinity for PIPs that probably directs Osh4p to the Golgi [84].
The full-length OSH proteins also contain at least two protein-binding domains. All three contain a FFAT motif which mediates an interaction with Scs2p, an ER-resident VAP homologue [96]. Osh1p and Osh2p also contain ankyrin repeats, which mediate protein-protein interactions for a diverse range of macromolecular complexes, and Osh3p has a ‘Golgi dynamics’ (GOLD) domain, which is found among a variety of proteins involved in Golgi function [97, 98]. The GOLD domain is typically found in proteins alongside other lipid and protein targeting motifs, such as the PH and FFAT of Osh3p, and may itself mediate protein-protein contacts although its full significance is still unclear [99]. The presence of multiple targeting signals in the full-length OSH proteins suggests that the Osh proteins may be capable of shuttling between organelles depending on conditions; targeting of mammalian OSBP may be influenced by the presence of oxysterol ligand [100], and the same could apply to the yeast homologues. The full-length yeast ORPs show complex patterns of localization depending on which targeting signals are present within the construct [93], but the physiological signals that might control the relative dominance of each targeting domain remain largely uncharacterized. Hence the purpose of any potential trafficking between organelles is unclear.
Recent insights into the full-length OSH proteins (Osh1p, Osh2p, Osh3p) have given weight to the idea that some ORPs function at regions of close apposition between different organelles. For instance, overexpression of Scs2p can effectively sequester the full-length Osh proteins due to its binding to their ankyrin repeats. But whereas overexpressed Scs2p is distributed over the entire ER membrane, Osh1p-3p localize only to very specific subdomains of the ER, suggesting that targeting signals besides the ankyrin repeats are also directing their localization [96]. This model of full-length OSH localization is supported by the emerging characterization of membrane contact sites, specialized domains mediating communication between organelles at sites of tight apposition [101, 102]. In yeast, certain ER subfractions may make extensive contacts with the mitochondria and plasma membrane [103, 104]. These domains are enriched in lipid biosynthetic enzymes, which in the case of the ER-PM sites include several members of the Erg family [104]. The ER-mitochondria contact site also appears to be the location of PS transfer from the ER for decarboxylation in the mitochondrion, marking it as a site where lipid transfer proteins may operate [105].
The most extensively characterized membrane contact site in yeast is the nucleus-vacuole junction (NVJ), a site of selective autophagy of nuclear material. This process, termed ‘piecemeal microautophagy of the nucleus’ (PMN), occurs during nutrient starvation and involves the budding and pinching-off of vesicles containing nuclear material into the vacuolar lumen [106]. The primary structural component of this junction consists of the outer nuclear membrane resident Nvj1p complexed with the vacuolar transmembrane protein Vac8p [107]. The ankyrin repeats at the N-terminus of Osh1p specifically target the protein to the NVJ via binding to a cytoplasmic tail of Nvj1p [93, 108]. Osh1p can be effectively sequestered to the NVJ by overexpression of NVJ1, which inhibits growth of trp1Δ cells in tryptophan-poor medium [109]. Growth can be restored by overexpression of the tryptophan permeases, suggesting that the problem is that the transporters are missorted away from the PM. As proper ergosterol distribution seems to be necessary for correct sorting of a tryptophan permease to the plasma membrane, as seen in certain ERG mutants, it is conceivable that sequestration of Osh1p affects intracellular sterol distribution in some manner [110]. It is interesting to note that osh4Δ and osh5Δ mutants are also defective in tryptophan uptake, and overexpression of OSH4 or OSH5 partially rescues the low-tryptophan growth defect in osh1 deletants [73]. It is possible that the shared function of the OSH proteins is sufficient for sorting of the Trp permease, but so far only Osh1p demonstrates a functional link to this process via its association with the NVJ.
7. OSH proteins as regulators of membrane trafficking
It is increasingly clear that certain yeast ORPs have a measurable impact on intracellular vesicular trafficking. For instance, Osh4p has been previously implicated in vesicular transport from the Golgi complex [84, 111]. These studies showed that Osh4p interacts genetically with Sec14p, an essential PITP in yeast necessary for secretory transport in the Golgi complex. Strains with conditional sec14 alleles are not viable at restrictive temperature. However, it is possible to isolate mutations that do not require, or “bypass,” Sec14p for viability. Some of these bypass mutants lack Osh4p [111]. Mutations in other Osh proteins do not bypass sec14, suggesting that Osh4p has a function in the TGN not shared with other Osh proteins. It has been proposed that Sec14p maintains a functionally important pool of DAG in the Golgi complex by regulating DAG consumption and synthesis [112, 113]. What role Osh4p plays in this process is not known, but it is not able to extract or transfer DAG from membranes [79]. It may act as a lipid sensor at the Golgi complex: for example, in response to binding sterols (or other lipids), it could regulate enzymes needed for maintaining DAG pools. It has also been proposed that Osh4p might directly bind and regulate a component of the vesicle budding machinery on the Golgi in response to binding a lipid ligand [86], though this component has not yet been identified. Alternatively, Osh4p might act as a lipid transfer protein at Golgi membranes. It could affect Golgi function indirectly by moving sterols to or from this organelle.
Another possibility is that PI(4,5)P2 transport or binding is the primary function of Osh4p at the Golgi complex. All sec14 bypass mutations require phospholipase D (PLD) [114, 115], an enzyme that hydrolyzes PC to PA, which can in turn be converted to DAG. It is thought that PLD, like Sec14p, helps maintain proper DAG levels in the TGN. Since this enzyme is specifically and potently activated by PI(4,5)P2 [116-118], modulating the amount of PI(4,5)P2 available to stimulate PLD could affect DAG levels in the Golgi complex. Thus, Osh4p might modulate PLD activity, and therefore Golgi DAG levels, by affecting the amount of PI(4,5)P2 available to stimulate PLD. Interestingly, PI(4,5)P2, PLD, and Osh4p have all been implicated in the proper function of some soluble NEM-sensitive factor receptors (SNAREs) at the PM in yeast [119]. These findings suggest that PI(4,5)P2 and Osh4p may affect PLD function and, indirectly, vesicular transport in a number of cellular membranes.
