Abstract
To evaluate and compare the extent of LD in cattle, 1536 SNPs, mostly localized on BTA03, were detected in silico from available sequence data using two different methods and genotyped on samples from 14 distinct breeds originating from Europe and Africa. Only 696 SNPs could be validated, confirming the importance of trace-quality information for the in silico detection. Most of the validated SNPs were informative in several breeds and were used for a detailed description of their genetic structure and relationships. Results obtained were in agreement with previous studies performed on microsatellite markers and using larger samples. In addition, the majority of the validated SNPs could be mapped precisely, reaching an average density of one marker every 311 kb. This allowed us to analyze the extent of LD in the different breeds. Decrease of LD with physical distance across breeds revealed footprints of ancestral LD at short distances (<10 kb). As suggested by the haplotype block structure, these ancestral blocks are organized, within a breed, into larger blocks of a few hundred kilobases. In practice, such a structure similar to that already reported in dogs makes it possible to develop a chip of <300,000 SNPs, which should be efficient for mapping purposes in most cattle breeds.
DOMESTIC cattle represent a major source of milk, meat, hides, and draft energy (Lenstra and Bradley 1997) with ∼800 different breeds found around the world and classified in two major morphological groups: the humpless taurine and the humped zebu types. Humpless cattle (Bos taurus) are the most common in regions with a temperate climate and include breeds reaching a high degree of specialization, such as the Holstein breed for milk production. Conversely, humped cattle (Bos indicus) are better adapted to dry and warm climates. Unravelling the genetic basis of phenotypic diversity among the numerous cattle breeds (Andersson and Georges 2004) contributes to the development of more efficient methodologies for genetic improvement. Until recently, genetic studies in domestic species have been hampered by the lack of detailed genomic resources. However, several studies have demonstrated the power of high-density genotyping in mapping disease or trait loci in cattle and the use of population linkage disequilibrium (LD) information has provided encouraging perspectives for increasing fine-mapping resolution (Meuwissen et al. 2002; Grisart et al. 2004; Olsen et al. 2005; Gautier et al. 2006). Such approaches are ultimately limited by the size of the haplotype segment remaining in the population and containing the causative allelic variant. Indeed, extensive studies in humans (Reich et al. 2001; Gabriel et al. 2002), dogs (Lindblad-Toh et al. 2005), and, more recently, cattle (Khatkar et al. 2007) have shown that the genome is mainly a mosaic of haplotype blocks (defined as regions with a high marker–marker LD and a low haplotype diversity) separated by short segments of very low LD. Several factors such as variable recombination and mutation rates and genetic hitchhiking explain this complex pattern (Ardlie et al. 2002; Reich et al. 2002). Thus, in the case of quantitative or other complex traits that are presumed to be controlled by common variants, genotyping only a fraction of the markers located inside haplotype blocks should decrease genotyping costs without altering mapping power (Cardon and Abecasis 2003). Furthermore, since evolutionary forces such as drift, inbreeding, or gene flow are expected to influence the structure of the whole genome in a similar fashion and are strongly related to the demographic history of the populations, the analysis of the extent of marker–marker LD provides valuable information (Hayes et al. 2003; Tenesa et al. 2007).
Nevertheless, characterizing the extent of LD requires that dense marker maps be available, which is still not the case for cattle. Additionally, until recently most studies have focused on bovine populations from developed countries where they are subjected to intensive breeding, such as the Holstein breed, and these studies have suggested that significant LD among markers extends over several megabases (Farnir et al. 2000; Tenesa et al. 2003; Khatkar et al. 2006; Thevenon et al. 2007). As part of the whole bovine genome sequencing project, significant efforts are currently being carried out to identify a large number of SNPs by comparing hundreds of thousands of random sequences originating from a small set of individuals belonging to different populations with a reference sequence. Furthermore, numerous bovine sequences (ESTs, BAC end sequences, shotgun reads) have been accumulating exponentially in databases since the beginning of the 1990s. Analyzing the redundancy offers a low-cost strategy for detecting SNPs in silico (Marth et al. 1999; Hawken et al. 2004; Pavy et al. 2006) but requires a validation step.
In this article, we report the detection and validation of 1536 SNPs identified in silico in 14 different cattle breeds, which represent various farming systems and origins. The SNPs were chosen to cover entirely bovine chromosome 3 (BTA03) and two segments of BTA01 and BTA15 to address three major topics:
Comparison of the efficiency of in silico SNP identification in the different breeds according to SNP detection methodology and sequence data used.
Analysis of the diversity within and relationships between the different breeds.
Comparison of the extent of LD within the different populations and its interpretation in terms of demographic history together with a description of the haplotype block structure.
MATERIALS AND METHODS
Animal material:
Table 1 summarizes information concerning breed sample size and origin, population size, group and subgroup affiliation (Fellius 1995), status, and main breeding purposes of the 1773 bovine individuals included in our study. According to Fellius's classification (Fellius 1995) based on geographical, historical, and morphological criteria, all but one (the North European polled and Celtic breeds) of the European and all the West African groups are represented. Holstein (HOL), Montbéliarde (MON), and Normande (NOR) are highly selected breeds, essentially for milk production, with a widespread use of artificial insemination (AI). MON and NOR are French regional breeds almost exclusively found close to their birthplace (eastern and northwestern France, respectively). The Charolaise (CHA) is one of the main French beef breeds now common in most French regions and also in other countries. Aubrac (AUB), Salers (SAL), Gasconne (GAS), and Maine-Anjou (MAJ) are French local breeds. The six West African cattle breeds are bred under more extensive farming systems and under tropical and subtropical conditions. They were sampled in three neighboring countries: Somba (SOM) samples were collected in the Atacora highlands (northwestern Benin/northeastern Togo), which is the birthplace of this breed, and Lagune (LAG), Borgou (BOR), and Sudanese Fulani zebu (SFU) samples were collected in Benin (in the Porto Novo region, the Borgou council, and the Malanville region, respectively). N'Dama (NDA) samples come from two experimental herds in Burkina-Faso where pure individuals originating from the breed birthplace in Fouta-Djallon (Guinea) had been introduced. Kuri (KUR) samples were collected in the area of Lake Chad.
TABLE 1.
Descriptions of samples
| Code | Breed name (species) | Sample size | Sample area | Population size (×1000)a | Groupb | Statusb | Purposeb |
|---|---|---|---|---|---|---|---|
| AUB | Aubrac (BTA) | 14 | Southwestern France | 105 | 4A | Regional | Beef |
| BOR | Borgou (BTA × BIN) | 15 | Benin | 600c | 11D | National | Dairy/work/beef |
| CHA | Charolais (BTA) | 132 | Eastern France | 1600 | 3F | National/global | Beef |
| GAS | Gasconne (BTA) | 15 | Southwestern France | 25 | 4B | Regional | Beef/work |
| HOL | French Holstein (BTA) | 1022 | France | 2800 | 2B | International | Dairy/beef |
| KUR | Kuri (BTA) | 15 | Chad | 120c | 11B | Regional | Dairy/work/beef |
| LAG | Lagune (BTA) | 16 | Benin | 37.5c | 11D | Regional/exported | Beef |
| MAJ | Maine-Anjou (BTA) | 16 | Northwestern France | 70 | 2F | Regional/international | Beef/dairy |
| MON | Montbéliarde (BTA) | 197 | Eastern France | 710 | 3E | Regional/international | Dairy/beef |
| NDA | N'Dama (BTA) | 11 | Guinea | 3760 | 11C | Imported | Dairy/work/beef |
| NOR | Normande (BTA) | 275 | Northwestern France | 810 | 2F | Regional/international | Dairy/beef |
| SAL | Salers (BTA) | 16 | Southwestern France | 180 | 4A | Regional/international | Beef/dairy |
| SFU | Sudanese Fulani (BIN) | 14 | Benin | ? | 12C | National | Dairy/work/beef |
| SOM | Somba (BTA) | 15 | Atacora highlands | 4.7c | 11D | Regional/exported | Beef/dairy |
| Saanen (CHI) | 40 | France | |||||
| Total | 1813 (1773 + 40) |
BTA, B. taurus; BIN, B. indicus; CHI, Capra hircus. Breed names for which a pedigree is available are in italics.
