Abstract
Background
The lancelet Asymmetron inferum (subphylum Cephalochordata) was recently discovered on the ocean floor off the southwest coast of Japan at a depth of 229 m, in an anaerobic and sulfide-rich environment caused by decomposing bodies of the sperm whale Physeter macrocephalus. This deep sulfide-rich habitat of A. inferum is unique among the lancelets. The distinguishing adaptation of this species to such an extraordinary habitat can be considered in a phylogenetic framework. As the first step of reconstruction of the evolutionary processes in this species, we investigated its phylogenetic position based on 11 whole mitochondrial genome sequences including the newly determined ones of the whale-fall lancelet A. inferum and two coral-reef congeners.
Results
Our phylogenetic analyses showed that extant lancelets are clustered into two major clades, the Asymmetron clade and the Epigonichthys + Branchiostoma clade. A. inferum was in the former and placed in the sister group to A. lucayanum complex. The divergence time between A. inferum and A. lucayanum complex was estimated to be 115 Mya using the penalized likelihood (PL) method or 97 Mya using the nonparametric rate smoothing (NPRS) method (the middle Cretaceous). These are far older than the first appearance of large whales (the middle Eocene, 40 Mya). We also discovered that A. inferum mitogenome (mitochondrial genome) has been subjected to large-scale gene rearrangements, one feature of rearrangements being unique among the lancelets and two features shared with A. lucayanum complex.
Conclusion
Our study supports the monophyly of genus Asymmetron assumed on the basis of the morphological characters. Furthermore, the features of the A. inferum mitogenome expand our knowledge of variation within cephalochordate mitogenomes, adding a new case of transposition and inversion of the trnQ gene. Our divergence time estimation suggests that A. inferum remained a member of the Mesozoic and the early Cenozoic large vertebrate-fall communities before shifting to become a whale-fall specialist.
Background
The subphylum Cephalochordata (lancelets), one of the basal groups of living chordates [1,2], appears to have maintained its basic body plan for several hundred million years [3,4]. Lancelets are widely distributed in tropical and temperate shallow seas and consist of three genera (Branchiostoma, Epigonichthys, and Asymmetron) with more than 30 known living species [5-8]. Adults are benthic, inhabiting sandy and shell/sand bottoms of clear seawater, while larvae are planktonic in both inshore and offshore areas (ca. 1.5 to 4 months duration) [5,9-11]. For a long time, it has been believed that the lancelets prefer such aerobic conditions, whereas recently a new lancelet Asymmetron inferum was discovered in an anaerobic and sulfide-rich bottom [7]. This species is only distinguishable morphologically from congeners by the number of myomeres [7].
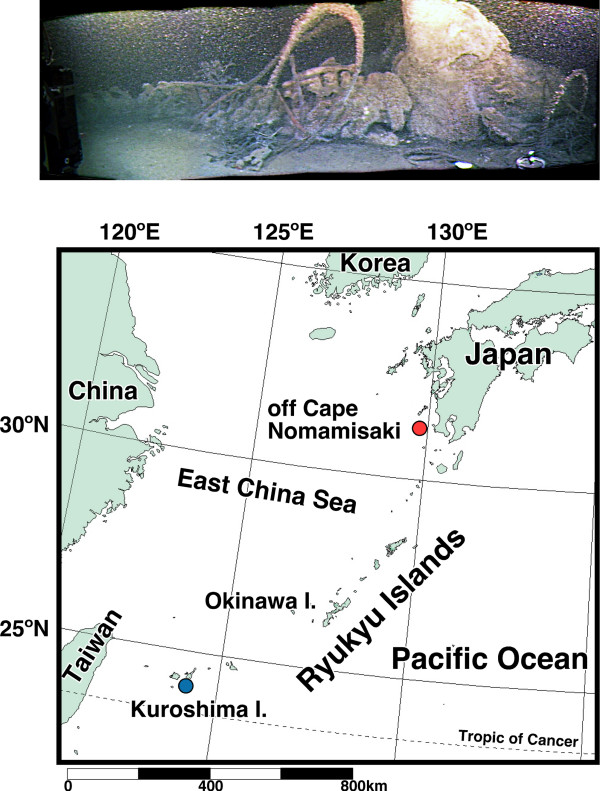
In July 2003, during the Hyper-Dolphin/Natsushima cruise of the Japan Marine Science and Technology Center (JAMSTEC), 26 specimens of A. inferum were collected from bottom sand subjacent to the decomposing bones of the sperm whale Physeter macrocephalus at a depth of 229 m, off Cape Nomamisaki, Kyushu Island, southwest Japan (Fig. 1). Twelve dead whales that had been stranded on the southwestern coast of Kyushu Island were submerged in February 2002, and then were observed to study the faunal composition and succession of the biological communities associated with whale-falls [12]. The submerged bodies of whales on the floor provided rich pickings that resulted in substantial amounts of organic material, lipids, and sulfides for dense biological community comprising of the mytillid mussels Adipicola pacifica and A. crypta [13,14] and the bone-eating marine worm Osedax japonicus [15]. A. inferum is also a member of this community [12].
Figure 1.
Submerged whale carcass (upper) and map of sampling sites of Asymmetron in Japan (lower). (Upper) Video still of skeletonized sperm whale carcass at a depth of 229 m in 2003. (Lower) Localities are color-coded: red, sampling site of A. inferum; blue, Asymmetron sp. A. The third examined species Asymmetron sp. C was collected in Bermuda, Atlantic Ocean (see [8], site not shown).
This deep (>200 m) and sulfide-rich habitat is unique to A. inferum in the lancelets. A sulfide-rich environment – including hydrogen sulfide – is generally toxic to macro-organisms. Therefore, the adaptation of this species to such a lethal environment is an intriguing phenomenon, but the phylogenetic framework for understanding its processes has not yet been established. Nohara et al. [16] conducted molecular phylogenetic analysis of the intra-relationships of some lancelets using whole mitochondrial genome (mitogenome) sequences, but A. inferum and two congeneric species of A. lucayanum complex [8] were not included. To know the phylogenetic position of the whale-fall lancelet A. inferum, we determined the whole mitogenome sequences from three species of the genus Asymmetron including this species (A. inferum, Asymmetron sp. A, and Asymmetron sp. C [=A. lucayanum sensu stricto] in A. lucayanum complex [8]) to compare with the published data for Nohara's A. lucayanum [16] (called here as Asymmetron sp. B in A. lucayanum complex [8]), Epigonichthys maldivensis, Branchiostoma belcheri, B. lanceolatum and B. floridae as well as three other deuterostomes as outgroups.