Deletion of other OSH proteins, including the Osh4p paralogue Osh5p/Hes1p, did not result in a ‘bypass Sec14p’ phenotype [111]. However, overexpression of OSH5 (normally expressed at significantly lower levels than OSH4) in osh4Δsec14ts mutants restores temperature sensitivity to the strain (Raychaudhuri and Prinz, unpublished observation). Several mammalian ORPs (ORP1S/L, ORP9S) also appear able to compensate for OSH4 deletions by restoring temperature sensitivity to sec14ts mutants [120, 121]. Determining whether these ORPs also bind and transport sterol or PI(4,5)P2 may indicate whether the function of Osh4p in the Sec14p pathway directly depends on this capability.
One recent study suggests that members of the OSH family may also regulate the vesicular transport required for polarized growth in yeast. Multicopy expression of any one OSH gene (except for OSH5 and OSH7) is able to compensate for temperature-sensitive CDC42 defects, permitting the establishment of polarization [122]. Cdc42p is a Rho family GTPase that establishes polarity by forming signaling complexes that define the bud site and induce localized changes in the actin cytoskeleton [123]. In turn, this polarity directs Rho1p-dependent trafficking of vesicles containing raw material for bud growth to the site where budding will occur. Mutants in which all seven OSH genes are disrupted accumulate undocked secretory vesicles that would normally be bound for the bud site (directed by the Rab GTPase Sec4p) [122]. As in the case of Sec14-bypass, it is not yet clear how Osh proteins affect this vesicular transport process. They could directly modulate proteins need for vesicle formation in response to binding lipid ligands or may affect these processes indirectly by affecting the lipid composition of cellular compartments. At present, there is no direct evidence for a physical link between any Osh protein and the relevant signaling complexes and, though five of the seven OSH proteins are able to compensate for the cdc42ts defect, only Osh2p has been seen to accumulate near the bud site, where Cdc42p works.
Osh6p and Osh7p have also been implicated in vesicular trafficking complexes, though their role is still unclear. Both can bind PIPs through their ORDs, and both possess putative C-terminal coiled-coil domains which appear to mediate an interaction with the AAA ATPase Vps4p [94, 124]. Vps4p induces disassembly of the endosomal sorting complex ESCRT-III, one of the final steps in delivery of cargo proteins to the multivesicular body (MVB) [125] The interaction between Osh7p and Vps4p is disrupted by ergosterol and requires an intact ORD [124]. GFP-Osh6p colocalizes with endocytic compartments in vps4Δ cells, and a lack of ATPase activity leads to both Osh6p and Osh7p partially pelleting with a membrane fraction. Deletions of other members of the ESCRT-III complex did not affect Osh6p/7p membrane association, and Osh6p/7p are dispensable for endocytosis [124]. The meaning of this association is yet unknown, but there are several possibilities. First, it has been suggested that Osh6p/7p function as sterol transfer proteins and Vps4p catalyzes their dissociation from membranes [126]. Alternatively, Osh6p/7p may regulate the activity of Vps4p in a ligand-sensitive fashion, thereby integrating sensitivity to local lipid concentrations with control of endosomal sorting. A third possibility is that Osh6p/7p act in concert with Vps4p to promote sterol transport from the MVB or vacuole. Correct sorting of the sterol-binding NPC1 homologue Ncr1p requires Vps4p [127], leading to the proposal of an intriguing functional link between two putative sterol-transport families, OSH and NPC [126]. The exact nature of this relationship in yeast has yet to be revealed, but in mammalian systems, the Vps4p orthologue SKD1 associates with NPC1 under conditions of cholesterol depletion, promoting the ubiquitination and subsequent degradation of NPC1 [128]. Osh6p/7p may therefore act as lipid sensors that, in the presence of ergosterol or some other lipid, bind Vps4p and regulate its association with Ncr1p.
