Abstract
Tumor necrosis factor α (TNFα) is a potent immunomodulator and proinflammatory cytokine that has been implicated in the pathogenesis of autoimmune and infectious diseases. For example, plasma levels of TNFα are positively correlated with severity and mortality in malaria and leishmaniasis. We have previously described a polymorphism at −308 in the TNFα promoter and shown that the rare allele, TNF2, lies on the extended haplotype HLA-A1-B8-DR3-DQ2, which is associated with autoimmunity and high TNFα production. Homozygosity for TNF2 carries a sevenfold increased risk of death from cerebral malaria. Here we demonstrate, with reporter genes under the control of the two allelic TNF promoters, that TNF2 is a much stronger transcriptional activator than the common allele (TNF1) in a human B cell line. Footprint analysis using DNase I and B cell nuclear extract showed the generation of a hypersensitive site at −308 and an adjacent area of protection. There was no difference in affinity of the DNA-binding protein(s) between the two alleles. These results show that this polymorphism has direct effects on TNFα gene regulation and may be responsible for the association of TNF2 with high TNFα phenotype and more severe disease in infections such as malaria and leishmaniasis.
Keywords: genetics, major histocompatibility complex, cytokine, gene regulation, autoimmune diseases
Tumor necrosis factor α (TNFα) is a potent cytokine with a wide range of proinflammatory activities (1). It is classically produced by monocytes/macrophages, although other cell types, such as T and B cells, also produce significant amounts. The TNFA gene lies in the class III region of the major histocompatibility complex (MHC), ≈250 kilobases centromeric of the HLA-B locus and 850 kilobases telomeric of HLA-DR. In view of its biological effects and gene location it has been speculated that polymorphism within this locus might contribute to MHC associations with autoimmune and infectious diseases (2), particularly those in which TNFα has been implicated in initiating or sustaining the inflammatory response, such as rheumatoid arthritis (3), or where increasing blood levels have been shown to be predictive of poorer outcome, such as malaria (4). This has been supported by the association of specific MHC haplotypes with different TNFα phenotypes: DR3 and DR4 haplotypes produce higher levels of TNFα (5, 6) while DR2 haplotypes are associated with low production (5, 7), suggesting that functional polymorphism might exist within regions that regulate the TNFA gene.
Studies in mice have implicated the TNF locus with disease phenotype. Polymorphisms have been described in the promoter region (8), the first intron, and 3′ untranslated region (9) of TNFA in different strains of mice. Further, variation in the production of TNFα from macrophages has been shown to vary between strains (10). Several of these polymorphisms have been correlated with TNFα production and with susceptibility to, or severity of, several diseases. The (NZB × NZW)F1 mouse develops a severe autoimmune disease that is very similar to systemic lupus erythematosus (SLE) in humans. A restriction fragment length polymorphism in the TNFA gene is correlated with low production of TNFα and with the development of lupus nephritis (11), and replacement therapy with recombinant TNFα delays the onset of nephritis with increased survival rate (12). Correlation of TNFα polymorphism with production of TNFα mRNA and resistance to development of murine Toxoplasma gondii encephalitis has also been demonstrated in the BALB/c strain (13). A study of Th2 cell-mediated local inflammatory responses has shown that the TNF dependence of this phenomenon is related to H2D haplotypes and corresponding TNFα production phenotypes, suggesting, at least in mice, that differential expression of TNFα from distinct alleles may influence the nature of an immune response (14).
TNF has potent biological actions, and control of its production is tightly regulated, occurring both at the transcriptional and posttranscriptional levels (15). In response to lipopolysaccharide stimulation of macrophages, TNF transcription increases 3-fold, TNF mRNA increases 50- to 100-fold, and protein secretion increases by a factor of ≈10,000-fold (16). Sequences within the 1100-bp stretch of DNA between the 3′ end of the lymphotoxin alpha gene and the 5′ end of TNFA have been shown to be central to the control of transcription (17, 18).
Recently we and others have described two polymorphisms in the human TNFA promoter at −308 (19) and −238 (20), both involving the substitution of guanine by adenosine in the uncommon alleles. We showed that the rare allele at −308 (TNF2) is part of an extended MHC haplotype HLA-A1-B8-DR3-DQ2 (21), which is associated with high TNFα production (5, 6). Studies in large populations have indicated that carriage of TNF2 is associated with a worse outcome in cerebral malaria (22) and in leishmaniasis (23).