Results
Features of A. inferum mitogenome
The nucleotide sequences from the mitogenomes of the three Asymmetron species have been deposited in DDBJ/EMBL/GenBank under the accession numbers of AP009352 for A. inferum, AP009353 for Asymmetron sp. A, and AP009354 for Asymmetron sp. C (=A. lucayanum sensu stricto). The total lengths of the A. inferum, Asymmetron sp. A, and Asymmetron sp. C mitogenomes were 15,084, 15,050, and 15,100 bp, respectively. Mitogenomes of the mentioned species of Asymmetron, each contained 37 genes for large- and small-subunit ribosomal RNAs (rrnL and rrnS, respectively), 22 transfer RNAs (trnX; X is the standard single-letter amino acid code), and 13 proteins (ATP synthetase subunits 6 and 8 [atp6 and atp8], cytochrome c oxidase subunits I-III [cox1-3], NADH dehydrogenase subunits 1–6 and 4L [nad1-6 and 4L], and cytochrome b apoenzyme [cob]), as is generally the case with metazoan mitogenomes (Fig. 2, Tables 1 and 2). The mitogenome organization of Asymmetron sp. A and Asymmetron sp. C was identical to that of Asymmetron sp. B (= Nohara's A. lucayanum [16]), while that of A. inferum was unique among the lancelets.
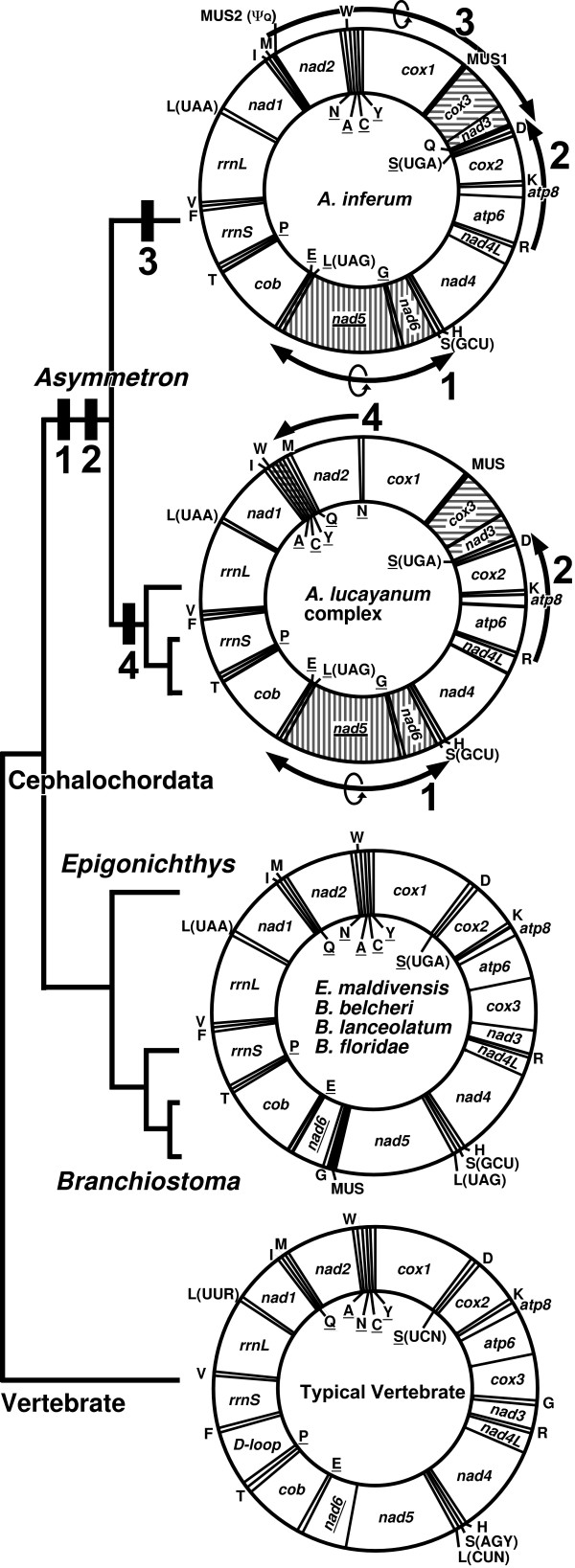
Figure 2.
Gene rearrangements found in the lancelets. Gene order rearrangement events of lancelet mitogenomes were mapped onto the phylogenetic tree. Bars (1–4) correspond to the unique gene order rearrangements as shown in right genome maps. Names of 13 mitochondrial protein genes, abbreviated as in text. Twenty-two tRNA genes, denoted by standard single letter amino acid code. MUS in the maps refers to major unassignable sequence in the mitogenome. Genes encoded on light strand of the mitogenome underlined.
Table 1.
Location of features in the mitogenome of Asymmetron inferum.
| Features | Position number | Size (bp) | Codon | Intergenic nucleotides | |||
| From | To | Start | Stop | anti-codon | |||
| cox1 | 1 | 1548 | 1548 | GTG | TAA | 45 | |
| cox3 | 1594 | 2381 | 788 | ATG | TA- | 0 | |
| nad3 | 2382 | 2735 | 354 | ATG | TAA | 8 | |
| trnQ | 2744 | 2812 | 69 | TTG | 3 | ||
| trnS(UGA) | 2816 | 2886 | 71 | TGA | 14 | ||
| trnD | 2901 | 2967 | 67 | GTC | 0 | ||
| cox2 | 2968 | 3658 | 691 | ATG | T-- | 0 | |
| trnK | 3659 | 3722 | 64 | TTT | 5 | ||
| atp8 | 3723 | 3896 | 174 | GTG | TAA | -7 | |
| atp6 | 3890 | 4574 | 685 | ATG | T-- | 0 | |
| trnR | 4575 | 4638 | 64 | TCG | 0 | ||
| nad4L | 4639 | 4913 | 275 | ATG | TA- | 0 | |
| nad4 | 4914 | 6272 | 1359 | ATG | TAA | 9 | |
| trnH | 6282 | 6346 | 65 | GTG | 0 | ||
| trnS(GCU) | 6347 | 6412 | 66 | GCT | 3 | ||
| nad6 | 6416 | 6916 | 501 | ATG | TAA | -15 | |
| trnG | 6902 | 6967 | 66 | TCC | 0 | ||
| nad5 | 6968 | 8759 | 1792 | GTG | T-- | 0 | |
| trnL(UAG) | 8760 | 8826 | 67 | TAG | 5 | ||
| trnE | 8832 | 8895 | 64 | TTC | 1 | ||
| cob | 8897 | 10039 | 1143 | ATG | TAA | 0 | |
| trnT | 10040 | 10104 | 65 | TGT | -1 | ||
| trnP | 10104 | 10168 | 65 | TGG | 0 | ||
| rrnS | 10169 | 11022 | 854 | 0 | |||
| trnF | 11023 | 11087 | 65 | GAA | 0 | ||
| trnV | 11088 | 11154 | 67 | TAC | 0 | ||
| rrnL | 11155 | 12514 | 1360 | 0 | |||
| trnL(UAA) | 12515 | 12584 | 70 | TAA | 0 | ||
| nad1 | 12585 | 13527 | 943 | ATG | T-- | 0 | |
| trnI | 13528 | 13593 | 66 | GAT | 1 | ||
| trnM | 13595 | 13661 | 67 | CAT | 48 | ||
| nad2 | 13710 | 14750 | 1041 | ATG | TAA | -8 | |
| trnN | 14743 | 14808 | 66 | GTT | 1 | ||
| trnW | 14810 | 14877 | 68 | TCA | 3 | ||
| trnA | 14881 | 14943 | 63 | TGC | 5 | ||
| trnC | 14949 | 15005 | 57 | GCA | 0 | ||
| trnY | 15006 | 15072 | 67 | GTA | 12 | ||
Genes encoded on light strand of the mitogenome underlined.
