Abstract
The relationship between animal mating system variation and patterns of protein polymorphism and divergence is poorly understood. Drosophila provides an excellent system for addressing this issue, as there is abundant interspecific mating system variation. For example, compared to D. melanogaster subgroup species, repleta group species have higher remating rates, delayed sexual maturity, and several other interesting differences. We previously showed that accessory gland protein genes (Acp's) of Drosophila mojavensis and D. arizonae evolve more rapidly than Acp's in the D. melanogaster subgroup and that adaptive Acp protein evolution is likely more common in D. mojavensis/D. arizonae than in D. melanogaster/D. simulans. These findings are consistent with the idea that greater postcopulatory selection results in more adaptive evolution of seminal fluid proteins in the repleta group flies. Here we report another interesting evolutionary difference between the repleta group and the D. melanogaster subgroup Acp's. Acp gene duplications are present in D. melanogaster, but their high sequence divergence indicates that the fixation rate of duplicated Acp's has been low in this lineage. Here we report that D. mojavensis and D. arizonae genomes contain several very young duplicated Acp's and that these Acp's have experienced very rapid, adaptive protein divergence. We propose that rapid remating of female desert Drosophila generates selection for continuous diversification of the male Acp complement to improve male fertilization potential. Thus, mating system variation may be associated with adaptive protein divergence as well as with duplication of Acp's in Drosophila.
POSTCOPULATORY conflict between males, in the form of sperm competition, can be an important component of male fitness in polyandrous species (Birkhead and Møller 1998). Numerous strategies have evolved to increase sperm competitive ability, often mediated by components of the seminal fluid (Birkhead and Møller 1998; Chapman 2001; Fry and Wilkinson 2004). Females also have an interest in paternity and can play an important role in deciding the outcome of sperm competition (Eberhard 1996; Birkhead and Pizzari 2002; Bernasconi et al. 2004). Thus, postcopulatory sexual selection may drive male adaptations to increase sperm competitive ability and female counter-adaptations to bias paternity, maintaining a state of antagonistic coevolution between the sexes (Rice 1996, 1998). Consistent with this hypothesis, proteins that mediate fertilization are known to evolve rapidly in many species (Vacquier 1998; Swanson and Vacquier 2002). Accordingly, postcopulatory interactions and the molecules behind them have drawn considerable attention for their potential role in generating reproductive isolation between populations (Parker and Partridge 1998; Rice 1998; Pitnick et al. 1999; Arnqvist et al. 2000; Gavrilets 2000; Knowles and Markow 2001).
In Drosophila, empirical studies suggest that there is abundant genetic variation affecting traits related to male–male and male–female postcopulatory interactions (Clark et al. 1995; Rice 1996; Clark and Begun 1998). Male accessory gland proteins (Acp's) of the Drosophila melanogaster subgroup have received most of the attention as potential molecular agents of male–male and male–female postcopulatory interactions in Drosophila. There are an estimated 70–106 Acp's in D. melanogaster (Mueller et al. 2005) that are transferred to females during mating as secreted seminal fluid peptides. Acp's have been shown to stimulate ovulation and increase egg-laying rates (Kalb et al. 1993; Herndon and Wolfner 1995; Heifetz et al. 2000), bind sperm and affect sperm storage (Neubaum and Wolfner 1999; Tram and Wolfner 1999), affect the outcome of sperm competition (Harshman and Prout 1994; Chapman et al. 2000), decrease female receptivity (Chen et al. 1988; Aigaki et al. 1991; Chapman et al. 2003; Liu and Kubli 2003), and decrease female life span (Chapman et al. 1993, 1995; Lung et al. 2002). Furthermore, Acp's evolve rapidly in the D. melanogaster subgroup (Begun et al. 2000; Swanson et al. 2001; Kern et al. 2004), in at least some cases as a result of directional selection (Tsaur and Wu 1997; Aguadé 1998, 1999; Tsaur et al. 1998; Begun et al. 2000; Kern et al. 2004).
Previous work has demonstrated that Acp's evolve more rapidly than most Drosophila genes (Begun et al. 2000; Swanson et al. 2001; Wagstaff and Begun 2005a) and that they evolve especially rapidly in desert Drosophila of the repleta group (Wagstaff and Begun 2005b). Elevated rates of Acp evolution in desert Drosophila may be due to differences between their mating system and that of flies from the D. melanogaster subgroup (Markow 1996, 2002). For example, desert Drosophila males take at least twice as long as D. melanogaster males to reach reproductive maturity (Pitnick et al. 1995). Male age at reproductive maturity is positively correlated with sperm size and the size of the female sperm-storage organ in Drosophila species (Pitnick et al. 1995, 1999). Moreover, sperm size and sperm-storage organ size are coevolving rapidly in D. mojavensis, with geographically distinct populations expressing different phenotypes for these correlated traits (Pitnick et al. 2003). Another difference relative to the D. melanogaster subgroup mating system is female remating, which occurs much more rapidly and often in desert Drosophila (Markow 2002). Higher remating rates in desert Drosophila could potentially increase selection on phenotypes related to postcopulatory male–male or male–female interactions (Markow 2002; Singh et al. 2002).
