Abstract
We have sequenced five distinct mitochondrial genomes in maize: two fertile cytotypes (NA and the previously reported NB) and three cytoplasmic-male-sterile cytotypes (CMS-C, CMS-S, and CMS-T). Their genome sizes range from 535,825 bp in CMS-T to 739,719 bp in CMS-C. Large duplications (0.5–120 kb) account for most of the size increases. Plastid DNA accounts for 2.3–4.6% of each mitochondrial genome. The genomes share a minimum set of 51 genes for 33 conserved proteins, three ribosomal RNAs, and 15 transfer RNAs. Numbers of duplicate genes and plastid-derived tRNAs vary among cytotypes. A high level of sequence conservation exists both within and outside of genes (1.65–7.04 substitutions/10 kb in pairwise comparisons). However, sequence losses and gains are common: integrated plastid and plasmid sequences, as well as noncoding “native” mitochondrial sequences, can be lost with no phenotypic consequence. The organization of the different maize mitochondrial genomes varies dramatically; even between the two fertile cytotypes, there are 16 rearrangements. Comparing the finished shotgun sequences of multiple mitochondrial genomes from the same species suggests which genes and open reading frames are potentially functional, including which chimeric ORFs are candidate genes for cytoplasmic male sterility. This method identified the known CMS-associated ORFs in CMS-S and CMS-T, but not in CMS-C.
PLANT mitochondrial genomes vary in size from 187 kb (Oda et al. 1992) to >2400 kb (Ward et al. 1981). It has long been known that the linear order of their genes can be highly variable among cytotypes even within a species (reviewed in Fauron et al. 1995a,b). However, it is not clear how plant mitochondrial genomes rearrange so readily or how their genome sizes can increase or decrease dramatically over relatively short evolutionary times.
Analysis of whole-plant mitochondrial genome sequences among divergent angiosperm species has shown that known protein-coding genes are highly conserved, but that intergenic spacer regions change rapidly (Kubo et al. 2000; Handa 2003; Clifton et al. 2004). Furthermore, the origin of this DNA is obscure. With two exceptions, each of which involved evaluation of two genomes (Satoh et al. 2004; Tian et al. 2006), comparisons have not been conducted among multiple mitochondrial genotypes (cytotypes) from within a single angiosperm species. Such studies are necessary to elucidate the mechanisms that underlie the rapid changes in intergenic sequences and in genome organization that occur in the mitochondrial genomes of angiosperms. To explore these issues, we have compared five mitochondrial genomes from Zea mays (maize), which provides an accessible system in which to clarify the dynamics of mitochondrial genomic change.
Five distinct maize mitochondrial genotypes have been identified (reviewed in Leaver et al. 1988; Fauron et al. 1995a). The fertile type NB is present in most commercial hybrids and has been the best studied. The other major mitochondrial genotype in male-fertile North American cultivated maize is termed NA. It was originally identified in the A188 inbred line (Fauron and Casper 1994) and is the cytoplasm present in most of the lines used to transform maize. The three major cytoplasmic-male-sterile groups (CMS)—CMS-C, CMS-T, and CMS-S—were originally classified on the basis of which nuclear restorer-of-fertility (Rf) genes would override their cytoplasmically determined pollen abortion (reviewed in Beckett 1971). Later, restriction enzyme analyses of mitochondrial DNAs showed distinctive patterns for each genotype (Levings and Pring 1976; Pring and Levings 1978). Cloning and mapping studies indicated that the mitochondrial genomes of the distinct cytotypes were rearranged relative to one another (reviewed in Fauron et al. 1995a). Other studies determined that, in two of the CMS genomes, sterility was due to rearrangements that yielded chimeric open reading frames (ORFs): T-urf13 in CMS-T (Dewey et al. 1987) and the cotranscribed orf355/orf77 in CMS-S (Zabala et al. 1997).
MATERIALS AND METHODS
Mitochondrial DNA Preparation:
The plants used for mitochondrial DNA (mtDNA) preparations were derived from B37 inbred lines with male-sterile CMS-T, CMS-C, and CMS-S cytoplasms and from the male-fertile inbred A188 (NA cytoplasm). The inbred lines were crossed by Mo17, and ear shoots of the F1 plants were harvested at the time of silk emergence. Mitochondria were prepared from the ear shoots using differential centrifugation; DNA was recovered from lysed mitochondria following phenol/chloroform extractions and ethanol precipitation as described previously (Newton 1994).
Library construction, sequencing, and finishing:
Full details were presented in Clifton et al. (2004). Briefly, the mtDNA was fragmented, linked to plasmid vector pOTMI, and transformed into and grown in Escherichia coli host strain DH10B-T1. Insert DNA was purified from each library and, following test sequencing to verify library quality, each library was sequenced at the Washington University Genome Sequencing Center. Chromatograms were processed using the Phred/Phrap package (Ewing and Green 1998; Ewing et al. 1998), and the Staden Package vector-clipping program (Staden 1996; Wendl et al. 1998) was used to remove vector sequences. All the genomes were shotgun sequenced to coverage depths of at least 10×. Sequences were assembled without reference to previous restriction mapping information. At each point where evidence for alternative conformations was obtained, the most common conformation was chosen. Following each sequence assembly, the database was verified in Consed (Gordon et al. 1998). Annotated sequences were deposited in GenBank under accession nos. DQ490951 (CMS-S), DQ490952 (NA), DQ490953 (CMS-T), and DQ645536 (CMS-C).
Annotation:
The primary database used for annotation was AceDB (http://www.acedb.org/). ORFs were initially identified using Artemis (Rutherford et al. 2000; http://www.sanger.ac.uk/Software/Artemis). The ORFs were used to query nonredundant databases using BLAST similarity searches, applying a cutoff of 70% sequence identity over at least 80% of the ORF length. To find putative homologs in other completed genomes, predicted proteins were searched for in the COG database (Tatusov et al. 2001), in the protein family database Pfam (Bateman et al. 2002; http://pfam.wustl.edu), and in the integrated view of the signature database Interpro (http://www.ebi.ac.uk/interpro). Additionally, to help define genes, ORF-Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), BLASTN, BLASTX (Altschul et al. 1990), and tRNAscan-SE (Lowe and Eddy 1997) were used. To improve the accuracy of the identifications, the mtDNA annotations were compared with other plant mitochondrial genome annotations, and all differences in coding predictions were reassessed on the basis of choice of start codon, length of conservation in plant mtDNA, and presence of identifiable motifs. In addition to known genes, ORFs of at least 150 nt (≥50 codons) were identified; only ORFs of at least 100 codons were annotated. ORFs containing or composed of plastid sequences were included if their upstream regions were not of plastid origin. All ORFs having upstream regions of plastid origin within 100 bp of the ATG were excluded from the analyses.
Chimeric genes were discovered by performing searches of all nongenic ORFs with a specific database of maize mitochondrial and plastid genes using BLASTN (E-value = 0.0001, G = 3, E = 1), as well as with public databases. Searches for plastid, plasmid, or known-gene DNA fragments used a lower limit of 16 bp, empirically determined by an inflection point in the number of BLAST hits (where E-values > 0.001). For inclusion in tables, fragments had an arbitrarily chosen lower size limit of 26 bp.
Data representation:
Linear genome representations were generated individually by an in-house drawing program that uses as input a spreadsheet containing the genome coordinates for genes and other features (available at http://mitolab.missouri.edu/newpage2/research/GenomeApplet).
Graphic genome-similarity alignments were generated using MultiPipMaker, a web-based tool for genomic sequence alignments (Schwartz et al. 2000, 2003; bio.cse.psu.edu/pipmaker). The annotated Z. mays NB mtDNA genomic sequence (Clifton et al. 2004) was used as the reference genome.
Pairwise graphic representations were generated with Miropeats (Parsons 1995) and manually modified using Adobe Illustrator.
Genome alignment:
The four mitochondrial genomes were aligned with the complete NB mitochondrial genome. Sequences >200 bp that are not present in the NB genome but are in the NA genome were then used as reference in an alignment appended to the NB alignment. Next, sequences present in CMS-C but not present in either NB or NA were used as a reference in a continuation of the combined alignment. Finally, alignments with sequences unique to CMS-T were appended to create a mega-alignment. There were no sequences unique to CMS-S.
Candidate homologs were obtained using BLASTN. Where large repeated sequences (>500 bp) were present, the larger of the repeats was used in the alignment. Paralogous sequences were detected using Multipip (Schwartz et al. 2000, 2003), which allows the rapid visual discrimination among sequences by pattern. Because of the abundant fragments of variable size and orientation in each of the genomes being aligned to NB, the alignment of the five sequences plus rice was done manually. Indels were arbitrarily limited to 200 bp to maintain the cohesion of the NB reference sequence; larger insertions were treated as unique sequence and placed in the appended alignments (described above).
Genome statistics:
In-house programs were used to generate statistics from the alignment, such as number of substitutions or indels, indel size, nucleotides in common between genomes, etc. Large searches for similar nucleotide or amino acid sequences were performed with BLAST (Altschul et al. 1990).
Analysis of short repeats:
Small dispersed repeats (SDRs) were defined as sequences of at least 20 bp (but <500 bp) that are present more than once in the genome, are at least 90% identical in sequence, and are of exactly the same length. Repeats were discovered using RepeatExtractor, an in-house script that is publicly available as part of BioExplorer (zeamtdna.missouri.edu/nlsap-gui.htm). See Clifton et al. (2004) for a detailed description of repeat extraction criteria.
Simple sequence repeats (SSRs) with repeat units of 1–5 bp were located and characterized using Sputnik (cbi.labri.fr/outils/Pise/sputnik.html; this program has an upper limit of 5 bp). Minimum acceptable total SSR length depended on repeat-unit size (1- and 2-bp repeat minimum length = 11 bp, 3 bp = 12 bp, 4 bp = 13 bp, and 5 bp = 14 bp). Tandem repeats with repeat units of 5–50 bp were located and characterized using Tandem Repeats Finder (Benson 1999); gap opening penalty = 5, minimum alignment score = 30, maximum period = 50, minimum overall identity = 95%.
Transmembrane-domain prediction:
SOSUI (Hirokawa et al. 1998) and TMBETA-NET (Gromiha et al. 2004) were used to determine transmembrane domains, transmembrane segment types (α- or β-strands), and residue positions of each transmembrane segment. SOSUI targets α-helical segments whereas TMBETA-NET targets β-strand segments. All predictions were made using default settings of the program parameters.