Table 2.
Location of features in the mitogenome of two species of Asymmetron lucayanum complex.
| A. lucayanum complex sp. A | A. lucayanum complex sp. C | |||||||||||||
| Position number | Size (bp) | Codon | anti-codon | Intergenic nucleotides | Position number | Size (bp) | Codon | anti-codon | Intergenic nucleotides | |||||
| Features | From | To | start | stop | From | To | start | stop | ||||||
| cox1 | 1 | 1548 | 1548 | GTG | TAA | 34 | 1 | 1548 | 1548 | GTG | TAA | 43 | ||
| cox3 | 1583 | 2371 | 789 | ATG | TAG | 15 | 1592 | 2380 | 789 | ATG | TAA | 13 | ||
| nad3 | 2387 | 2740 | 354 | ATG | TAG | 8 | 2394 | 2747 | 354 | ATG | TAG | 11 | ||
| trnS(UGA) | 2749 | 2819 | 71 | TGA | 11 | 2759 | 2829 | 71 | TGA | 14 | ||||
| trnD | 2831 | 2898 | 68 | GTC | 0 | 2844 | 2912 | 69 | GTC | 0 | ||||
| cox2 | 2899 | 3589 | 691 | ATG | T-- | 0 | 2913 | 3603 | 691 | ATG | T-- | 0 | ||
| trnK | 3590 | 3652 | 63 | TTT | 0 | 3604 | 3667 | 64 | TTT | 0 | ||||
| atp8 | 3653 | 3826 | 174 | GTG | TAA | -7 | 3668 | 3841 | 174 | GTG | TAA | -7 | ||
| atp6 | 3820 | 4504 | 685 | ATG | T-- | 0 | 3835 | 4519 | 685 | ATG | T-- | 0 | ||
| trnR | 4505 | 4568 | 64 | TCG | 0 | 4520 | 4583 | 64 | TCG | 0 | ||||
| nad4L | 4569 | 4843 | 275 | ATG | TA- | 0 | 4584 | 4858 | 275 | ATG | TA- | 0 | ||
| nad4 | 4844 | 6202 | 1359 | ATG | TAA | 1 | 4859 | 6217 | 1359 | ATG | TAA | 1 | ||
| trnH | 6204 | 6269 | 66 | GTG | 0 | 6219 | 6284 | 66 | GTG | 0 | ||||
| trnS(GCU) | 6270 | 6335 | 66 | GCT | 1 | 6285 | 6350 | 66 | GCT | 1 | ||||
| nad6 | 6337 | 6840 | 504 | ATG | TAA | -15 | 6352 | 6855 | 504 | ATG | TAA | -15 | ||
| trnG | 6826 | 6892 | 67 | TCC | 0 | 6841 | 6906 | 66 | TCC | 0 | ||||
| nad5 | 6893 | 8681 | 1789 | ATG | T-- | 0 | 6907 | 8695 | 1789 | GTG | T-- | 0 | ||
| trnL(UAG) | 8682 | 8749 | 68 | TAG | 6 | 8696 | 8762 | 67 | TAG | 9 | ||||
| trnE | 8756 | 8819 | 64 | TTC | 5 | 8772 | 8836 | 65 | TTC | 4 | ||||
| cob | 8825 | 9967 | 1143 | ATG | TAA | 0 | 8841 | 9983 | 1143 | ATG | TAG | 0 | ||
| trnT | 9968 | 10037 | 70 | TGT | 0 | 9984 | 10053 | 70 | TGT | 0 | ||||
| trnP | 10038 | 10101 | 64 | TGG | 0 | 10054 | 10116 | 63 | TGG | 0 | ||||
| rrnS | 10102 | 10943 | 842 | 0 | 10117 | 10964 | 848 | 0 | ||||||
| trnF | 10944 | 11006 | 63 | GAA | 0 | 10965 | 11027 | 63 | GAA | 0 | ||||
| trnV | 11007 | 11073 | 67 | TAC | 0 | 11028 | 11094 | 67 | TAC | 0 | ||||
| rrnL | 11074 | 12437 | 1364 | 0 | 11095 | 12460 | 1366 | 0 | ||||||
| trnL(UAA) | 12438 | 12506 | 69 | TAA | 0 | 12461 | 12530 | 70 | TAA | 0 | ||||
| nad1 | 12507 | 13449 | 943 | GTG | T-- | 0 | 12531 | 13473 | 943 | ATG | T-- | 1 | ||
| trnI | 13450 | 13515 | 66 | GAT | 15 | 13475 | 13540 | 66 | GAT | 20 | ||||
| trnW | 13531 | 13598 | 68 | TCA | 9 | 13561 | 13629 | 69 | TCA | 3 | ||||
| trnA | 13608 | 13670 | 63 | TGC | 3 | 13633 | 13695 | 63 | TGC | 2 | ||||
| trnC | 13674 | 13727 | 54 | GCA | 0 | 13698 | 13752 | 55 | GCA | 0 | ||||
| trnY | 13728 | 13792 | 65 | GTA | 12 | 13753 | 13818 | 66 | GTA | 42 | ||||
| trnM | 13805 | 13871 | 67 | CAT | -1 | 13861 | 13927 | 67 | CAT | -1 | ||||
| trnQ | 13871 | 13939 | 69 | TTG | 2 | 13927 | 13995 | 69 | TTG | 1 | ||||
| nad2 | 13942 | 14982 | 1041 | ATG | TAA | -8 | 13997 | 15037 | 1041 | ATG | TAA | -8 | ||
| trnN | 14975 | 15041 | 67 | GTT | 9 | 15030 | 15095 | 66 | GTT | 5 | ||||
Genes encoded on light strand of the mitogenome underlined.
The mitogenome of A. inferum contained 13 protein-coding genes, of which one reading-frame overlapped on the same strand (atp8 and atp6 shared 7 nucleotides). Ten protein-coding genes (atp6, cob, cox2, cox3, nad1-4, nad4L, and nad6) started with ATG, the other protein genes (atp8, cox1, and nad5) with GTG (Table 1). Stop codons of protein-coding genes were TAA in the atp8, cob, cox1, nad2-4, and nad6; TA in the cox3, and nad4L; and T in the atp6, cox2, nad1, and nad5. For those genes with an incomplete stop codon, the transcripts would be modified to form the complete termination signal UAA by polyadenylation after cleavage of the polycistronic RNA, as demonstrated for other metazoan mitogenomes [17]. There were 22 tRNA genes, which were clustered or individually scattered in the genome. The tRNA genes ranged in size from 57 to 71 nucleotides, large enough for the encoded tRNAs to fold into the cloverleaf secondary structure characteristic of tRNAs. The trnS(gcu) and trnC lacked the DHU arm, like Branchiostoma floridae [18]. The ribosomal genes (rrnS and rrnL) of A. inferum were 854 bp and 1360 bp, respectively. They were located, as in other lancelets, between the trnP and trnL(uaa) genes, being separated by the trnF and trnV genes. There were two major unassignable regions (MUS) longer than 40 bp in the A. inferum mitogenome. One of the regions (MUS1, 45 bp) was located at the identical position to MUS in the mitogenomes of the A. lucayanum complex (between the cox1 and cox3 genes) [16]. The other region (MUS2, 48 bp), located between the trnM and nad2 genes, was unique among the lancelet mitogenomes.