Additional differences between repleta group and D. melanogaster subgroup flies are evident in the short-term physiological response of females following copulation. Transfer of seminal fluid triggers an insemination reaction within the female reproductive tract of desert Drosophila (Patterson and Stone 1952) but is diminutive in D. melanogaster (Wheeler 1947; Markow and Ankney 1988). This insemination reaction, which is superficially similar to inflammation, results in a mass in the female reproductive tract. Remating does not occur during the several hours that it persists (Patterson 1947; Knowles and Markow 2001). The intensity of the insemination reaction is highly variable, with interspecific matings (e.g., D. arizonae and D. mojavensis) triggering an exaggerated and harder mass, which persists significantly longer than within-species insemination reactions (Patterson 1947). Interestingly, exaggerated insemination reactions are observed in some crosses between geographically distinct populations of D. mojavensis, suggesting that interpopulation postcopulatory incompatibilities may evolve quickly (Knowles and Markow 2001). Finally, ejaculate components of many repleta group species, including D. mojavensis, are incorporated into female somatic tissues, a phenomenon not known to occur in the D. melanogaster subgroup (Markow and Ankney 1984; Pitnick et al. 1997).
Our earlier results suggested that, although general patterns of protein variation in Acp's from desert Drosophila and D. melanogaster subgroup flies are similar, there are important quantitative differences between the groups. For example, we found faster rates of protein evolution and stronger evidence for directional selection in repleta group Acp's relative to D. melanogaster subgroup Acp's (Wagstaff and Begun 2005b). Analyses of annotated D. melanogaster Acp's show that, although several Acp duplicates exist, they tend to be relatively highly diverged at the nucleotide level (Holloway and Begun 2004; Mueller et al. 2005). Here we report the discovery of several recent Acp duplications in D. arizonae/D. mojavensis. Our analyses suggest that several of these recent duplications have diverged under directional selection, a phenomenon not observed in D. melanogaster (Holloway and Begun 2004). These data provide additional support for different evolutionary processes acting on Acp's in these lineages, perhaps as a result of mating system divergence.
MATERIALS AND METHODS
Stocks and DNA sequencing:
All fly stocks were acquired from the Drosophila Species Stock Center (Tucson, AZ). They included seven D. arizonae lines (15081-1271.00, 15081-1271.04, 15081-1271.05, 15081-1271.08, 15081-1271.12, 15081-1271.13, 15081-1271.14 from various locations in mainland Mexico) and seven D. mojavensis lines. Of the seven D. mojavensis stocks, four were D. mojavensis baja (15081-1351.03, 15081-1351.09, 15081-1351.12, 15081-1351.14 from various locations in Baja, Mexico), and three were D. mojavensis mojavensis (15081-1352.00, 15081-1352.01, 15081-1352.02 from various locations in southern California).
Most duplicate Acp's described here were accidentally amplified during our earlier survey (Wagstaff and Begun 2005b) as secondary PCR products from primers designed from D. mojavensis accessory gland ESTs. Sequence data from each putative duplicate Acp were used to design duplicate-specific PCR primers for amplifying additional copies. However, the very short length of some Acp's under investigation made it difficult to isolate duplicates from all of the fly lines in this survey. Expand high-fidelity polymerase (Roche Molecular Biochemicals) was used for PCR amplification. Single alleles for sequencing were isolated by TOPO vector (Invitrogen, San Diego) cloning of PCR products. PCR-amplified colony-PCR products and their associated sequences were obtained using M13 reverse and T7 vector primers. All sequencing was done on an Applied Biosystems 377 automated sequencer (ABI).
Organization of duplicated Acp's:
Patterns of sequence divergence (see below) in most cases provided unambiguous evidence that the Acp's in question are duplications rather than highly diverged alleles. However, we used molecular and further computational analysis to investigate the genomic organization of putative duplicate Acp's. Under the premise that recent duplications are often tandemly arranged, we designed PCR primers to amplify genomic DNA across the putative duplicates. We used LA-Taq long PCR polymerase (TaKaRa, Shiga, Japan) with an extension time of 10 min and cycling parameters according to the manufacturer's instructions. Successfully amplified fragments were end sequenced to confirm that the amplified product corresponded to the expected genomic sequence under the tandem duplication hypothesis. When the draft version of the whole-genome shotgun (WGS) D. mojavensis genome became available, we used BLAT (BLAST-like alignment tool, UCSC Genome Browser) analysis (Kent 2002) to confirm our observations and localize additional duplicate Acp's.
Molecular population genetic statistics and hypothesis tests of adaptive protein evolution:
Alleles and duplicate gene sequences were aligned and edited using the DNASTAR software package (Lasergene, Madison, WI). The DnaSP program of Rozas and Rozas (1999) was used to measure levels of polymorphism and divergence for duplicate genes represented by multiple alleles. Group averages (i.e., duplicates vs. other Acp's) were calculated by taking averages weighted according to sequence length.
Nucleotide distances were used to infer the topologies of duplicate family genealogies. Maximum-likelihood estimation of branch-specific dN and dS values used the free-ratio model (model 1) of the PAML computer program (Yang 1997). Outgroups were determined by pairwise distance estimates and corroborated by PAML branch length output. For genes sampled for multiple alleles, one random allele was chosen for PAML analyses. Alignments were generated using the DNASTAR software package (Lasergene) and manually adjusted where appropriate. Indel variation for codon positions that were gapped in >50% of the aligned sequences were omitted from the analyses. PAML tests for branch heterogeneity compared likelihood estimates from the free-ratio model to estimates from the one-ratio model (model 0). We then tested entire gene trees for significant evidence of dN/dS > 1 by comparing the one-ratio model to a one-ratio model with dN/dS = 1 (fix_ omega = 1; omega = 1). To test whether the dN value of a given branch significantly exceeds the dS value (Acp27 only), we used the two-ratio model (model 2) and set all background branches to have the same dN/dS value. The branch of interest either was allowed to be free or was fixed at dN/dS = 1. For likelihood-ratio tests, twice the log-likelihood difference was compared to a χ2 distribution with 1 d.f. [or (number of branches − 1) for the branch heterogeneity tests] to determine significance levels.