8. Concluding remarks
So long as yeast express a single OSH gene, all others are dispensable for survival. Yet it is increasingly apparent that most, if not all, Osh proteins possess a unique suite of structural characteristics: variability not just in primary sequence, but also in cellular function (as determined by genetic studies), the physiological ORD ligand, the specificity of its targeting domains, and so on. The seeming genetic redundancy of the Osh proteins might, perhaps, be based on a lingering, shared ancestral function based on lipid transport. At least two of the short Osh proteins (consisting primarily of the ORD) have been directly demonstrated to transport sterols in vitro. Since the ORD is highly conserved, it may be that the ORDs of all Osh family members can bind sterol ligands, albeit at widely diverging affinities, or requiring accessory proteins or other specific conditions. The elaborations seen in the extended Osh proteins (such as the PH domain or ankyrin repeats) may be stronger determinants of subcellular targeting and association with specific protein complexes, yet some remnant of an ancestral, shared ORD function may remain and be able to act in a more generalized capacity for lipid transfer. Since genetic screens may be complicated by redundancy in the context of genetic networks, strategies for teasing out the specific function of each Osh may therefore need to be based more on structural analyses: determining the protein and lipidic ligands of each domain, how these direct the subcellular localization of the Osh, and how the affinity for ligand or target organelle changes in response to external signals.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liscum L, Munn NJ. Intracellular cholesterol transport. Biochim Biophys Acta. 1999;1438:19–37. doi: 10.1016/s1388-1981(99)00043-8. [DOI] [PubMed] [Google Scholar]
- 2.Maxfield FR, Wustner D. Intracellular cholesterol transport. J Clin Invest. 2002;110:891–898. doi: 10.1172/JCI16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yeagle PL. The Membrane of Cells. Academic Press, Inc.; New York: 1993. [Google Scholar]
- 4.Incardona JP, Eaton S. Cholesterol in signal transduction. Curr Opin Cell Biol. 2000;12:193–203. doi: 10.1016/s0955-0674(99)00076-9. [DOI] [PubMed] [Google Scholar]
- 5.McIntosh TJ, Simon SA. Roles of bilayer material properties in function and distribution of membrane proteins. Annu Rev Biophys Biomol Struct. 2006;35:177–198. doi: 10.1146/annurev.biophys.35.040405.102022. [DOI] [PubMed] [Google Scholar]
- 6.Sturley SL, Patterson MC, Balch W, Liscum L. The pathophysiology and mechanisms of NP-C disease. Biochim Biophys Acta. 2004;1685:83–87. doi: 10.1016/j.bbalip.2004.08.014. [DOI] [PubMed] [Google Scholar]
- 7.Nofer JR, Remaley AT. Tangier disease: still more questions than answers. Cell Mol Life Sci. 2005;62:2150–2160. doi: 10.1007/s00018-005-5125-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orci L, Montesano R, Meda P, Malaisse-Lagae F, Brown D, Perrelet A, Vassalli P. Heterogeneous distribution of filipin--cholesterol complexes across the cisternae of the Golgi apparatus. Proc Natl Acad Sci U S A. 1981;78:293–297. doi: 10.1073/pnas.78.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 10.Coxey RA, Pentchev PG, Campbell G, Blanchette-Mackie EJ. Differential accumulation of cholesterol in Golgi compartments of normal and Niemann-Pick type C fibroblasts incubated with LDL: a cytochemical freeze-fracture study. J Lipid Res. 1993;34:1165–1176. [PubMed] [Google Scholar]
- 11.Holthuis JC, van Meer G, Huitema K. Lipid microdomains, lipid translocation and the organization of intracellular membrane transport (Review) Mol Membr Biol. 2003;20:231–241. [PubMed] [Google Scholar]
- 12.Brugger B, Sandhoff R, Wegehingel S, Gorgas K, Malsam J, Helms JB, Lehmann WD, Nickel W, Wieland FT. Evidence for segregation of sphingomyelin and cholesterol during formation of COPI-coated vesicles. J Cell Biol. 2000;151:507–518. doi: 10.1083/jcb.151.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maxfield FR, Menon AK. Intracellular sterol transport and distribution. Curr Opin Cell Biol. 2006;18:379–385. doi: 10.1016/j.ceb.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Chang TY, Chang CC, Ohgami N, Yamauchi Y. Cholesterol sensing, trafficking, and esterification. Annu Rev Cell Dev Biol. 2006;22:129–157. doi: 10.1146/annurev.cellbio.22.010305.104656. [DOI] [PubMed] [Google Scholar]
- 15.Soccio RE, Breslow JL. Intracellular Cholesterol Transport. Arterioscler Thromb Vasc Biol. 2004;4:1150–1160. doi: 10.1161/01.ATV.0000131264.66417.d5. [DOI] [PubMed] [Google Scholar]
- 16.Strauss JF, 3rd, Liu P, Christenson LK, Watari H. Sterols and intracellular vesicular trafficking: lessons from the study of NPC1. Steroids. 2002;67:947–951. doi: 10.1016/s0039-128x(02)00042-9. [DOI] [PubMed] [Google Scholar]
- 17.Mukherjee S, Maxfield FR. Lipid and cholesterol trafficking in NPC. Biochim Biophys Acta. 2004;1685:28–37. doi: 10.1016/j.bbalip.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 18.Sturley SL. Conservation of eukaryotic sterol homeostasis: new insights from studies in budding yeast. Biochim Biophys Acta. 2000;1529:155–163. doi: 10.1016/s1388-1981(00)00145-1. [DOI] [PubMed] [Google Scholar]