Phylogenetic relationships
Partitioned Bayesian inference (BI) phylogenetic analysis of the 11 mitogenomes from the concatenated nucleotide sequences from 13 protein-coding genes, 22 tRNA genes, plus 2 rRNA genes (dataset #1) under the general time reversible model with gamma correction and invariable-site assumption (GTR + I + Γ) [19] yielded a topology with resolution of the branching pattern among lancelets. All nodes were supported by higher Bayesian posterior probabilities (100%). BI analysis using dataset #2 (triplets converted amino acid sequences) under mtREV + I + Γ [20] (for protein-coding genes) and GTR + I + Γ (for tRNA and rRNA genes) models produced the same tree topology (Fig. 3). All nodes were supported by higher Bayesian posterior probabilities (100%). Maximum-likelihood (ML) analysis using dataset #1 under the transversional substitution model with gamma correction and invariable-site assumption (TVM + I + Γ) produced exactly the same tree topology as that found in the BI analyses (figure not shown). Most nodes were supported by high bootstrap probabilities (>98%), with the exception of a clade comprising B. lanceolatum and B. floridae (77%). Heuristic maximum parsimony (MP) analysis of the dataset #1 also produced the identical tree topology as in the above analyses (figure not shown). The MP analysis yielded the single most parsimonious tree, with a length of 23,178 steps (consistency index [CI], 0.605; retention index [RI], 0.455; and rescaled consistency index [RC], 0.275). Most nodes were supported by high bootstrap values (>92%), with the exception of the same clade (B. lanceolatum + B. floridae) as in ML analysis (56%).
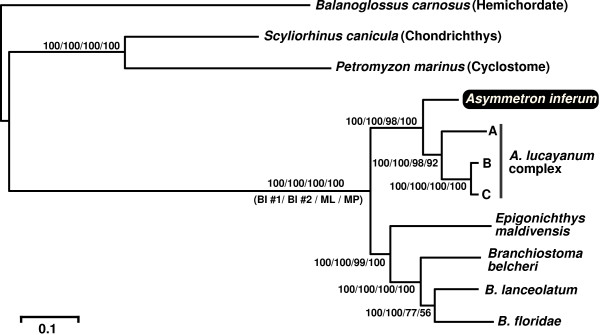
Figure 3.
Phylogenetic relationships of the lancelets. Bayesian inference (BI) tree for eight lancelets with three outgroups (one cyclostome, one chondrichthys, and one hemichordate) based on whole mitogenome sequences (dataset #2). Numbers on branches refer to BI posterior probabilities (dataset #1 and #2), ML bootstraps, and MP bootstraps, respectively.
Divergence time estimation
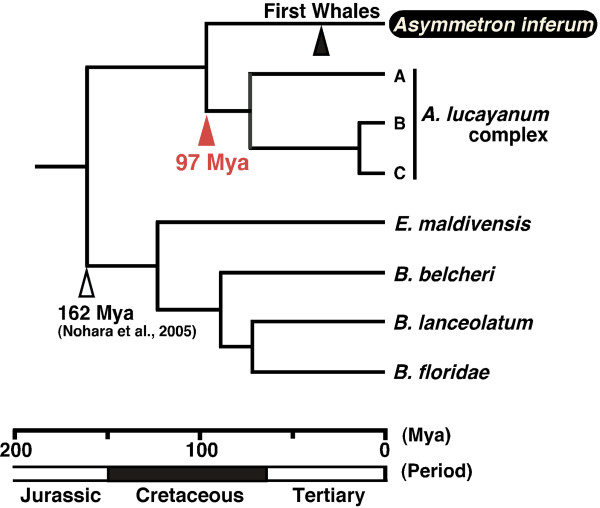
As a high rate of heterogeneity among lineages of lancelets was observed by the two-cluster test [21], we used two different molecular dating methods, the penalized likelihood (PL) [22]) based on the BI tree (dataset #2) and the nonparametric rate smoothing (NPRS) [23] based on the ML tree (dataset #1). The divergence time between A. inferum and A. lucayanum complex was estimated to be 115 Mya using PL method (Fig. 4) or 97 Mya using NPRS method (Fig. 5).
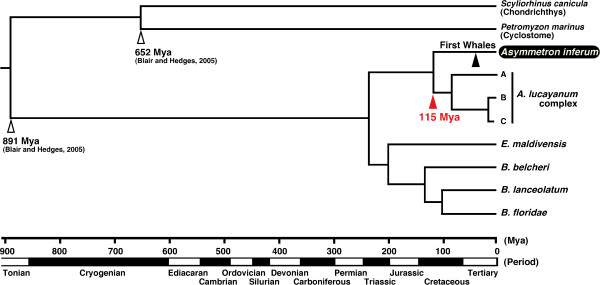
Figure 4.
Divergence time estimation of lancelets based on penalized likelihood (PL) method. Time tree from semiparametric rate smoothing (penalized likelihood, PL) based on the BI tree (Fig. 3). Scale bar shows time scale resulting from calibration using the divergence time between Cephalochordata and Vertebrata (+ Urochordata) (891 Mya) and between Agnatha (Cyclostome) and Gnathostomes (Chondrichthys) (652 Mya) [1]. Arrows indicate calibration point (open head), divergence time between A. inferum and A. lucayanum complex (red solid head), and first appearance of large whales (black solid head).
Figure 5.
Divergence time estimation of lancelets based on nonparametric rate smoothing (NPRS) method. Ultrametric tree based on nonparametric rate smoothing (NPRS) analysis of the ML tree. Scale bar shows time scale resulting from calibration using the divergence time between Asymmetron and the other genera (162 Mya) [16]. Arrows indicate calibration points (open head), divergence time between A. inferum and A. lucayanum complex (red solid head), and first appearance of large whales (black solid head).
Discussion
Phylogenetic position of A. inferum
Our phylogenetic analyses of mitogenome sequences for eight lancelets using Bayesian inference (BI), maximum likelihood (ML), and maximum parsimony (MP) methods show that extant lancelets are clustered into two major clades: the Asymmetron clade and the Epigonichthys + Branchiostoma clade (Fig. 3). This result is consistent with the findings of Nohara et al. [16]. The topology of the resulting trees implies that the asymmetrical arrangement of gonads seen paraphyletically in the genus Asymmetron and Epigonichthys can be regarded as a plesiomorphic feature, supporting the hypothesis that the gonadal symmetry in the genus Branchiostoma was derived from the asymmetric Epigonichthys-like ancestor [16]. Nishikawa [7] recovered the genus Asymmetron as a valid genus distinct from another valid genus Epigonichthys based on four morphological characters as follows: the elongated urostyloid process, marked metapleuran asymmetry, intercirral membrane with abrupt height change between lateral and ventral ones, and posterior shift of the cirral skeletal ring [24] with its anterodorsal extremity located at (or sometimes behind) the fourth myomere. The present molecular phylogeny shows that these morphological characters of the Asymmetron species are phylogenetically informative, supporting his claim. Although the A. lucayanum complex includes deep divergence (p-distance = 19% in cox1 of mitogenome [8]), A. inferum is placed on the sister group to this species complex, not nested within it (Fig. 3). This result indicates that the whale-fall lancelet A. inferum was diverged from the circumtropical lancelet A. lucayanum complex before the ancient separation in this complex [8].