RESULTS
Evidence of gene duplication:
In the course of our molecular population genetic analysis of 18 Acp's in D. arizonae and D. mojavensis (Wagstaff and Begun 2005b), sequence data from four genes revealed alleles that were unusually highly diverged from the majority of alleles sampled. These genes were clearly related to the target genes, but had levels of divergence that in most cases could be plausibly interpreted only as evidence of gene duplication. Table 1 provides a summary of alleles sampled and the number of fly lines that have been verified by PCR to carry particular putative duplicate gene copies.
TABLE 1.
Sample and distribution of duplicate genes
| Sample
|
Documented in the same fly line?
|
|||||
|---|---|---|---|---|---|---|
| Duplicate gene | ari | moj | a + b | a + c | b + c | a + b + c |
| Acp5a | 7 | 7 | ||||
| Acp5b | 3 | 1 | 3 ari, 1 moj | 1 moj | No | No |
| Acp5c | 0 | 1 | ||||
| Acp16a | 7 | 6 | ||||
| Acp16b | 7 | 4 | 7 ari, 3 moj | 2 moj | 1 moj | No |
| Acp16c | 0 | 3 | ||||
| Acp21a | 6 | 7 | No | — | — | — |
| Acp21b | 1 | 0 | ||||
| Acp27a | 7 | 7 | 5 moj | — | — | — |
| Acp27b | 0 | 5 | ||||
ari, D. arizonae; moj, D. mojavensis.
Under the assumption that recent duplicate Acp's likely originated through unequal crossing over and therefore were organized tandemly, we designed PCR primers to amplify intergenic sequence between putative paralogs. We successfully amplified intergenic sequences (data not shown) for Acp16a-b, Acp21a-b, and Acp27a-b, thereby confirming their duplicate status. BLAT analysis of the D. mojavensis assembly (UCSC Genome Browser) supports the proposition that the other highly diverged “alleles” isolated in our previous work (Wagstaff and Begun 2005b) are actually tandem duplicates. Table 2 shows the scaffold assembly positions of these paralogs. Thus, there is PCR evidence and/or support from genome assemblies for the duplicate status of the Acp's discussed here.
TABLE 2.
Duplicate Acp genomic organization inferred from D. mojavensis assembly
| Gene | Scaffold | Strand | CDS start | CDS stop | Intergenic sequence |
|---|---|---|---|---|---|
| Acp5a | 6540 | + | 3,094,356 | 3,094,529 | a–b: 22,390 |
| Acp5b | 6540 | − | 3,071,966 | 3,071,802 | b–c: 16,862 |
| Acp5c | 6540 | − | 3,054,940 | 3,054,758 | |
| Acp16a | — | — | — | — | — |
| Acp16b | 6680 | + | 18,985,243 | 18,985,461 | 2,761 |
| Acp16c | 6680 | + | 18,988,222 | 18,988,437 | |
| Acp21a | 6540 | + | 33,094,203 | 33,094,553 | — |
| Acp21b | — | — | — | — | |
| Acp27a | 6496 | − | 3,965,482 | 3,965,189 | — |
| Acp27b | — | — | — | — |
CDS, coding sequence.
Polymorphism and interspecific divergence of duplicate Acp's:
Polymorphism and interspecific orthologous divergence of several duplicate Acp's (Acp5a, Acp16a, Acp16b, Acp21a, and Acp27a) was previously reported in Wagstaff and Begun (2005b). We include these data in Table 3 along with the newly reported duplicates. Table 3 also shows estimates of average polymorphism and divergence for duplicate vs. single-copy Acp's in D. mojavensis and D. arizonae. Although synonymous and nonsynonymous heterozygosity shows considerable variation among duplicate genes, the small number of sites surveyed per gene precludes any speculation about heterogeneous forces. Overall, synonymous heterozygosity and divergence are slightly lower in duplicated D. arizonae and D. mojavensis Acp's compared to single-copy Acp's from these species. In contrast, nonsynonymous heterozygosity and divergence are higher for duplicated Acp's than for single-copy Acp's in D. arizonae and D. mojavensis, with nonsynonymous divergence marginally significantly higher for duplicate Acp's (Wagstaff and Begun 2005b, Mann–Whitney one-tailed U-test, P = 0.046). The average dN/dS ratio for duplicated Acp's is 2.123 (Table 3), which is significantly higher than the ratio for single-copy Acp's (0.761) from these species (Wagstaff and Begun 2005b, Mann–Whitney one-tailed U-test, P = 0.00679; one value was omitted from each group because dS = 0) and higher than the average ratio for Acp's in D. melanogaster vs. D. simulans comparisons (0.47) (Swanson et al. 2001).
TABLE 3.