RESULTS AND DISCUSSION
Genome maps:
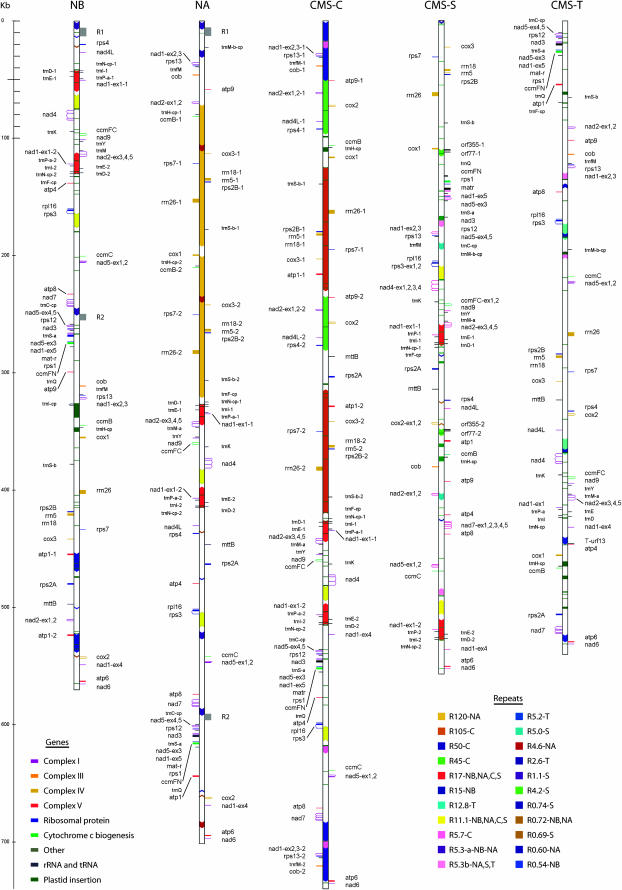
The NA, CMS-C, CMS-S, and CMS-T mitochondrial genomes were sequenced from highly purified mtDNA using the shotgun-sequencing method described previously for the NB maize mitochondrial genome (Clifton et al. 2004). The assemblies for NA, CMS-C, and CMS-T generated circular “master” genomes that are depicted in linearized form in Figure 1. Sequences were assembled without reference to an expected organization. Nevertheless, our maps were found to agree well with the maps generated from previous restriction mapping studies (reviewed in Fauron et al. 1995a).
Figure 1.—
Linear representations of maize mitochondrial genomes. The relative positions of genes are indicated by horizontal lines, color coded by function, as indicated in the key at the bottom left. Cis-spliced exons are connected by a caret. tRNA genes are indicated by short lines. Transcription direction is down for genes on the right and up for those on the left of each genome bar. For very closely spaced genes, spacing has been increased for visibility. Arrowed boxes within each genome bar indicate large repeats (>0.5 kb), color coded according to the key at the bottom right, with arrowheads indicating orientation. Genes in repeats contain the suffix -1 or -2. Rectangular green boxes/lines indicate transferred plastid sequences. Genes in plastid sequences contain the suffix -cp. Scale is in kilobases.
Circular master genome maps have been generated for most of the plant mitochondrial genomes sequenced to date (e.g., Fauron et al. 2004). We did not observe sets of shotgun clones with identical termini that would be suggestive of uniform linear ends. This implies that the genomes could exist as circular molecules, although not necessarily as master circles. Arguments have been made for the existence of a master circular form of a wheat mitochondrial genome after sequencing of gene-anchored mtDNA cosmids (Ogihara et al. 2005).
For the “circular” maize mitochondrial genomes, other conformations have been observed that are due to recombination across repeated sequences (reviewed in Lonsdale et al. 1984; Fauron et al. 1995a). Restriction enzyme analysis shows the existence of some recombination products (Lupold et al. 1999). Furthermore, Levy et al. (1991) identified a 120-kb subgenome in the mitochondrial genome of Black Mexican Sweet corn using pulsed-field gel electrophoresis. This subgenome did not correspond to any subgenome predicted by recombination across repeats, nor was it present in the mitochondrial genome of the inbred line (B73) most similar to the one used in this sequencing project (B37). The numbers of our sequencing reads for each genome were not sufficient to model subgenome configurations.
Despite the evidence for the existence of circular master maps from restriction mapping and genomic data, attempts to show their existence have been largely unsuccessful (Bendich 1996; Jacobs et al. 1996). Thus, it remains controversial whether circular plant mitochondrial master genomes exist in vivo.
In contrast to the other maize mitochondrial genomes, CMS-S assembled as a linear genome. That a majority of this genome is present in linear form had been shown by Schardl et al. (1984), who used digestion with the exonuclease Bal31 to show degradation of linear ends. Our sequence data resulted in multiple possible configurations, which was consistent with the restriction enzyme mapping results of Schardl et al. (1984). They predicted that a multiplicity of linear forms results from recombination between either of two abundant linear plasmids (the S1 and S2 plasmids) found within CMS-S mitochondria and short regions of identity that are present at two locations within an originally circular main mitochondrial genome. Supplemental Figure 1 (http://www.genetics.org/supplemental/) diagrams such a recombination event, which leads to a linearized S genome with an S-plasmid at one end and a plasmid terminal inverted repeat at the other end. Multiple genomic organizations are expected to exist for the linearized CMS-S mtDNAs because, theoretically, either plasmid can integrate via recombination in either the forward or the reverse orientation in either of the two places in the genome. Our sequencing read depth (10×) was not sufficient to estimate the proportions of individual conformations. We did find evidence for integration of the S1 plasmid into the main genome. We assembled a circular form of the CMS-S mitochondrial genome by computationally recombining out the S plasmid that linearized the genome. The genomic sequence that results (Figure 1) also concurs with data from Xiao et al. (2006).
Genome size and composition:
The four newly sequenced maize mitochondrial genomes vary 38% in size from smallest to largest: 535,825 bp in CMS-T, 557,162 bp in CMS-S, 701,046 bp in NA, and 739,719 bp in CMS-C. The sizes of the newly sequenced circular genomes agreed well with earlier restriction enzyme mapping estimates of 700 kb for NA (Fauron and Casper 1994) and 540 kb for CMS-T (Fauron and Havlik 1989). The GC content of each genome is ∼44% (NA, 43.82%; C, 43.97%; S, 43.91%; T, 44.06%), similar to that of NB (43.93%) and other plant mitochondrial genomes (Clifton et al. 2004).
Duplications are responsible for most of the size differences among the five genomes (Table 1). The difference in size between the smallest genome (CMS-T) and the largest (CMS-C) is 203,894 bp (38% larger). However, if genomic complexities are compared—i.e., only one copy of each repeat (>0.5 kb) is considered—the differences are much reduced. The resulting genome complexities range from 506,760 bp (CMS-C) to 537,180 bp (NA), a difference of only 30,420 bp (6%). Thus, the genome complexity of CMS-T is actually larger than that of CMS-C. Large duplications do not, however, account for the greater sizes of the maize mitochondrial genomes relative to other plant mitochondrial genomes, such as rice (Oryza sativa). Repeated sequences (>0.5 kb) account for 28% of the 491-kb rice genome (Notsu et al. 2002), yet it is 45 kb smaller than the smallest maize genome, CMS-T, which has only 5.3% of its genome as duplications.
TABLE 1.
Portions of maize mitochondrial genomes present as genes and ORFs
| Genome
|
|||||
|---|---|---|---|---|---|
| Genome feature | NB | NA | CMS-C | CMS-S | CMS-T |
| Genomes | |||||
| Total genome size | 569,630 | 701,046 | 739,719 | 557,162 | 535,825 |
| Total repeated sequencea | 49,436 | 163,866 | 232,959 | 44,987 | 28,469 |
| Genome complexityb | 520,194 | 537,180 | 506,760 | 512,175 | 507,356 |
| Genes | |||||
| Protein genescd | 34,541 | 36,137 | 41,933 | 35,624 | 32,892 |
| Single-copyb protein genesc | 32,628 | 32,634 | 32,730 | 33,936 | 32,892 |
| cis introns | 23,135 | 23,137 | 26,156 | 23,150 | 23,145 |
| Single-copybcis introns | 23,135 | 23,137 | 23,147 | 23,150 | 23,145 |
| rRNA genes | 5,646 | 11,292 | 11,298 | 5,651 | 5,651 |
| Single-copyb rRNA genes | 5,646 | 5,646 | 5,649 | 5,651 | 5,651 |
| tRNA genesd | 1,573 | 1,735 | 1,662 | 1,574 | 1,207 |
| Single-copyb tRNA genesc | 1,206 | 1,353 | 1,134 | 1,207 | 1,207 |
| Pseudogenes and pseudo-exons | 1,487 | 1,416 | 1,335 | 1,488 | 663 |
| Single-copyb pseudogenes/exons | 1,487 | 1,335 | 1,335 | 1,488 | 663 |
| Coding totals | |||||
| Total known genescd | 41,760 | 49,164 | 54,893 | 42,849 | 39,840 |
| Total single-copyb genesc | 39,480 | 39,633 | 39,513 | 40,794 | 39,840 |
| Total genes, % of total genome | 7.33 | 7.01 | 7.42 | 7.69 | 7.44 |
| Single-copyc genes, % of complexity | 7.59 | 7.38 | 7.80 | 7.96 | 7.85 |
| ORFs | |||||
| Total ORFs | 53,343 | 68,199 | 62,754 | 51,846 | 41,973 |
| Single-copyb ORFs | 49,644 | 56,058 | 44,916 | 47,493 | 39,912 |
| ORFs % of total genome | 9.36 | 9.73 | 8.48 | 9.31 | 7.83 |
| Single-copyb ORFs, % of complexity | 9.54 | 10.44 | 8.86 | 9.27 | 7.87 |
Sizes are given in number of nucleotides.
Repeats >0.5 kb.
Second (and third) copies on repeats are not included.
Coding regions only.
Includes both exon and gene duplications for all genes listed in Table 2, but does not include introns, pseudogenes, or pseudo-exons.
Conserved genes:
All four of the newly sequenced maize mitochondrial genomes contain the same basic suite of 51 functional genes as does NB (Clifton et al. 2004), although the copy number of individual genes or exons varies among cytotypes due to the presence of duplications (Figure 1; Table 2). The conserved genes code for 33 known proteins. Eighteen of the mitochondrially encoded proteins are components of the ATP-generating electron transport chain: nine subunits of complex I (NAD 1, 2, 3, 4, 4L, 5, 6, 7, 9), apocytochrome b (COB) of complex III, three subunits of complex IV (COX 1, 2, 3), and five subunits of complex V (ATP 1, 4, 6, 8, 9). Four additional proteins participate in the biogenesis of cytochrome c (CCM B, C, FN, and FC). Nine genes encode eight ribosomal proteins (RPS 1, 2, 3, 4, 7, 12, 13, and RPL16). In all five maize mitochondrial genomes, two distinct rps2 genes have been retained (rps2A and rps2B), although previous studies suggested that rps2A is the major active gene (Perrotta et al. 2002). All the maize mitochondrial genomes carry a conserved gene for a putative maturase (mat-r) located within the fourth intron of nad1 (Wahleithner et al. 1990), as well as the mttB/tatC gene (Bonnard and Grienenberger 1995; Bogsch et al. 1998), which encodes a protein translocase (Weiner et al. 1998).
TABLE 2.
