Abstract
Biological factors involved in reproductive barriers between two divergent races of Ceratopteris richardii were investigated. We used a combination of spore germination rates, QTL analysis of spore germination rates, and transmission ratio distortion (TRD) of 729 RFLPs, AFLPs, and isozyme markers distributed across the genome on the basis of hybrid populations of 488 doubled haploid lines (DHLs) and 168 F2's. Substantial reproductive barriers were found between the parental races, predominantly in the form of spore inviability (23.7% F1 spore viability). Intrinsic genetic factors such as Bateson–Dobzhansky–Muller (BDM) incompatibilities involving both nuclear–nuclear and nuclear–cytoplasmic factors and chromosomal rearrangements appear to contribute to intrinsic postzygotic isolation. The genomewide distribution patterns of TRD loci support the hypothesis that reproductive barriers are a byproduct of divergence in allopatry and that the strong reproductive barriers are attributable to a small number of genetic elements scattered throughout the genome.
DIVERGENT populations often exhibit various types of reproductive barriers. These range from ecological isolation such as habitat differentiation to reduced courtship arising from differences in mating behavior to complete hybrid sterility, ultimately resulting in separate species. Reproductive isolation has long been thought to accumulate gradually following population separation (Stebbins 1950; Dobzhansky 1951; Mayr 1963; Coyne and Orr 1989), but an understanding of reproductive isolation at the genetic and molecular levels did not emerge until recently. Genetic mapping of hybrid incompatibilities has provided insights into the number, location, and effect of the genetic factors underlying intrinsic postzygotic reproductive barriers (Naveira 1992; True et al. 1996; Coyne et al. 1998; Presgraves 2003; Tao et al. 2003a,b; Moyle and Graham 2005; Fishman and Willis 2006; Sweigart et al. 2006). Likewise, studying transmission ratio distortion (TRD) of marker loci in segregating hybrid populations has proven to be a sensitive assay for loci underlying reproductive barriers between parental populations (e.g., Fishman et al. 2001a; Harushima et al. 2002; Hall and Willis 2005). Determining the identity of such genetic factors, their interactions, and their evolutionary patterns will advance understanding of the process of speciation.
Ceratopteris, a homosporous fern genus comprising several herbaceous species distributed throughout the tropics and subtropics, typically found in aquatic to semi-aquatic habitats such as river banks and ponds (Lloyd 1974), is among the few ferns with truly nonperennial life cycles. Our study system, C. richardii Brongn., occurs in the tropics from Central to South America (Lloyd 1974) and is a diploid species with 39 chromosomes. The species has recently been promoted as a model system in ferns (Hickok et al. 1987) and its genetic linkage map was recently described (Nakazato et al. 2006).
Although ferns have received extensive taxonomic and phylogenetic study and are the second most diverse group of vascular plants after seed plants, the mechanisms driving speciation in ferns are largely unknown. Here, we use segregating hybrid populations between genetically divergent allopatric populations of C. richardii to provide the first genetic study of factors contributing to reproductive barriers in ferns. We investigated reproductive barriers between these populations at three levels: spore germination rates as a measure of hybrid fertility, QTL analysis of spore germination rates, and TRD of marker loci distributed across the genome. This multilevel approach permits inferences of the likely genetic mechanisms and developmental phases involved in reproductive isolation. In addition, we took advantage of the fact that C. richardii gametophytes can self-fertilize to form completely homozygous doubled haploid lines (DHLs). We compared spore germination rates and TRD between DHLs and F2's to understand differences in reproductive barriers in completely homozygous vs. partially heterozygous progeny.
MATERIALS AND METHODS
Sample preparation and genetic linkage map construction:
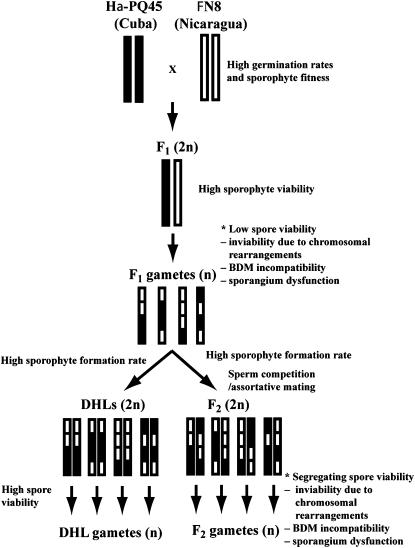
A genetic linkage map for C. richardii was constructed using 368 RFLPs, 358 AFLPs, and 3 isozyme markers on the basis of a mapping population of 488 DHLs. The parental strains of the mapping population, Hα-PQ45 from Cuba (Killip 44595, GH) and ΦN8 from Nicaragua (Nichols 1719, GH), were kindly provided by Leslie Hickok (University of Tennessee) and were crossed using Hα-PQ45 as the maternal parent to generate our mapping population (Figure 1). DHLs were generated by isolating F1 gametophytes and self-fertilized in the presence of distilled water (intragametophytic selfing). The methods of plant culture, genotyping, and genetic linkage map construction have been detailed elsewhere (Nakazato and Gastony 2006; Nakazato et al. 2006). In addition to DHLs, 168 F2 sporophytes were generated by random mating among F1 gametes (intergametophytic selfing) upon flooding mature F1 gametophytes on petri dishes with water and sporophytes were cultured using the same methods employed for the DHLs (Figure 1). These F2's were subjected to RFLP genotyping along with DHLs, but only codominant markers were used for the TRD analysis.