Polymorphism and orthologous divergence of duplicate Acp's
| No. of sites
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene | No. of alleles | Sample | Synonymous | Nonsynonymous | θsyn | θrep | dS | dN | dN/dS |
| Acp5aa | 7 | ari | 27 | 72 | 0.0151 | 0.0057 | 0.111 | 0.110 | 0.990 |
| Acp5aa | 7 | moj | 27 | 72 | 0.0000 | 0.0170 | — | — | — |
| Acp5b | 3 | ari | 27 | 69 | 0.0000 | 0.0000 | 0.000 | 0.112 | dN > dS |
| Acp16aa | 7 | ari | 38 | 103 | 0.0000 | 0.0159 | 0.060 | 0.132 | 2.205 |
| Acp16aa | 6 | moj | 38 | 103 | 0.0000 | 0.0299 | — | — | — |
| Acp16ba | 7 | ari | 49 | 155 | 0.0251 | 0.0184 | 0.062 | 0.05 | 0.808 |
| Acp16ba | 4 | moj | 49 | 155 | 0.0336 | 0.0070 | — | — | — |
| Acp16c | 3 | moj | 45 | 156 | 0.0000 | 0.0086 | — | — | — |
| Acp21aa | 6 | ari | 48 | 132 | 0.0092 | 0.0066 | 0.055 | 0.227 | 4.121 |
| Acp21aa | 7 | moj | 48 | 132 | 0.0086 | 0.0278 | — | — | — |
| Acp27aa | 7 | ari | 68 | 214 | 0.0000 | 0.0019 | 0.006 | 0.013 | 2.138 |
| Acp27aa | 7 | moj | 68 | 214 | 0.0120 | 0.0076 | — | — | — |
| Acp27b | 5 | moj | 71 | 208 | 0.0068 | 0.0115 | — | — | — |
| All duplicates | ari | 257 | 745 | 0.0080 | 0.0083 | 0.044 | 0.094 | 2.123 | |
| moj | 346 | 1040 | 0.0097 | 0.0139 | — | — | — | ||
| Other Acp'sa | ari | 712 | 2336 | 0.0149 | 0.0058 | 0.068 | 0.052 | 0.761 | |
| moj | 712 | 2336 | 0.0166 | 0.0075 | — | — | — | ||
ari, D. arizonae; moj, D. mojavensis; θsyn, synonymous heterozygosity; θrep, nonsynonymous heterozygosity.
Data are from Wagstaff and Begun (2005b).
Contingency table comparison of synonymous to nonsynonymous polymorphisms (Table 4) provides further evidence that different evolutionary forces are acting on duplicate vs. other Acp's in D. mojavensis (P = 0.012).The heterogeneity can be interpreted as either a deficit of synonymous or an excess of nonsynonymous polymorphisms. Comparison to non-Acp D. mojavensis polymorphisms from our previous survey on the same fly lines (Wagstaff and Begun 2005b) suggests that the heterogeneity is primarily attributable to an excess of nonsynonymous polymorphisms, although a smaller deficit of synonymous polymorphisms also contributes to the pattern. A comparable contingency table from D. arizonae shows a similar trend in the same direction, although it is not significantly heterogeneous (P = 0.071).
TABLE 4.
Contingency analysis of duplicate vs. single-copy Acp polymorphism
| Synonymous | Nonsynonymous | ||
|---|---|---|---|
| D. mojavensis | |||
| Duplicates | 7 | 32 | G = 6.350 |
| Other Acp's | 29 | 42 | P = 0.012 |
| D. arizonae | |||
| Duplicates | 5 | 17 | G = 3.250 |
| Other Acp's | 26 | 33 | P = 0.071 |
Paralogous dN/dS ratios:
Paralogous dN/dS estimates are >1 for all pairwise comparisons and are extraordinarily high in some cases (Table 5). For example, dN/dS is >4 in at least one pairwise comparison for three of the four Acp families. For the Acp16 family, the highest value is for D. arizonae Acp16a-b (1.934). These extremely high dN/dS estimates seem particularly noteworthy, given that only 3 of 14 putative single-copy Acp's investigated in D. arizonae/D. mojavensis (Wagstaff and Begun 2005b) have a dN/dS >1.
TABLE 5.
Paralogous divergence of duplicate Acp's
| No. of alleles
|
No. of sites
|
||||||
|---|---|---|---|---|---|---|---|
| Gene pair | First gene | Second gene | Synonymous | Nonsynonymous | dS | dN | dN/dS |
| Acp5 | |||||||
| ari (a:b) | 7 | 3 | 27 | 69 | 0.199 | 0.272 | 1.370 |
| moj (a:b) | 7 | 1 | 24 | 63 | 0.043 | 0.205 | 4.799 |
| moj (a:c) | 7 | 1 | 25 | 65 | 0.124 | 0.474 | 3.817 |
| moj (b:c) | 1 | 1 | 25 | 68 | 0.157 | 0.434 | 2.757 |
| Acp16 | |||||||
| ari (a:b) | 7 | 7 | 40 | 116 | 0.229 | 0.442 | 1.934 |
| moj (a:b) | 6 | 4 | 40 | 113 | 0.247 | 0.461 | 1.867 |
| moj (a:c) | 6 | 3 | 40 | 116 | 0.196 | 0.314 | 1.599 |
| moj (b:c) | 4 | 3 | 46 | 149 | 0.313 | 0.378 | 1.209 |
| Acp21 | |||||||
| ari (a:b) | 6 | 1 | 49 | 137 | 0.014 | 0.134 | 9.734 |
| Acp27 | |||||||
| moj (a:b) | 7 | 5 | 65 | 196 | 0.021 | 0.103 | 4.899 |
ari, D. arizonae; moj, D. mojavensis.