Known functional genes in maize mitochondrial genomes
| Product group | Gene | NB | NA | CMS-C | CMS-S | CMS-T |
|---|---|---|---|---|---|---|
| Complex I | nad1a | +b | +b | +bc | +b | + |
| nad2a | +d | +d | +de | +bd | + | |
| nad3 | + | + | + | + | + | |
| nad4 | + | + | + | + | + | |
| nad4L | + | + | 2+ | + | + | |
| nad5a | + | + | + | + | + | |
| nad6 | + | + | + | + | + | |
| nad7 | + | + | + | + | + | |
| nad9 | + | + | + | + | + | |
| Complex III | cob | + | + | 2+ | + | + |
| Complex IV | cox1 | + | + | + | + | + |
| cox2 | + | + | 2+ | + | + | |
| cox3 | + | 2+ | 2+ | + | + | |
| Complex V | atp1 | 2+ | + | 2+ | 2+ | + |
| atp4 | + | + | + | + | + | |
| atp6 | + | + | + | + | + | |
| atp8 | + | + | + | + | + | |
| atp9 | + | + | 2+ | + | + | |
| Cytochrome c biogenesis | ccmB | + | 2+ | + | + | + |
| ccmC | + | + | + | + | + | |
| ccmFN | + | + | + | + | + | |
| ccmFC | + | + | + | + | + | |
| Ribosome | rps1 | + | + | + | + | + |
| rps2A | + | + | + | + | + | |
| rps2Bf | + | 2+ | 2+ | + | + | |
| rps3 | +b | +b | +b | +b | +g | |
| rps4 | + | + | 2+ | + | + | |
| rps7 | + | 2+ | 2+ | + | + | |
| rps12 | + | + | + | + | + | |
| rps13 | + | + | 2+ | + | + | |
| rpl16 | + | + | + | + | + | |
| Other proteins | mat-r | + | + | + | + | + |
| mttB | + | + | + | + | + | |
| T-urf13h | − | − | − | − | + | |
| orf355/orf77h | − | − | − | + | − | |
| tRNA | ||||||
| Asparagine | trnN | 2 cp | 2 cp | 2 cp | 2 cp | cp |
| Aspartic acid | trnD | 2 mt | 2 mt | 2 mt | 2 mt | mt |
| Cysteine | trnC | cp | cp | cp | cp | cp |
| Glutamic acid | trnE | 2 mt | 2 mt | 2 mt | 2 mt | mt |
| Glutamine | trnQ | mt | mt | mt | mt | mt |
| Histidine | trnH | cp | 2 cp | cp | cp | cp |
| Isoleucine | trnI | 2 mt, cp | 2 mt | 2 mt | 2 mt | mt |
| Lysine | trnK | mt | mt | mt | mt | mt |
| Methionine | trnM | mt | mt-a,i cp-b | mt | mt-a,i cp-b | mt-a,i cp-b |
| f Methionine | trnfM | mt | mt | 2 mt | mt | mt |
| Phenylalanine | trnF | cp | cp | cp | cp | cp |
| Proline | trnP | 2 mt | 2 mt | 2 mt | 2 mt | mt |
| Serine | trnS | mt-a,i mt-b | mt-a,i mt-b | 2 mt-a,i mt-b | mt-a,i mt-b | mt-a,i mt-b |
| Tryptophan | trnW | cpj | cpj | cpj | cpj | cpj |
| Tyrosine | trnY | mt | mt | mt | mt | mt |
| rRNA | rrn5 | + | 2+ | 2+ | + | + |
| rrn18 | + | 2+ | 2+ | + | + | |
| rrn26 | + | 2+ | 2+ | + | + |
+, present; −, absent. Whole-gene copy numbers >1 are given.
trans-spliced.
Two copies of exon 1.
Two copies of exons 2 and 3.
Two copies of exons 4 and 5.
Two copies of exons 1 and 2.
May not be functional.
Three copies of exon 1.
Involved in cytoplasmic male sterility.
“a” and “b” represent different anticodons. “mt” and “cp” denote mitochondrial and plastid origin, respectively.
Present on the mitochondrial 2.3-kb linear plasmid (2.1 kb in NA and CMS-T).
Three genes do not contain a DNA-encoded AUG start codon by comparison with other genomes. In two instances (nad1, nad4L), an ACG codon is assumed to be RNA edited to AUG (Chapdelaine and Bonen 1991; Handa 2003), and in the last instance (mat-r), an AGA has been taken to be the start codon in maize mitochondrial genomes by comparison with other organisms in the public sequence databases. The stop codon is UAA for 15 genes, including atp6 that is RNA edited from CAA (Kumar and Levings 1993), UAG for 9 genes, and UGA for 9 genes, including ccmFC that is RNA edited from CGA (Handa 2003).
All of the maize mitochondrial genomes contain three ribosomal RNA genes (5S, 18S, and 26S). Each of the genome complexities also possesses 15 nonduplicate tRNA genes for the same 14 amino acids (Table 2). In addition, the functional tryptophan tRNA gene is located on a small linear plasmid that was not sequenced in this project (2.1 kb in NA and CMS-T, 2.3 kb in the other cytotypes; Bedinger et al. 1986; Bogsch et al. 1998). Of the 16 tRNA genes, 11 are “native” mitochondrial genes, including two distinct tRNA-Ser genes. The tRNAs for 5 amino acids are encoded solely by genes of plastid origin. For two tRNA genes, some genomes have only a “native” mitochondrial version, while other genomes have an additional plastid-derived version. These include the tRNA-Met genes in the NA, CMS-S, and CMS-T mitochondrial genomes, and an intron-containing tRNA-Ile gene in NB. The transfer RNAs for 6 amino acids (alanine, arginine, glycine, leucine, threonine, and valine) are not encoded in the mitochondrion and are presumed to be encoded in the nuclear genome and imported into the mitochondrion (Kumar et al. 1996). There are also several tRNA pseudogenes in each genome.
Most of the protein-coding genes are identical in both amino acid and nucleotide sequence in all five maize cytotypes. Among the 11 polymorphic genes (Table 3), 3 are notable. First, the CMS-C atp6 gene contains 481 nucleotides at its 5′ end that are unique, as noted by Dewey et al. (1991). Second, the atp4 gene of CMS-T has 10 nucleotide substitutions, compared with the other four cytotypes, and another substitution shared only with CMS-S. These nucleotide differences predict six amino acid changes. Third, relative to NB and NA, rps2A has several nonsynonymous substitutions in its 3′ end: 2 in CMS-C, 5 in CMS-S, and 3 in CMS-T. This region is part of a large, grass-specific C-terminal extension of unknown function that is proteolytically cleaved to yield the mature protein (Perrotta et al. 2002). In the genes other than atp4 and rps2A, there is a total of 12 substitutions, 8 of which are nonsynonymous. Relative to NB, there are few transitions compared to transversions within the coding regions; for NA, there are 0 transitions:1 transversion; for CMS-C, 2:2; for CMS-S, 1:9; and for CMS-T, 5:14 (1:8 if atp4 is excluded).
TABLE 3.
Substitutions and indels within maize mitochondrial genes, relative to cytotype NB
| NB vs.:
|
||||
|---|---|---|---|---|
| NB gene | NA | CMS-C | CMS-S | CMS-T |
| atp4 (660 bp) | 28-36 | |||
| GGTaTaaaT → AAAaGaaaA | ||||
| (GIN → KRK) | ||||
| 227 tTc → tGc (F → C) | ||||
| 287 cGg → cAg (R → Q) | ||||
| 348 tgT → tgC (C) | ||||
| 498 ttA → ttT (L → F) | ||||
| 513 acA → acT (T) | ||||
| 603 ctC → ctA (L) | 603 ctC → ctA (L) | |||
| 634 acc → aAGAGATcc | 634 acc → aAGAGATcc | 634 acc → aAGAGATcc | 634 acc → aAGAGATcc | |
| (T → KRS) | (T → KRS) | (T → KRS) | (T → KRS) | |
| atp6 (1233 bp) | P | First 481 bp unique | P | P |
| 537 ggcaag → ggTGag | ||||
| (GK → GE) | ||||
| ccmFN (1863 bp) | P | 1485 aGa → aTa (R → I) | ||
| 1577 ttG → ttT (L → F) | ||||
| nad4L (303 bp) | 234 gcC → gcA (A) | P | 210 gcT → gcA (A) | |
| nad7 exon 3 (467 bp) | P | P | P | 56 Ttg → Gtg (L → V) |
| nad9 (573 bp) | P | P | P | 402 acT → acG (T) |
| rps2A (1596 bp) | P | 225 ttA → ttC (L → F) | ||
| 691 Tgg → Ggg (W → G) | 691 Tgg → Ggg (W → G) | 691 Tgg → Ggg (W → G) | ||
| 815 gGc → gAc (G → D) | 815 gGc → gAc (G → D) | |||
| 835 Agt → Tgt (S → C) | 835 Agt → Tgt (S → C) | 835 Agt → Tgt (S → C) | ||
| 1243 Caa → Aaa (Q → K) | ||||
| rps2B (1251 bp) | P | P | 231 ttA → ttC (L → F) | |
| 465 gtT → gtG (V) | ||||
| 750 gaA → gaT (E → D) | ||||
| rps3 exon 2 (1606 bp) | P | P | 651 tTt → tAt (F → Y) | P |
| rps13 (351 bp) | P | 349 at → cGGAAAtcreates 34 bp 3′ extension | P | P |
| rrn18 (1968 bp) | P | 890 ct → cCTACGt | P | P |
| 1342 cCACGGAGa → ca | ||||
| rrn26 (3552 bp) | P | 398 cc → cGGGCGc | 2882 at → aTCATTt | |
| 1898 cg → cGTTAGg | ||||
Altered nucleotides are in capital letters, flanking nucleotides in lowercase. Numbers indicate nucleotide position of change, relative to the start site of the coding region (protein-coding genes) or mature RNA (rRNA genes). For protein-coding genes, the codon containing the change is given. Letters within parentheses are single-letter amino acid codes. “P” indicates that the gene is present in identical form. The number of base pairs in each coding region (whole gene or exon) is given.
All but 4 of the 28 substitutions and all seven indels within genes are unique to a single cytotype, including 15 substitutions and one indel in CMS-T. The exceptions are three of the differences in rps2A and one in atp4. Because the genes in Table 3 are being compared to the NB versions, the apparent insertion in atp4 at 634 in four genomes is actually a deletion only in NB. Five of the indels are in the 18S and 26S rRNA genes.
In addition to the conserved set of functional genes, the CMS-S and CMS-T genomes each have expressed ORFs that are known to be associated with cytoplasmic male sterility (T-urf13 in T and orf355/orf77 in S; Dewey et al. 1987; Zabala et al. 1997).
Because of the large duplications, a twofold variation in copy number occurs for some mitochondrial genes, yet it does not appear to pose a gene-regulation problem. Even though the two male-fertile genomes, NB and NA, differ substantially in the number of repeated genes, as do cytoplasmic-male-sterile CMS-C (which has 13) and CMS-T (which has none), there is no detectable phenotype that distinguishes the genomes except the phenotypes associated with the CMS genes themselves (Laughnan and Gabay-Laughnan 1983). Two- and even threefold variation occurs for individual exons of mitochondrial genes among the cytotypes, yet this also does not cause a detectable problem for the plant. Mitochondrial gene expression has been shown to be regulated at both transcriptional and post-transcriptional levels in maize (Mulligan et al. 1991) and mainly at the post-transcriptional level in Arabidopsis (Giege et al. 2000). Post-transcriptional regulation may account for the lack of dosage effects in maize mitochondrial genomes.