Figure 1.—
Illustration of the crossing design for construction of the C. richardii mapping population. Bars with solid and open segments indicate genomic content of Hα-PQ45 and ΦN8 parents, respectively. Reproductive barriers and their possible causes are summarized at asterisks. Sporangium dysfunction refers to problems of sporangium tissues to nurture spores.
TRD analysis:
χ2-tests were conducted to determine the significance of TRD for each marker at the α = 0.05 level. For DHLs, equal representation of the parental genotypes was tested. For F2's, in addition to equal representation of the parental genotypes, deviation of heterozygote frequencies from the Hardy–Weinberg expectation was tested on the basis of the frequency of the parental genotypes. Gradients of TRD along linkage groups were estimated on the basis of available markers using the “Negative Exponential” or “Bisquare” smoothing function implemented in SigmaPlot (Systat Software, San Jose, CA). To determine the likelihood of each linkage group hosting at least one TRDL, a multivariate statistical test incorporating the dependencies between loci was developed and implemented separately for each linkage group. For a given chromosome with m markers, let X = (X1, X2, … , Xm) be the number of Hα-PQ45 alleles observed at each marker. Let EX be the number of Hα-PQ45 alleles expected at each marker that would be half the total number scored. The covariance matrix G = (g(i, j)) is given by g(i, j) = n(i, j)(0.25 − 0.5r(i, j)), where n(i, j) is the number of spores scored at both markers i and j and r(i, j) is the recombination frequency between markers i and j calculated using the Kosambi map function from the genetic distance between markers. Under the null hypothesis of no distortion, (X − EX)G−1(X − EX)t follows a χ2-distribution with m d.f. If the χ2-statistic is significant, then the linkage group as a whole demonstrates some segregation distortion. This distortion can be due to an over- or underrepresentation of Hα-PQ45 alleles at a single locus or adjacent loci or the distortion can be due to an abrupt change in the frequencies in Hα-PQ45 alleles between adjacent loci. All statistical analyses in this study were conducted using either Microsoft Excel or SPSS v.12.0 (SPSS, Chicago).
Spore-germination-rate estimation and QTL analysis:
Spore-germination rates were estimated by averaging the proportion of spores germinated in four different areas on petri dishes in the fern medium described in the C-Fern Manual (Hickok et al. 1998). QTL analyses of spore germination rates among the DHLs were performed using Windows QTL Cartographer V2.5 (Wang et al. 2006, http://statgen.ncsu.edu/qtlcart/WQTLCart.htm). Because of the highly skewed distribution observed, spore germination rates were arcsine-transformed prior to analysis. Permutation tests (1000 times) implemented in the program were used to estimate the threshold level. Using the estimated threshold score, composite interval mapping (CIM) was conducted using the “forward regression method” with the window size of 10 cM, five background markers, and a walk speed of 2 cM. Two-way epistatic interactions on spore germination rates were tested between a pair of markers on different linkage groups. For each pair, the mean difference between homoallelic (AABB, aabb) and heteroallelic (AAbb, aaBB) DHLs was tested by single-factor ANOVA implemented in Microsoft Visual Basic.
RESULTS
TRD among DHLs:
A total of 729 markers were scored and analyzed for TRD among DHLs. The transmission ratio of Hα-PQ45 alleles for the studied markers followed a roughly normal distribution, but the mean ratio (49.5%) deviated significantly (two-tailed t-test, P = 0.023) from the neutral expectation of 50%, indicating that ΦN8 genotypes are slightly but significantly overrepresented among hybrid individuals. A substantial proportion (26.89%) of markers showed a significant deviation from the neutral expectation of 50% at the α = 0.05 level, with a maximum deviation of 65.3% (82.7:17.3% Hα-PQ45:ΦN8 genotypic ratio). Of these, 78 and 118 deviated toward Hα-PQ45 and ΦN8, respectively (χ2-test, P = 0.004), again indicating a significant overrepresentation of ΦN8 genotypes.