- 19.Henneberry AL, Sturley SL. Sterol homeostasis in the budding yeast, Saccharomyces cerevisiae. Semin Cell Dev Biol. 2005;16:155–161. doi: 10.1016/j.semcdb.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 21.Daum G, Lees ND, Bard M, Dickson R. Biochemistry, cell biology and molecular biology of lipids of Saccharomyces cerevisiae. Yeast. 1998;14:1471–1510. doi: 10.1002/(SICI)1097-0061(199812)14:16<1471::AID-YEA353>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 22.Edqvist J, Blomqvist K. Fusion and fission, the evolution of sterol carrier protein-2. J Mol Evol. 2006;62:292–306. doi: 10.1007/s00239-005-0086-3. [DOI] [PubMed] [Google Scholar]
- 23.Bagnat M, Keranen S, Shevchenko A, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proc Natl Acad Sci U S A. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu X, Bittman R, Duportail G, Heissler D, Vilcheze C, London E. Effect of the structure of natural sterols and sphingolipids on the formation of ordered sphingolipid/sterol domains (rafts). Comparison of cholesterol to plant, fungal, and disease-associated sterols and comparison of sphingomyelin, cerebrosides, and ceramide. J Biol Chem. 2001;276:33540–33546. doi: 10.1074/jbc.M104776200. [DOI] [PubMed] [Google Scholar]
- 25.Bard M, Woods RA, Barton DH, Corrie JE, Widdowson DA. Sterol mutants of Saccharomyces cerevisiae: chromatographic analyses. Lipids. 1977;12:645–654. doi: 10.1007/BF02533759. [DOI] [PubMed] [Google Scholar]
- 26.Gaber RF, Copple DM, Kennedy BK, Vidal M, Bard M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol Cell Biol. 1989;9:3447–3456. doi: 10.1128/mcb.9.8.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lees ND, Bard M, Kemple MD, Haak RA, Kleinhans FW. ESR determination of membrane order parameter in yeast sterol mutants. Biochim Biophys Acta. 1979;553:469–475. doi: 10.1016/0005-2736(79)90302-x. [DOI] [PubMed] [Google Scholar]
- 28.Emter R, Heese-Peck A, Kralli A. ERG6 and PDR5 regulate small lipophilic drug accumulation in yeast cells via distinct mechanisms. FEBS Lett. 2002;521:57–61. doi: 10.1016/s0014-5793(02)02818-1. [DOI] [PubMed] [Google Scholar]
- 29.Kleinhans FW, Lees ND, Bard M, Haak RA, Woods RA. ESR determinations of membrane permeability in a yeast sterol mutant. Chem Phys Lipids. 1979;23:143–154. doi: 10.1016/0009-3084(79)90042-2. [DOI] [PubMed] [Google Scholar]
- 30.Heese-Peck A, Pichler H, Zanolari B, Watanabe R, Daum G, Riezman H. Multiple functions of sterols in yeast endocytosis. Mol Biol Cell. 2002;13:2664–2680. doi: 10.1091/mbc.E02-04-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grossmann G, Opekarova M, Novakova L, Stolz J, Tanner W. Lipid raft-based membrane compartmentation of a plant transport protein expressed in Saccharomyces cerevisiae. Eukaryot Cell. 2006;5:945–953. doi: 10.1128/EC.00206-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Munn AL, Heese-Peck A, Stevenson BJ, Pichler H, Riezman H. Specific sterols required for the internalization step of endocytosis in yeast. Mol Biol Cell. 1999;10:3943–3957. doi: 10.1091/mbc.10.11.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Proszynski TJ, Klemm RW, Gravert M, Hsu PP, Gloor Y, Wagner J, Kozak K, Grabner H, Walzer K, Bagnat M, Simons K, Walch-Solimena C. A genome-wide visual screen reveals a role for sphingolipids and ergosterol in cell surface delivery in yeast. Proc Natl Acad Sci U S A. 2005;102:17981–17986. doi: 10.1073/pnas.0509107102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kato M, Wickner W. Ergosterol is required for the Sec18/ATP-dependent priming step of homotypic vacuole fusion. Embo J. 2001;20:4035–4040. doi: 10.1093/emboj/20.15.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lange Y, Matthies HJ. Transfer of cholesterol from its site of synthesis to the plasma membrane. J Biol Chem. 1984;259:14624–14630. [PubMed] [Google Scholar]
- 36.Kaplan MR, Simoni RD. Transport of cholesterol from the endoplasmic reticulum to the plasma membrane. J Cell Biol. 1985;101:446–453. doi: 10.1083/jcb.101.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urbani L, Simoni RD. Cholesterol and vesicular stomatitis virus G protein take separate routes from the endoplasmic reticulum to the plasma membrane. J Biol Chem. 1990;265:1919–1923. [PubMed] [Google Scholar]
- 38.Field FJ, Born E, Murthy S, Mathur SN. Transport of cholesterol from the endoplasmic reticulum to the plasma membrane is constitutive in CaCo-2 cells and differs from the transport of plasma membrane cholesterol to the endoplasmic reticulum. J Lipid Res. 1998;39:333–343. [PubMed] [Google Scholar]
- 39.Baumann NA, Sullivan DP, Ohvo-Rekila H, Simonot C, Pottekat A, Klaassen Z, Beh CT, Menon AK. Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via nonvesicular equilibration. Biochemistry. 2005;44:5816–5826. doi: 10.1021/bi048296z. [DOI] [PubMed] [Google Scholar]
- 40.Schnabl M, Daum G, Pichler H. Multiple lipid transport pathways to the plasma membrane in yeast. Biochim Biophys Acta. 2005;1687:130–140. doi: 10.1016/j.bbalip.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 41.Juschke C, Wachter A, Schwappach B, Seedorf M. SEC18/NSF-independent, protein-sorting pathway from the yeast cortical ER to the plasma membrane. J Cell Biol. 2005;169:613–622. doi: 10.1083/jcb.200503033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Athenstaedt K, Daum G. The life cycle of neutral lipids: synthesis, storage and degradation. Cell Mol Life Sci. 2006;63:1355–1369. doi: 10.1007/s00018-006-6016-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zinser E, Paltauf F, Daum G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J Bacteriol. 1993;175:2853–2858. doi: 10.1128/jb.175.10.2853-2858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Milla P, Athenstaedt K, Viola F, Oliaro-Bosso S, Kohlwein SD, Daum G, Balliano G. Yeast oxidosqualene cyclase (Erg7p) is a major component of lipid particles. J Biol Chem. 2002;277:2406–2412. doi: 10.1074/jbc.M104195200. [DOI] [PubMed] [Google Scholar]