Open reading frames:
A complete list of ORFs (minimum size, 100 codons) that did not overlap known genes was generated for each genome. Each of the maize mitochondrial genomes contains between 95 and 116 different ORFs that are not thought to be associated with CMS (supplemental Tables 1–3 at http://www.genetics.org/supplemental/). The percentage of each genome's complexity occupied by such ORFs (7.87–10.44%) was found to be higher than the percentage occupied by known exon sequences (7.38–7.96%; Table 1).
The largest ORFs found in the maize mitochondrial genomes (orf1159, orf917, and orf911; supplemental Tables 1–3) are larger than any of the known genes. The largest ORFs appear to be degenerate copies of all or part of the DNA polymerase (orf917, orf911, and orf734) or RNA polymerase (orf1159, orf653, orf487, and orf417) genes derived from either the S plasmids of CMS-S maize or the homologous R plasmids of RU maize (Weissinger et al. 1982). These plasmids are the source of sequences that yield other ORFs throughout the genomes as well. For example, 10 of the 105 nonduplicate ORFs in NB are derived from degenerate segments of R/S plasmid genes.
With the exception of ORFs that confer cytoplasmic male sterility, mitochondrial ORFs that were later identified as functional genes were found to be conserved across a range of taxa. They include the ccm genes (Handa et al. 1996) and the previously designated orfX (now mttB/tatC) (Bonnard and Grienenberger 1995; Bogsch et al. 1998), orf25 (now atp4), and orfB (now atp8) (Heazlewood et al. 2003). However, only 54 of the 105 such ORFs found in the NB mitochondrial genome complexity (Clifton et al. 2004) were present as open reading frames in all of the other genomes (supplemental Table 1). Fifty ORFs from the NB genome are missing in at least one of the other four maize mitochondrial genomes (supplemental Table 2), and many ORFs were identified in the other genomes that were not open reading frames in NB (supplemental Table 3). Just one of the ORFs, orf99-a, is also present as an ORF in the rice mitochondrial genome (Notsu et al. 2002), but it is not present as an ORF in the wheat mitochondrial genome (Ogihara et al. 2005) or any other plant mitochondrial genome. Most of the mitochondrial ORF sequences were not found to be expressed as abundant stable RNAs in Arabidopsis (Giege et al. 1998) or NB maize (Meyer 2004). Thus few, if any, of the unknown ORFs are likely to be functional.
Chimeric ORFs:
Rearrangement of the mitochondrial DNA can join pieces of genes, ORFs, or noncoding sequences to produce novel, chimeric open reading frames. Chimeric ORFs have been implicated in CMS in many taxa (reviewed by Hanson and Bentolila 2004); in maize, they include T-urf13 in CMS-T and orf355/orf77 in CMS-S. Recently, a small ORF, orf79, was shown to be the cause of CMS in Boro II rice (Wang et al. 2006), and orf77 is a candidate ORF in CMS-S (Zabala et al. 1997). Therefore, ORFs of at least 70 codons were characterized for chimerism using nucleotide BLAST (Table 4). Only ORFs containing gene fragments of at least 26 bp are included in Table 4. For such ORFs, gene fragments of 16–25 bp are also shown. Some gene sequences are parts of two chimeric ORFs; e.g., in the CMS-T genome, 35 bp of a plastid-derived trnR is present in orf149-a and 31 bp in orf118-b. The chimeric ORFs that have already been associated with cytoplasmic male sterility in CMS-S and CMS-T mitochondrial genomes are included in the gene table (Table 2).
TABLE 4.
Chimeric ORFs (≥70 codons) present in the five maize mitochondrial genomes
| Genome
|
|||||
|---|---|---|---|---|---|
| NB | NA | CMS-C | CMS-S | CMS-T | Gene fragments present |
| orf73 | — | — | — | — | 28 bp atp9 |
| orf75 | orf75 | — | orf75 | orf75 | 29 bp atp9 |
| orf70 | orf70 | orf70 | orf70 | orf70 | 42 bp nad7 exon 4 |
| — | — | — | orf72 | — | 153 bp nad2 pseudo-exon 1 |
| orf73-a | — | — | — | — | 219 bp nad2 pseudo-exon 4 |
| orf73-b | — | — | — | — | 51 bp nad2 pseudo-exon 4 |
| orf86 | orf86 | orf86 | orf86 | — | 67 bp atp6 |
| orf90 | — | orf90 | orf90 | orf90 | 31 bp rps3 exon 1 |
| orf89 | orf89 | orf89 | orf89 | orf89 | 28 bp plastid rrn23 |
| orf197 | — | — | orf221 | — | 573 bp nad2 pseudo-exon 4 |
| orf191a | — | orf191a | orf191a | orf191a | 30 bp rps3 exon 1 |
| — | orf130-bb | — | — | — | 30 bp rps3 exon 1 |
| orf159-a | orf159-a | orf163-b | orf163-b | orf159-a | 264 bp rps3 (exon 1 + 190 bp of intron 1); 209 bp rps12 |
| orf101-c | orf101-c | orf101-c | orf101-c | — | 26 bp rps3 exon 2 |
| orf186 | orf186 | orf186 | orf186 | — | 80 bp cox2 exon 1; 28 bp rps2B |
| — | orf179-b | — | — | orf179-b | 21, 43 bp cox2 exon 1 |
| orf248 | orf248 | orf246 | — | orf248 | 26, 24, 23, 29 bp atp6-CMS-C; 21 bp rps3 exon 2 |
| orf248b | orf248b | orf246b | orf189 | orf248b | 26, 24 bp atp6-CMS-C |
| orf396 | orf364 | orf410 | orf462 | — | 112 bp atp6-NBc |
| – | orf105-h | — | — | orf105-h | 47 bp atp9 |
| orf147-b | orf147-b | orf147-b | orf147-b | orf190 | 24 bp atp4; 39 bp atp9; 19 bp cox1 |
| orf149-a | orf149-a | orf149-a | orf134-b | orf149-a | 35 bp cp trnR |
| — | — | — | — | orf118-b | 31 bp cp trnRd |
| 8 | 4 | 4 | 6 | 4 | Chimeric ORFs 70–99 codons |
| 9 | 10 | 8 | 9 | 8 | Chimeric ORFs ≥100 codons |
| 296 | 360 | 388 | 279 | 280 | Total ORFs 70–99 codons |
| 105 | 116 | 103 | 107 | 95 | Total ORFs ≥100 codons |
| 2.7% | 1.1% | 1.0% | 2.2% | 1.4% | Chimeric ORFs 70–99 codons as percentage of all ORFs 70–99 codons |
| 8.6% | 8.6% | 7.8% | 8.4% | 8.4% | Chimeric ORFs ≥100 codons as percentage of all ORFs ≥100 codons |
ORFs of 70–99 codons are in the top portion of the list, and ORFs of ≥100 codons are in the bottom portion of the list. Identified CMS-related ORFs (T-urf13, orf355, orf77) and ORFs within sequences of plastid origin (see materials and methods) are not included. ORF totals are for genome complexity. ORFs must have at least one gene fragment ≥26 bp, but additional fragments of 16–25 bp are shown if present.
The orf130-b sequence lies within the orf191 sequence and contains the same gene fragment, but is in the opposite orientation.
Contains the fragments listed plus additional fragments listed in the line above. See Figure 2 for a graphic representation of some of the chimeric ORFs or supplemental Table 1 at http://www.genetics.org/supplemental/ for all of them.
This atp6 version is also the version in NA, CMS-S, and CMS-T. This 112-bp segment is also found 7 bp upstream of the cox2 ATG in all the genomes except CMS-C.
Part of the fragment on the line above it.
Eight to 10 chimeric ORFs of ≥100 codons (the minimum size annotated; Table 4, supplemental Figure 2 at http://www.genetics.org/supplemental/) are present within the genome complexities of each of the five mitochondrial types, accounting for <10% of all ORFs of that size range in each genome (supplemental Tables 1–3). An additional 4–8 chimeric ORFs of 70–99 codons were identified in each genome (Table 4).
Three main types of chimeric ORFs are present (supplemental Figure 2 at http://www.genetics.org/supplemental/). The first type is the same length in all the mitochondrial genomes in which it is found (e.g., the 186-amino-acid orf186). The second type has different lengths in different cytotypes due to a changed start or stop site (e.g., orf147/orf190, which contains pieces of three genes). The third type has both a different start or stop and additional gene segments in some of the ORFs. This type is represented by orf189 (CMS-S), orf246 (CMS-C), and orf248 (NB, NA, CMS-T), which all have the same start site and the first two gene segments; the two longer versions have three additional gene segments.
CMS-associated sequences:
Finding the genes responsible for CMS has often been a tedious task, and attempts to identify them by comparing a single CMS genome with a single fertile genome have not been generally successful (Hanson and Bentolila 2004). However, by comparing the complete sequences of multiple, closely related mitochondrial genomes, the list of chimeric ORFs that are unique to a given CMS cytotype, and thus are candidates for causing CMS, is reduced.
Using a 70-codon minimum size, only two chimeric ORFs were found to be present uniquely in the CMS-T genome. One of them, orf115, is T-urf13, the gene known to cause T-type cytoplasmic male sterility (Dewey et al. 1986, 1987; Wise et al. 1987). It encodes a 13-kDa membrane-spanning polypeptide, and oligomers of this protein can depolarize the mitochondrion, leading to cell death (Rhoads et al. 1994). In contrast, the other ORF, orf118-b, is predicted to be a soluble protein with no membrane-spanning domains (data not shown). Because CMS-associated genes usually contain membrane-spanning domains, orf118-b was not considered to be a candidate gene for T-type male sterility.
The only chimeric ORF ≥70 codons that was found uniquely in the CMS-S genome sequence is orf77, which is composed largely of sequences from atp9 (Zabala et al. 1997). It lies on the same strand as, and is located just 11 bp downstream from, orf355. orf355 is composed of sequences from several genomic locations: the 5′ third is composed of sequences otherwise found only in NB, the middle third only in NB and NA, and the final third and most of orf77 only in NA and CMS-T. The latter two-thirds are separated by 33 bp of unknown origin. Because orf355 does not contain recognizable pieces of known genes, it is not considered to be a chimeric ORF by our definition. The cotranscribed orf355/orf77 region has been suggested as the cause of CMS-S (Zabala et al. 1997). CMS-associated genes are often located very near another gene or ORF (Schnable and Wise 1998). For example, T-urf13 is just 78 bp upstream of atp4. As sequencing costs are further reduced and analytical tools are improved, comparative sequencing of related mitochondrial genomes may become the preferred first step in the process of identifying other CMS-causing genes.