Analysis of duplicate Acp gene trees:
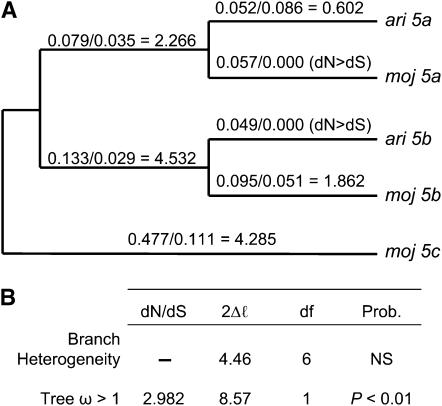
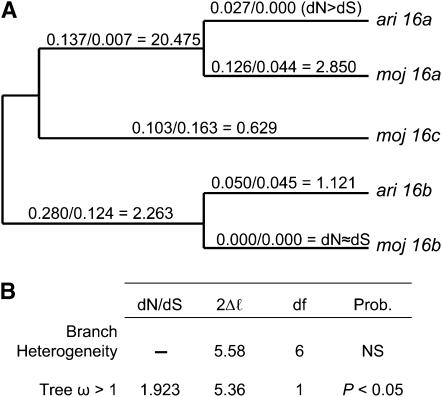
Maximum-likelihood analysis of the Acp5 duplicate gene family reveals very high rates of protein evolution along most gene-tree branches, with average dN/dS = 2.982 (Figure 1B). The complete gene tree has a dN/dS ratio that is significantly >1 (Figure 1B; P < 0.01) and provides no evidence of branch heterogeneity. We report individual branch dN/dS estimates as a guideline for future investigations (Figure 1A). However, the lack of evidence for branch heterogeneity precludes individual branch hypothesis testing. Similarly, the Acp16 gene tree shows high nonsynonymous divergence along most branches (Figure 2A), significant evidence of adaptive evolution with an average dN/dS of 1.923 (Figure 2B; P < 0.05), and no statistical evidence for branch heterogeneity (Figure 2B). Note that, although our PCR and sequencing efforts identified only D. mojavensis alleles for both Acp5c and Acp16c, paralogous synonymous and nonsynonymous divergence associated with both duplicates (Table 5) greatly exceeded average levels of orthologous divergence (Table 3). Thus, our failure to identify D. arizonae alleles might be explained by interspecific divergence that was too great to amplify D. arizonae alleles using primers designed from D. mojavensis DNA sequences or by loss of duplications in D. arizonae. As the genome assembly becomes more complete, a strategy of long PCR, cloning, and sequencing should reveal the full complement of Acp5 and Acp16 duplicates in D. arizonae and D. mojavensis.
Figure 1.—
Phylogeny of Acp5 duplicate genes. (A) Evolution along each branch is shown as dN/dS ratios. (B) Maximum-likelihood analyses of the complete gene tree. There is no significant evidence of branch heterogeneity; however, the complete Acp5 gene tree has dN/dS > 1 (P < 0.01). ω, dN/dS ratio; 2Δℓ, likelihood-ratio test; NS, not significant.
Figure 2.—
Phylogeny of Acp16 duplicate genes. (A) dN/dS values are shown for each branch. (B) Maximum-likelihood analyses of the complete gene tree. There is no significant evidence of branch heterogeneity; however, the complete Acp16 gene tree has dN/dS > 1 (P < 0.05). ω, dN/dS ratio; 2Δℓ, likelihood-ratio test; NS, not significant.
Divergence estimates and tests of branch heterogeneity for Acp21 and Acp27 are shown in Table 6. Both gene families generally show little synonymous divergence and very high levels of nonsynonymous divergence. However, in many cases, an informative estimate of dN/dS cannot be reported because synonymous divergence is too close to zero. The Acp21 gene family shows dramatically higher levels of nonsynonymous vs. synonymous divergence, with dN/dS ratios significantly >1 for the complete gene tree (P < 0.001), and all three branches are individually estimated at dN > dS. Because there is no significant evidence of Acp21 branch heterogeneity (Table 6), we do not report individual branch likelihood-ratio tests. Even so, note that the D. mojavensis Acp21a branch has estimated dN at severalfold higher than dN along the D. arizonae Acp21a and Acp21b branches.
TABLE 6.
Divergence and branch heterogeneity of Acp21 and Acp27 duplicate families
| Gene family | dN | dS | ω | 2Δla | Significance |
|---|---|---|---|---|---|
| Acp21 heterogeneity | — | — | — | 0.41 | NS |
| Acp21 complete tree | — | — | dN > dS | 18.52 | P < 0.001 |
| ari a branch | 0.090 | 0.000 | dN > dS | — | — |
| ari b branch | 0.060 | 0.000 | dN > dS | — | — |
| moj a branch | 0.180 | 0.014 | 12.857 | — | — |
| Acp27 heterogeneity | — | — | — | 6.40 | P < 0.05 |
| Acp27 complete tree | — | — | 8.348 | 8.24 | P < 0.01 |
| ari a branch | 0.014 | 0.000 | dN > dS | 0.97 | NS |
| moj a branch | 0.000 | 0.013 | 0.000 | — | — |
| moj b branch | 0.110 | 0.000 | dN > dS | 12.12 | P < 0.01 |
w, dN/dS ratio (dN > dS is shown when dS = 0); NS, not significant.