- 45.Mullner H, Zweytick D, Leber R, Turnowsky F, Daum G. Targeting of proteins involved in sterol biosynthesis to lipid particles of the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2004;1663:9–13. doi: 10.1016/j.bbamem.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 46.Leber R, Landl K, Zinser E, Ahorn H, Spok A, Kohlwein SD, Turnowsky F, Daum G. Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol Biol Cell. 1998;9:375–386. doi: 10.1091/mbc.9.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mo C, Milla P, Athenstaedt K, Ott R, Balliano G, Daum G, Bard M. In yeast sterol biosynthesis the 3-keto reductase protein (Erg27p) is required for oxidosqualene cyclase (Erg7p) activity. Biochim Biophys Acta. 2003;1633:68–74. doi: 10.1016/s1388-1981(03)00088-x. [DOI] [PubMed] [Google Scholar]
- 48.Ott RG, Athenstaedt K, Hrastnik C, Leitner E, Bergler H, Daum G. Flux of sterol intermediates in a yeast strain deleted of the lanosterol C-14 demethylase Erg11p. Biochim Biophys Acta. 2005;1735:111–118. doi: 10.1016/j.bbalip.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Ness F, Achstetter T, Duport C, Karst F, Spagnoli R, Degryse E. Sterol uptake in Saccharomyces cerevisiae heme auxotrophic mutants is affected by ergosterol and oleate but not by palmitoleate or by sterol esterification. J Bacteriol. 1998;180:1913–1919. doi: 10.1128/jb.180.7.1913-1919.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis TL, Keesler GA, Fenner GP, Parks LW. Pleiotropic mutations in Saccharomyces cerevisiae affecting sterol uptake and metabolism. Yeast. 1988;4:93–106. doi: 10.1002/yea.320040203. [DOI] [PubMed] [Google Scholar]
- 51.Arthington-Skaggs BA, Crowell DN, Yang H, Sturley SL, Bard M. Positive and negative regulation of a sterol biosynthetic gene (ERG3) in the post-squalene portion of the yeast ergosterol pathway. FEBS Lett. 1996;392:161–165. doi: 10.1016/0014-5793(96)00807-1. [DOI] [PubMed] [Google Scholar]
- 52.Wilcox LJ, Balderes DA, Wharton B, Tinkelenberg AH, Rao G, Sturley SL. Transcriptional profiling identifies two members of the ATP-binding cassette transporter superfamily required for sterol uptake in yeast. J Biol Chem. 2002;277:32466–32472. doi: 10.1074/jbc.M204707200. [DOI] [PubMed] [Google Scholar]
- 53.Bourot S, Karst F. Isolation and characterization of the Saccharomyces cerevisiae SUT1 gene involved in sterol uptake. Gene. 1995;165:97–102. doi: 10.1016/0378-1119(95)00478-o. [DOI] [PubMed] [Google Scholar]
- 54.Alimardani P, Regnacq M, Moreau-Vauzelle C, Ferreira T, Rossignol T, Blondin B, Berges T. SUT1-promoted sterol uptake involves the ABC transporter Aus1 and the mannoprotein Dan1 whose synergistic action is sufficient for this process. Biochem J. 2004;381:195–202. doi: 10.1042/BJ20040297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li Y, Prinz WA. ABC-transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the ER. J Biol Chem. 2004;279:45226–45234. doi: 10.1074/jbc.M407600200. [DOI] [PubMed] [Google Scholar]
- 56.Kaminski WE, Piehler A, Wenzel JJ. ABC A-subfamily transporters: structure, function and disease. Biochim Biophys Acta. 2006;1762:510–524. doi: 10.1016/j.bbadis.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 57.Reiner S, Micolod D, Zellnig G, Schneiter R. A genomewide screen reveals a role of mitochondria in anaerobic uptake of sterols in yeast. Mol Biol Cell. 2006;17:90–103. doi: 10.1091/mbc.E05-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sesaki H, Jensen RE. Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion. J Biol Chem. 2004;279:28298–28303. doi: 10.1074/jbc.M401363200. [DOI] [PubMed] [Google Scholar]
- 59.Kishimoto T, Yamamoto T, Tanaka K. Defects in structural integrity of ergosterol and the Cdc50p-Drs2p putative phospholipid translocase cause accumulation of endocytic membranes, onto which actin patches are assembled in yeast. Mol Biol Cell. 2005;16:5592–5609. doi: 10.1091/mbc.E05-05-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heiniger HJ, Kandutsch AA, Chen HW. Depletion of L-cell sterol depresses endocytosis. Nature. 1976;263:515–517. doi: 10.1038/263515a0. [DOI] [PubMed] [Google Scholar]