Source for European breeds is at http://www.inapg.inra.fr/dsa/especes/bovins/ (reported values correspond to the French population) and for African breeds at http://dad.fao.org/.
From Fellius (1995). The group is a numeral and subgroups are denoted by a letter. The underlined terms correspond to the main breeding purpose.
Reliability unknown.
Pedigree information was available for 4 breeds: HOL, NOR, MON, and CHA. Of the 1022 HOL individuals, 973 are AI bulls organized in 17 half-sib families (30–117 individuals/family) and 49 belong to a complex pedigree in which the causal polymorphism for syndactyly segregates (Duchesne et al. 2006). For NOR and MON, the samples consist of 275 and 197 AI bulls, respectively, organized in six and four half-sib families (18–66 and 10–65 bulls/family). For CHA, the 132 individuals belong to a complex five-generation pedigree. For the other 10 breeds, samples are composed of unrelated individuals collected in France and different West African countries (Moazami-Goudarzi et al. 1997, 2002; Quéval et al. 1998; Souvenir Zafindrajaona et al. 1999). DNA was available from previous studies except for HOL, MON, and NOR AI bulls for which DNA was extracted according to standard procedures (Jeanpierre 1987) from the semen sample bank maintained at the Institut National de la Recherche Agronomique with the help of the French AI industry. Finally, 40 goat samples with known pedigree relationships were also included in the study to evaluate the rate of interspecific success of the genotyping procedure and to deduce the potential ancestral allelic state.
SNP genotyping:
SNP detection methodologies:
We chose to detect SNPs in silico on the basis of the sequences available in public databases. However, one drawback of this approach is that, for most of the bovine sequence data, no trace-quality information is available and at the time this study was carried out, all available SNP detection software strongly relied on trace-quality values. Thus, we had to develop our own bioinformatic solutions, and SNPs were detected using two different strategies. The first approach aimed at detecting SNPs from available EST data. A set of 1,000,000 bovine ESTs available in dbEST in January 2006 was downloaded and clustered according to their similarities with human transcripts provided by the ensembl database (http://www.ensembl.org). The sequences were subsequently assembled using the Cap3 software. A position was considered polymorphic if it satisfied the following criteria: the position had to be included in a multiple alignment containing at least five EST sequences showing at most two different residues in the corresponding column with the rare variant observed at least twice. In addition, the five adjacent left and right columns of this candidate SNP position should not show any discrepancy among sequences. The results of this SNP detection strategy on EST data for bovine and a few other species are available at http://www.bioinfo.genopole-prd.fr/Iccare. The second approach was based on whole-genome shotgun data produced by the Baylor College of Medicine (BCM) for five different breeds (see ftp://ftp.hgsc.bcm.tmc.edu/pub/data/Btaurus/snp/Btau20050310/README for details). We have implemented our own in silico detection method, which is essentially the same as the BCM method. The shotgun reads were masked, using RepeatMasker (http://www.repeatmasker.org), for known bovine repeats and aligned to the Hereford bovine Btau20050310-freeze assembly using BLAST. Only the reads, which could be confidently assigned to a contig were retained (read–contig alignment >300 bp with at least 97% of identity). A position was defined as polymorphic when the read at that position differed from the nucleotide in the assembly while having a good trace-quality value (>60) as estimated by the phred quality score (Ewing and Green 1998). In addition, we required that the four nucleotides surrounding the candidate position be identical to those in the assembly with a high supporting phred quality score (>30).
SNP selection and genotyping:
Overall, 1536 SNPs, 931 resulting from the strategy using EST data and 605 from that using shotgun data, were selected from among all the available in silico-detected SNPs (details are given in supplemental Table 1 at http://www.genetics.org/supplemental/). The selection strategy aimed at providing a dense coverage of the complete BTA03 chromosome and two small regions of BTA01 and BTA15. To that end, predicted locations were obtained on the basis of sequence similarities with the human genome (hg18 whole-genome sequence assembly) and state-of-the-art comparative maps (Everts-Van Der Wind et al. 2004). In total, 1373 SNPs were chosen to cover BTA03 and were conserved with three different regions of the human genome spanning ∼120 Mb (from positions 35 to 120 Mb and 142 to 166 Mb on HSA01 and from 232 to 242 Mb on HSA02). A total of 96 and 67 SNPs anchoring, respectively, to HSA21 (from positions 29 to 46 Mb) and to HSA11 (from positions 43 to 46 Mb) were chosen to cover the centromeric region of BTA01 and the telomeric region of BTA15. Genotyping of the 1536 SNPs was performed at the French National Genotyping Center according to standard procedures using a high-throughput GoldenGate assay provided by Illumina (http://www.illumina.com; Illumina, San Diego).
Map construction:
All the markers were mapped to bovine contig sequences of the currently available whole-genome assembly (Btau 3.1) and anchored to the most recent version of the human genome assembly (hg18) using the BLAST program (Altschul et al. 1997). Linkage maps were then constructed using the Multimap/Crimap software suite (Matise et al. 1994). Twenty-five half-sib families (17, 5, and 3 belonging, respectively, to HOL, NOR, and MON breeds) with >30 offspring were used, providing a pedigree of 1381 individuals (from 31 to 114 individuals/family). At first, we considered only the most informative marker-per-contig sequence. When the linkage map order derived from the whole-genome assembly was challenged, we observed inconsistencies, confirming discrepancies among the Btau 2.0 assembly, published radiation hybrid (RH) maps, and the latest Btau 3.1 bovine assembly. Therefore, we decided to build a linkage map from scratch. We started by identifying the three expected linkage groups (one for each chromosome) and then constructed framework maps at different LOD-score thresholds. This allowed us to identify and confirm, independently, blocks of conserved synteny identified from dense RH maps (Everts-Van Der Wind et al. 2004) in the bovine regions of interest, taking the human genome as reference. On the basis of this comparative mapping information, we produced comprehensive maps, which were challenged using the “flips” option. Unlikely double crossovers were finally identified using the “chrompic” option.
Physical map distances between markers belonging to the same chromosome were estimated according to their position on the human genome. Distances between SNPs within identical bovine sequence contigs from the most recent bovine genome assembly were in good agreement with their respective human counterpart. Finally, the physical distances separating contiguous blocks of conserved synteny on bovine chromosomal regions were estimated from the genetic distance obtained, considering 1 cM as equivalent to 1 Mb. Within blocks, the average observed centimorgan-to-megabase ratio was 0.930 and thus in good agreement with this latter approximation.
LD and genetic diversity analysis:
Genotyping data:
Since individuals from 10 of the 14 breeds considered in our study are unrelated, analyses were performed using diplotypic data. For the remaining four breeds (CHA, HOL, MON, and NOR), we selected only a small subset of individuals from the available pedigrees: i.e., for the CHA breed, 25 founder individuals (with no parental information), and for the HOL, MON, and NOR breeds, two to four individuals/half-sib family without any common ancestor for at least three generations on the maternal side. For HOL, 6 founder individuals from the pedigree segregating the syndactyly causal mutation (Duchesne et al. 2006) were also included in the sample. Finally, 39, 36, and 33 individuals were selected for the HOL, MON, and NOR breeds, respectively.
Across-breed LD was investigated by artificially constructing several composite populations of 56 individuals (4 individuals randomly drawn per breed). To limit sampling biases, results from 30 samples were averaged.