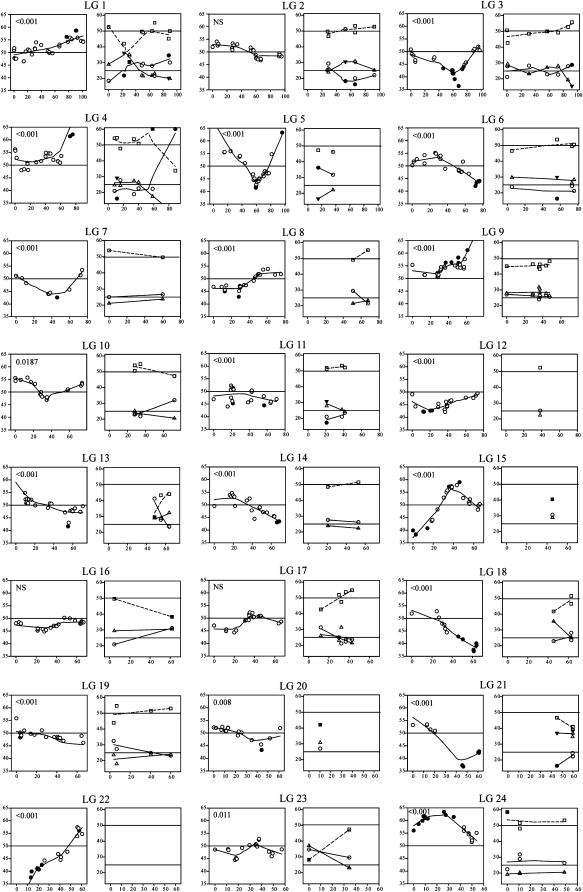
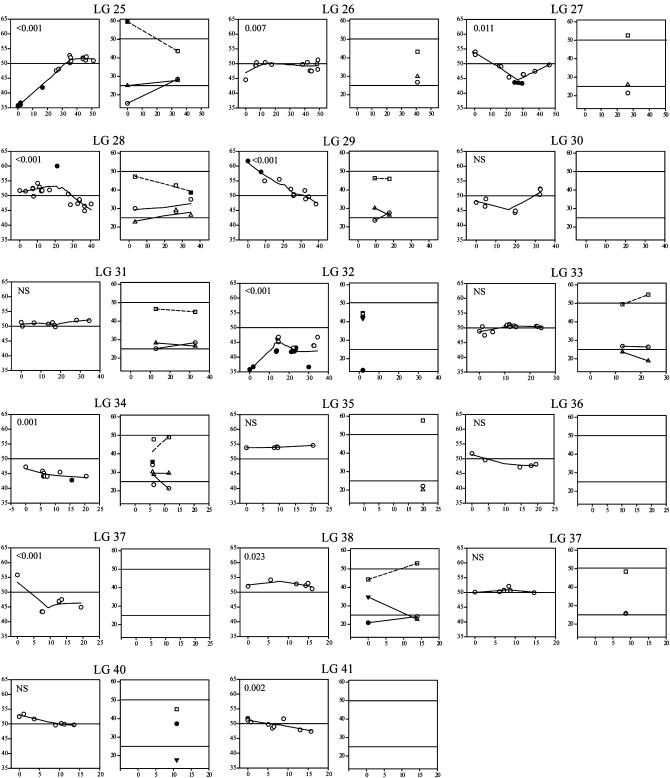
TRD along linkage groups (Figure 2) generally followed smooth gradients around local peaks, consistent with the presence of loci under selection and genetic hitchhiking at the linked loci. On a visual basis, we detected ∼30 TRD loci (TRDLs) across the genome. Most TRDLs had only modest effects (10–20% deviation), but loci under extreme selection pressure are fairly common (e.g., LG4, -5, -9, -22, -25, and -32). Interestingly, the average percentage of TRD effect for Hα-PQ45 (18.9%) was significantly greater (two-tailed t-test, P = 0.035) than that for ΦN8 (16.2%). Therefore, at the genome level, TRD toward Hα-PQ45 was less frequent, but stronger compared to that toward ΦN8. Statistical analysis of linkage-group-wide TRD patterns indicated that 31 of 41 linkage groups (75.61%) showed evidence of at least one TRDL at the α = 0.05 level (Figure 2), indicating that genetic factors contributing to reproductive barriers are common and scattered throughout the genome. In some cases, the entire linkage group showed TRD (e.g., LG9, -12, -24, -32, and -34), suggesting linkage of neutral markers to strongly selected loci and/or the presence of multiple TRDLs in the same direction (Chevin and Hospital 2006).
Figure 2.—
TRD along linkage groups for DHLs (left graph of each linkage group) and F2's (right graph of each linkage group). Each data point represents a marker reflecting the percentage of Hα-PQ45 genotypes (circles), ΦN8 genotypes (triangles), and heterozygotes (squares) along the linkage group (x-axis) in centimorgans of genetic distance with significant (solid symbols) or nonsignificant (open symbols) deviation from the expected segregation ratio (horizontal lines) at the α = 0.05. For F2's, the expected Hardy–Weinberg frequency of heterozygotes deviates from 50% depending on the frequencies of the parental genotypes. The likelihood that each linkage group hosts at least one TRD locus (TRDL) is indicated in the left graphs.
TRD among F2's:
A total of 108 codominant RFLP markers among F2's were scored and analyzed for TRD. As for DHLs, Hα-PQ45 genotypes were underrepresented in F2's (49.3%), but this was not significant (two-tailed t-test, P > 0.1). A much smaller proportion of markers (16; 14.81%) showed a significant deficiency (α = 0.05) of one of the parental genotypes compared to that among DHLs (26.89%), although a direct comparison is difficult because of much lower statistical power among F2's. Nonetheless, the proportions of markers with distorted parental genotypic ratios among DHLs and F2's are comparable to that of some intraspecific crosses [13−18% (Zamir and Tadmor 1986; Jenczewski et al. 1997; Lu et al. 2002), but much lower than that of some interspecific crosses; 80% (Devicente and Tanksley 1993); 49% (Fishman et al. 2001a)]. Of 16 markers with distorted parental genotypic ratios among F2's, 11 showed overrepresentation of ΦN8 genotypes, again consistent with the trend among DHLs. Seven of these 16 markers also exhibited TRD among DHLs. As expected, in all cases the direction of deviation was consistent across the two progeny types, indicating that common selection often applies to different crossing types. However, more than half of the distorted markers (9/16) showed distortion only among F2's, despite its lower statistical power. A clear example is the marker on LG40, where the Hα-PQ45 genotype is overrepresented among F2's, but the same marker showed no such bias among DHLs. This suggests that some markers experience different selection pressures between DHL and F2 progeny types, presumably because of differences in levels of heterozygosity (see discussion).