To test whether the −308 polymorphism has a functional significance, we have investigated its effects on transcription using reporter gene assays. Our results show that the TNF2 allele is a much more powerful transcriptional activator than the common allele. Although we can demonstrate specific binding of a nuclear protein and DNase I hypersensitivity at the polymorphic site, there was no obvious difference in affinity of the protein(s) for the two alleles. These results indicate that this polymorphism may have a direct effect on transcriptional activity and may underlie the association of the HLA-A1-B8-DR3 haplotype with high TNFα production and be directly responsible for the poorer outcome reported in malaria and leishmaniasis with carriage of the TNF2 allele.
MATERIALS AND METHODS
Generation and Cloning of TNFα Promoter Fragments.
A fragment of 691 bp (−585 to +106) of the TNFA gene was amplified by PCR using primers 5′-GCTTGTCCCTGCTACCCGC-3′ and 5′-GTCAGGGGATGTGGCGTCT-3′, and cycles as described (19). The fragments were cloned into the TA vector and used to transform Escherichia coli (strain INVF′α) (Invitrogen, United States Biochemical). Following selection and propagation, pure plasmid DNA was prepared by standard methods (24). The TNF promoter alleles were removed from the TA vector by restriction enzyme digestion with HindIII and XbaI (Promega) to allow for directional cloning into the pBLCAT3 expression vector (25). The sequences of the inserts were checked by the dideoxy chain termination method using Sequenase (United States Biochemical).
Transfection of Human B Cells.
Experiments were performed using the human Raji B cell line. Cells were cultured in 1× RPMI 1640 medium adjusted to pH 7.4 with 1 M NaOH, buffered with 7.5% (vol/vol) sodium bicarbonate, and supplemented with 5% (vol/vol) fetal calf serum (GIBCO/BRL), penicillin (100 units per ml), streptomycin (100 μg/ml), and glutamine (2 mM) (Northumbria Biologicals, Northumberland, England). Cultures were incubated at 37°C in a humidified 5% CO2/95% air atmosphere. Raji cells, which were maintained in rapid growth phase by change of medium and passaged 1:10 every 3 days, were centrifuged at 400 × g for 5 min. The cells were washed once with medium, centrifuged, and then resuspended at a concentration of 1 × 107 cells per ml of medium; 800 μl of this was used for each transfection. In each experiment three different plasmids were transfected: (i) pBLCAT3, (ii) TNF1-pBLCAT3, and (iii) TNF2-pBLCAT3. Each time 80 μg of DNA was used. Electroporation was performed with a single pulse from a gene pulser apparatus (Bio-Rad) with a capacitance extender unit at settings of 300 V and 960 μFd. Cells were incubated at room temperature for 10 min before and after electroporation. The cell suspension was added to 19.2 ml of medium and incubated for 24 hr. Each cell culture was then split in half and again resuspended in 20 ml of medium. Phorbol 12-myristate 13-acetate (PMA) (Sigma) was added to one culture from each duplicate to a final concentration of 50 ng/ml and the cultures incubated for a further 48 hr. The cells were then harvested by centrifugation, and the pellets were resuspended in 0.25 M Tris·HCl (pH 8.0) and stored at −70°C. The cell suspensions were subjected to three episodes of rapid freezing/thawing to obtain lysates.
Quantification and Normalization of Chloramphenicol Acetyltransferase (CAT) Expression.
Measurement of CAT protein production in transfected cells was performed using a commercially available enzyme-linked immunosorbant assay (Boehringer Mannheim). The lower detection limit was 100 pg/ml CAT. Quantification of protein concentrations in cellular extracts was determined using a modification of the micro-Lowry technique (protein assay kit; Sigma).
To determine transfection efficiencies, a dot-blotting procedure was followed (26) using a β-counter (Tri-Carb 260-DU, Packard). The probe used was isolated from pBLCAT3 by digesting the plasmid with EcoRI. The fragment, ≈1500 bp in length, spanning the CAT cDNA, was randomly labeled with [α-32P]dCTP [3000 Ci/mmol (1 Ci = 37 GBq); Amersham] using a T7 Quickprime kit according to the manufacturer’s instructions (Pharmacia).
Extraction of DNA-Binding Proteins from Raji Cells.