- 61.Blanchette-Mackie EJ, Dwyer NK, Amende LM, Kruth HS, Butler JD, Sokol J, Comly ME, Vanier MT, August JT, Brady RO, et al. Type-C Niemann-Pick disease: low density lipoprotein uptake is associated with premature cholesterol accumulation in the Golgi complex and excessive cholesterol storage in lysosomes. Proc Natl Acad Sci U S A. 1988;85:8022–8026. doi: 10.1073/pnas.85.21.8022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sleat DE, Wiseman JA, El-Banna M, Price SM, Verot L, Shen MM, Tint GS, Vanier MT, Walkley SU, Lobel P. Genetic evidence for nonredundant functional cooperativity between NPC1 and NPC2 in lipid transport. Proc Natl Acad Sci U S A. 2004;101:5886–5891. doi: 10.1073/pnas.0308456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Malathi K, Higaki K, Tinkelenberg AH, Balderes DA, Almanzar-Paramio D, Wilcox LJ, Erdeniz N, Redican F, Padamsee M, Liu Y, Khan S, Alcantara F, Carstea ED, Morris JA, Sturley SL. Mutagenesis of the putative sterol-sensing domain of yeast Niemann Pick C-related protein reveals a primordial role in subcellular sphingolipid distribution. J Cell Biol. 2004;164:547–556. doi: 10.1083/jcb.200310046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Berger AC, Vanderford TH, Gernert KM, Nichols JW, Faundez V, Corbett AH. Saccharomyces cerevisiae Npc2p is a functionally conserved homologue of the human Niemann-Pick disease type C 2 protein, hNPC2. Eukaryot Cell. 2005;4:1851–1862. doi: 10.1128/EC.4.11.1851-1862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Berger AC, Hanson PK, Wylie Nichols J, Corbett AH. A yeast model system for functional analysis of the Niemann-Pick type C protein 1 homolog, Ncr1p. Traffic. 2005;6:907–917. doi: 10.1111/j.1600-0854.2005.00327.x. [DOI] [PubMed] [Google Scholar]
- 66.Cheruku SR, Xu Z, Dutia R, Lobel P, Storch J. Mechanism of cholesterol transfer from the Niemann-Pick type C2 protein to model membranes supports a role in lysosomal cholesterol transport. J Biol Chem. 2006;281:31594–31604. doi: 10.1074/jbc.M602765200. [DOI] [PubMed] [Google Scholar]
- 67.Ko DC, Binkley J, Sidow A, Scott MP. The integrity of a cholesterol-binding pocket in Niemann-Pick C2 protein is necessary to control lysosome cholesterol levels. Proc Natl Acad Sci U S A. 2003;100:2518–2525. doi: 10.1073/pnas.0530027100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alpy F, Stoeckel ME, Dierich A, Escola JM, Wendling C, Chenard MP, Vanier MT, Gruenberg J, Tomasetto C, Rio MC. The steroidogenic acute regulatory protein homolog MLN64, a late endosomal cholesterol-binding protein. J Biol Chem. 2001;276:4261–4269. doi: 10.1074/jbc.M006279200. [DOI] [PubMed] [Google Scholar]
- 69.Alpy F, Tomasetto C. Give lipids a START: the StAR-related lipid transfer (START) domain in mammals. J Cell Sci. 2005;118:2791–2801. doi: 10.1242/jcs.02485. [DOI] [PubMed] [Google Scholar]
- 70.Tsujishita Y, Hurley JH. Structure and lipid transport mechanism of a StAR-related domain. Nat Struct Biol. 2000;7:408–414. doi: 10.1038/75192. [DOI] [PubMed] [Google Scholar]
- 71.Kishida T, Kostetskii I, Zhang Z, Martinez F, Liu P, Walkley SU, Dwyer NK, Blanchette-Mackie EJ, Radice GL, Strauss JF., 3rd Targeted mutation of the MLN64 START domain causes only modest alterations in cellular sterol metabolism. J Biol Chem. 2004;279:19276–19285. doi: 10.1074/jbc.M400717200. [DOI] [PubMed] [Google Scholar]
- 72.Hanada K, Kumagai K, Yasuda S, Miura Y, Kawano M, Fukasawa M, Nishijima M. Molecular machinery for non-vesicular trafficking of ceramide. Nature. 2003;426:803–809. doi: 10.1038/nature02188. [DOI] [PubMed] [Google Scholar]
- 73.Jiang B, Brown JL, Sheraton J, Fortin N, Bussey H. A new family of yeast genes implicated in ergosterol synthesis is related to the human oxysterol binding protein. Yeast. 1994;10:341–353. doi: 10.1002/yea.320100307. [DOI] [PubMed] [Google Scholar]
- 74.Beh CT, Cool L, Phillips J, Rine J. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 2001;157:1117–1140. doi: 10.1093/genetics/157.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olkkonen VM, Lehto M. Oxysterols and oxysterol binding proteins: role in lipid metabolism and atherosclerosis. Ann Med. 2004;36:562–572. doi: 10.1080/07853890410018907. [DOI] [PubMed] [Google Scholar]
- 76.Olkkonen VM, Johansson M, Suchanek M, Yan D, Hynynen R, Ehnholm C, Jauhiainen M, Thiele C, Lehto M. The OSBP-related proteins (ORPs): global sterol sensors for co-ordination of cellular lipid metabolism, membrane trafficking and signalling processes? Biochem Soc Trans. 2006;34:389–391. doi: 10.1042/BST0340389. [DOI] [PubMed] [Google Scholar]
- 77.Wang PY, Weng J, Anderson RGW. OSBP Is a Cholesterol-Regulated Scaffolding Protein in Control of ERK1/2 Activation. Science. 2005;307:1472–1476. doi: 10.1126/science.1107710. [DOI] [PubMed] [Google Scholar]
- 78.Im Y, Raychaudhuri J,S, Prinz WA, Hurley JH. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 2005;437:154–158. doi: 10.1038/nature03923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raychaudhuri S, Im YJ, Hurley JH, Prinz WA. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein-related proteins and phosphoinositides. J Cell Biol. 2006;173:107–119. doi: 10.1083/jcb.200510084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sullivan DP, Ohvo-Rekila H, Baumann NA, Beh CT, Menon AK. Sterol trafficking between the endoplasmic reticulum and plasma membrane in yeast. Biochem Soc Trans. 2006;34:356–358. doi: 10.1042/BST0340356. [DOI] [PubMed] [Google Scholar]
- 81.Beh CT, Rine J. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J Cell Sci. 2004;117:2983–2996. doi: 10.1242/jcs.01157. [DOI] [PubMed] [Google Scholar]
- 82.Voelker DR. Bridging gaps in phospholipid transport. Trends Biochem Sci. 2005;30:396–404. doi: 10.1016/j.tibs.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 83.Audhya A, Emr SD. Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell. 2002;2:593–605. doi: 10.1016/s1534-5807(02)00168-5. [DOI] [PubMed] [Google Scholar]