TRD among F2's allows us to estimate the relative contribution of haploid and diploid phases to reproductive barriers. Like TRD among DHLs, the relative frequency of the two homozygous classes among F2's most likely reflects the relative fitness of each parental allele in the haploid phase. On the other hand, deviation of the heterozygote frequency from the Hardy–Weinberg expectation indicates selection on heterozygotes at the diploid phase. In this study, 12 of 108 codominant RFLP markers (11.11%) showed a significant deviation (α = 0.05) of heterozygote frequency from the neutral Hardy–Weinberg frequency (maximum percentage deviation of 43.55%). Of those, 9 were underdominant and 3 were overdominant, indicating that selection against rather than for heterozygotes is a more prevalent consequence of hybridization between divergent populations. The fact that we detected similar numbers of loci with parental genotype frequency deviation (16) and heterozygote frequency deviation (12) among F2's suggests that reproductive barriers in haploid and diploid phases are equally common. This observation is consistent with a comparative mapping of TRDLs among three interspecific crosses of rice (Harushima et al. 2002), where they found 15–19 gametophytic barriers and 17–18 sporophytic barriers.
Viability of spores and sporophytes:
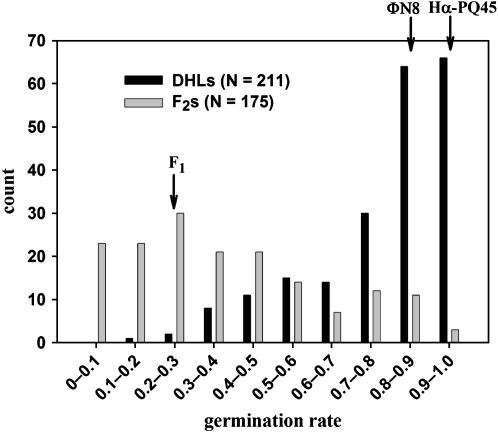
For both parental strains, we observed relatively high spore-germination rates (Hα-PQ45, 93.2%; ΦN8, 82.8%; Figure 3) and viable sporophyte formation rates (Hα-PQ45, 86.2%; ΦN8, 82.0%), indicating that the environmental conditions of our study had little influence on their fitness, although Hα-PQ45 seems to have slightly higher fitness. The proportion of Hα-PQ45 gametophytes that formed viable F1 hybrid sporophytes upon fertilization with ΦN8 male gametes (81.7%) was comparable with that of the parents, indicating that there is little, if any, incompatibility during fertilization. However, the germination rate of spores from the F1 sporophyte was low (27.3%), typical of intrinsic postzygotic isolation. Observation of hybrid spores with a microscope revealed that the reduced hybrid fertility was commonly caused by spore abortion, evident by collapsed shape instead of the tetrahedral shape of viable spores. The proportions of F1 gametophytes that formed viable DHL sporophytes and F2 sporophytes were 78.6 and 81.1%, respectively, again indicating that fertilization contributes little to reproductive isolation.
Figure 3.—
Histogram of spore germination rates for DHLs (solid) and F2's (shaded). Arrows indicate the categories of the mean spore germination rates for the parents and the F1.
The viability of DHL spores was modestly reduced relative to those of the parental populations, ranging from 16.1 to 100% with an average of 78.4% (Figure 3). The distribution was highly skewed toward high values with the mode of 93.3%, which was equivalent to the value of the parents. High viability of DHL spores was expected because these spores are in theory genetically identical to their respective DHL parents, which had successfully germinated. The facts that DHL spores were not always viable and that spore abortion was also observed for these spores indicate that factors other than spore genotype contribute to DHL fertility, such as the ability of the sporangium (spore-forming organ) to nurture spores (see discussion).
Viability of F2 spores exhibited transgressive segregation, with viabilities ranging from 0 to 100%. Average F2 spore viability (35.2%) was substantially lower than that of the parents and DHLs, but higher than the F1 (23.7%; Figure 3), again a pattern commonly observed in plant hybrids.
QTL for spore viability:
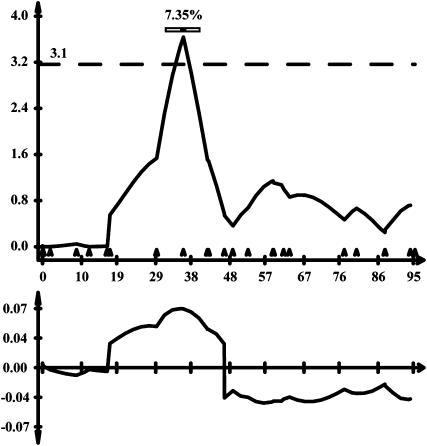
A single significant QTL for spore viability among DHLs was detected on LG4 (Figure 4). The QTL effect was modest with a percentage of variance explained (PVE) of 7.35% and was toward higher viability of spores carrying ΦN8 alleles. No other genomic regions showed association with spore viability close to the threshold.
Figure 4.—
Significant QTL for spore germination rate on LG4. Top: LOD score along the linkage group with the threshold level indicated by the dashed line. The horizontal bar on top shows the QTL within a 1-LOD margin with percentage of variance explained (PVE) at the peak position indicated on top. Triangles on the x-axis indicate the marker locations. Bottom: Additive genetic variance of the ΦN8 parental allele on the trait along the linkage group.