Nuclear proteins were extracted from Raji cells as described (27). Prior to use, the protein concentration was determined.
Generation of Radiolabeled TNFA Promoter Fragment.
A 119-bp fragment (−345 to −226) of the TNFα promoter was amplified by PCR from TNF1 and TNF2 homozygous individuals using primers 5′-CAAAAGAAATGGAGGCAAT-3′ and 5′-TCCTCCCTGCTCCGATTCCG-3′. The two allelic fragments were then cloned into the TA vector, and the sequences were confirmed as above. Probe was prepared by restriction digestion using NsiI and HindIII, [α-32P]dCTP end-labeled using the Klenow fragment (Promega), and then purified with a Sephadex G-50 NICK column (Pharmacia). The specific activity of the probe was measured with a β-counter.
Electrophoretic Mobility Shift Assay (EMSA).
Nuclear protein extract (10 μg) was incubated with 2 μg of poly(dI-dC) (Pharmacia) in 25 μl of buffer composed of 10 mM Tris·HCl (pH 7.5), 75 mM KCl, 5 mM MgCl2, 1 mM DTT, 1 mM EDTA, 12.5% glycerol, and 0.1% (vol/vol) Triton X-100 at room temperature for 30 min. After the addition of 2.5 ng of labeled probe and incubation for a further 30 min, 2.5 μl of loading buffer consisting of 250 mM Tris·HCl (pH 7.5), 0.2% bromophenol blue, 0.2% xylene cyanol, and 40% glycerol was added and the samples were electrophoresed in a 0.25 × TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3)/4% nondenaturing polyacrylamide gel. Visualization of bands was by autoradiography. Quantification of competitor DNA concentrations was by measurement of optical density at 260 nm. This was checked by comparing band intensities following agarose gel electrophoresis and ethidium bromide staining.
DNase I Footprint.
Initially 80 ng of Raji nuclear extract was incubated at 37°C for 20 min in 50 μl of buffer containing 25 mM Hepes (pH 7.8), 50 mM KCl, 0.05 mM EDTA, 0.5 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, 5% glycerol, and 100 ng poly(dI-dC). Labeled probe (40,000 cpm) was then added and incubated for a further 20 min. Digestion was performed at 0°C with 0.01 units of DNase I (Boehringer Mannheim) for 1 min following the addition of 1 mM CaCl2 and 5 mM MgCl2. The reactions were then terminated with 100 μl stop solution consisting of 0.375% (wt/vol) SDS, 15 mM EDTA, 100 mM NaCl, and 100 mM Tris·HCl (pH 7.6). The products were then incubated at 37°C for 15 min with 0.18 mg proteinase K and 10 μg of tRNA in a final volume of 163 μl, and phenol/chloroform extracted and ethanol precipitated. At the same time a Maxam–Gilbert guanidine ladder was generated as described (28). The fragments were separated on a 6% denaturing polyacrylamide gel. The gel was then dried and visualized by autoradiography.
RESULTS
Induction of CAT Protein by TNFA Promoter Fragments.
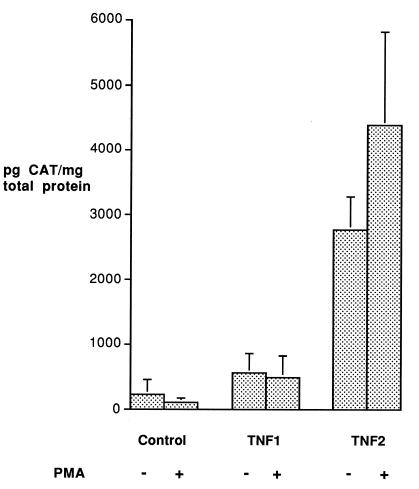
Raji cells were transfected with three plasmids: a negative control consisting of the pBLCAT3 vector alone and plasmids containing either of the two TNF allelic promoter fragments. The experiments were performed four times using DNA from different plasmid preparations. Efficiency of transfection was assessed by Southern blot analysis of the CAT gene and quantified by β-counting. Total protein was also measured in cellular lysates. The CAT protein concentration was corrected using these results to minimize differences due to transfection efficiency and cell numbers and was expressed in picograms of CAT protein per milligram of total protein. The results of this experiment are shown in Fig. 1. As expected, the negative control produced very low levels of CAT protein in both the unstimulated and PMA-stimulated cells. There was a significantly higher production of CAT from the TNF2-CAT plasmid compared with TNF1-CAT in both the unstimulated and stimulated cells. The TNF1-CAT plasmid showed no evidence of inducible production of CAT. Although there appeared to be some inducibility of the TNF2-CAT plasmid by PMA, this was not significant. This result was replicated four times with different batches of Raji cells.