Likelihood-ratio tests vs. the null model (dN/dS = 1 or dN/dS is constant for branch heterogeneity tests).
Unlike Acp21, Acp27 does show significant evidence of branch heterogeneity (P < 0.05) that is clearly attributable to most of the nonsynonymous divergence occurring along the D. mojavensis Acp27b branch. Both this branch individually and the complete gene tree have dN/dS significantly >1 (P < 0.01; Table 6). Furthermore, all 17 nonsynonymous differences between D. mojavensis Acp27a and Acp27b fall on the Acp27b branch when D. arizonae Acp27a is used as the outgroup. Note that levels of divergence for D. arizonae Acp21b and D. mojavensis Acp27b are low enough to be consistent with postspeciation duplication events. Further sequence analysis should confirm whether or not this is the case.
DISCUSSION
Sequence analysis of Acp genes from the D. melanogaster subgroup has demonstrated that seminal fluid proteins evolve rapidly relative to other classes of genes (Begun et al. 2000; Swanson et al. 2001; Kern et al. 2004). This rapid evolution is often interpreted as evidence of natural selection, which is thought to play an important role in sperm competition and male–female postcopulatory interactions (Rice 1996; Swanson and Vacquier 2002). We have previously shown that single-copy Acp genes in D. arizonae and D. mojavensis evolve more rapidly than the D. melanogaster subgroup Acp's (Wagstaff and Begun 2005b), an observation that is consistent with expectations based on their dramatically different mating systems (Markow 1996, 2002). Here we have documented that desert Drosophila Acp's differ from the D. melanogaster subgroup Acp's in other important ways. In contrast to the D. melanogaster subgroup Acp's (Saudan et al. 2002; Holloway and Begun 2004; Mueller et al. 2005), Acp duplications in D. mojavensis/D. arizonae are very recent (Table 5), including two (D. arizonae Acp21b and D. mojavensis Acp27b) that potentially originated after the D. mojavensis/D. arizonae speciation event.
The four D. arizonae/D. mojavensis Acp gene families investigated here evolve more rapidly than putative single-copy Acp's, with evidence of adaptive evolution in all four families. These results are consistent with observations suggesting that gene duplication can facilitate adaptive protein evolution (Ohno 1970; Ohta 1994; Li 1995). Interspecific dN/dS ratios for all duplicate Acp's varied from 0.808 to 4.121, significantly exceeding the distribution of dN/dS ratios for putative single-copy Acp's. Moreover, paralogous dN/dS ratios were even higher, demonstrating a broad time frame for adaptive evolution since most duplication events clearly predate D. arizonae/D. mojavensis speciation. Our maximum-likelihood analyses show that 16 of 20 duplicate gene tree branches have dN/dS ratios >1. The complete gene tree for each duplicate gene family significantly exceeds dN/dS = 1.
Duplicated Acp's also show higher levels of nonsynonymous polymorphism compared to single-copy Acp's. The high rate of adaptive protein evolution at these loci and the evidence for significant geographical variation in postcopulatory D. mojavensis phenotypes (Knowles and Markow 2001; Pitnick et al. 2003; Reed and Markow 2004) suggest that some nonsynonymous polymorphism in Acp gene families might be due to divergent selection between geographically isolated populations. Alternatively, the high level of nonsynonymous polymorphism could be due to selected amino acid polymorphisms during their sojourn through the population. The lower synonymous heterozyogsity in duplicated vs. single-copy Acp's is consistent with this scenario. Additional population genetics and functional data comparing intra- and interpopulation dynamics between conspecific desert Drosophila populations are needed to resolve this question.
Acknowledgments
We thank A. Holloway, S. Schaeffer, and two anonymous reviewers for useful comments. This work was supported by a National Institutes of Health grant (GM55298 to D.J.B.), a National Science Foundation grant (NSF) (DEB-0327049 to D.J.B.), and an NSF doctoral dissertation improvement grant.
References
- Aguadé, M., 1998. Different forces drive the evolution of the Acp26Aa and Acp26Ab accessory gland genes in the Drosophila melanogaster species complex. Genetics 150: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguadé, M., 1999. Positive selection drives the evolution of the Acp29AB accessory gland protein in Drosophila. Genetics 152: 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aigaki, T., I. Fleischmann, P. S. Chen and E. Kubli, 1991. Ectopic expression of sex peptide alters reproductive behavior of female D. melanogaster. Neuron 4: 557–563. [DOI] [PubMed] [Google Scholar]
- Arnqvist, G., M. Edvardsson, U. Friberg and T. Nilsson, 2000. Sexual conflict promotes speciation in insects. Proc. Natl. Acad. Sci. USA 97: 10460–10464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begun, D. J., P. Whitley, B. L. Todd, H. M. Waldrip-Dail and A. G. Clark, 2000. Molecular population genetics of male accessory gland proteins in Drosophila. Genetics 156: 1879–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernasconi, G., T. L. Ashman, T. R. Birkhead, J. D. Bishop, U. Grossniklaus et al. 2004. Evolutionary ecology of the prezygotic stage. Science 303: 971–975. [DOI] [PubMed] [Google Scholar]
- Birkhead, T. R., and A. P. Møller, 1998. Sperm Competition and Sexual Selection. Academic Press, London.