Although spore viability QTL acting additively are rare, we detected several inter-linkage-group QTL combinations showing significant (ANOVA, P < 10−4) two-way epistatic interactions on the trait among DHLs (Table 1). One of the QTL combinations involved the additive QTL on LG4 (Figure 4) and it is not clear at this time whether this involves significant epistasis or is largely a result of additive effects on LG4. However, other QTL combinations do not involve loci with significant additive effects and therefore likely result from genuine interactions between unlinked genetic elements. Interestingly, although three QTL combinations showed negative effects on spore germination rates, expected from BDM incompatibilities (see discussion), the majority of significant QTL interactions resulted in higher spore viability of heteroallelic DHLs (AAbb, aaBB) compared to the homoallelic ones (AABB, aabb), suggesting that hybridization promotes fertility in many cases.
TABLE 1.
QTL pairs with significant epistasis affecting DHL spore germination rates
| QTL 1
|
QTL 2
|
||||
|---|---|---|---|---|---|
| LG | cM position | LG | cM position | P (ANOVA) | Directiona |
| 2 | 7.9 | 4 | 0 | 5.85E-05 | − |
| 3 | 46 | 15 | 2.8 | 8.60E-05 | + |
| 4b | 34.3 | 9 | 74.1 | 1.95E-05 | + |
| 5 | 64.2 | 10 | 28.4 | 5.28E-06 | + |
| 14 | 66.6 | 32 | 13.5 | 4.29E-05 | − |
| 15 | 2.8 | 16 | 3 | 8.28E-05 | + |
| 15 | 0 | 32 | 13.7 | 9.34E-05 | + |
| 22 | 57.7 | 37 | 12.7 | 2.91E-05 | + |
| 24 | 53.9 | 31 | 13.8 | 6.79E-05 | + |
| 28 | 32.7 | 33 | 24.4 | 4.61E-05 | + |
| 30 | 4.5 | 40 | 1.1 | 2.04E-06 | − |
Higher (+) or lower (−) spore germination rates of heteroallelic DHLs compared to homoallelic ones.
QTL with significant association with germination rates by itself.
QTL analysis on spore germination rates among F2's detected no significant QTL, presumably because of low marker coverage and small sample size. More rigorous comparison of fertility QTL between F2's and DHLs in future studies will permit comparisons of the reproductive isolation mechanisms resulting from different mating types (i.e., intra- vs. inter-gametophytic selfing) and may reveal reproductive isolation mechanisms unique to ferns.
DISCUSSION
Mechanisms of reproductive isolation:
We detected strong but incomplete reproductive isolation between divergent populations of C. richardii, consistent with the low F1 spore viability reported for the same cross (∼30%) in a previous study (Hickok et al. 1995). Although multiple developmental stages likely contribute to reproductive isolation, spore inviability appears to be the dominant reproductive barrier in Ceratopteris (Figure 1). High spore viability and sporophyte formation rates of the parents indicate that extrinsic factors such as differential fitness of the parental alleles or their combination under our growing conditions contributed little to reproductive isolation, although extrinsic factors may play an important role in nature (Arnold and Hodges 1995). Rather, the low F1 spore viability and segregating F2 spore viability suggest that most of the observed reproductive barriers were likely due to intrinsic factors, namely interactions of the parental genomes. Although inbreeding depression could contribute to reduced fitness of the later generation hybrids, complete homozygosity of the parents makes it unlikely in our experiment.
Two of the potential mechanisms causing intrinsic reproductive barriers between divergent populations are BDM incompatibilities and chromosomal rearrangements. The BDM model hypothesizes that intrinsic postzygotic isolation can result from incompatible epistatic interactions of alleles from divergent populations brought together upon hybridization (reviewed in Orr 1996). The chromosomal rearrangement hypothesis, on the other hand, proposes that intrinsic postzygotic isolation could result from karyotypic differences such as inversions and translocations. This is because crossovers during meiosis in hybrids heterozygous for the rearrangements may generate gametes that lack essential genetic elements within the rearranged segments.
If reproductive isolation is largely caused by chromosomal rearrangements or BDM incompatibility acting at the haploid phase, fertility should be lowest in F1 hybrids where heterozygosity is highest and the fertility of the subsequent hybrid generations should never be lower than the F1 generation's. On the other hand, if fertility reduction is caused mainly by BDM incompatibility acting at the diploid phase, lower fertility of F2 hybrids than F1's is possible, depending on the fitness of the hybrid genotypes. Fishman and Willis (2001b) observed significantly lower F2 than F1 fertility and concluded that reproductive isolation between two populations of Mimulus was caused at least partly by BDM incompatibilities acting at the diploid phase. In our study, mean F2 spore viability was substantially higher than that of F1's supporting both postzygotic isolation by chromosomal rearrangements and BDM incompatibility at the haploid phase. However, many F2's also had lower fertility than F1's, suggesting that BDM incompatibilities acting at the diploid phase are partly responsible for the hybrid fertility reduction. Therefore, we failed to detect a dominant mechanism of postzygotic isolation as Fishman and Willis (2001b) did, and different intrinsic mechanisms are likely responsible for postzygotic isolation in Ceratopteris.