- 84.Li X, Rivas MP, Fang M, Marchena J, Mehrotra B, Chaudhary A, Feng L, Prestwich GD, Bankaitis VA. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J Cell Biol. 2002;157:63–77. doi: 10.1083/jcb.200201037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perry RJ, Ridgway ND. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol Biol Cell. 2006;17:2604–2616. doi: 10.1091/mbc.E06-01-0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bankaitis VA, Phillips S, Yanagisawa L, Li X, Routt S, Xie Z. Phosphatidylinositol transfer protein function in the yeast Saccharomyces cerevisiae. Adv Enzyme Regul. 2005;45:155–170. doi: 10.1016/j.advenzreg.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 87.Park YU, Hwang O, Kim J. Two-hybrid cloning and characterization of OSH3, a yeast oxysterol-binding protein homolog. Biochem Biophys Res Commun. 2002;293:733–740. doi: 10.1016/S0006-291X(02)00288-7. [DOI] [PubMed] [Google Scholar]
- 88.Tinkelenberg AH, Liu Y, Alcantara F, Khan S, Guo Z, Bard M, Sturley SL. Mutations in yeast ARV1 alter intracellular sterol distribution and are complemented by human ARV1. J Biol Chem. 2000;275:40667–40670. doi: 10.1074/jbc.C000710200. [DOI] [PubMed] [Google Scholar]
- 89.Swain E, Baudry K, Stukey J, McDonough V, Germann M, Nickels JT., Jr. Sterol-dependent regulation of sphingolipid metabolism in Saccharomyces cerevisiae. J Biol Chem. 2002;277:26177–26184. doi: 10.1074/jbc.M204115200. [DOI] [PubMed] [Google Scholar]
- 90.Daum G, Tuller G, Nemec T, Hrastnik C, Balliano G, Cattel L, Milla P, Rocco F, Conzelmann A, Vionnet C, Kelly DE, Kelly S, Schweizer E, Schuller HJ, Hojad U, Greiner E, Finger K. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast. 1999;15:601–614. doi: 10.1002/(SICI)1097-0061(199905)15:7<601::AID-YEA390>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 91.Yano T, Inukai M, Isono F. Deletion of OSH3 gene confers resistance against ISP-1 in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2004;315:228–234. doi: 10.1016/j.bbrc.2004.01.039. [DOI] [PubMed] [Google Scholar]
- 92.Sano T, Kihara A, Kurotsu F, Iwaki S, Igarashi Y. Regulation of the sphingoid long-chain base kinase Lcb4p by ergosterol and heme: studies in phytosphingosine-resistant mutants. J Biol Chem. 2005;280:36674–36682. doi: 10.1074/jbc.M503147200. [DOI] [PubMed] [Google Scholar]
- 93.Levine TP, Munro S. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. 2001;12(6):1633–44. doi: 10.1091/mbc.12.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang P, Duan W, Munn AL, Yang H. Molecular characterization of Osh6p, an oxysterol binding protein homolog in the yeast Saccharomyces cerevisiae. FEBS J. 2005;272:4703–4715. doi: 10.1111/j.1742-4658.2005.04886.x. [DOI] [PubMed] [Google Scholar]
- 95.Levine TP, Munro S. The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr Biol. 1998;8:729–739. doi: 10.1016/s0960-9822(98)70296-9. [DOI] [PubMed] [Google Scholar]
- 96.Loewen CJ, Roy A, Levine TP. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO. 2003;22:2025–2035. doi: 10.1093/emboj/cdg201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lehto M, Olkkonen VM. The OSBP-related proteins: a novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signalling. Biochim Biophys Acta. 2003;1631:1–11. doi: 10.1016/s1388-1981(02)00364-5. [DOI] [PubMed] [Google Scholar]
- 98.Schmalix WA, Bandlow W. SWH1 from yeast encodes a candidate nuclear factor containing ankyrin repeats and showing homology to mammalian oxysterol-binding protein. Biochim Biophys Acta. 1994;1219:205–210. doi: 10.1016/0167-4781(94)90273-9. [DOI] [PubMed] [Google Scholar]
- 99.Anantharaman V, Aravind L. The GOLD domain, a novel protein module involved in Golgi function and secretion. Genome Biol. 2002;3:research0023. doi: 10.1186/gb-2002-3-5-research0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ridgway ND, Dawson PA, Ho YK, Brown MS, Goldstein JL. Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J Cell Biol. 1992;116:307–319. doi: 10.1083/jcb.116.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Levine T, Loewen C. Inter-organelle membrane contact sites: through a glass, darkly. Curr Opin Cell Biol. 2006;18:371–378. doi: 10.1016/j.ceb.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 102.Voeltz GK, Rolls MM, Rapoport TA. Structural organization of the endoplasmic reticulum. EMBO Rep. 2002;3:944–950. doi: 10.1093/embo-reports/kvf202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gaigg B, Simbeni R, Hrastnik C, Paltauf F, Daum G. Characterization of a microsomal subfraction associated with mitochondria of the yeast, Saccharomyces cerevisiae. Involvement in synthesis and import of phospholipids into mitochondria. Biochim Biophys Acta. 1995;1234:214–220. doi: 10.1016/0005-2736(94)00287-y. [DOI] [PubMed] [Google Scholar]
- 104.Pichler H, Gaigg B, Hrastnik C, Achleitner G, Kohlwein SD, Zellnig G, Perktold A, Daum G. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur J Biochem. 2001;268:2351–2361. doi: 10.1046/j.1432-1327.2001.02116.x. [DOI] [PubMed] [Google Scholar]
- 105.Achleitner G, Gaigg B, Krasser A, Kainersdorfer E, Kohlwein SD, Perktold A, Zellnig G, Daum G. Association between the endoplasmic reticulum and mitochondria of yeast facilitates interorganelle transport of phospholipids through membrane contact. Eur J Biochem. 1999;264:545–553. doi: 10.1046/j.1432-1327.1999.00658.x. [DOI] [PubMed] [Google Scholar]