- Birkhead, T. R., and T. Pizzari, 2002. Postcopulatory sexual selection. Nat. Rev. Genet. 3: 262–273. [DOI] [PubMed] [Google Scholar]
- Chapman, T., 2001. Seminal fluid-mediated fitness traits in Drosophila. Heredity 87: 511–521. [DOI] [PubMed] [Google Scholar]
- Chapman, T., J. Hutchings and L. Partridge, 1993. No reduction in the cost of mating for Drosophila melanogaster females mating with spermless males. Proc. R. Soc. Lond. B. Biol. Sci. 253: 211–217. [DOI] [PubMed] [Google Scholar]
- Chapman, T., L. F. Liddle, J. M. Kalb, M. F. Wolfner and L. Partridge, 1995. Cost of mating in Drosophila melanogaster females is mediated by male accessory gland products. Nature 373: 241–244. [DOI] [PubMed] [Google Scholar]
- Chapman, T., D. M. Neubaum, M. F. Wolfner and L. Partridge, 2000. The role of male accessory gland protein Acp36DE in sperm competition in Drosophila melanogaster. Proc. R. Soc. Lond. B. Biol. Sci. 267: 1097–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman, T., J. Bangham, G. Vinti, B. Seifried, O. Lung et al., 2003. The sex peptide of Drosophila melanogaster: female post-mating responses analyzed by using RNA interference. Proc. Natl. Acad. Sci. USA 100: 9923–9928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, P. S., E. Stumm-Zollinger, T. Aigaki, J. Balmer, M. Bienz et al., 1988. A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster. Cell 54: 291–298. [DOI] [PubMed] [Google Scholar]
- Clark, A. G., and D. J. Begun, 1998. Female genotypes affect sperm displacement in Drosophila. Genetics 149: 1487–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. G., M. Aguadé, T. Prout, L. G. Harshman and C. H. Langley, 1995. Variation in sperm displacement and its association with accessory gland protein loci in Drosophila melanogaster. Genetics 139: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard, W. G., 1996. Female Control: Sexual Selection by Cryptic Female Choice. Princeton University Press, Princeton, NJ.
- Fry, C. L., and G. S. Wilkinson, 2004. Sperm survival in female stalk-eyed flies depends on seminal fluid and meiotic drive. Evolution Int. J. Org. Evolution 58: 1622–1626. [DOI] [PubMed] [Google Scholar]
- Gavrilets, S., 2000. Rapid evolution of reproductive barriers driven by sexual conflict. Nature 403: 886–889. [DOI] [PubMed] [Google Scholar]
- Harshman, L. G., and T. Prout, 1994. Sperm displacement without sperm transfer in Drosophila melanogaster. Evolution 48: 758–766. [DOI] [PubMed] [Google Scholar]
- Heifetz, Y., O. Lung, E. A. Frongillo, Jr. and M. F. Wolfner, 2000. The Drosophila seminal fluid protein Acp26Aa stimulates release of oocytes by the ovary. Curr. Biol. 10: 99–102. [DOI] [PubMed] [Google Scholar]
- Herndon, L. A., and M. F. Wolfner, 1995. A Drosophila seminal fluid protein, Acp26Aa, stimulates egg laying in females for 1 day after mating. Proc. Natl. Acad. Sci. USA 92: 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holloway, A., and D. J. Begun, 2004. Molecular evolution and population genetics of duplicated accessory gland protein genes in Drosophila. Mol. Biol. Evol. 21: 1625–1628. [DOI] [PubMed] [Google Scholar]
- Kalb, J. M., A. J. DiBenedetto and M. F. Wolfner, 1993. Probing the function of Drosophila melanogaster accessory glands by directed cell ablation. Proc. Natl. Acad. Sci. USA 90: 8093–8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent, W. J., 2002. BLAT: the BLAST-like alignment tool. Genome Res. 12: 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern, A. D., C. D. Jones and D. J. Begun, 2004. Molecular population genetics of male accessory gland proteins in the Drosophila simulans complex. Genetics 167: 725–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles, L. L., and T. A. Markow, 2001. Sexually antagonistic coevolution of a postmating-prezygotic reproductive character in desert Drosophila. Proc. Natl. Acad. Sci. USA 98: 8692–8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W., 1995. Molecular Evolution. Sinauer Associates, Sunderland, MA.
- Liu, H., and E. Kubli, 2003. Sex-peptide is the molecular basis of the sperm effect in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100: 9929–9933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung, O., U. Tram, C. M. Finnerty, M. A. Eipper-Mains, J. M. Kalb et al., 2002. The Drosophila melanogaster seminal fluid protein Acp62F is a protease inhibitor that is toxic upon ectopic expression. Genetics 160: 211–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markow, T. A., 1996. Evolution of Drosophila mating systems. Evol. Biol. 29: 73–106. [Google Scholar]
- Markow, T. A., 2002. Perspective: female remating, operational sex ratio, and the arena of sexual selection in Drosophila species. Evolution 56: 1725–1734. [DOI] [PubMed] [Google Scholar]
- Markow, T. A., and P. F. Ankney, 1984. Drosophila males contribute to oogenesis in a multiple mating species. Science 224: 302–303. [DOI] [PubMed] [Google Scholar]
- Markow, T. A., and P. F. Ankney, 1988. Insemination reaction in Drosophila: found in species whose males contribute material to oocytes before fertilization. Evolution 42: 1097–1101. [DOI] [PubMed] [Google Scholar]
- Mueller, J. L., K. Ravi Ram, L. A. McGraw, M. C. Bloch Qazi, E. D. Siggia et al., 2005. Cross-species comparison of Drosophila male accessory gland protein genes. Genetics 171: 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubaum, D. M., and M. F. Wolfner, 1999. Mated Drosophila melanogaster females require a seminal fluid protein, Acp36DE, to store sperm efficiently. Genetics 153: 845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno, S., 1970. Evolution by Gene Duplication. Springer-Verlag, Berlin.