Genetic diversity analysis:
SNP allele frequencies, the mean number of alleles (MNA), and unbiased estimates of gene diversity (Nei 1978) were determined across the different breeds using the program GENETIX 4.05 (Belkhir et al. 2004). Fisher's exact test for Hardy–Weinberg equilibrium (HWE) was performed for each marker using the R genetics package (http://cran.r-project.org/src/contrib/Descriptions/genetics.html).
Measuring pairwise LD:
Due to the small size of the samples, SNPs were rejected if their minor allele frequency (MAF) was <0.05 or their P-value for HWE test was <0.01. The r2 and other classical LD measures were computed with the R genetics package. To evaluate how far the same marker phase is likely to persist across pairs of breeds (the extent of ancestral LD), we calculated, for different distance ranges, the correlation coefficient between the mean pairwise r defined as the square root of r2 (Goddard et al. 2006). The sign of r in each population was given so that the 2 × 2 contingency tables (haplotype phase combination) used to calculate LD were the same across populations.
Inferring population demographic history from LD:
For autosomal loci and considering both experimental and evolutionary sampling effects, the expected r2 between neutral markers can be related to genetic distance c (in centimorgans), effective population size Ne, and experimental chromosomal sample size n according to the formula E(r2) = 1/(α + 4Nec) + 1/n, where α = 1 (α ≈ 2) if mutation is (not) taken into account (Hill 1975; Sved 1971; Tenesa et al. 2007). Assuming a linear population growth and without considering mutation (α = 1) in the model, the (chromosome) effective population size Ne, [1/2c] generations ago, can then be estimated provided c and E(r2) are known (Hayes et al. 2003; Tenesa et al. 2007). Simulation studies revealed that estimates of past effective population sizes were not greatly affected by departure from the assumption of a linear population growth (Hayes et al. 2003). For our different populations, marker-pair r2 values adjusted for chromosome sample size (Tenesa et al. 2007) were averaged for different distance ranges to give an estimate of E(r2) for a distance c (midpoint of the corresponding range). Since our linkage map was not sufficiently resolute for small distances, genetic distances were obtained from physical distances, assuming 1 cM is equivalent to 1 Mb (see above).
Population haplotype block structure:
Haploview 4.0 software (Barrett et al. 2005) was used to identify haplotype block boundaries and to estimate within-block haplotype diversity using the so-called four-gamete rule. In this approach, the population frequencies of the four possible two-marker haplotypes are computed. If all four are observed with a frequency of at least 0.01, a recombination is deemed to have taken place. Blocks are then formed by consecutive markers where only three gametes are observed. SNPs were rejected if their P-value for the HWE test was <0.01 or their MAF was <0.1.
Genetic structure and relationships:
The F-statistics (Wright 1965) FIT, FST, and FIS were estimated, respectively, in the form of F, θ, and f (Weir and Cockerham 1984) using the program GENETIX 4.05 (Belkhir et al. 2004). Significance and variance of the F-statistics were determined from permutation tests (1000 permutations) and jack-knife over loci. GENETIX 4.05 was also used to compute FST statistics among pairs of breeds, within-breed FIS, and respective statistical significances (1000 permutations). The Nei's genetic distances (Nei 1978) between the different pairs of breeds were estimated using PHYLIP 3.65 package (Felsenstein 1989). These were further used for dendogram construction according to the neighbor-joining (NJ) algorithm (Saitou and Nei 1987) implemented in the PHYLIP package (Felsenstein 1989). The reliability of each node was estimated from 10,000 random bootstrap resamplings of the data.
RESULTS
SNP validation:
Among the 1536 SNPs genotyped, 111 failed to give any genotype and 524 were found to be completely monomorphic across the full cattle sample (Table 2). Among the 901 SNPs (∼60%) polymorphic in at least one of the 14 breeds, 196 were discarded because of their low genotyping success rate (<90%). Nine additional SNPs were discarded because of a high genotyping error rate identified when analyzing segregation within available pedigrees (CHA, HOL, MON, NOR) or because of other discrepancies (either only heterozygous or both homozygous genotypes present in at least one population).Thus, 696 SNPs were retained for further analysis.
TABLE 2.
Number of markers as a function of the in silico detection method
| Identification method | First approach (EST) | Second approach (shotgun reads) | Total |
|---|---|---|---|
| All markers | 931 | 605 | 1536 |
| Failed | 97 (10)a | 14 (2)a | 111 (7)a |
| Monomorphic | 497 (53)a | 27 (4)a | 524 (34)a |
| Low genotyping success rate or other problems | 120 (13)a | 85 (14)a | 203 (13)a |
| Conserved for further analysis | 217 (23)a | 479 (79)a | 696 (45)a |
| MAF > 0.05 in 0 breed | 10 (5)b | 4 (0.1)b | 14 (2)b |
| MAF > 0.05 in 1 breed | 16 (7)b | 19 (4)b | 35 (5)b |
| MAF > 0.05 in 2–5 breeds | 18 (8)b | 58 (12)b | 76 (11)b |
| MAF > 0.05 in 6–10 breeds | 44 (20)b | 121 (25)b | 165 (24)b |
| MAF > 0.05 in 11–13 breeds | 73 (33)b | 152 (31)b | 225 (32)b |
| MAF > 0.05 in 14 breeds | 56 (26)b | 125 (26)b | 181 (26)b |
| Worked on goat (genotype success rate >0.8) | 114 (52)b | 187 (39)b | 301 (43)b |
Numbers within parentheses are percentages.
Calculated from the corresponding total number of SNPs.
Calculated from the corresponding number of SNPs conserved for further analysis.
Overall, there is a clear difference between the two SNP prediction methods regarding their ability to detect true polymorphic sites and their validation rate (Table 2). The most striking differences between the two approaches reside in the higher rate of failure and monomorphic proportion of markers. Only ∼25% of the SNPs detected from EST data were retained, while 79% were retained in the 605 SNPs detected on shotgun reads. For the two methods, a similar proportion of SNPs was discarded because of low genotyping success rate. Interestingly, among the 696 SNPs finally retained, 303 had a genotyping rate success >80% in the 40 goat individuals tested (Table 2), of which 7 appeared polymorphic within the goat group. Four of these latter SNPs displayed a clear deviation from Mendelian inheritance expectations (P < 0.001) and another one harbored a genotyping error. Thus, only 2 (<1%) SNPs (rstoul_bta3_snp_460 and rstoul_bta3_snp_602) of the 303 bovine SNPs working in goat were found to be polymorphic in this latter species. SNPs derived from EST sequences tended to work better on the goat sample (Table 2).
SNP polymorphism across the different breeds:
As shown in Table 3, the LAG breed is the least variable with <50% of the 696 SNPs displaying a MAF <0.05. For the other breeds, a moderate-to-high proportion of SNPs are informative: the proportion of SNPs with a MAF >0.05 varies from 63.6% (NDA) to 82.9% (HOL). When considering previous work based on the same populations but with other marker types (microsatellite, blood protein loci, or blood group systems) (Moazami-Goudarzi et al. 1997; Quéval et al. 1998; Souvenir Zafindrajaona et al. 1999), there is an unexpected lower observed variability in African compared to European breeds. While 94.6% of the SNPs are polymorphic (MAF > 0.05) in at least one European breed, only 81.8% are polymorphic in at least one African breed. Part of this observation might be explained by the small size of the sample. Indeed, 93.5% of the SNPs are polymorphic in CHA, HOL, MON, or NOR breeds while 87.5% of the SNPs are polymorphic in at least one of the four remaining European breeds, which have sample sizes similar to those of the African breeds. Nevertheless, the ascertainment bias in SNP discovery most probably originates from the overrepresentation of sequences from European cattle origin in sequence databases. Hence, among the 577 SNPs polymorphic in HOL, only 71.1% are polymorphic in at least one African breed while 78.0% are polymorphic in at least one of the four European breeds with small sample sizes (AUB, GAS, MAJ, and SAL) and 77.2% in at least one of the three other European breeds (CHA, MON, and NOR). Conversely, only 3.3% of the SNPs, which are polymorphic in at least one African breed, are not polymorphic in any other European breed (6% when considering only AUB, GAS, MAJ, and SAL). As a consequence, the unbiased gene diversity (He) on average is 0.25 (from 0.188 in LAG to 0.279 in BOR) for African breeds and 0.30 (from 0.282 in NOR to 0.322 in HOL) for European breeds. Likewise, the MNA over the 696 SNPs on average is 1.73 (from 1.54 in LAG to 1.80 in BOR) in African breeds and 1.85 (from 1.82 in SAL to 1.90 in HOL) in European breeds.