Gene rearrangement of A. inferum
We discovered that the A. inferum mitogenome has been subjected to large-scale gene rearrangements. To elucidate the relative timing of these gene rearrangements, we mapped gene orders of the lancelet mitogenome onto the phylogenetic tree (Fig. 2). Gene orders of Epigonichthys and Branchiostoma are similar to that of typical vertebrate mitogenomes [25], with the exception of slight differences in the location of four tRNA genes [18]. Therefore, it is reasonable to assume that the ancestral lancelet gene order is almost the same as that of the Epigonichthys and Branchiostoma. The A. inferum mitogenome has three novel gene positions: (1) an inversion extending from the trnL(uag) to nad6 genes; (2) transposition of the cox3 and nad3 genes from between the atp6 and the trnR genes to between the cox1 and trnS(uga) genes; and (3) an inversion of the trnQ gene and its transposition from between the trnM and nad2 genes to between the nad3 and the trnS(uga) genes. The gene order of this mitogenome can be parsimoniously explained by the following steps: events (1) and (2) occurred in the ancestor of all Asymmetron species, event (3) subsequently occurred in the lineage of A. inferum, and event (4) (see [16]) occurred in the ancestor of A. lucayanum complex (Fig. 2). In other words, novel gene positions (1) and (2) represent molecular synapomorphies shared among the Asymmetron species, and novel gene position (4) is the synapomorphy of A. lucayanum species complex.
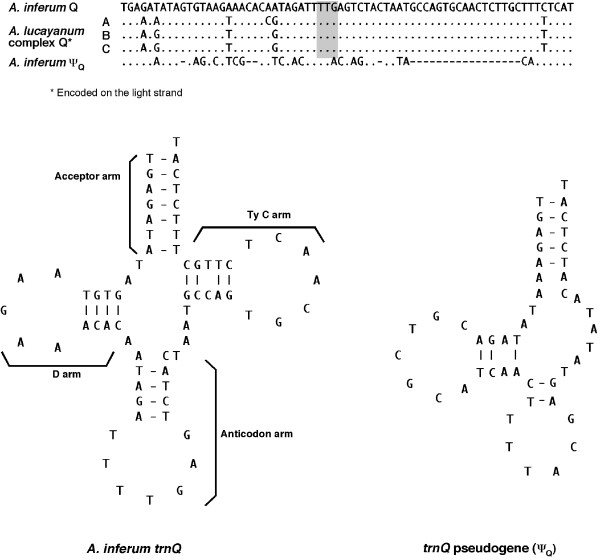
Gene position (3) and existence of MUS2 are unique to A. inferum. MUS2, located at the identical position of trnQ in the mitogenomes of A. lucayanum complex, appears to be a trnQ pseudogene because of its sequence similarity to the acceptor and anticodon arm regions of trnQ of A. inferum and complement sequences of that of A. lucayanum complex (Fig. 6). Therefore, gene arrangement pattern (3) may have resulted from two events as follows: the inversion of trnQ gene occurred at the original position by recombination of the mitogenome as a possible cause for pattern (1) [16], followed by transposition, involving a tandem genomic duplication and subsequent random deletions of the duplicated part, as invoked for many cases of gene rearrangement in animal mitogenomes ([25] and references therein).
Figure 6.
Aligned sequences (upper) and potential secondary structures (lower) of trnQ gene and putative trnQ pseudogene. (Upper) Aligned sequences of the four trnQ genes (Q) of four species Asymmetron and a putative trnQ pseudogene (ψQ) from A. inferum. Dots indicate sequence identity with the first sequences (trnQ gene of A. inferum), and dashes indicate alignment gaps. Sequence in grey box indicates anticodon. (Lower) Potential secondary structures of trnQ gene and putative trnQ pseudogene (ψQ) in A. inferum.
When and how did A. inferum become a whale-fall specialist?
A. inferum is considered as whale-fall specialist. This species has been found only in the whale-fall community at the “sulfophilic stage” (fueled by anaerobic breakdown of bone lipids) and has never been found elsewhere including shallow waters, wood-falls, cold seeps, or hydrothermal vents [7,15]. Therefore, it is natural to suppose that this lancelet may make use of the lipid-and-sulfide-rich or the organic material as a food source like other community members [26]. So far as the gross anatomy is concerned, there are no significant differences between A. inferum and its shallow-water congeners [7], which indicates that A. inferum may be a filter-feeder like its congeners. The mechanism of feeding of this whale-fall lancelet still remains as an open question.
When did A. inferum adapt to sulfide-rich environment? To examine the timing of A. inferum lineage, we estimated the divergence time between A. inferum and the other species of Asymmetron inhabiting coral reefs. With a slight difference between two resultant estimations of 115 Mya (Fig. 4) and 97 Mya (Fig. 5) (the middle Cretaceous), these appear far older than the first appearance of large whales, represented by the inshore archeocetacean Basilosaulus (40 Mya, the middle Eocene) [27]. Therefore, an A. inferum-like ancestor seems to have become a member of the whale-fall community after more than 57 million years of the emergence of A. inferum lineage (Figs. 4 and 5). Prior to the appearance of large whales, there were the Cenozoic or Mesozoic large fishes (e.g., the giant fossil shark Carcharocles, 10–20 m long; the ichthyodectid Xiphactinus, 4.2 m long) and the Mesozoic reptiles (e.g. ichthyosaurus [<16 m long] and plesiosaurus [<14 m long]) in the sea [4]. These large fishes and reptiles in the Mesozoic may have played an ecological role similar to the whales on the sea floor, as suggested by Martill et al. [28] and Hogler [29]. A. inferum may have been a member of these large vertebrate-fall communities before becoming a whale-fall specialist.
Another whale-fall specialist, the bone-eating marine worm Osedax also shows ancient divergence time (> 100 Mya) from its sister group (vestimentiferans + Sclerolinum) (inferred from [30]); however, the divergence time within Osedax (<42 Mya) [15,30,31] coincides with the first appearance of the large whales, based on the molecular clock. In the whale-fall bathymodiolin bivalves of the genera Adipicola and Idas, the divergence time (<30 Mya, the middle Oligocene) is synchronous with the major radiation of unequivocally oceanic whales [32,33], based on molecular age estimation (inferred from [26,34]) and the fossil records [35]. This synchronicity suggests that high sulfide-rich habitats derived from large whales with oil-rich bones [36] may play an important role in the divergences of Osedax, Adipicola, and Idas. Their tolerance and utilization of sulfide may have already been developed at the time of their invasion into whale-fall communities, because their sister groups are chemosymbiotic [26,30,34], provided actually or potentially with these abilities.