In contrast to the relative ease of identifying candidate sterility-causing genes in CMS-T and CMS-S, no chimeric ORF of at least 70 codons was found that was both unique to CMS-C and did not overlap a known gene. It may be that an even smaller chimeric ORF is responsible for this sterility. Thus, the lower size cutoff was reduced to 50 codons. CMS-C contains 752 ORFs of 50–69 codons, of which only 11 are chimeric (containing known-gene fragments of at least 26 bp; supplemental Table 4 at http://www.genetics.org/supplemental/). Three are unique to CMS-C, none of which is predicted to have a membrane-spanning region. The smallest, orf59-b, has only a short sequence from atp9, predicting an out-of-frame 9-amino-acid segment. orf62 contains three fragments, from rps3 and atp6, in the reverse orientation relative to their original genes. Twenty-one base pairs from rps3 exon 2 are followed by 29 and 23 bp from atp6. The two atp6 segments are apparently derived from a single segment that has undergone a 60-bp deletion. orf61 is also unique to CMS-C, but it is closely related to CMS-T orf54. Both contain two large gene fragments: 56 bp from atp4 and ∼70 bp from atp1. CMS-C orf61 and orf62 are candidates for further analyses.
Another possibility is that CMS-C is caused by one or more of the three rearranged functional genes—atp6, atp9, and cox2—identified by Dewey et al. (1991). We paid particular attention to the chimeric atp6 gene in CMS-C because (1) it is present in a single copy and (2) some CMS-associated ORFs are cotranscribed with atp genes (Hanson and Bentolila 2004; Wang et al. 2006). The CMS-C atp6 gene contains a 482-nucleotide leader that has a completely different sequence from the 424-nucleotide leader in the other four maize mitochondrial genomes. The extreme 5′ end of the C-atp6 leader sequence is derived from the atp9 gene, including the first 39 bp of the coding region and all of its upstream regulatory region (see supplemental Figure 3 at http://www.genetics.org/supplemental/). The rest of the CMS-C atp6 leader sequence is not present anywhere in the other maize mitochondrial genomes (the sequences of “unknown origin” of Dewey et al. 1991). However, a segment 93% identical to nucleotides 41–478 of the C-atp6 leader, plus the first 131 bp of the atp6 core, is found immediately upstream of the cox2 start codon in Z. perennis (Newton et al. 1995). Although no other homologs were readily apparent in the nucleotide databases, the predicted amino acid sequence of the “unknown origin” region identified atp6 leader sequences in the mitochondrial genomes of tobacco, pepper, potato, petunia, male-sterile radish, and Arabidopsis (Bland et al. 1987; Loessl et al. 1987; Makaroff et al. 1989; Marienfeld et al. 1996; Lu and Hanson 1999; Kim and Kim 2006), but not in other eudicots studied so far. The eudicot leader sequences are highly diverged from the maize C-atp6 leader, although the atp6 core sequence is highly conserved.
An atp6 leader is associated with CMS in the Owen type of CMS in sugar beet, where a 387-codon leader precedes the core atp6 sequence (Yamamoto et al. 2005). After the gene is translated, the protein is processed to yield the core ATP6 plus a 35-kDa membrane protein that is associated with CMS. The sugar beet CMS-atp6 leader shares no sequence similarity with the maize CMS-C atp6 leader. Furthermore, the sugar beet CMS atp6 leader sequence is predicted to encode 17 transmembrane segments, whereas the maize CMS-C atp6 leader (as well as the loosely related atp6 leaders from both male-sterile and male-fertile eudicots) is predicted to have none. This suggests that the distinctive atp6 leader sequence in CMS-C maize is probably not directly responsible for the male sterility trait.
The upstream region of cox2 is rearranged (supplemental Figure 3 at http://www.genetics.org/supplemental/). In all five maize mitochondrial genomes, a region of similarity to the atp6 coding region lies upstream of the cox2 start codon. In the non-CMS-C genomes, an in-frame 112-bp atp6 core sequence (nucleotides 440–551) begins 7 bp upstream of the cox2 ATG. In CMS-C, the atp6 sequence starts at the same location, but continues upstream for >5 kb, thereby including 118 bp of core and the entire normal (e.g., NB) atp6 leader sequence (from its start codon to nucleotide 551). Nevertheless, the mature COXII protein is the same size in all maize mitochondria, leading to the proposal that the transcription start site for the CMS-C cox2 gene may be within the translocated atp6 coding region (Dewey et al. 1991). The identity of COXII in size and sequence to all other maize mitochondrial COXII proteins argues against the protein being the cause of CMS-C. However, if transcription in CMS-C starts at the atp6 promoter and the truncated atp6 protein sequence is post-translationally cleaved, the leader product may interfere with normal ATP6 function.
We found that there are two distinct forms of the atp9 gene in CMS-C. One of them, designated atp9-1, is the same as is found in the other four genomes and thus is expected to be a completely functional copy. The other copy, designated atp9-2 and first described by Dewey et al. (1991), is also complete. However, relative to the other four genomes, 113 bp upstream of its start codon, rearrangements introduced a 199-bp segment that is also present 611 bp upstream of the nad4L start codon (supplemental Figure 3 at http://www.genetics.org/supplemental/).
Only one other major difference in a protein-coding gene distinguishes CMS-C from the other maize cytoplasms. A 5-bp insertion immediately upstream of the stop codon of rps13 changes the reading frame and leads to a 13-amino-acid addition to the gene. It is not clear how such a mutation might be related to CMS. Indeed, if this change caused a malfunctioning ribosomal protein to be made, one might expect to see a constitutively defective growth phenotype (Newton et al. 1996) rather than CMS.
Even after extensive sequence comparisons, we lack a clear candidate gene for CMS-C. This illustrates that the use of multiple genomes to identify CMS-associated regions will not always be successful. Additional techniques will be necessary to distinguish among candidates or to confirm the designation of a CMS gene. These include testing tissue-specific expression of candidate CMS genes in the presence or absence of nuclear restorer alleles, and experimentally inducing CMS following introduction of candidate CMS genes into male-fertile plants (reviewed in Hanson and Bentolila 2004).
Plastid sequences in the mitochondrial genomes:
The discovery of plastid sequences in maize mitochondrial genomes was first reported by Stern and Lonsdale (1982), and it is now known that sequences of plastid origin are widespread among plant mitochondrial genomes. What is evident from our analysis is how fluid the content of such exogenous sequences in the main mitochondrial genome can be. Maize NB was reported to have 25,281 bp of transferred plastid DNA (cpDNA: Clifton et al. 2004), or 4.44% of the genome complexity. The genomes reported here all contain substantially differing amounts. In the non-NB mtDNAs, sequences of plastid origin account for between 2.98% (CMS-C) and 4.42% (CMS-T) of genome complexity (Table 5). If the proportions of all five mtDNAs are calculated on the basis of total genome size, so that plastid insertions on repeats are included, the range is between 2.29 and 4.61% (NB). The additional maize mitochondrial genome sequences allowed the retrospective identification of four additional, small cpDNA insertions in the NB genome, and thus the amount and percentage of cpDNA presented here are slightly larger than reported in Clifton et al. (2004). Of the 45 regions of the plastid genome that are present in at least one of the maize mitochondrial genomes, 35 are found in all of them. Most of the inserts are short: 78% are <500 bp and 51% are <100 bp (supplemental Table 5 at http://www.genetics.org/supplemental/).
TABLE 5.
Sequences of plastid or plasmid origin integrated into the maize mitochondrial genomes
| Genome | Total nucleotides | cpDNA nt | % of genome | Plasmid nt | % of genome |
|---|---|---|---|---|---|
| NB | 569,630 (520,194) | 26,239 (25,453) | 4.61 (4.89) | 12,102 (12,102) | 2.12 (2.33) |
| NA | 701,046 (537,180) | 29,470 (20,854) | 4.20 (3.88) | 11,377 (11,377) | 1.62 (2.12) |
| CMS-C | 739,719 (506,760) | 16,929 (15,084) | 2.29 (2.98) | 0 | 0 |
| CMS-S | 557,162 (512,175) | 20,780 (20,433) | 3.73 (3.99) | 374a (374) | 0.07 (0.07) |
| CMS-T | 535,825 (509,912) | 23,669 (22,423) | 4.42 (4.40) | 0 | 0 |
Numbers in parentheses do not include nucleotides in large repeats (>0.5 kb); i.e., they are genome complexity numbers. Minimum sequence identity is 80%.
The linearizing S1 and S2 plasmids are not included.
Three major differences account for much of the variation in cpDNA among the newly sequenced genomes. First, in NB mtDNA, a 12,592-bp segment from the plastid inverted repeat accounts for almost half of its total cpDNA (Clifton et al. 2004). Clifton et al. (2004) hypothesized that this insert in NB maize was actually part of a 23-kb insert that had been fragmented by normal mitochondrial recombination, followed by the loss of some of the resulting fragments. The other genomes have much smaller versions of the 12.6-kb insert: NA, CMS-S, and CMS-T have only 3206 bp and CMS-C has 1464 bp (supplemental Table 5 at http://www.genetics.org/supplemental/). The most parsimonious explanation is that these genomes lost more DNA from a larger plastid DNA segment. The second major difference is that NA, CMS-S, and CMS-T (but not CMS-C) contain an additional 3756 bp of cpDNA from the middle of the large single copy (LSC) region of the plastid genome that is not present in NB. Third, CMS-T is unique in having 2682 bp from yet another part of the LSC region.
It appears that plastid DNA can be gained and lost rapidly from the mtDNA. Whereas the maize mitochondrial genomes that we examined differ in complexity (only one copy of each repeat is included) by a maximum of 6%, the amount of plastid DNA differs by up to 69%. However, the variation in plastid amount is due to only 10 of the 45 segments of plastid origin (supplemental Table 5 at http://www.genetics.org/supplemental/). In general, the variable segments are larger than the nonvariable ones, in keeping with the idea that large plastid segments are acquired and then fragmented, after which some of the fragments may be lost (Clifton et al. 2004).
All five of the maize mitochondrial genomes contain the previously described 4.1-kb plastid DNA insert whose unique organization and sequence suggested that recent plastid DNA transfers have copy corrected part of the insert (Clifton et al. 2004).
Integrated plasmid sequences:
Several linear and circular plasmids are present in maize mitochondria, and portions of some of them are found in the main mitochondrial genomes (Table 5). The 6397-bp S1 (Paillard et al. 1985) and 5453-bp S2 (Levings and Sederoff 1983) linear plasmids are uniquely associated with S-type cytoplasmic male sterility. Some of the male-fertile South American RU cytoplasms have related plasmids designated R1 and R2 (Weissinger et al. 1982). The 7.4-kb R1 plasmid contains 4.5 kb that are nearly identical in sequence to the left end of S1, whereas R2 appears to be identical to S2. Slightly >2% of the NB and NA genome complexities are present as integrated S/R-plasmid sequences (Table 5). The NB mitochondrial genome possesses an integrated homolog of R1 (Houchins et al. 1986) that is missing 230 bp from its left end and an R2 that is missing 320 bp from its right end. The NA genome has the same R2 segment, but the integrated R1 is missing the right 603 bp. Neither CMS-C nor CMS-T contains sequences homologous to the S or R plasmids >55 bp that are not highly divergent.
The linear conformations of CMS-S that we observed contain integrated complete copies of S1; their sequences are identical to the free plasmid sequences. We did not detect integrated S2 plasmids. Previous data suggested that integrated copies of S2 do exist (Schardl et al. 1984, 1985).