TABLE 3.
Genetic variability within the different breeds across 696 SNPs
| Breed | % of markers genotyped on >90% of the sample | % of markers with MAF > 0.05 | % of markers with MAF > 0.01 | Expected heterozygosity (SD) | He (SD) | MNA | % in HWE (P > 0.01) |
|---|---|---|---|---|---|---|---|
| AUB | 99.1 | 75.0 | 81.3 | 0.287 (0.190) | 0.298 (0.187) | 1.84 | 81.2 |
| BOR | 97.4 | 71.0 | 77.7 | 0.269 (0.187) | 0.279 (0.194) | 1.80 | 77.4 |
| CHA | 99.4 | 79.3 | 85.8 | 0.312 (0.171) | 0.319 (0.175) | 1.89 | 87.1 |
| GAS | 98.9 | 74.9 | 81.2 | 0.296 (0.193) | 0.307 (0.189) | 1.83 | 80.7 |
| HOL | 100 | 82.9 | 90.1 | 0.318 (0.168) | 0.322 (0.170) | 1.90 | 87.9 |
| KUR | 97.0 | 68.7 | 77.2 | 0.258 (0.188) | 0.268 (0.193) | 1.79 | 76.7 |
| LAG | 98.7 | 47.4 | 52.7 | 0.182 (0.203) | 0.188 (0.207) | 1.54 | 52.4 |
| MAJ | 99.7 | 78.2 | 84.1 | 0.287 (0.179) | 0.298 (0.186) | 1.85 | 82.0 |
| MON | 100 | 74.1 | 85.1 | 0.280 (0.181) | 0.284 (0.183) | 1.86 | 83.0 |
| NDA | 99.1 | 63.6 | 72.7 | 0.242 (0.188) | 0.254 (0.197) | 1.74 | 72.4 |
| NOR | 100 | 74.9 | 83.2 | 0.278 (0.181) | 0.282 (0.184) | 1.84 | 81.6 |
| SAL | 98.6 | 76.1 | 80.6 | 0.291 (0.185) | 0.301 (0.190) | 1.82 | 79.6 |
| SFU | 97.1 | 64.7 | 74.3 | 0.242 (0.188) | 0.254 (0.197) | 1.75 | 73.4 |
| SOM | 98.4 | 64.4 | 69.8 | 0.238 (0.194) | 0.247 (0.199) | 1.75 | 69.8 |
Yet, most SNPs are polymorphic in several breeds (Table 2), probably because of their ancient origin. In particular, 181 SNPs (26%) display a MAF >0.05 in all 14 breeds, and 546 SNPs (78%) are polymorphic in at least one European and one African breed. Finally, no significant departures from the HWE were observed among the polymorphic markers in any breed since only 0.14% (respectively 0.4%) of the HWE tests performed had a P-value <0.1% (respectively 1%). This corresponds to the proportions expected from the type I error rate at a 0.001 (respectively 0.01) threshold.
Map construction:
The 696 SNPs retained were anchored to 506 different sequence contigs (from 1 to 6 markers/contig and 1.4 on average) from the most recent Btau3.1 whole bovine genome assembly. Thirty-one (including 44 markers), 342 (including 487 markers), and 19 (including 22 markers) of these contigs were assigned to BTA01, BTA03, and BTA15, respectively, on the assembly while 63 contigs (including 87 markers) were unassigned. Among the 87 SNPs unassigned to a chromosome on the assembly, 5, 76, and 6 were expected on BTA01, BTA03, and BTA15, respectively, from comparative mapping results. Conversely, 56 SNPs (anchoring to 51 different contigs) were assigned to a chromosome different from the one initially targeted. As the order of contigs and scaffolds is suboptimal on the Btau 3.1 assembly, we decided to construct a genetic map for the three chromosomes targeted using available pedigrees in HOL, NOR, and MON. Among the 696 SNPs, 90 SNPs had no or <50 informative meioses and were not considered to build the genetic maps. The remaining 606 SNPs had an average of 244 (from 54 to 476) informative meioses. Forty-one, 471, and 27 SNPs (anchoring to 31, 349, and 21 different contigs) were assigned or expected on BTA01, BTA03, and BTA15, respectively. At a LOD-score threshold of 6, two linkage groups were identified for BTA03 and a LOD-3 framework map containing 44 SNPs was further constructed. The LOD-3 framework map was anchored on the human hg18 genome assembly, allowing the identification of three blocks of conserved synteny, which confirmed previous results (Everts-Van Der Wind et al. 2004). On the basis of comparative map information, we finally obtained a 460-SNP comprehensive map extending the block boundaries somewhat (position 165 to 142 Mb and position 120 to 35 Mb on HSA01 and from 234 to 242 Mb on HSA02). With a similar strategy, we constructed linkage maps for the regions mapping to BTA01 and BTA15. Details of the maps are given in Table 4.
TABLE 4.
Size, marker number, and density of the genetic maps
| Marker no. | Size
|
Marker density
|
Marker spacing mean (min–max)
|
|||||
|---|---|---|---|---|---|---|---|---|
| Region | BTA | In cM | In Mb | Per cM | Per Mb | In cM | In Mb | |
| 1 | BTA01 | 20 | 15.6 | 12 | 1.28 | 1.67 | 0.822 (0.0–7.3) | 0.631 (0.0–7.3) |
| 2 | BTA01 | 23 | 13.3 | 13.1 | 1.73 | 1.76 | 0.607 (0.0–4.1) | 0.596 (0.0–3.0) |
| 3 | BTA03 | 460 | 127 | 125 | 3.62 | 3.68 | 0.277 (0.0–5.3) | 0.273 (0.0–3.0) |
| 4 | BTA15 | 23 | 6.32 | 4.99 | 3.64 | 4.61 | 0.287 (0.0–1.9) | 0.222 (0.0–0.67) |
| All | 526 | 162 | 155 | 3.24 | 3.39 | 0.297 (0.0–7.3) | 0.311 (0.0–7.3) | |
Extent of LD with marker distance:
The 526 SNPs with confirmed mapping information were used to evaluate the extent of pairwise LD with physical distance. In addition, 56 SNPs belonging to 24 sequence contigs of the Btau 3.1 assembly containing at least two markers were included in the analysis. The number of marker pairs available (with the two SNPs satisfying a MAF >0.05) for different distance ranges is detailed in Figure 1A. The small differences observed among African and European cattle are mostly related to the number of available SNPs satisfying the condition on the MAF (see above). As expected, the level of pairwise LD as measured by r2 decreases within each breed with marker distance (Figure 1B). The decrease is more or less pronounced across the different breeds until a rather high average value (>0.1) at large distances (>1 Mb). Interestingly, r2 mean values across the different breeds and the average composite population samples are very high (>0.5) at distances <10 kb. This high LD signal at small physical distances is almost completely eroded when considering markers >500 kb apart: for the 500 kb to 1 Mb distance range, the mean r2 is <0.05 for the average composite population samples while always >0.1 within each breed.