Figure 1.
Induction of CAT protein from reporter gene constructs transfected into Raji cells. Raji cells (0.8 × 107) were transfected with the pBLCAT3 vector containing either no insert, which served as a negative control, or 691 bp of each allelic promoter. After 24 hr the cells were split in two, and one flask of each duplicate was stimulated with PMA (50 ng/ml). After a further 48 hr incubation the cells were harvested. Results have been corrected for transfection efficiency by Southern blot analysis of CAT DNA and also for total cell numbers by measuring total protein. The experiments were performed four times and means and standard errors are shown.
EMSA of TNFα Fragments.
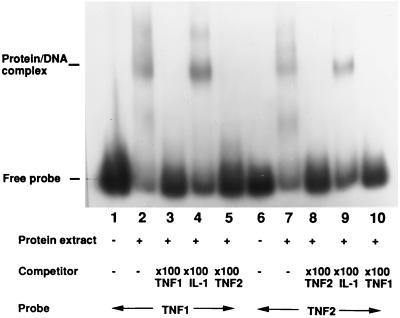
To investigate protein–DNA interactions in the vicinity of the polymorphism we performed EMSAs using extract from Raji cells and a 119-bp TNF fragment. The results of this experiment are shown in Fig. 2. Binding of at least one protein is demonstrated with both allelic fragments (lanes 2 and 7). The specific nature of binding is demonstrated by the disappearance of both DNA–protein complexes using competition with a 100-fold excess of unlabeled TNF1 probe (lanes 3 and 8) or TNF2 probe (lanes 5 and 10), but not when a similar-sized fragment of the interleukin 1A (IL-1A) promoter was used (lanes 4 and 9).
Figure 2.
Specific binding of the TNF promoter fragment by a nuclear protein. Nuclear extract (10 μg) from Raji cells was incubated with each of the two TNF promoter fragments with (lanes 3–5 and 8–10) or without (lanes 1, 2, 6, 7) 100-fold molar excess of unlabeled probe. The gel retardation complex is indicated by “Protein/DNA complex”. Lanes 4 and 9, competition with unlabeled IL-1 DNA fragment did not disrupt the complex.
DNase I Footprint of TNFα Promoter Region Fragment.
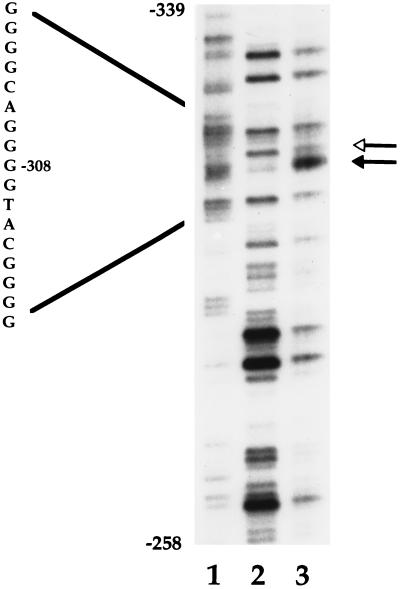
The exact site of DNA–protein interaction around the polymorphic site was defined in a footprint assay. A hypersensitive site was induced at −308 with an adjacent protected region (Fig. 3). No other evidence of interaction was seen.
Figure 3.
Detection of a hypersensitive site at −308. In vitro DNase I footprint analysis of TNF1 allele (coding strand) is shown. The unfilled arrowhead indicates an area of protection, and the filled arrowhead a DNase I hypersensitive site. Lanes: 1, Maxam–Gilbert guanidine ladder; 2, naked DNA control; 3, plus Raji cell nuclear extract.
Competitive EMSA.
To examine whether the difference in transcriptional activity was due to a difference in affinity of the two alleles for the binding protein, a competition EMSA was performed using labeled TNF1 and increasing excesses of cold oligonucleotides. There was no evidence of a major difference in affinity between the two alleles (Fig. 4).
Figure 4.