It is unlikely that the TRD detected in the DHLs is caused by chromosomal rearrangements because it is the heterozygous class, rather than parental markers, that should be under-represented if rearrangements are involved. However, TRD caused by BDM incompatibilities can be detected in both DHLs and F2's. In order for BDM incompatibilities among nuclear genes to result in TRD, there must be asymmetric fitness of interacting locus combinations. In the simplest case, a two-locus interaction involved in the haploid phase, there exist four haplotypes among F1 gametes: AB, Ab, aB, and ab, derived from the parents (AABB and aabb). Assuming equally high fitness of the parental haplotypes, higher fitness of Ab relative to the aB haplotype, for example, results in TRD. Complete lethality of aB haplotype results in 66.7% AA genotype (and bb) and 33.3% aa (and BB) among DHLs. Among F2's, the same fitness scheme results in 44.4% AA (and bb), 44.4% Aa, and 11.2% aa (and BB). In fact, they are the maximum TRD effects that can be achieved by nuclear–nuclear BDM incompatibility, because higher-order interactions and lower penetrance on lethality result in weaker TRD. The fact that the majority of the significant TRDs (73.2%) we observed among DHLs had only moderate effects (40–60% Hα-PQ45 genotype) suggests that nuclear–nuclear BDM incompatibility may be a primary mechanism of TRD. However, some genomic regions showed TRDs clearly outside of this range, namely LG4 (82.7% Hα-PQ45 allele), LG5 (74.3%), LG9 (75.1%), LG13 (70.2%), and LG22 (24.1%) among DHLs and LG4 (60.2%) among F2's, indicating that they were caused by factors other than nuclear–nuclear BDM incompatibility or possibly by tightly linked multiple BDM incompatibility loci.
One potential mechanism that could be responsible for such strong TRD is cytonuclear BDM incompatibility, which results from epistasis between nuclear and cytoplasmic genes. Given the strong interdependency between genes in these genomes during cellular processes (Rand et al. 2004), incompatibility among these genes has a great potential to result in postzygotic barriers. Because nuclear and cytoplasmic genomes have conflicting evolutionary interests, such incompatibility may evolve very rapidly between divergent populations (Levin 2003), as exemplified by cytoplasmic male sterility. Unlike the conventional nuclear–nuclear BDM incompatibility, which has a limit to TRD magnitude (see above), cytonuclear BDM incompatibility can result in up to 100% TRD of parental alleles in a segregating population when uniparentally inherited cytoplasmic alleles show deleterious epistasis with segregating nuclear alleles from the other parent. Because Ceratopteris likely has maternal cytoplasmic inheritance based on a study of closely related cheilanthoid ferns (Gastony and Yatskievych 1992), and Hα-PQ45 was used as a maternal parent of the cross, cytonuclear BDM incompatibility can result in complete TRD of Hα-PQ45, but not ΦN8 alleles, if single nuclear factors are involved. Consistent with this idea, all but one of the strongest TRDLs among DHLs and F2's (LG4, -5, -9, and -13; see above) showed overrepresentation of Hα-PQ45 alleles, perhaps each involving a single nuclear factor or a few. Furthermore, the fact that the mean TRD magnitude for Hα-PQ45 (18.9%) over all markers was significantly greater than that for ΦN8 (16.2%) among DHLs (see results) may indicate that cytonuclear BDM incompatibility is common throughout the genome. In Mimulus, reciprocal crossings and genetic linkage mapping revealed that anther sterility involved in hybrids with M. guttatus cytoplasm but not in the reciprocal cross is caused by cytonuclear BDM incompatibility involving a single nuclear QTL (Fishman and Willis 2006). Similar study is desirable in Ceratopteris in the future to seek more definitive evidence of cytonuclear BDM and its roles in speciation.
Our results suggest that intrinsic factors such as BMD incompatibility and chromosomal rearrangements acting in the gametophytic genomes play a major role in spore inviability. However, the common occurrence of spore abortion among DHLs, where no genetic incompatibility is expected, suggests that the genetic makeup of hybrid spores is not the only factor responsible for intrinsic postzygotic isolation. Instead, the genotype of spore-bearing sporophytes may also be a significant contributor; some hybrid genotypes may have problems nurturing spores within sporangia, resulting in many aborted spores regardless of the spore haplotypes. It is generally believed that the tapetal layer in the sporangia is important in depositing the outer layer (perispore or perine) on fern spores, and dysfunction of the tapetal layer cells in the hybrids may have contributed to spore abortion in this study. In angiosperms, it has been shown that pollen quality and initial performance of male gametophytes are highly dependent on the resources provided by anther tissue (Vasil 1974; Mulcahy and Mulcahy 1982; Wetzel and Jensen 1992) and a number of studies have demonstrated that environmental conditions of parental plants affect pollen quality and performance (reviewed in Delph et al. 1997). In other words, these studies indicate that gametophyte fitness is at least partly a parental trait.
The potential role of the sporangium genotype on hybrid fertility is further supported by a puzzling observation of similar numbers of sporophytic barriers (12 TRDLs) and gametophytic barriers (16 TRDLs), despite the high sporophyte viability among the parents and their hybrids. These cryptic sporophytic barriers may result from intrinsic reproductive barriers involving genes that are expressed during spore formation in sporangium tissues. Alternatively, sporophytic barriers may result from sexual selection due to preferential fertilization by sperm carrying one of the parental haplotypes, although competition among free-swimming fern sperm is probably less intense than pollen competition in flowering plants. Finally, assortative mating of gametes could lead to a deficiency of heterozygotes, without any selection against the heterozygotes themselves.