Unlike these whale-fall specialists, the A. inferum-like ancestor might have had a very low sulfide tolerance, because its sister group, A. lucayanum complex, and all the remaining lancelets inhabit aerobic environments [37]. This does not necessarily deny our hypothesis that A. inferum lineage may have started its life-mode as a vertebrate-fall specialist in the Mesozoic, because, unlike modern whale-falls with oil-rich bones, the Mesozoic large vertebrate-falls may possibly have lacked sulfophilic stages during decomposition [35,36], resulting in a low concentration of sulfide. Subsequently, the A. inferum lineage may have come to gain the sulfide tolerance to become a whale-fall specialist.
Conclusion
The trees based on the whole mitogenome sequences show that the whale-fall specialist A. inferum is the sister group to the coral reef lancelet A. lucayanum complex, supporting the monophyly of the genus Asymmetron, assumed on the basis of the morphological characters. Furthermore, the features of the A. inferum mitogenome expand our knowledge of variation within cephalochordate mitogenomes, adding new cases of inversion and transposition of the trnQ gene. Novel gene orders in Asymmetron were found to be useful for phylogenetic inference.
The divergence time between A. inferum and its sister group A. lucayanum complex is estimated to be 115 Mya or 97 Mya (the middle Cretaceous), which is older than the first appearance of large whales (40 Mya, the middle Eocene). Our results raise the possibility that A. inferum lineage was initially a member of the Mesozoic and the early Cenozoic large vertebrate-fall communities, and subsequently shifting to become a whale-fall specialist.
Methods
Specimens and DNA extraction
Specimens of A. inferum were collected from the sediments underneath the bones of a sperm whale off Cape Nomamisaki, Kagoshima Prefecture, Japan, 229 m deep (Fig. 1) during the Hyper-Dolphin/Natsushima cruise of JAMSTEC (NT03-08 leg1). Comparative specimens of two species of A. lucayanum complex were collected with the aid of SCUBA from coral sand 10–20 m deep off the northwest coast of Kuroshima Island, Yaeyama Islands, Japan, and Castle Harbour in Bermuda [8]. These collected specimens were fixed and preserved in 99.5% ethanol. The whole body of a specimen from each species was used to extract total DNA, which was performed using the DNeasy Tissue Kit (QIAGEN), according to manufacturer's protocols.
PCR and Sequencing
The mitogenomes of the three Asymmetron species were amplified in their entirety using a long PCR technique. Four lancelet-versatile long-PCR primers [see Additional File 1] were used to amplify the entire mitogenome in two reactions. The long-PCR products were diluted with TE buffer (1:19) for subsequent use as PCR templates. Forty-three lancelet-versatile, 39 fish-versatile, and 23 species-specific PCR primers [see Additional File 1] were used in various combinations to amplify contiguous, overlapping segments of the entire mitogenome. Fifty species-specific primers were designed in cases where no appropriate lancelet-versatile primers were available for A. inferum. Long PCR and subsequent nested PCR were performed as previously described [16]. Double-stranded PCR products, purified using a Pre-Sequencing Kit (USB), were subsequently used for direct cycle sequencing with dye-labeled terminators (Applied Biosystems). Primers used were the same as those for PCR. All sequencing reactions were performed according to the manufacturer's instructions. Labeled fragments were analyzed on a Model 3100 DNA sequencer (Applied Biosystems).
Alignments
Four Asymmetron, an Epigonichthys, and three Branchiostoma species, including whole mitogenomes of known species (Asymmetron sp. B, AB110092[16]; E. maldivensis, AB110093[16]; B. floridae, AF098298[18]; B. lanceolatum, AB194383 [38]; B. belcheri, AB083384 [Matsuzaki et al., unpublished data]) were phylogenetically analyzed, based on surveyed mtDNA sequence data. An enteropneust Balanoglossus carnosus (AF051097[39]), a cyclostome Petromyzon marinus (U11880 [40]), and a small-spotted catshark Scyliorhinus canicula (X16067[41]) were chosen as outgroups. Urochordates were not included in the present analysis because of peculiarity of their mitogenome sequences that was remarkably different from those of other chordates, supposedly because of rapid evolutionary rate in the mitogenome [42,43].
The DNA sequences for the 11 species were edited and analyzed with EditView ver. 1.0.1, AutoAssembler ver. 2.1 (Applied Biosystems), and DNASIS ver. 3.2 (Hitachi Software Engineering Co. Ltd.). Amino acids were used for alignments of the protein-coding genes, and secondary structure models were used for the alignment of tRNA genes. Since strictly secondary-structure-based alignment for the two rRNA genes was impractical for the large dataset, we employed machine alignment instead, which would minimize erroneous assessment of the positional homology of the rRNA molecules. The two rRNA gene (rrnL and rrnS) sequences were initially aligned using CLUSTAL X, ver. 1.81 [44]. Each primary aligned sequence was realigned using ProAlign ver. 0.5 [45] and those regions with posterior probabilities ≥70% used in the phylogenetic analyses. These probabilities seemed to effectively remove all ambiguously aligned regions. Ambiguous alignment regions, such as the 5' and 3' ends of several protein-coding genes and loop regions of several tRNA genes, were excluded, leaving a total of 12,497 available nucleotide positions (10,059, 1,275, and 1,163 positions for protein-coding, tRNA, and rRNA genes, respectively) for phylogenetic analyses. Two datasets were used in our analyses: dataset #1, concatenated nucleotide sequences from 13 protein-coding, 22 tRNA, and two rRNA genes (total position 12,497); dataset #2, concatenated amino acid sequences from 13 protein-coding genes plus nucleotide sequences from 22 tRNA and two rRNA genes (5,791).
Phylogenetic analysis
Maximum-likelihood (ML) analysis for dataset #1 using PAUP* 4.0b10 [46] was performed under a transversional substitution model with gamma correction and invariable-site assumption (TVM + I + Γ), which was chosen as the most fit for the present case based on hierarchical likelihood tests by Modeltest 3.6 [47]. The base frequencies were estimated to be A = 0.2940, C = 0.2233, G = 0.1598, and T = 0.3230. The substitution rates were A-C = 0.9657, A-G = 8.4537, A-T = 1.3911, C-G = 1.6808, C-T = 8.4537, and G-T = 1.0000. Assumed proportion of invariable sites was 0.1312. Gamma distribution shape parameter was 0.4086. Heuristic search option of PAUP* was chosen for obtaining the ML tree. Robustness of each internal branch of the ML tree estimated was evaluated with 100 bootstrap replications [48].
Partitioned Bayesian inference (BI) phylogenetic analysis was performed with MrBayes version 3.1.2 [49,50]. Five (dataset #1) and three (dataset #2) partitions were set (1st, 2nd, 3rd codon positions, tRNA genes, and rRNA genes; and amino acid sequences of 13 protein-coding genes, tRNA genes, and rRNA genes, respectively). The general time reversible (GTR) model with gamma correction and invariable-site assumption was used in the analysis for dataset #1, and for tRNA and rRNA genes of dataset #2. As mentioned above, TVM + I + Γ was chosen as the best fitted for the present case. However, the TVM model is a special case of the GTR model and is not yet implemented in MrBayes. Therefore, the GTR model (GTR + I + Γ) was used in the analyses. The mtREV [20] model with gamma correction and invariable-site assumption (mtREV + I + Γ) was used in the analysis for the protein-coding genes of dataset #2. This model was selected as the best-fit model of amino acid substitution by MrBayes. Model parameter values were treated as unknown and were estimated for each analysis. Random starting trees were used, and analyses were run for one million generations, sampling every 100 generations. Bayesian posterior probabilities were then calculated from the sample points after the Markov Chain Monte Carlo (MCMC) algorithm began to converge. To ensure that our analyses were not trapped in local optima, four independent MCMC runs were performed. Topologies and posterior clade probabilities from different runs were compared for congruence.