- Ohta, T., 1994. Further examples of evolution by gene duplication revealed through DNA sequence comparisons. Genetics 138: 1331–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, G. A., and L. Partridge, 1998. Sexual conflict and speciation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 353: 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, J. T., 1947. The insemination reaction and its bearing on the problem of speciation in the mulleri subgroup. Univ. Texas Publ. 4720: 41–77. [Google Scholar]
- Patterson, J. T., and W. S. Stone, 1952. Evolution in the Genus Drosophila. Macmillan, New York.
- Pitnick, S., T. A. Markow and G. S. Spicer, 1995. Delayed male maturity is a cost of producing large sperm in Drosophila. Proc. Natl. Acad. Sci. USA 92: 10614–10618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitnick, S., G. S. Spicer and T. A. Markow, 1997. Phylogenetic examination of female incorporation of ejaculate in Drosophila. Evolution 51: 833–845. [DOI] [PubMed] [Google Scholar]
- Pitnick, S., T. A. Markow and G. S. Spicer, 1999. Evolution of multiple kinds of female sperm-storage organs in Drosophila. Evolution 53: 1804–1822. [DOI] [PubMed] [Google Scholar]
- Pitnick, S., G. T. Miller, K. Schneider and T. A. Markow, 2003. Ejaculate-female coevolution in Drosophila mojavensis. Proc. Biol. Sci. 270: 1507–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, L. K., and T. A. Markow, 2004. Early events in speciation: polymorphism for hybrid male sterility in Drosophila. Proc. Natl. Acad. Sci. USA 101: 9009–9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice, W. R., 1996. Sexually antagonistic male adaptation triggered by experimental arrest of female evolution. Nature 381: 232–234. [DOI] [PubMed] [Google Scholar]
- Rice, W. R., 1998. Intergenomic conflict, interlocus antagonistic coevolution, and the evolution of reproductive isolation, pp. 261–270 in Endless Forms: Species and Speciation, edited by D. J. Howard and S. H. Berlocher. Oxford University Press, New York.
- Rozas, J., and R. Rozas, 1999. DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15: 174–175. [DOI] [PubMed] [Google Scholar]
- Saudan, P., K. Hauck, M. Soller, Y. Choffat, M. Ottiger et al., 2002. Ductus ejaculatorius peptide 99B (DUP99B), a novel Drosophila melanogaster sex-peptide pheromone. Eur. J. Biochem. 269: 989–997. [DOI] [PubMed] [Google Scholar]
- Singh, S. R., B. N. Singh and H. F. Hoenigsberg, 2002. Female remating, sperm competition and sexual selection in Drosophila. Genet. Mol. Res. 1: 178–215. [PubMed] [Google Scholar]
- Swanson, W. J., and V. D. Vacquier, 2002. The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3: 137–144. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., A. G. Clark, H. M. Waldrip-Dail, M. F. Wolfner and C. F. Aquadro, 2001. Evolutionary EST analysis identifies rapidly evolving male reproductive proteins in Drosophila. Proc. Natl. Acad. Sci. USA 98: 7375–7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tram, U., and M. F. Wolfner, 1999. Male seminal fluid proteins are essential for sperm storage in Drosophila melanogaster. Genetics 153: 837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsaur, S. C., and C. I. Wu, 1997. Positive selection and the molecular evolution of a gene of male reproduction, Acp26Aa, of Drosophila. Mol. Biol. Evol. 14: 544–549. [DOI] [PubMed] [Google Scholar]
- Tsaur, S. C., C. T. Ting and C. I. Wu, 1998. Positive selection driving the evolution of a gene of male reproduction, Acp26Aa, of Drosophila. II. Divergence versus polymorphism. Mol. Biol. Evol. 15: 1040–1046. [DOI] [PubMed] [Google Scholar]
- Vacquier, V. D., 1998. Evolution of gamete recognition proteins. Science 281: 1995–1998. [DOI] [PubMed] [Google Scholar]
- Wagstaff, B. J., and D. J. Begun, 2005. a Comparative genomics of accessory gland protein genes in Drosophila melanogaster and D. pseudoobscura. Mol. Biol. Evol. 22: 818–832. [DOI] [PubMed] [Google Scholar]
- Wagstaff, B. J., and D. J. Begun, 2005. b Molecular population genetics of accessory gland protein genes and testis-expressed genes in Drosophila mojavensis and D. arizonae. Genetics 171: 1083–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler, M. R., 1947. The insemination reaction in intraspecific matings of Drosophila. University of Texas Publication 4720, pp. 78–115.
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. (http://abacus.gene.ucl.ac.uk/software/paml.html). [DOI] [PubMed] [Google Scholar]