Modes of speciation:
The facts that TRDLs are scattered throughout the genome and selection against both parental genotypes is common in Ceratopteris are consistent with the view that genetic factors underlying intrinsic postzygotic isolation accumulate gradually across diverging parental genomes following geographic separation (Stebbins 1950; Dobzhansky 1951; Mayr 1963; Coyne and Orr 1989). The generality of such a conclusion is supported by similar distribution patterns of TRDLs observed in other organisms (e.g., Fishman et al. 2001a; Harushima et al. 2001, 2002; Hall and Willis 2005). In addition, Harushima et al. (2002) showed that hybridization of different cultivars of rice generated unique TRDLs, indicating that genetic factors involved in reproductive barriers have evolved independently and fairly rapidly.
Although the genetic factors involved in reproductive barriers are widespread in the Ceratopteris genome, occurring in most linkage groups, the number of such factors appears to be small compared to the number of genetic elements in the entire genome (>104) that can potentially be selected. Hence, despite substantial reproductive isolation between the parental populations (∼80% fertility reduction), it seems likely that much of the genome could be freely exchanged between the divergent lineages if they were to come into contact. Indeed, substantial genomewide gene flow was found between divergent sunflower species showing nearly complete reproductive isolation (Yatabe et al. 2007).
The most unexpected finding of this study was the small magnitude of the single QTL underlying spore viability. Indeed, QTL size (7.35% PVE) was at the detection limit despite the large size of the QTL population. This contrasts with recent studies in Drosophila, where several genes have recently been cloned that cause almost complete reproductive isolation (Ting et al. 1998; Barbash et al. 2003; Presgraves 2003; Masly et al. 2006). Likewise, major QTL for sterility are frequently detected in flowering plants (Moyle and Graham 2005; Fishman and Willis 2006; Sweigart et al. 2006). The lack of major QTL for spore viability in ferns might be due to the use of DHLs for QTL mapping. A possible biological explanation is that homosporous ferns may be less subject to sexual selection than heterosporous flowering plants or animals. In the latter, sexual selection has been suggested as a possible cause of rapid evolution of genes causing postzygotic isolation (Ting et al. 1998).
Acknowledgments
We are grateful to two anonymous reviewers for insightful comments and suggestions and to Troy Wood for fruitful discussions. This material is based upon work supported by the National Science Foundation under grant no. 0128926.
References
- Arnold, M. L., and S. A. Hodges, 1995. Are natural hybrids fit or unfit relative to their parents. Trends Ecol. Evol. 10: 67–71. [DOI] [PubMed] [Google Scholar]
- Barbash, D. A., D. F. Siino, A. M. Tarone and J. Roote, 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. USA 100: 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevin, L. M., and F. Hospital, 2006. The hitchhiking effect of an autosomal meiotic drive gene. Genetics 173: 1829–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne, J. A., and H. A. Orr, 1989. Two rules of speciation, pp. 180–207 in Speciation and Its Consequences, edited by D. Otte and J. A. Endler. Sinauer Associates, Sunderland, MA.
- Coyne, J. A., S. Simeonidis and P. Rooney, 1998. Relative paucity of genes causing inviability in hybrids between Drosophila melanogaster and D. simulans. Genetics 150: 1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delph, L. F., M. H. Johannsson and A. G. Stephenson, 1997. How environmental factors affect pollen performance: ecological and evolutionary perspectives. Ecology 78: 1632–1639. [Google Scholar]
- Devicente, M. C., and S. D. Tanksley, 1993. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 134: 585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky, T. G., 1951. Genetics and the Origin of Species. Columbia University Press, New York.