Figure 1.—
Pairwise LD analysis within the different populations and across the corresponding average composite population (30 replicates of a composite population consisting of 56 individuals with 4 randomly drawn within each of the 14 breeds). (A) Number of SNP pairs available for each range of marker distance considered (measured in megabases). (B) Mean r2 value for each range of marker distance considered. (C) Estimates of the effective population size (Ne) from r2 at different times in the past (measured in number of generations). Further details are given in supplemental Table 3 at http://www.genetics.org/supplemental/.
In addition, for markers <10 kb apart (supplemental Table 2 at http://www.genetics.org/supplemental/), r values are in general always highly correlated even for distantly related breeds. On average, the correlation coefficient is equal to 0.77 (±0.1) among pairs of European breeds (from 0.54 between AUB and GAS to 0.93 between HOL, CHA, and MON) to 0.71 (±0.13) among pairs of African breeds (from 0.44 between BOR and LAG to 0.86 between LAG and SOM) and to 0.65 (±0.13) among pairs of African and European breeds (from 0.32 between BOR and GAS to 0.84 between NDA and CHA). The correlation coefficient then drops quickly to <0.5 when considering larger SNP distances (>50 kb). On average, for the 50–500 kb distance range, it is equal to 0.48 (±0.06) among pairs of European breeds, 0.42 (±0.05) among pairs of African breeds, and 0.41 (±0.07) among pairs of African and European breeds.
Estimation and evolution of ancestral Ne:
The observed decrease of r2 with physical distance from a high value (>0.5) suggests a decline of the overall population size as illustrated for the different populations in Figure 1C. Interestingly, the pattern is similar for the different breeds, which have been subjected to different constraints in their recent history. The most striking bottleneck appeared ∼1500 generations ago, which corresponds roughly to the beginning of the domestication process, assuming a generation time of six to seven years. A more recent event (50–100 generations ago) might correspond to an intensification of the population isolation (breed formation in Europe corresponding to an extreme). Finally, estimation from long-range LD of the current (fewer than five generations ago) effective population size (Figure 1C and supplemental Table 3 at http://www.genetics.org/supplemental/) gave very low values for the different populations with an average of 35 (from 22 in NDA to 46 in CHA). Nevertheless, at large physical distances, these estimates might be somewhat downwardly biased by low sample sizes.
Haplotype block structure:
As shown in Table 5, from 53 (for LAG) to 97 (for SAL) haplotype blocks were identified with an average of 81.5, which is above the value (70.8) observed for the average simulated composite population. The corresponding mean block size varies from 298 kb (for CHA) to 766 kb (for LAG in which far fewer SNPs are informative) with an average of 427 kb, i.e., three times more than for the average composite population (171 kb). The within-block haplotype variability is quite similar among the different breeds with on average 3.2 (from 2.90 in CHA to 3.43 in SFU) common haplotypes segregating for blocks defined on average by 2.7 SNPs (from 2.47 in CHA to 2.85 in LAG). Assuming by definition that no recombination occurred in the history of the block, three (respectively four) haplotypes at most must be observed when considering a block of two (respectively three) SNPs. Thus, the observed haplotype variability is in the range imposed by the method used. Nevertheless, the chromosome coverage of the haplotype blocks is still limited for the different breeds (from 20.7% for NOR to 30.1% for BOR), suggesting that a higher SNP density might be necessary to draw a more precise picture of the haplotype block structure among the different breeds.
TABLE 5.
Haplotype block structure identified using the four-gamete rule for the BTA03 chromosome
| Breed | No. of blocks | No. of markers within blocks | Mean no. of markers per block (min–max) | Mean block size in kb (min–max) | % chromosome coverage | Mean no. of haplotypes per blocks (min–max) | Mean frequency of the most frequent haplotype (min–max) |
|---|---|---|---|---|---|---|---|
| AUB | 91 | 243 | 2.67 (2–6) | 417 (0.2–2740) | 29.9 | 3.19 (2–6) | 0.53 (0.29–0.86) |
| BOR | 78 | 208 | 2.67 (2–8) | 464 (0.3–2565) | 28.5 | 3.29 (2–8) | 0.52 (0.21–0.82) |
| CHA | 94 | 232 | 2.47 (2–7) | 298 (0.22–2195) | 22.0 | 2.9 (2–5) | 0.56 (0.30–0.88) |
| GAS | 93 | 249 | 2.68 (2–6) | 392 (0.24–2696) | 28.7 | 3.23 (2–7) | 0.53 (0.29–0.79) |
| HOL | 94 | 268 | 2.85 (2–9) | 345 (0.22–2696) | 25.5 | 3.22 (2–6) | 0.54 (0.22–0.90) |
| KUR | 79 | 207 | 2.62 (2–6) | 438 (0.3–4623) | 27.2 | 3.27 (2–7) | 0.54 (0.31–0.89) |
| LAG | 53 | 151 | 2.85 (2–6) | 766 (0.22–5930) | 31.9 | 3.37 (2–10) | 0.54 (0.34–0.84) |
| MAJ | 89 | 245 | 2.75 (2–8) | 387 (0.2–3174) | 27.1 | 3.14 (2–5) | 0.54 (0.32–0.86) |
| MON | 83 | 217 | 2.61 (2–7) | 369 (0.299–3061) | 24.1 | 3.07 (2–5) | 0.53 (0.27–0.77) |
| NDA | 75 | 194 | 2.59 (2–6) | 436 (0.22–2610) | 25.7 | 3.09 (2–6) | 0.54 (0.27–0.86) |
| NOR | 81 | 220 | 2.72 (2–5) | 325 (0.3–1894) | 20.7 | 3.06 (2–5) | 0.54 (0.35–0.89) |
| SAL | 97 | 255 | 2.63 (2–7) | 395 (0.2–2841) | 30.1 | 3.14 (2–8) | 0.55 (0.27–0.90) |
| SFU | 68 | 190 | 2.79 (2–6) | 516 (0.4–2565) | 27.6 | 3.43 (2–7) | 0.51 (0.25–0.86) |
| SOM | 67 | 176 | 2.63 (2–7) | 435 (0.2–1459) | 22.9 | 3.24 (2–7) | 0.53 (0.31–0.89) |
| All | 70.8 (±3.2) | 164.1 (±6.9) | 2.32 (±0.05) | 171 (±26) | 9.50 | 2.76 (±0.04) | 0.595 (±0.007) |
The whole population (All) values correspond to the average across 30 samples (standard deviations are given in parentheses) of a composite simulated population constructed after randomly drawing four samples per breed.