- 106.Roberts P, Moshitch-Moshkovitz S, Kvam E, O'Toole E, Winey M, Goldfarb DS. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:129–141. doi: 10.1091/mbc.E02-08-0483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pan X, Roberts P, Chen Y, Kvam E, Shulga N, Huang K, Lemmon S, Goldfarb DS. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol Biol Cell. 2000;11:2445–2457. doi: 10.1091/mbc.11.7.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kvam E, Goldfarb DS. Nvj1p is the outer-nuclear-membrane receptor for oxysterol-binding protein homolog Osh1p in Saccharomyces cerevisiae. J Cell Sci. 2004;117:4959–4968. doi: 10.1242/jcs.01372. [DOI] [PubMed] [Google Scholar]
- 109.Kvam E, Goldfarb DS. Structure and function of nucleus-vacuole junctions: outer-nuclear-membrane targeting of Nvj1p and a role in tryptophan uptake. J Cell Sci. 2006;119:3622–3633. doi: 10.1242/jcs.03093. [DOI] [PubMed] [Google Scholar]
- 110.Umebayashi K, Nakano A. Ergosterol is required for targeting of tryptophan permease to the yeast plasma membrane. J Cell Biol. 2003;161:1117–1131. doi: 10.1083/jcb.200303088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fang M, Kearns BG, Gedvilaite A, Kagiwada S, Kearns M, Fung MK, Bankaitis VA. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO. 1996;15:6447–6459. [PMC free article] [PubMed] [Google Scholar]
- 112.Kearns BG, Alb JGJ, Bankaitis V. Phosphatidylinositol transfer proteins: the long and winding road to physiological function. Trends Cell Biol. 1998;8:276–282. doi: 10.1016/s0962-8924(98)01281-1. [DOI] [PubMed] [Google Scholar]
- 113.Routt SM, Bankaitis VA. Biological functions of phosphatidylinositol transfer proteins. Biochem Cell Biol. 2004;82:254–262. doi: 10.1139/o03-089. [DOI] [PubMed] [Google Scholar]
- 114.Xie Z, Fang M, Rivas MP, Faulkner AJ, Sternweis PC, Engebrecht JA, Bankaitis VA. Phospholipase D activity is required for suppression of yeast phosphatidylinositol transfer protein defects. Proc Natl Acad Sci U S A. 1989;95:12346–12351. doi: 10.1073/pnas.95.21.12346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sreenivas A, Patton-Vogt JL, Bruno V, Griac P, Henry SA. A role for phospholipase D (Pld1p) in growth, secretion, and regulation of membrane lipid synthesis in yeast. J Biol Chem. 1998;273:16635–16638. doi: 10.1074/jbc.273.27.16635. [DOI] [PubMed] [Google Scholar]
- 116.Liscovitch M, Chalifa V, Pertile P, Chen CS, Cantley LC. Novel function of phosphatidylinositol 4,5-bisphosphate as a cofactor for brain membrane phospholipase D. J Biol Chem. 1994;269:21403–21406. [PubMed] [Google Scholar]
- 117.Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J. Phospholipase D signaling is essential for meiosis. Proc Natl Acad Sci U S A. 1995;92:12151–12155. doi: 10.1073/pnas.92.26.12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sciorra VA, Rudge SA, Wang J, McLaughlin S, Engebrecht J, Morris AJ. Dual role for phosphoinositides in regulation of yeast and mammalian phospholipase D enzymes. J Cell Biol. 2002;159:1039–1049. doi: 10.1083/jcb.200205056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Coluccio A, Malzone M, Neiman AM. Genetic evidence of a role for membrane lipid composition in the regulation of soluble NEM-sensitive factor receptor function in Saccharomyces cerevisiae. Genetics. 2004;166:89–97. doi: 10.1534/genetics.166.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fairn GD, McMaster CR. The roles of the human lipid-binding proteins ORP9S and ORP10S in vesicular transport. Biochem Cell Biol. 2005;83:631–636. doi: 10.1139/o05-064. [DOI] [PubMed] [Google Scholar]
- 121.Fairn GD, McMaster CR. Identification and assessment of the role of a nominal phospholipid binding region of ORP1S (oxysterol-binding-protein-related protein 1 short) in the regulation of vesicular transport. Biochem J. 2005;387:889–896. doi: 10.1042/BJ20041915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kozminski KG, Alfaro G, Dighe S, Beh CT. Homologues of oxysterol-binding proteins affect Cdc42p- and Rho1p-mediated cell polarization in Saccharomyces cerevisiae. Traffic. 2006;7:1224–1242. doi: 10.1111/j.1600-0854.2006.00467.x. [DOI] [PubMed] [Google Scholar]
- 123.Pruyne D, Bretscher A. Polarization of cell growth in yeast. J Cell Sci. 2000;113(Pt 4):571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- 124.Wang P, Zhang Y, Li H, Chieu HK, Munn AL, Yang H. AAA ATPases regulate membrane association of yeast oxysterol binding proteins and sterol metabolism. EMBO. 2005;24:2989–2999. doi: 10.1038/sj.emboj.7600764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Babst M, Katzmann DJ, Snyder WB, Wendland B, Emr SD. Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell. 2002;3:283–289. doi: 10.1016/s1534-5807(02)00219-8. [DOI] [PubMed] [Google Scholar]
- 126.Yang H. Nonvesicular sterol transport: two protein families and a sterol sensor? Trends Cell Biol. 2006;16:427–432. doi: 10.1016/j.tcb.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 127.Zhang S, Ren J, Li H, Zhang Q, Armstrong JS, Munn AL, Yang H. Ncr1p, the yeast ortholog of mammalian Niemann Pick C1 protein, is dispensable for endocytic transport. Traffic. 2004;5:1017–1030. doi: 10.1111/j.1600-0854.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- 128.Ohsaki Y, Sugimoto Y, Suzuki M, Hosokawa H, Yoshimori T, Davies JP, Ioannou YA, Vanier MT, Ohno K, Ninomiya H. Cholesterol depletion facilitates ubiquitylation of NPC1 and its association with SKD1/Vps4. J Cell Sci. 2006;119:2643–2653. doi: 10.1242/jcs.02993. [DOI] [PubMed] [Google Scholar]