Competitive EMSA using allelic TNF promoter fragments and Raji nuclear extracts. Labeled TNF2 was used. Lanes: 1 and 7, no competitor; lanes 2–6 and 7–12, increasing excess of unlabeled TNF2 and TNF1, respectively, as competitor. No difference in affinity for nuclear proteins between TNF alleles was apparent.
DISCUSSION
Several features make it difficult to determine whether a particular DNA variant within the MHC is directly responsible for a disease association. This is the most polymorphic region of the genome and contains many genes that encode proteins that are involved in inflammatory and immune responses. Another important feature is the strong linkage disequilibrium between alleles across the MHC. Thus, for example, the haplotype HLA-A1-B8-DR3-DQ2 occurs much more frequently than the product of the individual allelic frequencies would suggest. Therefore, the association of MHC haplotypes with TNFα phenotypes might not be due to polymorphism within the TNF gene itself, but rather to variation in a linked gene that regulates expression of this cytokine. A possible example is the recent cloning of an IkB-like gene within 90 kilobases of the TNF locus (29). There are at least three NFkB consensus sequences within the TNFα promoter and these have each been demonstrated specifically to bind a B cell nuclear protein (17). Therefore, it is important to show a direct functional effect of any polymorphism, because the association with a disease may be entirely due to linkage disequilibrium with the true etiological gene.
Our results demonstrate that the polymorphism at −308 has a significant effect on transcriptional activity in reporter gene assays and that this could explain the association between the high TNFα phenotype and the DR3 haplotype. The molecular mechanism of this difference is not completely clear because there was no evidence of a major difference in affinity of the DNA-binding protein(s) to the two allelic forms of the TNFA promoter, at least in Raji cells. This should have shown up in the competition experiment if based on protein–DNA interactions. Perhaps as a result of difference in the DNA/chromatin structure at the polymorphic site, the interaction of transcription factors is enhanced leading to stronger transactivation of the TNFα gene. Our results demonstrating a lack of inducibility of both TNFα promoter allelic fragments in human B cells are in keeping with a previous study which demonstrated that the minimum promoter fragment required for PMA responsiveness in the 729–6 B cell line extended to −1105 bp with a high basal activity and poor inducibility compared with a T cell and monocytic cell line (30), and with the finding of high constitutive levels of TNFα mRNA in Raji cells (18). Although the polymorphic site lies in a consensus sequence for AP2 we found no evidence, using recombinant human protein in a gel retardation assay, of AP2 binding to the polymorphic site (data not shown). Interestingly, a homologous sequence in the TNFA promoter (−254 to −230) has been shown to bind a transcriptional repressor and not AP2 (31). It may be, therefore, that a novel protein binds to the polymorphic TNFA −308 site. A number of other groups have studied the effects of this polymorphism on gene expression. Results similar to ours have been found in one study in both Jurkat cells or U937 cells with evidence of binding of a protein only to the TNF2 allele (32). Two other groups have been unable to demonstrate a difference in transcriptional activity between the promoter alleles. This may be because of differences in cell types, stimulants, and reporter gene constructs used (33, 34). For example, we found that TNFA fragments shorter than those described here were inactive in reporter gene assays (data not shown). However, a recent study of TNFα production from peripheral blood mononuclear cells stimulated with anti-CD3 and anti-CD28 has shown a higher TNFα production phenotype in TNF2 carriers, and of two TNF haplotypes, differing only at −308, the TNF2 +ve haplotype produced significantly more TNFα (35). There is, therefore, evidence that the TNF2 genotype is associated with increased TNFα productions in vitro.
An interesting observation is the low incidence of SLE in areas of West Africa in which malaria is endemic (36), in contrast to the high incidence in Afro-American populations who are mostly of West African descent (37). Furthermore infection of the (NZB × NZW)F1 mouse with Plasmodium Berghei leads to protection from the spontaneous lupus-like disease (38). In humans with malaria, the highest levels of TNF are seen in fatal cases of cerebral malaria (4). A study of TNF genotypes in West African malaria patients has shown that homozygosity for TNF2 is associated with a 7-fold increased risk of death or severe neurological complications due to cerebral malaria (22). The TNF2 allele may therefore be responsible for the lower incidence of SLE in Africa as a consequence of endemic malaria causing higher levels of TNF production, while the absence of this stimulant of TNF production in the United States allows for the increased incidence of lupus seen in Afro-Americans. Despite the adverse effects of homozygosity in malaria, TNF2 is maintained at similar levels in West African and Northern European populations, suggesting that compensatory pressures in Africa exist to maintain the allele. Perhaps it has beneficial effects in other major infectious diseases, such as measles, meningococcal disease, leprosy, or tuberculosis. There may also be heterozygous advantages.