Genetic structure and relationships among 14 breeds:
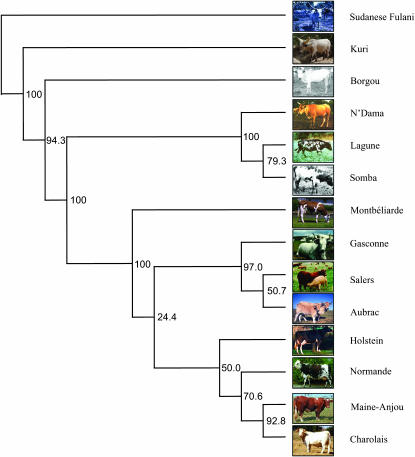
Among the 696 available SNPs, 526 (75%) were unambiguously included in a linkage map with a mean average spacing equal to 311 kb (Table 4). At such a distance, small mean r2 values between markers are observed (Figure 1B). We thus chose to consider all the validated SNPs genotyped in at least 90% of each breed sample, leading to a total of 632 SNPs. Population differentiation was first examined by the fixation indices FIT, FIS and FST across all loci. FIS appears nonsignificant in African breeds while slightly negative in European breeds, confirming only a small departure from the HWE (see above). Stronger differences in FIS values appear within the different breeds (supplemental Table 4 at http://www.genetics.org/supplemental/) but the high locus heterogeneity suggests that they originate more likely from small sampling biases. The average genetic differentiation among breeds, measured as FST value, was 15.5%. According to geographic origin, we obtained average values of 11.9% for African breeds and 9.9% for European breeds. All FST values between pairs of breeds were significantly different from zero (supplemental Table 4). Among pairs of European breeds, FST varies from 0.035 (SAL–AUB) to 0.132 (HOL–NOR) with a mean value of 0.090. Genetic differentiation is slightly higher among pairs of African breeds ranging from 0.031 (KUR–BOR) to 0.277 (KUR–LAG) with a mean value of 0.12. As expected, the differentiation among pairs of European and African breeds remains important, varying from 0.162 (CHA–BOR) to 0.315 (MAJ–LAG) with a mean value of 0.21. Genetic differentiation is confirmed when looking at the Nei's genetic distances calculated among the different breeds (supplemental Table 4). Indeed, average distances between European and African breeds (0.110 ±0.02) are higher than within African (0.040 ± 0.03) and European (0.040 ± 0.01) breeds. Finally, Figure 2 shows the unrooted consensus tree obtained for the 14 cattle breeds, using the NJ clustering method with Nei's distance matrix. This tree clearly separates European breeds from African breeds. As indicated by the high bootstrap values (half of the nodes overcome the 95% confidence level), three main groups are separated: the European taurine cluster, the African taurine cluster, and the group formed by KUR, BOR, and SFU.
Figure 2.—
Unrooted consensus tree showing the genetic relationships among the 14 breeds considered using the neighbor-joining method and the unbiased Nei's genetic distance. Numbers at the nodes are the reliabilities in percentages estimated after 10,000 bootstrap resamplings.
DISCUSSION
Efficiency of in silico SNP detection methods and SNP polymorphism and influence of sequence data:
Genotyping of the 1536 SNPs detected in silico from a large panel of individuals from several different breeds allowed us to draw a posteriori conclusions on the efficiency of the different detection methods. The different validation values (true positive rate) of the SNP prediction methods vary mainly according to the nature of the sequence data. Indeed, we discarded 55% of the 1536 genotyped SNPs, which corresponded to 75% of the SNPs detected from EST data (with no trace-quality values) and to only 21% of the SNPs detected using shotgun reads. When considering EST data, the validation rate has been shown to vary from as little as 24% in the absence of trace-quality values (Huntley et al. 2006) to as high as 99% when SNPs are detected in sequences from PCR-amplified diploid samples. An intermediate 80% validation rate was observed when using PolyBayes software on EST data with associated trace-quality values (Pavy et al. 2006). Our results fall within the range corresponding to methods using the same kind of information. Given the true positive rate values in the studied population, we tried to improve our SNP detection method for EST data by adjusting several parameters (cluster depth, size of dissimilarity-less neighborhood, a priori rate of difference between paralogous sequences). We were able to increase the true positive rate only up to 60% (data not shown). Overall, this underlines that trace-quality information and the availability of a complete genome sequence are of primary importance for SNP detection and that the method of choice, in this high-throughput genomic era, is the exploitation of shotgun reads produced in one or a few large-scale projects and a reference assembly.
Most of the bovine sequence data available in databases are from individuals belonging to European cattle breeds (Hereford and Holstein). This might explain why the observed polymorphism of our SNP data set was highest in the Holstein breed, although differences in sample sizes also affect the observed SNP ascertainment bias in our study (see results). Nevertheless, 60% of the SNPs analyzed had a MAF >0.05 in >10 of the 14 breeds studied. Most of the SNPs identified might in fact be old relative to the very recent formation of breeds (∼200 years ago). This has attractive implications for SNP detection programs, since most of the SNPs detected in one breed with a sufficiently high polymorphism are expected to be polymorphic in several other breeds, even if distantly related. Finally, due to sequence similarities with the goat genome, SNP genotyping was effective in our goat sample for >40% of the SNPs, with a slightly better score when considering those derived from EST sequences, as expected from a better conservation of coding sequences. Two of these were found to be polymorphic (<1%) in goat; the remaining ones were monomorphic, which gave insights into the ancestral status of the cattle allele. Even if these results need to be taken with caution since some of the amplified sequences might not be strictly orthologous and only two of the bovine expected alleles can be detected using our genotyping methodology, they are not surprising because the divergence time between goat and cattle corresponds to that of the bovids, i.e., ∼18.5 million years (Vrba and Schaller 2000). Nevertheless, genotyping individuals from other Bos species more closely related to cattle, such as buffalo (Bos bubalis), bison (Bos bison) or yak (Bos grunniens), might be a more straightforward way to determine ancestral SNP alleles.
Relationships between the different breeds:
On average, genetic differentiation (FST) among breeds was 15.5, 9.9, and 11.9% for European and African breeds, respectively. When considering European breeds, similar values of genetic differentiation (FST = 9.9%) have been obtained using microsatellite data: 11.2% for 7 European breeds (MacHugh et al. 1998), 10.7% for 20 northern European breeds (Kantanen et al. 2000), and 6.8% for 18 southwestern European cattle breeds (Jordana et al. 2003). In our study, genetic differentiation among the 6 African breeds was slightly higher than in the European breeds (11.9% vs. 9.9%), the value obtained being almost identical to that (11.4%) obtained using microsatellite data available for 4 of them (Moazami-Goudarzi et al. 2002). As expected, the NJ tree (Figure 2) shows a clear separation between African and European breeds. Within African breeds, two groups were distinguished: (i) the African taurine group (LAG, NDA, and SOM living in regions where the tsetse fly is endemic) and (ii) the KUR, BOR, and SFU group. These findings are in agreement with previous and more documented studies that demonstrated the influence of historical and ecological factors in hybridization events in Africa between the two subspecies of cattle (B. taurus and B. indicus) (Hanotte et al. 2002; Freeman et al. 2004, 2006). Similarly, although less robustly, relationships among European cattle breeds remain concordant with previous breed classifications according to geographical, morphological, and historical criterions (Fellius 1995). A notable exception is represented by CHA, which appears closer to the group represented by NOR and MAJ than the group represented by MON, as expected. To improve meat quality, infusion of the British Durham breed is known to have occurred at a significant level in the NOR, MAJ, and CHA breeds during the 19th century, probably contributing to positioning of these three breeds in the same group. Nevertheless, previous results have tended to minimize such an influence of the Durham breed (Grosclaude et al. 1990).
Extent of LD and haplotype block structure:
Most of the SNPs genotyped in our study were included in a linkage map constructed on the basis of pedigree and comparative mapping information. On the basis of this information we were able to study and compare the extent of LD across different breeds. Interestingly, a similar pattern was observed irrespective of the breed origin. In particular, a high level of LD was described at short distances (<10 kb), which was >0.6 on average when considering r2 measures. Recently, similar values were reported for the Angus and Holstein breeds (Goddard et al. 2006). At such small distances, our observations are not consistent with the model considering mutation, for which the theoretical limit is 0.5 when c tends toward 0 (see materials and methods), and suggest a decreasing trend in the effective population sizes. In addition, for SNPs <10 kb apart, we also found a high correlation among r values across the different breeds and even between European and African breeds. This strong LD signal most probably reflects the ancestral one, which might originate from the domestication period that started ∼10,000 years ago. In addition, estimates of different past effective population sizes from the decrease of LD with marker distance (Hayes et al. 2003; Tenesa et al. 2007) suggest an exponentially decreasing trend for the various breeds, which began roughly at that time. From an average effective population size of 2000–5000 individuals, a first clear decrease was indeed observed in our study ∼1500 generations ago (equivalent to 10,000 years ago, assuming a generation time in cattle of 6–7 years). A second and more recent inflection seems also to have occurred ∼50–100 generations ago and might thus correspond to several events related to the isolation of different populations, which recently reached an extreme for European breeds after breed formation (∼200 years ago). For these latter breeds and in particular for the Holstein breed, the recent increase in population size was not accompanied by an increase in effective population size due to enhancement of selection and intensive use of AI. Because of the increased bias in estimating r2 over large distances for small samples when considering diplotypic data, it was not possible to provide a precise estimate of the current effective population size. However, values <50 for the HOL, MON, and NOR breeds are quite consistent with previous estimations from pedigree data (Boichard et al. 1996). Overall, the exponential decrease in the different cattle population sizes corresponds tightly to the exponential increase of the human population size during the same period (Tenesa et al. 2007). The development of human populations has been conditioned by the possibility of getting better food and field work supply, a significant part of which was provided by cattle. In that regard, improvement of selection methods together with the adaptation to different agro-ecological constraints have been necessary and might have had a direct consequence on the population structure of cattle.