S1 and R1 carry a gene for a viral-like DNA polymerase (S1 ORF3; Kuzmin and Levchenko 1987) and S2/R2 carries a gene for a viral-like RNA polymerase (S2 ORF1; Kuzmin et al. 1988). In other taxa, including Brassica napus, Podospora anserina, and Claviceps purpurea (a phytopathogenic fungus), similar ORFs for DNA and RNA polymerases are located on single mitochondrial plasmids (Oeser and Tudzynski 1989; Hermanns and Osiewacz 1992; Handa et al. 2002). It has been suggested that the plasmid polymerase genes are used only for plasmid maintenance and expression (Oeser and Tudzynski 1989). In contrast, the genes for the polymerases involved in maintaining and expressing the normal set of genes in the main mitochondrial genome are coded for in the nucleus (Chang et al. 1999).
Diverged copies of plasmid ORFs are the basis for the largest ORFs in the five mitochondrial genomes. ORFs that have >90% similarity to the plasmid polymerase ORFs are present in two of the maize cytotypes (NB and NA). The maize ORFs that are most closely related to the S1-DNA polymerase (orf917, orf911, and orf734) show similarities (∼40% identical and 60% similar) to several ORFs in the main mitochondrial genomes of Beta vulgaris, Daucus carota, and Secale cereale. Most of the non-maize ORFs contain 100- to 500-codon regions with similarities to the S1-DNA polymerase. The maize ORFs that are most closely related at the protein level to the RNA polymerase of the S2 plasmid (orf1159, orf653, orf487, and orf417) also show similarity (∼30% identity and 50% similarity) to several ORFs in the main mitochondrial genomes of B. vulgaris and D. carota. Again, most of the ORFs contain 100- to 500-codon regions with similarities to the plasmid ORFs.
All five maize mitochondrial genomes also contain other highly divergent (<80% sequence identity) copies of R/S plasmid fragments that appear to have become integrated at multiple times prior to the divergence of the genomes. None of the genomes contains integrated portions that are >34 bp (>80% sequence identity) from the short mitochondrial plasmids (2312-bp linear plasmid in NB, CMS-C, and CMS-S; 2176-bp linear plasmid in NA and CMS-T; 1913-bp circular plasmid in all except CMS-S; and 1445-bp circular plasmid present sporadically) (Ludwig et al. 1985; Bedinger et al. 1986; Smith and Pring 1987).
Genome conservation:
Considering that all the maize mitochondrial genomes have similar genome complexity, a comparative analysis was conducted to determine the proportion of the sequences that each has in common with the others (Table 6). One of the most surprising outcomes of our analyses is how rapidly sequence segments are gained and lost. Even comparing the two most similar genomes, NA is missing 1.97% of the sequences present in NB.
TABLE 6.
Percentage of maize mitochondrial genome complexity that is absent in other genomes
| Reference genome
|
|||||
|---|---|---|---|---|---|
| vs. | NB | NA | CMS-C | CMS-S | CMS-T |
| NB | — | 5.11 | 2.66 | 5.09 | 7.42 |
| NA | 1.97 | — | 0.80 | 0.48 | 3.11 |
| CMS-C | 4.97 | 6.26 | — | 5.24 | 6.73 |
| CMS-S | 6.05 | 4.61 | 3.92 | — | 6.60 |
| CMS-T | 7.07 | 5.96 | 4.12 | 5.31 | — |
Small deletions (<200 bp) are not considered to be missing segments. Percentages for the NA genome, which shares the most complexity with each of the other genomes, are in italics.
Each of the three cytoplasmic-male-sterile genomes is missing substantial amounts of sequence that are present in the male-fertile genomes. Over 7% of the NB genome is not present in the CMS-T genome (Table 6), and CMS genome pairs are missing from 3.92 to 6.73% of each other. The NA genome might be considered the most complete, in that it contains the greatest proportion of each of the other genomes, including 99.20% of the sequences present in CMS-C and 99.52% of those in CMS-S.
Reciprocity is generally not seen in any comparison, even between the two male-fertile genomes; the NA genome is missing 1.97% of the NB genome, but the NB genome is missing 5.11% of the NA genome (Table 6). The difference is more striking when comparing NA with CMS-C: NA is missing only 0.80% of the CMS-C genome, but CMS-C does not possess 6.26% of the NA genome. Reciprocal differences were also seen in which sequences are present in male-fertile vs. CMS sugar beet mitochondrial genomes (Satoh et al. 2004).
Especially in the comparisons between the fertile and CMS maize mitochondrial genomes, a large proportion of the missing sequences are exogenous, originating either from the plastid genome or from mitochondrial plasmids. In fact, between the two most closely related cytotypes, NB and NA, all but 313 bp of the difference is the result of the presumed loss in the NA genome of a 9.4-kb segment of the 12.6-kb NB plastid insert. Even for the 5% of the NB genome that is not present in CMS-C, missing plastid and plasmid inserts together account for 97% of the difference. Thus, acquired exogenous sequences can play a major role in size and compositional evolution of plant mitochondrial genome complexity on a subspecific timescale. In fact, the genome with the least integrated plastid and plasmid DNA, CMS-C, is also the genome with the smallest complexity.
“Native” mitochondrial sequences can also be present in only some genomes. For example, 75% of the sequence that is present in NA but not present in NB cannot be attributed to a difference in plastid or plasmid DNA. Among the five mitochondrial genomes, all of the missing “native” DNA segments are <10 kb, and most are <1 kb. Some of these missing mitochondrial sequences may have been lost by simple stochastic processes following looping out or double recombination events (see Fauron et al. 1995b).
Some of the sequences are found only in one of the genomes and may be newly acquired. The unique sequences include a 313-bp segment in NB, 426- and 131-bp segments in NA, and 288- and 442-bp segments in CMS-C. CMS-T has 10 unique segments ranging from 359 to 3353 bp in length, comprising a total of 12,576 bp. (Indels of <50 bp were not included.) None of these unique sequences has matches in the public databases. In addition, 9249 bp of the 12.6-kb plastid insertion in NB mtDNA is not present in any of the other four genomes.
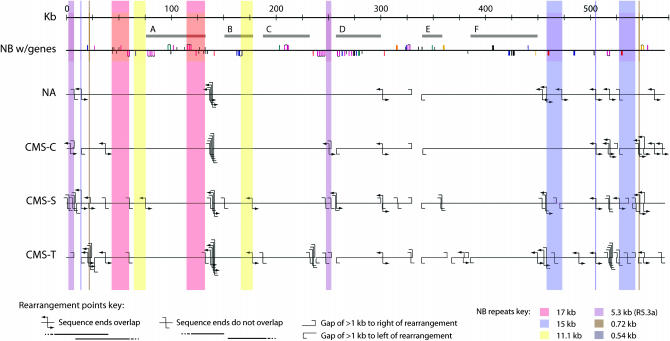
Genome rearrangement:
Despite the phylogenetic closeness of the five maize mitochondrial genomes (Doebley 1990), regions of synteny are generally short, due to the abundant rearrangements among them. There are six regions (not including repeats) of at least 15 kb that are syntenic in all five genomes, the largest of which is 66 kb (“F” in Figure 2). When the genomes are compared pairwise, the largest contiguous region is 161 kb, found in NB and NA (NB, 138,491–299,194; NA, 479,468–640,246). In the 10 pairwise genome comparisons, there are an additional seven common segments of >100 kb (139-kb NAxC, 123-kb NBxNA, 114-kb NBxNA, 110-kb NAxC, 109-kb NAxS, 109-kb NBxC, and 100-kb NBxC). An additional 17 segments of >50 kb are present. The smallest syntenic segment observed (above our arbitrary 25-bp lower limit) is 27 bp when NB is compared to CMS-T. The next smallest is a 32-bp segment in the four non-NB cytotypes relative to NB.
Figure 2.—
Multiple rearrangements lead to small syntenic regions. The NA, C, S, and T mitochondrial genomes are arranged relative to the linearized NB map. NB gene positions are indicated by vertical lines colored as in Figure 1. Horizontal gray bars (A–F) indicate regions of synteny (15–66 kb) among all five maize mitochondrial genomes. Colored boxes indicate regions that are present in NB as repeats of >500 bp, using the color scheme of Figure 1. Some very closely spaced rearrangements may be even closer than indicated due to line separation requirements for visibility.
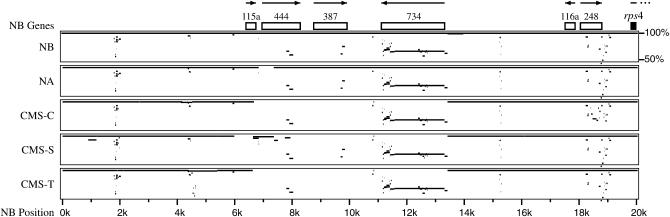
The numerous rearrangement points, repeats, and insertions and deletions among the five maize mitochondrial genomes are not evenly distributed across the genomes. For instance, a large number of rearrangements occurred near NB coordinate 140 kb, but almost none occurred in the 80-kb interval preceding it (Figure 2). No clear explanation for this hot spot has been discerned. Another region involving both rearrangement and deletion occurs in the first 20 kb of the NB genome and is shown in more detail in Figure 3 (see also Clifton et al. 2004). These differences are almost exclusively in the R1-plasmid-homlogous region (coordinates 6638–13,429). A comparison to the entire NB genome is shown in supplemental Figure 4 at http://www.genetics.org/supplemental/.
Figure 3.—
Conservation of maize mitochondrial genomic sequences. Multipip representation of the first 20 kb of the NB map. The region is typical in that sequences are either present and highly conserved or absent. This region is unusual in that large portions of the NB genome are not present in three of the genomes. Multipip (Schwartz et al. 2000, 2003) aligns the sequences of each taxon to the reference (NB), irrespective of the locations of those sequences in the other taxa, and plots them according to the percentage of identity to the reference genome (vertical scale). The solid block indicates a gene, open blocks indicate ORFs, and arrows indicate the direction of transcription.
Figure 1 clearly illustrates the differences in the order of genes that result from rearrangement. Unlike plastid genomes, which always include ribosomal RNA genes on their large inverted repeats, the duplications in maize mtDNA occur throughout the genome, so that different members from each of the functional groups of protein-coding genes may be duplicated. There does not appear to be a bias for which genes are present on duplications in the maize mitochondrial genomes.
Relatively gene-rich and gene-poor regions can each show evidence of numerous rearrangements. Except for the leader sequence of atp6 in CMS-C, the intercytotype rearrangements do not delete parts of genes. Rearrangements that greatly affect gene function, such as those of the NCS mutants (Newton et al. 2004), would be expected to be strongly selected against. Two rearrangements occur within introns. In four of the mitochondrial genomes, one end of the 17-kb repeat lies within the 1905-bp intron 3 of nad2. In all the genomes, one end of the 11.1-kb repeat (12.8 kb in CMS-T) lies within the 1842-bp intron of rps3. In each case, the normal gene is preserved, but supernumerary exons are created on the duplicated segments.