Maximum parsimony (MP) analysis for dataset #1 was performed using PAUP* 4.0b10 [46]. Heuristic MP analyses were conducted with TBR (tree bisection-reconnection) branch swapping and 100 random addition sequences. All phylogenetically uninformative sites were ignored. Robustness of each internal branch of the MP tree estimated was evaluated with 1,000 bootstrap replications [48].
Divergence time estimation
The analyses of divergence time were conducted with the penalized likelihood (PL) [22] and the nonparametric rate smoothing (NPRS) [23] methods. Molecular clock approaches were not used because a high rate of heterogeneity among lineages of lancelets was observed by the two-cluster test (LINTREE [21]). The previous analyses based on molecules were referred to the calibration points for our dating because of the absence of a useful fossil record in the lancelets. PL approach based on the BI tree (dataset #2) was performed by r8s 1.71 [51]. All r8s analyses utilized the truncated Newton (TN) algorithm and the additive rate penalty function. All analyses were reoptimized 1,000 times (set_num_restarts = 1,000) to avoid entrapment on a local solution optimum. The optimal smoothing parameter (121) was estimated using cross-validation. The divergence times between Cephalochordata and Vertebrata (+ Urochordata) (891 Mya) and between Agnatha (Cyclostome) and Gnathostomes (Chondrichthys) (652 Mya) [1] were used for the age of two calibration points. NPRS approach based on the ML tree was performed by TreeEdit 1.0 [52]. As a reference point for dating, the divergence time between Asymmetron and the other genera (162 Mya) was used for the age of root node [16].
Authors' contributions
TK, MNi, and TN conceived and designed the research; MNo, YF, and TN collected materials; TK and MNo performed experiments; TK, MNo, and YY analyzed data, TK, MNi, and TN wrote the paper. All authors read and approved the final manuscript.
Supplementary Material
List of primers. List of primers used in the PCR and sequencing for all species of the genus Asymmetron.
Acknowledgments
Acknowledgements
Our cordial thanks are due to Dr. J. G. Inoue (SCS, Florida State University) for his helpful discussion regarding phylogenetic analyses, and Dr. T. P. Satoh (ORI, University of Tokyo) for his helpful discussion regarding gene rearrangements. This study was financially supported by Grants-in-Aid from JSPS (Nos. 13440253, 15380131, 16370044, and 12NP0201).
Contributor Information
Takeshi Kon, Email: kon@ori.u-tokyo.ac.jp.
Masahiro Nohara, Email: nhrmasa@mar.email.ne.jp.
Yusuke Yamanoue, Email: ayyamano@mail.ecc.u-tokyo.ac.jp.
Yoshihiro Fujiwara, Email: fujiwara@jamstec.go.jp.
Mutsumi Nishida, Email: mnishida@ori.u-tokyo.ac.jp.
Teruaki Nishikawa, Email: nishikawa@num.nagoya-u.ac.jp.
References
- Blair JE, Hedges SB. Molecular phylogeny and divergence times of deuterostome animals. Mol Biol Evol. 2005;22:2275–2284. doi: 10.1093/molbev/msi225. [DOI] [PubMed] [Google Scholar]
- Bourlat SJ, Juliusdottir T, Lowe CJ, Freeman R, Aronowicz J, Kirschner M, Lander ES, Thorndyke M, Nakano H, Kohn AB, et al. Deuterostome phylogeny reveals monophyletic chordates and the new phylum Xenoturbellida. Nature. 2006;444:85–88. doi: 10.1038/nature05241. [DOI] [PubMed] [Google Scholar]
- Holland ND, Chen JY. Origin and early evolution of the vertebrates: new insights from advances in molecular biology, anatomy, and palaeontology. Bioessays. 2001;23:142–151. doi: 10.1002/1521-1878(200102)23:2<142::AID-BIES1021>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Benton MJ. Vertebrate Palaeontology. 3. Oxford, UK: Blackwell Publishing; 2005. [Google Scholar]
- Poss SG, Boschung HT. Lancelets (Cephalochordata: Branchiostomatidae): How many species are valid? Israel J Zool. 1996;42:S13–S66. [Google Scholar]
- Nishikawa T, Nishida M. Problems in lancelet systematics. In: Kawashima S, Kikuyama S, editor. Advances in comparative endocrinology. Vol. 1. Bolonga: Monduzzi Editore; 1997. pp. 241–246. [Google Scholar]
- Nishikawa T. A new deep-water lancelet (Cephalochordata) from off Cape Nomamisaki, SW Japan, with a proposal of the revised system recovering the genus Asymmetron. Zool Sci. 2004;21:1131–1136. doi: 10.2108/zsj.21.1131. [DOI] [PubMed] [Google Scholar]
- Kon T, Nohara M, Nishida M, Sterrer W, Nishikawa T. Hidden ancient diversification in the circumtropical lancelet Asymmetron lucayanum complex. Mar Biol. 2006;149:875–883. doi: 10.1007/s00227-006-0271-y. [DOI] [Google Scholar]
- Whickstead JH. Chordata: Acrania (Cephalochordata) In: Giese AC, Pearse JS, editor. Reproduction of Marine Invertebrate. Vol. 2. New York: Academic Press; 1975. pp. 283–319. [Google Scholar]
- Wu X, Zhang S, Wang Y, Zhang B, Qu Y, Jiang X. Laboratory observation on spawning, fecundity and larval development of amphioxus (Branchiostoma belcheri tsingtaunese) Chinese J Oceanol Limnology. 1994;12:289–294. [Google Scholar]
- Ueda H, Kamakura H. Synchronous recruitment and growth pattern of planktonic larvae of the amphioxus Branchiostoma belcheri in the Seto Inland Sea, Japan. Mar Biol. 2006;148:1263–1271. doi: 10.1007/s00227-005-0176-1. [DOI] [Google Scholar]
- Fujiwara Y, Kawato M, Yamamoto T, Yamanaka T, Sato-Okoshi W, Noda C, Tsuchida S, Komai T, Cubelio SS, Sasaki T, et al. Three-year investigations into sperm whale-fall ecosystems in Japan. Mar Ecol. 2007;28:219–232. doi: 10.1111/j.1439-0485.2007.00150.x. [DOI] [Google Scholar]
- Okutani T, Fujiwara Y, Fujikura K, Miyake H, Kawano M. A mass aggregation of the mussel Adipicola pacifica (Bivalvia: Mytylidae) on submerged whale bones. Venus. 2003;63:61–64. [Google Scholar]
- Iwasaki H, Kyuno A, Shintaku M, Fujita Y, Fujiwara Y, Fujikura K, Hashimoto J, Martins LD, Gebruk A, Miyazaki JI. Evolutionary relationships of deep-sea mussels inferred by mitochondrial DNA sequences. Mar Biol. 2006;149:1111–1122. doi: 10.1007/s00227-006-0268-6. [DOI] [Google Scholar]
- Fujikura K, Fujiwara Y, Kawato M. A new species of Osedax (Annelida: Siboglinidae) associated with whale carcasses off Kyushu, Japan. Zool Sci. 2006;23:733–740. doi: 10.2108/zsj.23.733. [DOI] [PubMed] [Google Scholar]