To further compare the effect of the demographic history at the genomic level, we tried to describe the haplotype block structure of the different breeds. Using the four-gamete rule, we identified haplotype blocks covering 20–30% of the BTA03 chromosome with an average size concordant across the different considered breeds (except for the LAG breed, which presented a marked reduced gene diversity). The size range of 300–500 kb was found to be similar to that observed for dogs using the same methods (Lindblad-Toh et al. 2005). In addition and as suggested by the extent of across-breed LD (see above), the haplotype block structure in cattle might be strongly similar to that reported in dogs. Within breed, the genome might be composed of haplotype blocks of a few hundred kilobases, each of these blocks corresponding to a mosaic of smaller blocks (<10 kb long) from a more ancient origin (before domestication).
Practical implications for mapping purposes:
These results are promising for achieving a rather high resolution in mapping experiments when using new generation mapping methodologies such as those exploiting within-population LD (Meuwissen et al. 2002). QTL are likely to each be determined by a small number of causal polymorphisms at an intermediate population frequency and thus embedded in common haplotypes. Thus, the average length of haplotype blocks, previously defined, represents a higher bound of the expected mapping resolution. This is in good agreement with recent findings in the Holstein breed for which QTL affecting milk production traits have been mapped in intervals only a few hundred kilobases long (Olsen et al. 2005; Gautier et al. 2006). Recently, Khatkar et al. (2007) proposed that genotyping one tag SNP every 30–50 kb (<100,000 SNPs genomewide) would be sufficient to capture most of the LD information within the different cattle breeds. As suggested in our study, most of the SNPs are expected to be segregating in several populations. Thus, a substantial gain in mapping resolution (up to 10 kb) would still be obtained by considering several breeds since the allelic association reflecting ancestral LD structure is preserved only at very small distances across breeds. Designing a common set of 300,000 SNPs (one tag every 10 kb) for all the different breeds might thus be a straightforward approach.
Acknowledgments
We thank Tom Druet and Sébastien Fritz for their help in collecting semen samples for the Holstein, Normande, and Montbéliarde breeds and Hélène Hayes for English correction of the manuscript. We also acknowledge the assistance of the respective breeder associations in the collection of Aubrac, Charolais, Gasconne, Maine-Anjou, and Salers cattle samples and the following persons for their help in planning and conducting the sampling missions for African samples: V. Codja (Bénin), N. T. Kouagou (Togo), I. Sidibé (Burkina-Faso), and P. Souvenir Zafindrajaona (Chad). Finally, we thank the two anonymous reviewers for their helpful suggestions and corrections.
References
- Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang et al., 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, L., and M. Georges, 2004. Domestic-animal genomics: deciphering the genetics of complex traits. Nat. Rev. Genet. 5: 202–212. [DOI] [PubMed] [Google Scholar]
- Ardlie, K. G., L. Kruglyak and M. Seielstad, 2002. Patterns of linkage disequilibrium in the human genome. Nat. Rev. Genet. 3: 299–309. [DOI] [PubMed] [Google Scholar]
- Barrett, J. C., B. Fry, J. Maller and M. J. Daly, 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21: 263–265. [DOI] [PubMed] [Google Scholar]
- Belkhir, K., P. Borsa, L. Chikhi, N. Raufaste and F. Bonhomme, 2004. GENETIX, logiciel sous WindowsTM pour la génétique des populations. Université de Montpellier II, Montpellier, France.
- Boichard, D., L. Maignel and E. Verrier, 1996. Analyse généalogique des races bovines laitières françaises. INRA Prod. Anim. 9: 323–335. [Google Scholar]
- Cardon, L. R., and G. R. Abecasis, 2003. Using haplotype blocks to map human complex trait loci. Trends Genet. 19: 135–140. [DOI] [PubMed] [Google Scholar]
- Duchesne, A., M. Gautier, S. Chadi, C. Grohs, S. Floriot et al., 2006. Identification of a doublet missense substitution in the bovine LRP4 gene as a candidate causal mutation for syndactyly in Holstein cattle. Genomics 88: 610–621. [DOI] [PubMed] [Google Scholar]
- Everts-van der Wind, A., S. R. Kata, M. R. Band, M. Rebeiz, D. M. Larkin et al., 2004. A 1463 gene cattle-human comparative map with anchor points defined by human genome sequence coordinates. Genome Res. 14: 1424–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Farnir, F., W. Coppieters, J. J. Arranz, P. Berzi, N. Cambisano et al., 2000. Extensive genome-wide linkage disequilibrium in cattle. Genome Res. 10: 220–227. [DOI] [PubMed] [Google Scholar]
- Fellius, M., 1995. Cattle Breeds: An Encyclopedia. Misset, The Hague, The Netherlands.
- Felsenstein, J., 1989. PHYLIP: Phylogeny Inference Package (Version 3.2). Cladistics 5: 164–166. [Google Scholar]
- Freeman, A. R., C. M. Meghen, D. E. Machugh, R. T. Loftus, M. D. Achukwi et al., 2004. Admixture and diversity in West African cattle populations. Mol. Ecol. 13: 3477–3487. [DOI] [PubMed] [Google Scholar]
- Freeman, A. R., C. J. Hoggart, O. Hanotte and D. G. Bradley, 2006. Assessing the relative ages of admixture in the bovine hybrid zones of Africa and the Near East using X chromosome haplotype mosaicism. Genetics 173: 1503–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel, S. B., S. F. Schaffner, H. Nguyen, J. M. Moore, J. Roy et al., 2002. The structure of haplotype blocks in the human genome. Science 296: 2225–2229. [DOI] [PubMed] [Google Scholar]
- Gautier, M., R. R. Barcelona, S. Fritz, C. Grohs, T. Druet et al., 2006. Fine mapping and physical characterization of two linked quantitative trait loci affecting milk fat yield in dairy cattle on BTA26. Genetics 172: 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard, M. E., B. Hayes, A. Chamberlain and H. McPartlan, 2006. Can the same markers be used in multiple breeds? 8th World Congress on Genetics Applied to Livestock Products, Belo Horizonte, Brazil, Communication 22–16.