Examination of the region within 1500 bp of the start or end of a coding region revealed six upstream and six downstream rearrangement sites (supplemental Table 6 at http://www.genetics.org/supplemental/). Ten of these 12 rearrangements occur near the one cob, three cox, and five atp genes. In the maize mitochondrial genomes, there are 51 genes in 59 transcriptional units (because of trans-splicing), and thus five-sixths of the rearrangements near genes occur adjacent to fewer than one-sixth of the transcriptional units. We have not determined why the regions surrounding these genes are more recombinationally active.
Relatively fewer rearrangements occur near genes than in the genomes overall. Within 1.5 kb upstream and downstream of coding regions (a total of ∼725 kb in all five genomes combined), the frequency of rearrangements (1/60 kb) is about one-fifth of that at which they occur in the genomes in total (198 rearrangements in 2586 kb = 1/13 kb). However, the frequency of rearrangements near chimeric ORFs (8 rearrangements in 108 kb = 1/13 kb) is no different from the overall rearrangement frequency. This suggests that rearrangements near genes are selected against or that regions adjacent to most of the genes are not recombinationally competent. In contrast, recombination in regions adjacent to chimeric ORFs appears not to be selected against. This is a further argument that unidentified chimeric ORFs are not functional.
Approximately half of the rearrangement sites have fragment ends that overlap (double arrows in Figure 2); i.e., the rearrangement sites are repeated sequences, with sizes that range from a single nucleotide to >4500 bp. The overlaps containing very small numbers of nucleotides were detected by aligning adjacent homologous segments onto the NB genome. These overlaps are distinct from SDRs and SSRs. Among the 35 rearrangement sites that have overlapping fragments, only 4 include SDRs and none have SSRs, although three SDRs and one SSR are immediately adjacent to the breakpoint, and one of each type is within 20 bp of the breakpoint.
Some of the large duplications in one maize mitochondrial genome are composed of sequences that are adjacent to deletions in another genome. For example, sequences adjacent to one end of the recombinationally active R5.3a repeat in NB and NA are absent in CMS-C, CMS-S, and CMS-T (Figure 2). Rearrangements involving small repeats can lead to the loss of genomic sequences (Small et al. 1989; Hanson and Folkerts 1992), sometimes creating duplicate-deficient genomes, which we did not observe. We were unable to definitively explain the repeat-based reorganizations in terms of the proposed models. The rearrangements that do not appear to involve identified repeats may be the result of chromosome breakage and rejoining.
Repeated sequences:
The largest repeat within the five maize mitochondrial genomes is 120 kb in NA (Table 7, Figure 1); its two copies account for 34.1% of the genome. This and the three other very large repeats in CMS-C (105, 50, and 45 kb) are present in only a single cytotype. Repeats <11 kb are present only sporadically. In contrast, the 11.1-kb repeat (12.8 kb in CMS-T) is present in all cytotypes, and the 17-kb repeat (R17) is present in all but CMS-T (Table 7). Four of the smaller repeats are present in three copies (R4.6 in NA, R5.7 in CMS-C, R1.1 in CMS-S, and R2.6 in CMS-T; Table 7), with two of the three copies located within a larger repeat (Figure 1). Each triplication occurs in only one genome.
TABLE 7.
Large repeats (>05 kb) in maize mitochondrial genomes
| Genomes
|
|||||
|---|---|---|---|---|---|
| Repeats | NB | NA | CMS-C | CMS-S | CMS-T |
| R120 | − | +, R4.6 | −, R105, R50 | − | − |
| R105 | − | −, R15, R4.6, R0.6 | +, R15 | −, R4.2, R0.74 | − |
| R50 | −, R5.3a | −, R5.3a | +, R5.7 | −, R5.0, R1.1 | − |
| R45 | −, R0.7 | −, R0.7 | +, R0.7 | −, R5.0, R0.7 | −, R5.2, R0.7 |
| R17 | + | + | + | + | − |
| R15 | + | −, R0.6 | − | −, R4.2, R0.74 | − |
| R12.8 | −, R11 | −, R11 | −, R11 | −, R11 | +, R2.6 |
| R11.1 | + | + | + | + | −, R2.6 |
| R5.7 | − | −, R5.3b | +, 3× | −, R5.3b | −, R5.3b |
| R5.3a | + | + | − | −, R5.0, R1.1 | − |
| R5.3b | − | + | − | + | + |
| R5.2 | − | − | − | − | + |
| R5.0 | − | − | − | +, R1.1 | − |
| R4.6 | − | +, 3× | − | − | − |
| R4.2 | − | − | − | + | − |
| R2.6 | − | − | − | − | +, 3× |
| R1.1 | − | − | − | +, 3× | − |
| R0.74 | − | −, R0.60 | − | + | − |
| R0.72 | + | + | + | + | − |
| R0.69 | − | − | − | + | − |
| R0.60 | − | + | − | − | − |
| R0.54 | + | − | − | − | − |
Repeats (R) are indicated by numbers indicating lengths (in kilobases) as in Figure 1. +, complete repeats present; −, complete repeats not present. Where smaller repeats are entirely contained within a larger repeat, they are specified; italics indicate that only parts of the smaller repeats are present within the larger repeat. “3×” indicates that sequences are present three times.
Most of the duplicated regions are related to at least one other repeat. The terminal 86 kb of the 105-kb repeat (R105) in CMS-C is also the terminal 86 kb of the 120-kb repeat (R120) in NA (supplemental Figure 5 at http://www.genetics.org/supplemental/). The other terminus of R105 is composed of what is the 15-kb repeat (R15) in the NB genome. Only 4 kb of R105 is not part of a repeat in another maize mitochondrial genome. This sharing of repeated sequences is a common theme among the genomes: (1) 258 kb (45%) of the 564 kb of total genome complexity present in the five genomes is part of a repeat in at least one genome; (2) of that 258 kb, 141 kb (55%) is present in >1 repeat; (3) only 2 repeats (R17 and R0.54) do not share sequences with another of the 22 repeats; (4) 11 complete repeats constitute parts of larger repeats; and (5) 7 repeats share a terminus with one or more larger repeats.
Parsimony leads us to conclude that smaller repeats were derived from larger repeats. In this scenario, a large repeat was formed and then either (1) a segment of one of the two copies was lost, shortening or dividing it, or (2) a rearrangement in one of the copies cleaved it, creating two separate repeats, one pair of which retained the back-to-back structure. A good example of the latter is found in CMS-C, where one copy of each of R50 and R45 are contiguous, but the second copies are separated by hundreds of kilobases. The less parsimonious scenario is that a region of the genome lying immediately adjacent to an existing repeat also became repeated, lengthening the existing repeat. In either case, the larger a repeat, the more likely it is that its sequence will include a region that is duplicated by a separate event in another genome, or even the same genome, in which case a triplication will be produced. Ends of repeats tend to be shared, suggesting that repeat termini are more prone to duplication than other sequences (supplemental Figure 5 at http://www.genetics.org/supplemental/).
The ends of large repeats (>0.5 kb) are also more commonly associated with SDRs and SSRs. There are only 37 unique repeat ends among the 22 repeats because of common ends (supplemental Figure 5). Nine of the 37 unique ends contain members of SDR families; 6 of the SDRs are present many times in each of the genomes. Four repeat ends contain SSRs.
The two copies of each of the largest repeats do not appear to be randomly distributed. For each of the nine repeats of ≥15 kb, the two copies lie within <10% of genome complexity of each other (i.e., within ∼50 kb of each other, not including other intervening repeats; Figure 1). For example, in the circularized CMS-C genome, the R50 repeats are 8563 bp apart (1.14% of genome size, 1.67% of genome complexity). The lone exception is the 17-kb repeat (R17) in CMS-S.
Sequence evolution:
Despite their differences in organization and DNA content, the five maize genomes are very closely related at the nucleotide level. The most similar genomes are NB and NA. There are only 92 nucleotide substitutions in the 559,015 nt in common in the alignment of NB with NA, which corresponds to 1.65 substitutions/10,000 bp (Table 8). For CMS-C, the value is 2.31 substitutions/10,000 bp, for CMS-T it is 4.32, and for CMS-S it is 5.36. The least similar genomes are CMS-S and CMS-T, which have 7.04 nucleotide differences/10,000 bp of their common sequence (supplemental Table 7 at http://www.genetics.org/supplemental/). The relative levels of indels are roughly the same as they are for substitutions, except that, whereas CMS-S has the most substitutions relative to NB, NA, and CMS-C, CMS-T has the most indels relative to those three genomes.
TABLE 8.
Nucleotide differences relative to the NB mitochondrial genome
| Total genome
|
Genes
|
|||||
|---|---|---|---|---|---|---|
| Genome | Lengtha | Nucleotide substitutions | Nucleotide substitutions/10 kb | Lengthb | No. of nucleotide substitutions | Nucleotide substitutions/10 kb |
| NA | 559,015 | 92 | 1.65 | 32,670 | 1 | 0.31 |
| CMS-C | 540,991 | 125 | 2.31 | 32,246 | 4 | 1.24 |
| CMS-S | 537,139 | 288 | 5.36 | 32,670 | 9 | 2.75 |
| CMS-T | 521,063 | 225 | 4.32 | 32,670 | 19 (9)c | 5.82 (2.75)c |
| Rice | 153,419 | 2842 | 185 | 29,501 | 401 | 136 |
Nucleotides in common with NB for each of the genomes; genes are included, indels are excluded.
Number of common nucleotides in single copies of all of the protein-coding genes.
Numbers in parentheses exclude atp4 (see text).
The nucleotide substitution frequency between the mitochondrial genomes of the rice indica and japonica subspecies is 1.93/10,000 bp (Tian et al. 2006), which is similar to that among the three most similar maize mitochondrial genomes. On the other hand, the male-fertile and cytoplasmic-male-sterile sugar beet mitochondrial genomes differed by 9.06/10,000 bp (Satoh et al. 2004), which is more than the maximum for any maize pair (7.04 in CMS-S vs. CMS-T). Putting these divergence levels into context will require the establishment of reliable estimates of divergence time.
Within the maize mitochondrial genomes, sequences are highly conserved, and coding regions of known genes are approximately twice as conserved as noncoding regions. In contrast, two types of sequences are more variable: integrated plasmid sequences and short dispersed repeats. The level of divergence among maize genomes for ORFs showing similarity to S/R-plasmid ORFs is greater than that for any functional gene comparison within higher plants. In fact, NB orf1159 alone contains over one-third of all of the nucleotide substitutions in the 559,015 nucleotides in common between the NB and NA genomes.
Small repeats (<30 bp) also appear to be rapidly diverging. In Multipip representations, many areas containing “vertical scatter” occur (e.g., at 2 kb in Figure 3; see also supplemental Figure 4 at http://www.genetics.org/supplemental/). Each cluster represents small, dispersed repeats that have varying levels of sequence identity. Their presumed degradation may be a step toward the loss of the repeats since, despite these clusters, repeats <30 bp are less common in the NB maize mitochondrial genome (the reference genome in Figure 3 and supplemental Figure 4) than would be expected by chance (Clifton et al. 2004).