- Fishman, L., A. J. Kelly, E. Morgan and J. H. Willis, 2001. a A genetic map in the Mimulus guttatus species complex reveals transmission ratio distortion due to heterospecific interactions. Genetics 159: 1701–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman, L., and J. H. Willis, 2001. b Evidence for Dobzhansky-Muller incompatibilites contributing to the sterility of hybrids between Mimulus guttatus and M. nasutus. Evolution 55: 1932–1942. [DOI] [PubMed] [Google Scholar]
- Fishman, L., and J. H. Willis, 2006. A cytonuclear incompatibility causes anther sterility in Mimulus hybrids. Evolution 60: 1372–1381. [DOI] [PubMed] [Google Scholar]
- Gastony, G. J., and G. Yatskievych, 1992. Maternal inheritance of the chloroplast and mitochondrial genomes in cheilanthoid ferns. Am. J. Bot. 79: 716–722. [Google Scholar]
- Hall, M. C., and J. H. Willis, 2005. Transmission ratio distortion in intraspecific hybrids of Mimulus guttatus: implications for genomic divergence. Genetics 170: 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., M. Nakagahra, M. Yano, T. Sasaki and N. Kurata, 2001. A genomewide survey of reproductive barriers in an intraspecific hybrid. Genetics 159: 883–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., M. Nakagahra, M. Yano, T. Sasaki and N. Kurata, 2002. Diverse variation of reproductive barriers in three intraspecific rice crosses. Genetics 160: 313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickok, L. G., T. R. Warne, S. L. Baxter and C. T. Melear, 1998. Sex and the C-Fern: not just another life cycle. Bioscience 48: 1031–1037. [Google Scholar]
- Hickok, L. G., T. R. Warne and R. S. Fribourg, 1995. The biology of the fern Ceratopteris and its use as a model system. Int. J. Plant Sci. 156: 332–345. [Google Scholar]
- Hickok, L. G., T. R. Warne and M. K. Slocum, 1987. Ceratopteris richardii: applications for experimental plant biology. Am. J. Bot. 74: 1304–1316. [Google Scholar]
- Jenczewski, E., M. Gherardi, I. Bonnin, J. M. Prosperi, I. Olivieri et al., 1997. Insight on segregation distortions in two intraspecific crosses between annual species of Medicago (Leguminosae). Theor. Appl. Genet. 94: 682–691. [Google Scholar]
- Levin, D. A., 2003. The cytoplasmic factor in plant speciation. Syst. Bot. 28: 5–11. [Google Scholar]
- Lloyd, R. M., 1974. Systematics of the genus Ceratopteris Brongn. (Parkeriaceae) II. Taxonomy. Brittonia 26: 139–160. [Google Scholar]
- Lu, H., J. Romero-Severson and R. Bernardo, 2002. Chromosomal regions associated with segregation distortion in maize. Theor. Appl. Genet. 105: 622–628. [DOI] [PubMed] [Google Scholar]
- Masly, J. P., C. D. Jones, M. A. F. Noor, J. Locke and H. A. Orr, 2006. Gene transposition as a cause of hybrid sterility in Drosophila. Science 313: 1448–1450. [DOI] [PubMed] [Google Scholar]
- Mayr, E., 1963. Animal Species and Evolution. Belknap Press, Cambridge, MA.
- Moyle, L. C., and E. B. Graham, 2005. Genetics of hybrid incompatibility between Lycopersicon esculentum and L. hirsutum. Genetics 169: 355–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy, G. B., and D. L. Mulcahy, 1982. The two phases of growth of Petunia hybrida pollen tubes through compatible styles. J. Palynol. 18: 1–3. [Google Scholar]
- Nakazato, T., and G. J. Gastony, 2006. High-throughput RFLP genotyping method for large genomes based on a chemiluminescent detection system. Plant Mol. Biol. Rep. 24: 245a–245f. [Google Scholar]
- Nakazato, T., M.-K. Jung, E. A. Housworth, L. H. Rieseberg and G. J. Gastony, 2006. Genetic map-based analysis of genome structure in the homosporous fern Ceratopteris richardii. Genetics 173: 1585–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveira, H. F., 1992. Location of X-linked polygenic effects causing sterility in male hybrids of Drosophila simulans and D. mauritiana. Heredity 68: 211–217. [DOI] [PubMed] [Google Scholar]
- Orr, H. A., 1996. Dobzhansky, Bateson, and the genetics of speciation. Genetics 144: 1331–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves, D. C., 2003. A fine-scale genetic analysis of hybrid incompatibilities in Drosophila. Genetics 163: 955–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand, D. M., R. A. Haney and A. J. Fry, 2004. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol. Evol. 19: 645–653. [DOI] [PubMed] [Google Scholar]
- Stebbins, G. L., 1950. Variation and Evolution in Plants. Columbia University Press, New York.
- Sweigart, A. L., L. Fishman and J. H. Willis, 2006. A simple genetic incompatibility causes hybrid male sterility in mimulus. Genetics 172: 2465–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., S. N. Xhen, D. L. Hartl and C. C. Laurie, 2003. a Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. I. Differential accumulation of hybrid male sterility effects on the X and autosomes. Genetics 164: 1383–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, Y., Z. B. Zeng, J. Li, D. L. Hartl and C. C. Laurie, 2003. b Genetic dissection of hybrid incompatibilities between Drosophila simulans and D. mauritiana. II. Mapping hybrid male sterility loci on the third chromosome. Genetics 164: 1399–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, C. T., S. C. Tsaur, M. L. Wu and C. I. Wu, 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504. [DOI] [PubMed] [Google Scholar]
- True, J. R., B. S. Weir and C. C. Laurie, 1996. A genomewide survey of hybrid incompatibility factors by the introgression of marked segments of Drosophila mauritiana chromosomes into Drosophila simulans. Genetics 142: 819–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil, J. K., 1974. The histology and physiology of pollen germination and pollen growth on the stigma and in the style, pp. 105–118 in Fertilization in Higher Plants, edited by H. F. Linskens. North-Holland Publishing, New York.
- Wang, S., C. J. Basten and Z. B. Zeng, 2006. Windows QTL Cartographer 2.5. North Carolina University, Raleigh, NC.
- Wetzel, C. L. R., and W. A. Jensen, 1992. Studies of pollen maturation in cotton: the storage reserve accumulation phase. Sex. Plant Reprod. 5: 117–127. [Google Scholar]
- Yatabe, Y., N. C. Kane, C. Scotti-Saintagne and L. H. Rieseberg, 2007. Rampant gene exchange across a strong reproductive barrier between the annual sunflowers, Helianthus annuus and H. petiolaris. Genetics 175: 1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamir, D., and Y. Tadmor, 1986. Unequal segregation of nuclear genes in plants. Bot. Gaz. 147: 355–358. [Google Scholar]