- Nohara M, Nishida M, Miya M, Nishikawa T. Evolution of the mitochondrial genome in Cephalochordata as inferred from complete nucleotide sequences from two Epigonichthys species. J Mol Evol. 2005;60:526–537. doi: 10.1007/s00239-004-0238-x. [DOI] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- Boore JL, Daehler LL, Brown WM. Complete sequence, gene arrangement, and genetic code of mitochondrial DNA of the cephalochordate Branchiostoma floridae (Amphioxus) Mol Biol Evol. 1999;16:410–418. doi: 10.1093/oxfordjournals.molbev.a026122. [DOI] [PubMed] [Google Scholar]
- Yang Z. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: Approximate methods. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- Adachi J, Hasegawa M. Model of amino acid substitution in proteins encoded by mitochondrial DNA. J Mol Evol. 1996;42:459–468. doi: 10.1007/BF02498640. [DOI] [PubMed] [Google Scholar]
- Takezaki N, Rzhetsky A, Nei M. Phylogenetic test of the molecular clock and linearized trees. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol Biol Evol. 2002;19:101–109. doi: 10.1093/oxfordjournals.molbev.a003974. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. A nonparametric approach to estimating divergence times in the absence of rate constancy. Mol Biol Evol. 1997;14:1218–1231. [Google Scholar]
- Ruppert EE. Cephalochordata (Acrania) In: Harrison FW, Ruppert EE, editor. Microscopic Anatomy of Invertebrates, Hemichordata, Chaetognatha, and the Invertebrate Chordates. Vol. 15. New York: Wiley-Liss, Inc; 1997. pp. 349–504. [Google Scholar]
- Boore JL. Animal mitochondrial genomes. Nucleic Acids Res. 1999;27:1767–1780. doi: 10.1093/nar/27.8.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CR, Baco AR. Ecology of whale falls at the deep-sea floor. Oceanogr Mar Biol. 2003;41:311–354. [Google Scholar]
- Gingerich PD, Smith BH, Simons EL. Hind limbs of Eocene Basilosaurus – Evidence of feet in whales. Science. 1990;249:154–157. doi: 10.1126/science.249.4965.154. [DOI] [PubMed] [Google Scholar]
- Martill DM, Cruickshank ARI, Taylor MA. Dispersal via whale bones. Nature. 1991;351:193. doi: 10.1038/351193a0. [DOI] [Google Scholar]
- Hogler JA. Speculations on the role of marine reptile deadfalls In Mesozoic deep-sea paleoecology. Palaios. 1994;9:42–47. doi: 10.2307/3515077. [DOI] [Google Scholar]
- Rouse GW, Goffredi SK, Vrijenhoek RC. Osedax: Bone-eating marine worms with dwarf males. Science. 2004;305:668–671. doi: 10.1126/science.1098650. [DOI] [PubMed] [Google Scholar]
- Glover A, Källström B, Smith C, Dahlgren T. World-wide whale worms? A new species of Osedax from the shallow north Atlantic. Proc Royal Soc B: Biol Sci. 2005;272:2587–2592. doi: 10.1098/rspb.2005.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fordyce RE. Cetacean evolution. In: Perrin WF, Würsig B, Thewissem JG, editor. Encyclopedia of Marine Mammals. San Diego: Academic Press; 2002. pp. 214–220. [Google Scholar]
- Sasaki T, Nikaido M, Hamilton H, Goto M, Kato H, Kanda N, Pastene LA, Cao Y, Fordyce RE, Hasegawa M, et al. Mitochondrial phylogenetics and evolution of mysticete whales. Syst Biol. 2005;54:77–90. doi: 10.1080/10635150590905939. [DOI] [PubMed] [Google Scholar]
- Jones WJ, Won YJ, Maas PAY, Smith PJ, Lutz RA, Vrijenhoek RC. Evolution of habitat use by deep-sea mussels. Mar Biol. 2006;148:841–851. doi: 10.1007/s00227-005-0115-1. [DOI] [Google Scholar]
- Amano K. Recent and fossil whale-fall communities – with special remarks on molluscs. Fossils (The Palaeontological Society of Japan) 2006;80:5–16. (in Japanese with English abstract) [Google Scholar]
- Kiel S, Goedert J. Deep-sea food bonanzas: early Cenozoic whale-fall communities resemble wood-fall rather than seep communities. Proc Royal Soc B: Biol Sci. 2006;273:2625–2631. doi: 10.1098/rspb.2006.3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasui K, Kubokawa K. Biology of Cephalochordate Lancelet. Tokyo, Japan: University of Tokyo Press; 2005. (in Japanese) [Google Scholar]
- Nohara M, Nishida M, Nishikawa T. New complete mitochondrial DNA sequence of the lancelet Branchiostoma lanceolatum (Cephalochordata) and the identity of this species' sequences. Zool Sci. 2005;22:671–674. doi: 10.2108/zsj.22.671. [DOI] [PubMed] [Google Scholar]
- Castresana J, Feldmaier-Fuchs G, Yokobori S, Satoh N, Pääbo S. The mitochondrial genome of the hemichordate Balanoglossus carnosus and the evolution of deuterostome mitochondria. Genetics. 1998;150:1115–1123. doi: 10.1093/genetics/150.3.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WJ, Kocher TD. Complete sequence of a sea lamprey (Petromyzon marinus) mitochondrial genome – Early establishment of the vertebrate genome Organization. Genetics. 1995;139:873–887. doi: 10.1093/genetics/139.2.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delarbre C, Spruyt N, Delmarre C, Gallut C, Barriel V, Janvier P, Laudet V, Gachelin G. The complete nucleotide sequence of the mitochondrial DNA of the dogfish, Scyliorhinus canicula. Genetics. 1998;150:331–344. doi: 10.1093/genetics/150.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori S, Ueda T, Feldmaier-Fuchs G, Pääbo S, Ueshima R, Kondow A, Nishikawa K, Watanabe K. Complete DNA sequence of the mitochondrial genome of the ascidian Halocynthia roretzi (Chordata, Urochordata) Genetics. 1999;153:1851–1862. doi: 10.1093/genetics/153.4.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori S, Watanabe Y, Oshima T. Mitochondrial genome of Ciona savignyi (Urochordata, Ascidiacea, Enterogona): Comparison of gene arrangement and tRNA genes with Halocynthia roretzi mitochondrial genome. J Mol Evol. 2003;57:574–587. doi: 10.1007/s00239-003-2511-9. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A, Milinkovitch MC. A hidden Markov model for progressive multiple alignment. Bioinformatics. 2003;19:1505–1513. doi: 10.1093/bioinformatics/btg193. [DOI] [PubMed] [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4. Sunderland: Sinauer Associates; 2002. [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sanderson MJ. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Charleston M. TreeEdit Version 10. Oxford: University of Oxford; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of primers. List of primers used in the PCR and sequencing for all species of the genus Asymmetron.