- Grisart, B., F. Farnir, L. Karim, N. Cambisano, J. J. Kim et al., 2004. Genetic and functional confirmation of the causality of the DGAT1 K232A quantitative trait nucleotide in affecting milk yield and composition. Proc. Natl. Acad. Sci. USA 101: 2398–2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosclaude, F., R. Y. Aupetit, J. Lefebvre and J. C. Mériaux, 1990. Essai d'analyse des relations génétiques entre les races bovines françaises à l'aide du polymorphisme biochimique. Genet. Sel. Evol. 22: 317–338. [Google Scholar]
- Hanotte, O., D. G. Bradley, J. W. Ochieng, Y. Verjee, E. W. Hill et al., 2002. African pastoralism: genetic imprints of origins and migrations. Science 296: 336–339. [DOI] [PubMed] [Google Scholar]
- Hawken, R. J., W. C. Barris, S. M. McWilliam and B. P. Dalrymple, 2004. An interactive bovine in silico SNP database (IBISS). Mamm. Genome 15: 819–827. [DOI] [PubMed] [Google Scholar]
- Hayes, B. J., P. M. Visscher, H. C. McPartlan and M. E. Goddard, 2003. Novel multilocus measure of linkage disequilibrium to estimate past effective population size. Genome Res. 13: 635–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, W. G., 1975. Linkage disequilibrium among multiple neutral alleles produced by mutation in finite population. Theor. Popul. Biol. 8: 117–126. [DOI] [PubMed] [Google Scholar]
- Huntley, D., A. Baldo, S. Johri and M. Sergot, 2006. SEAN: SNP prediction and display program utilizing EST sequence clusters. Bioinformatics 22: 495–496. [DOI] [PubMed] [Google Scholar]
- Jeanpierre, M., 1987. A rapid method for the purification of DNA from blood. Nucleic Acids Res. 15: 9611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordana, J., P. Alexandrino, A. Beja-Pereira, I. Bessa, J. Canon et al., 2003. Genetic structure of eighteen local south European beef cattle breeds by comparative F-statistics analysis. J. Anim. Breed. Genet. 120: 73–87. [Google Scholar]
- Kantanen, J., I. Olsaker, L. E. Holm, S. Lien, J. Vilkki et al., 2000. Genetic diversity and population structure of 20 North European cattle breeds. J. Hered. 91: 446–457. [DOI] [PubMed] [Google Scholar]
- Khatkar, M. S., A. Collins, J. A. Cavanagh, R. J. Hawken, M. Hobbs et al., 2006. A first-generation metric linkage disequilibrium map of bovine chromosome 6. Genetics 174: 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatkar, M. S., K. R. Zenger, M. Hobbs, R. J. Hawken, J. A. Cavanagh et al., 2007. A primary assembly of a bovine haplotype block map based on a 15,036-single-nucleotide polymorphism panel genotyped in Holstein-Friesian cattle. Genetics 176: 763–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra, J. A., and D. G. Bradley, 1997. Systematics and phylogeny of cattle, pp. 1–14 in The Genetics of Cattle, edited by R. Fries and A. Ruvinsky. CABI Publishing, Oxon, UK.
- Lindblad-Toh, K., C. M. Wade, T. S. Mikkelsen, E. K. Karlsson, D. B. Jaffe et al., 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438: 803–819. [DOI] [PubMed] [Google Scholar]
- MacHugh, D. E., R. T. Loftus, P. Cunningham and D. G. Bradley, 1998. Genetic structure of seven European cattle breeds assessed using 20 microsatellite markers. Anim. Genet. 29: 333–340. [DOI] [PubMed] [Google Scholar]
- Marth, G. T., I. Korf, M. D. Yandell, R. T. Yeh, Z. Gu et al., 1999. A general approach to single-nucleotide polymorphism discovery. Nat. Genet. 23: 452–456. [DOI] [PubMed] [Google Scholar]
- Matise, T. C., M. Perlin and A. Chakravarti, 1994. Automated construction of genetic linkage maps using an expert system (MultiMap): a human genome linkage map. Nat. Genet. 6: 384–390. [DOI] [PubMed] [Google Scholar]
- Meuwissen, T. H., A. Karlsen, S. Lien, I. Olsaker and M. E. Goddard, 2002. Fine mapping of a quantitative trait locus for twinning rate using combined linkage and linkage disequilibrium mapping. Genetics 161: 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazami-Goudarzi, K., D. Laloe, J. P. Furet and F. Grosclaude, 1997. Analysis of genetic relationships between 10 cattle breeds with 17 microsatellites. Anim. Genet. 28: 338–345. [DOI] [PubMed] [Google Scholar]
- Moazami-Goudarzi, K., D. M. A. Belemsaga, G. Ceriotti, D. Laloe, F. Fagbohoun et al., 2002. Caractérisation de la race bovine Somba à l'aide de marqueurs moléculaires. Rev. Elev. Méd. Vét. Pays Trop. 54: 129–138. [Google Scholar]
- Nei, M., 1978. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen, H. G., S. Lien, M. Gautier, H. Nilsen, A. Roseth et al., 2005. Mapping of a milk production quantitative trait locus to a 420-kb region on bovine chromosome 6. Genetics 169: 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavy, N., L. S. Parsons, C. Paule, J. MacKay and J. Bousquet, 2006. Automated SNP detection from a large collection of white spruce expressed sequences: contributing factors and approaches for the categorization of SNPs. BMC Genomics 7: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quéval, R., K. Moazami-Goudarzi, D. Laloe, J. C. Mériaux and F. Grosclaude, 1998. Relations génétiques entre populations de taurins ou zébus d'Afrique de l'Ouest et taurins Européens. Genet. Sel. Evol. 30: 367–383. [Google Scholar]
- Reich, D. E., M. Cargill, S. Bolk, J. Ireland, P. C. Sabeti et al., 2001. Linkage disequilibrium in the human genome. Nature 411: 199–204. [DOI] [PubMed] [Google Scholar]
- Reich, D. E., S. F. Schaffner, M. J. Daly, G. McVean, J. C. Mullikin et al., 2002. Human genome sequence variation and the influence of gene history, mutation and recombination. Nat. Genet. 32: 135–142. [DOI] [PubMed] [Google Scholar]
- Saitou, N., and M. Nei, 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4: 406–425. [DOI] [PubMed] [Google Scholar]
- Souvenir Zafindrajaona, P., V. Zeuh, K. Moazami-Goudarzi, D. Laloë, D. Bourzat et al., 1999. Etude du satut phylogénétique du bovin Kouri du lac Tchad à l'aide de marqueurs moléculaires. Rev. Elev. Méd. Vét. Pays Trop. 52: 155–162. [Google Scholar]
- Sved, J. A., 1971. Linkage disequilibrium and homozygosity of chromosome segments in finite populations. Theor. Popul. Biol. 2: 125–141. [DOI] [PubMed] [Google Scholar]
- Tenesa, A., S. A. Knott, D. Ward, D. Smith, J. L. Williams et al., 2003. Estimation of linkage disequilibrium in a sample of the United Kingdom dairy cattle population using unphased genotypes. J. Anim. Sci. 81: 617–623. [DOI] [PubMed] [Google Scholar]
- Tenesa, A., P. Navarro, B. J. Hayes, D. L. Duffy, G. M. Clarke et al., 2007. Recent human effective population size estimated from linkage disequilibrium. Genome Res. 17: 520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenon, S., G. K. Dayo, S. Sylla, I. Sidibe, D. Berthier et al., 2007. The extent of linkage disequilibrium in a large cattle population of western Africa and its consequences for association studies. Anim. Genet. 38: 277–286. [DOI] [PubMed] [Google Scholar]
- Vrba, E. S., and G. B. Schaller, 2000. Phylogeny of Bovidae based on behavior, glands, skulls and postcrania, pp. 203–222 in Antelopes, Deer, and Relatives: Fossil Record, Behavorial Ecology, Systematics, and Conservation, edited by E. S. Vrba and G. B. Schaller. Yale University Press, New Haven, CT.
- Weir, B. S., and C. C. Cockerham, 1984. Estimating F-statistics for the analysis of population structure. Evolution 19: 395–420. [DOI] [PubMed] [Google Scholar]
- Wright, S., 1965. Interpretation of population structure by F-statistics with special regard to system of mating. Evolution 19: 395–420. [Google Scholar]