The associations of HLA-DR4 and -DR2 with high and low TNFα production, respectively, has not been explained. It seems reasonable to speculate that polymorphism, which may exist in other regulatory regions of the TNFA gene or in linked genes, plays a role in TNFα production. The most important region in the regulation of gene expression seems to be the 3′ untranslated region, which contains a tandem repeat of an octameric sequence, TTATTTAT (39). The corresponding UA-rich sequence in mRNA binds an inducible cytoplasmic factor (40), resulting in mRNA instability and translational blockade (41). It may be that DNA variants in this region are in linkage disequilibrium with HLA-DR4 or -DR2. Microsatellite alleles in the TNF locus can be used to subdivide DR4 haplotypes into high and low TNF phenotypes, and this has suggested that functionally important DNA variants do indeed exist within, or close to, this locus. TNFα is believed to be one of the key mediators of the chronic inflammatory response seen in rheumatoid arthritis (42). Severe forms of this disease are associated with homozygosity of the DRB1*0401 allele (43). It will therefore be interesting to see if this subtype of DR4 is associated with a high TNFα phenotype.
A study of the −238 TNFA promoter polymorphism has demonstrated strong linkage disequilibrium of the rare allele with two extended MHC haplotypes: B18-DR3 and B57-DR7. However, no genotype–phenotype association could be seen, and the high TNFα production of the B18-DR3 haplotype could not be further differentiated by typing this variant (35), suggesting that it does not have a direct effect on gene expression. Polymorphisms may of course also exist in the coding region of the gene, as has been found in the nearby lymphotoxin alpha gene (44). With regard to −308 TNFA2, however, there is evidence that it is overrepresented in diseases where TNFα levels are associated with poor prognosis. TNFα production from this allele has been reported to be higher in vitro, and now we have shown an increased transcriptional rate from the TNFA2 promoter in reporter gene assays. These observations begin to make a strong case that the −308 variation is of biological significance.
Polymorphisms within the human IL-1 gene cluster on chromosome 2 have been characterized, and associations with a number of autoimmune diseases have been described (45–47). An allele of a TaqI restriction fragment-length polymorphism in the IL-1β gene is associated with a high IL-1β production phenotype and psoriasis (F.S. di Giovine, personal communication). Different cytokine genotypes may exist in the population as a result of the selective pressure of infectious diseases. It may be that specific cytokine genotypes are beneficial in the eradication of infectious diseases, but by creating a “proinflammatory” phenotype, they predispose to chronic inflammatory diseases or to a more severe form of inflammatory disease with a worse clinical outcome, irrespective of whether the initial triggering event is an infectious agent, autoimmunity, or, indeed, any cause sufficient to stimulate an inflammatory response. This possibility is currently being tested in a wide range of human diseases.
Acknowledgments
We thank Gerald Crabtree, Francesco di Giovine, James Lorens, and Martin Nicklin for advice and discussion. This work was supported by an Arthritis and Rheumatism Council Copeman Travelling Fellowship (A.G.W.), a program grant from the Arthritis and Rheumatism Council (G.W.D.), a grant from Rhône-Poulenc Rorer, and a grant from the National Institute of Diabetes, Digestive, and Kidney Diseases (H.O.M.).
ABBREVIATIONS
- TNFα
tumor necrosis factor α
- TNFA
gene for TNFα
- MHC
major histocompatibility complex
- SLE
systemic lupus erythematosus
- CAT
chloramphenicol acetyltransferase
- EMSA
electrophoretic mobility-shift assay
- IL
interleukin
- PMA
phorbol 12-myristate 13-acetate
References
- 1.Vassalli P. Annu Rev Immunol. 1992;10:411–452. doi: 10.1146/annurev.iy.10.040192.002211. [DOI] [PubMed] [Google Scholar]
- 2.Jacob C O. Immunol Today. 1992;13:122–125. doi: 10.1016/0167-5699(92)90107-i. [DOI] [PubMed] [Google Scholar]
- 3.di Giovine F S, Nuki G, Duff G W. Ann Rheum Dis. 1988;47:768–772. doi: 10.1136/ard.47.9.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwiatkowski D, Hill A V S, Sambou I, Twumasi P, Castracane J, Manogue K R, Cerami A, Brewster D R, Greenwood B M. Lancet. 1990;336:1201–1204. doi: 10.1016/0140-6736(90)92827-5. [DOI] [PubMed] [Google Scholar]
- 5.Jacob C O, Fronek Z, Lewis G D, Koo M, Hansen J A, McDevitt H O. Proc Natl Acad Sci USA. 1990;87:1233–1237. doi: 10.1073/pnas.87.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abraham L J, French M A H, Dawkins R L. Clin Exp Immunol. 1993;92:14–18. doi: 10.1111/j.1365-2249.1993.tb05940.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bendtzen K, Morling N, Fomsgaard A, Svenson M, Jakobsen B, Ødum N, Svejgaard A. Scand J Immunol. 1988;28:599–606. doi: 10.1111/j.1365-3083.1988.tb01492.x. [DOI] [PubMed] [Google Scholar]
- 8.Jongeneel C V, Acha-Orbea H, Blankenstein T. J Exp Med. 1990;171:2141–2146. doi: 10.1084/jem.171.6.2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beutler B, Brown T. Gene. 1993;129:279–283. doi: 10.1016/0378-1119(93)90280-g. [DOI] [PubMed] [Google Scholar]
- 10.Jacob C O, Hwang F, Lewis G D, Stall A M. Cytokine. 1991;3:551–556. doi: 10.1016/1043-4666(91)90481-r. [DOI] [PubMed] [Google Scholar]
- 11.Jacob C O, McDevitt H O. Nature (London) 1988;331:356–357. doi: 10.1038/331356a0. [DOI] [PubMed] [Google Scholar]
- 12.Gordon C, Ranges G E, Greenspan J S, Wofsy D. Clin Immunol Immunopathol. 1989;52:421–434. doi: 10.1016/0090-1229(89)90157-8. [DOI] [PubMed] [Google Scholar]
- 13.Freund Y R, Sgarlato G, Jacob C O, Suzuki Y, Remington J S. J Exp Med. 1992;175:683–688. doi: 10.1084/jem.175.3.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Müller K M, Lisby S, Arrighi J F, Grau G E, Saurat J H, Hauser C. J Immunol. 1994;153:316–324. [PubMed] [Google Scholar]
- 15.Sariban E, Imamura K, Leubbers R, Kufe D. J Clin Invest. 1988;81:1506–1510. doi: 10.1172/JCI113482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beutler B, Cerami A. Annu Rev Immunol. 1989;7:625–655. doi: 10.1146/annurev.iy.07.040189.003205. [DOI] [PubMed] [Google Scholar]
- 17.Goldfeld A E, Doyle C, Maniatis T. Proc Natl Acad Sci USA. 1990;87:9769–9773. doi: 10.1073/pnas.87.24.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldfeld A E, Strominger J L, Doyle C. J Exp Med. 1991;174:73–81. doi: 10.1084/jem.174.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson A G, di Giovine F S, Blakemore A I F, Duff G W. Hum Mol Genet. 1992;1:353. doi: 10.1093/hmg/1.5.353. [DOI] [PubMed] [Google Scholar]
- 20.D’Alfonso S, Momigliano Richiardi P. Immunogenetics. 1994;39:150–154. doi: 10.1007/BF00188619. [DOI] [PubMed] [Google Scholar]
- 21.Wilson A G, de Vries N, Pociot F, di Giovine F S, van de Putte L B A, Duff G W. J Exp Med. 1993;177:557–560. doi: 10.1084/jem.177.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGuire W, Hill A V S, Allsopp C E M, Greenwood B M, Kwiatkowski D. Nature (London) 1994;371:508–511. doi: 10.1038/371508a0. [DOI] [PubMed] [Google Scholar]
- 23.Cabrera M, Shaw M A, Sharpes C, Williams H, Castes M, Convit J, Blackwell J M. J Exp Med. 1995;182:1259–1264. doi: 10.1084/jem.182.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 25.Lucklow B, Schütz G. Nucleic Acids Res. 1987;15:5490. doi: 10.1093/nar/15.13.5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abken H, Reifenrath B. Nucleic Acids Res. 1992;20:3527. doi: 10.1093/nar/20.13.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohlsson H, Edlund T. Cell. 1986;45:35–44. doi: 10.1016/0092-8674(86)90535-0. [DOI] [PubMed] [Google Scholar]
- 28.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J D, Smith J A, Struhl K. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D P, Seidman J G, Smith J A, Struhl K, editors. New York: Wiley; 1994. pp. 12.1–12.10. [Google Scholar]
- 29.Albertella M R, Campbell R D. Hum Mol Genet. 1994;3:793–799. doi: 10.1093/hmg/3.5.793. [DOI] [PubMed] [Google Scholar]
- 30.Rhoades K L, Golub S H, Economou J S. J Biol Chem. 1992;267:22102–22107. [PubMed] [Google Scholar]
- 31.Fong C L, Siddiqui A H, Mark D F. Nucleic Acids Res. 1994;22:1108–1114. doi: 10.1093/nar/22.6.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abraham L J, Kroeger K M. Eur Cytokine Netw. 1996;7:183. [Google Scholar]
- 33.Brinkman B M N, Zuijdgeest D, Kaijzel E L, Breedveld F C, Verweij C L. J Inflamm. 1996;46:32–41. [PubMed] [Google Scholar]
- 34.Stuber F, Udalova I A, Book M, Drutskaya L N, Kuprash D V, Turetskaya R L, Schade F U, Nedospasov S A. J Inflamm. 1996;46:42–50. [PubMed] [Google Scholar]
- 35.Bouma G, Crusius J B A, Oudkerk Pool M, Kolkman J J, Von Blomberg B M E, Kostense P J, Giphart M J, Schreuder G M T, Meuwissen S G M, Peña A S. Scand J Immunol. 1996;43:456–463. doi: 10.1046/j.1365-3083.1996.d01-65.x. [DOI] [PubMed] [Google Scholar]
- 36.Butcher G A. J R Soc Med. 1991;84:451–453. doi: 10.1177/014107689108400802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ballou S P, Kahn M, Kusher A. Arthritis Rheum. 1982;25:55–60. doi: 10.1002/art.1780250109. [DOI] [PubMed] [Google Scholar]
- 38.Greenwood B M, Herrick E M, Voller A. Nature (London) 1970;226:266–267. doi: 10.1038/226266a0. [DOI] [PubMed] [Google Scholar]
- 39.Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. Proc Natl Acad Sci USA. 1986;83:1670–1674. doi: 10.1073/pnas.83.6.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bohjanen P R, Petryniak B, June C H, Thompson C B, Lindsen T. Mol Cell Biol. 1991;11:3288–3295. doi: 10.1128/mcb.11.6.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kruys V, Marinx O, Shaw G, Deschamps J, Huez G. Science. 1989;245:852–855. doi: 10.1126/science.2672333. [DOI] [PubMed] [Google Scholar]
- 42.Elliott M J, Maini R N, Feldmann M, Long-Fox A, Charles P, Katsikis P, Brennan F M, Walker J, Bijl H, Ghrayeb J, Woody J N. Arthritis Rheum. 1993;36:1681–1690. doi: 10.1002/art.1780361206. [DOI] [PubMed] [Google Scholar]
- 43.Weyand C M, Xie C, Goronzy J J. J Clin Invest. 1992;89:2033–2039. doi: 10.1172/JCI115814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Messer G, Spengler U, Jung M C, Honold G, Blömer K, Pape G R, Riethmüller G, Weiss E H. J Exp Med. 1991;173:209–219. doi: 10.1084/jem.173.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blakemore A I F, Tarlow J K, Cork M J, Gordon C, Emery P, Duff G W. Arthritis Rheum. 1994;37:1380–1385. doi: 10.1002/art.1780370917. [DOI] [PubMed] [Google Scholar]
- 46.Tarlow J K, Clay F E, Cork M J, Blakemore A I F, McDonagh A J G, Messenger A G, Duff G W. J Invest Dermatol. 1994;103:387–389. doi: 10.1111/1523-1747.ep12395398. [DOI] [PubMed] [Google Scholar]
- 47.McDowell T L, Symons J A, Ploski R, Førre Ø, Duff G W. Arthritis Rheum. 1995;38:221–228. doi: 10.1002/art.1780380210. [DOI] [PubMed] [Google Scholar]