Elimination of small repeats reduces the potential for recombination. Small repeats appear to have been involved in several of the events that led to major rearrangement of the maize mitochondrial genomes. The participation of small repeats in recombination and genome rearrangement was also found in the wheat mitochondrial genome (Ogihara et al. 2005). Small repeats are known to be involved in the generation of deleterious mutations, such as the NCS mutants in maize. For example, NCS2 is the result of a rare recombination event between two imperfect 16-bp sequences within introns of nad4 and nad7 that led to the creation of a nonfunctional, chimeric nad4/nad7 gene (Marienfeld and Newton 1994).
Among the maize mitochondrial genomes, there is a twofold range in the prevalence of indels (≤200 bp) relative to substitutions. The CMS-C and CMS-T genomes have the highest ratio of indels to substitutions: each has 1.45 indels/substitution relative to NB (supplemental Table 7 at http://www.genetics.org/supplemental/). The smallest ratio is 0.73 indel/substitution for NB vs. CMS-S. The ratio of insertions to deletions also shows roughly twofold variation (from 2.42 insertions:1 deletion in NA compared with NB to 1.03 insertions:1 deletion in NA relative to CMS-C). Since an insertion in a comparison in one direction is a deletion in the comparison in the opposite direction, we used the following hierarchy in computing the 10 possible comparisons: NB was the first genome used as reference (with respect to the four other genomes); NA was then used as the reference for the remaining three genomes (i.e., not NB); then CMS-C was used for the remaining two, and CMS-S was used for CMS-T.
In general, the frequency of indels in plant mitochondrial genomes is thought to be lower than the frequency of substitutions (e.g., Freudenstein and Chase 2001). For example, the ratio of indels to substitutions is 0.26 for the mitochondrial genomes of the two male-fertile rice subspecies indica and japonica (Tian et al. 2006), the closest relatives for which whole-genome data are available. The ratio for the two fertile maize mitochondrial genomes (NB vs. NA) is three times the rice ratio, and the ratios among the male-sterile maize genomes are five times the rice ratio (supplemental Table 7 at http://www.genetics.org/supplemental/). However, in comparisons of the NB, CMS-C, and CMS-T genomes, indels are in fact more common than substitutions. The cause of this difference may be that maize substitution frequencies are unusually low for grasses or that their indel frequencies are particularly high.
In all five of the genomes, approximately half of the indels have a length of five nucleotides (supplemental Figure 6 at http://www.genetics.org/supplemental/); relative to NB, 41–57% of insertions and 29–44% of deletions (i.e., 41–52% of indels) are 5 bp long. There is no discernible sequence motif involved. The next-largest classes are 4- and 6-bp indels, each of which accounts for ∼10% of the indels (5% of insertions and 5% of deletions). Indels of 2 and 3 nucleotides are very rare, in aggregate accounting for only 1.4% of all indels, as are indels of >12 nucleotides. Relative to NB, the NA genome does not have any indels of >12 bp, whereas the other three genomes each have indels of at least 35 bp. The largest in all comparisons is 36 bp in CMS-S relative to NB.
Length variation in small tandem repeats, including SSRs, is a subset of the indels. In the genome complexities, between 273 (NB) and 292 (NA) SSRs were found, >97% of which have repeat units of ≤20 bp, and >93% are of ≤10 bp (not shown). The length distribution of the repeat units is similar to the indel distributions, especially in NA and CMS-C, although 5-bp repeat units account for only 17–24% of the SSR indel total, and 6-bp repeats make up 14–19%.
The most similar genomes in terms of the number of substitutions, indels, or common nucleotides are NB and NA (the two male-fertile cytotypes). The most divergent cytotypes are CMS-S and CMS-T. Thus, those two male-sterile cytotypes are most different not from the male-fertile cytotypes, but rather from each other. They are also quite divergent from the fertile types. This is consistent with the separate origins of the male-sterile cytotypes from closely related fertile ancestors, followed by the more recent divergence of the two fertile cytotypes.
The early divergence of CMS-S is supported by the observation that a mitochondrial genome identical to the maize CMS-S mitochondrial genome at the restriction-length-polymorphism level has been observed in populations of the teosinte Z. mays ssp. mexicana, from which it may have introgressed into local maize populations (Weissinger et al. 1983; Doebley and Sisco 1989). Interestingly, although the Latin American maize is male sterile, the teosinte itself is not male sterile. Furthermore, introgressing the teosinte cytoplasm into maize does not make the maize male sterile (Allen 2005). The level of sequence divergence and the number of indels in the CMS-T mitochondrial genome suggest that it diverged as long ago as did CMS-S. However, although the CMS-T cytotype and T-type cytoplasmic male sterility have been observed in Latin American maize land races (Weissinger et al. 1983), the CMS-T cytoplasm has not yet been reported in any teosinte accession.
Conclusions:
Our analyses indicate that NB and NA are the most closely related sequenced mitochondrial genomes. Relative to NB and NA, the most closely related genome is CMS-C, followed by CMS-S and CMS-T. All of the genomes have the same set of 51 genes in their main genomes, plus one carried on a linear plasmid. Gene sequences are approximately twice as conserved as nongenic sequences.
Comparative genomics was helpful at narrowing the search for CMS-associated chimeric ORFs in two of the three maize CMS genomes. Therefore, whole-genome sequence comparisons among multiple cytotypes within a species may be the preferable first step in looking for CMS-associated genes in other systems, especially as sequencing costs continue to decline.
Rearrangements are very common in the maize mitochondrial genomes; the largest syntenic region among the five genomes is only 66 kb long. Rearrangements occur less frequently near genes (transcriptional units) than in the genomes in general. In contrast, rearrangements occur at the same frequency near unidentified ORFs as they do in the genomes overall. None of the unidentified ORFs is likely to be functional based on this fact and their lack of conservation among maize and other grass genomes.
Substantial size differences exist among the five maize mitochondrial genomes, with the largest (CMS-C) being 38% larger than the smallest (CMS-T). Large repeats (>0.5 kb) are the main factor in the difference in overall genome size; the largest genome complexity (NA) is only 6% larger than the smallest (CMS-C). However, large repeats do not account for size differences between the mitochondrial genomes of maize and those of other plants.
Our comparative analyses of the maize mitochondrial genomes shows that their DNA sequences are generally either highly conserved or absent. As much as 7% of the genome complexity of one maize mitochondrial genome is missing in another maize mitochondrial genome. DNA of exogenous origin is a major source of the differences in the genome complexities. Despite the fact that ∼80% of the genome complexity has no known function, the majority of the sequences missing in several of the comparisons derive from integrated plastid or plasmid sequences, each of which constitutes <5% of the genome. It is not clear if the plastid or plasmid DNA is passively lost or if it is actively eliminated from the main mitochondrial genomes. Nevertheless, one of the most surprising outcomes of our comparative analyses is how rapidly mitochondrial sequence segments are gained and lost within a single subspecies.
Acknowledgments
We thank Suman Kanuganti, Claude dePamphilis, and John Spieth for helpful suggestions on data analyses and Jeff Palmer and a reviewer for helpful comments on the manuscript. This work was funded by the National Science Foundation Plant Genome Research Program (DBI-0110168).
References
- Allen, J. O., 2005. Effect of teosinte cytoplasmic genomes on maize phenotype. Genetics 169: 863–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers and D. J. Lipman, 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller et al., 2002. The Pfam protein families database. Nucleic Acids Res. 30: 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett, J. B., 1971. Classification of male-sterile cytoplasms in maize (Zea mays L.). Crop Sci. 11: 724–727. [Google Scholar]
- Bedinger, P., E. L. de Hostos, P. Leon and V. Walbot, 1986. Cloning and characterization of a linear 2.3 kb mitochondrial plasmid of maize. Mol. Gen. Genet. 205: 206–212. [DOI] [PubMed] [Google Scholar]
- Bendich, A. J., 1996. Structural analysis of mitochondrial DNA molecules from fungi and plants using moving pictures and pulsed-field gel electrophoresis. J. Mol. Biol. 255: 564–588. [DOI] [PubMed] [Google Scholar]
- Benson, G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland, M. M., C. S. I. Levings and D. F. Matzinger, 1987. The ATPase subunit 6 gene of tobacco mitochondria contains an unusual sequence. Curr. Genet. 12: 475–481. [DOI] [PubMed] [Google Scholar]
- Bogsch, E. G., F. Sargent, N. R. Stanley, B. C. Berks, C. Robinson et al., 1998. An essential component of a novel bacterial protein export system with homologues in plastids and mitochondria. J. Biol. Chem. 273: 18003–18006. [DOI] [PubMed] [Google Scholar]
- Bonnard, G., and J. M. Grienenberger, 1995. A gene proposed to encode a transmembrane domain of an ABC transporter is expressed in wheat mitochondria. Mol. Gen. Genet. 246: 91–99. [DOI] [PubMed] [Google Scholar]
- Chang, C. C., J. Sheen, M. Bligny, Y. Niwa, S. Lerbs-Mache et al., 1999. Functional analysis of two maize cDNAs encoding T7-like RNA polymerases. Plant Cell 11: 911–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapdelaine, Y., and L. Bonen, 1991. The wheat mitochondrial gene for subunit I of the NADH dehydrogenase complex: a trans-splicing model for this gene-in-pieces. Cell 65: 465–472. [DOI] [PubMed] [Google Scholar]
- Clifton, S. W., P. Minx, C. M. Fauron, M. Gibson, J. O. Allen et al., 2004. Sequence and comparative analysis of the maize NB mitochondrial genome. Plant Physiol. 136: 3486–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey, R. E., C. S. Levings, III and D. H. Timothy, 1986. Novel recombinations in the maize mitochondrial genome produce a unique transcriptional unit in the Texas male-sterile cytoplasm. Cell 44: 439–449. [DOI] [PubMed] [Google Scholar]
- Dewey, R. E., D. H. Timothy and C. S. Levings, 1987. A mitochondrial protein associated with cytoplasmic male sterility in the T cytoplasm of maize. Proc. Natl. Acad. Sci. USA 84: 5374–5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey, R. E., D. H. Timothy and C. Levings, III, 1991. Chimeric mitochondrial genes expressed in the C male-sterile cytoplasm of maize. Curr. Genet. 20: 475–482. [DOI] [PubMed] [Google Scholar]
- Doebley, J., 1990. Molecular systematics of Zea (Gramineae). Maydica 35: 143–150. [Google Scholar]
- Doebley, J., and P. Sisco, 1989. On the origin of the maize male sterile cytoplasms: it's completely unimportant, that's why it's so interesting. Maize Genet. Coop. News. Lett. 63: 108–109. [Google Scholar]
- Ewing, B., and P. Green, 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8: 186–194. [PubMed] [Google Scholar]
- Ewing, B., L. Hillier, M. C. Wendl and P. Green, 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8: 175–185. [DOI] [PubMed] [Google Scholar]
- Fauron, C. M., and M. Casper, 1994. A second type of normal maize mitochondrial genome: an evolutionary link. Genetics 137: 875–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauron, C., and M. Havlik, 1989. The maize mitochondrial genome of the normal type and the male sterile type T have very different organization. Curr. Genet. 15: 149–154. [Google Scholar]
- Fauron, C., M. Casper, Y. Gao and B. Moore, 1995. a The maize mitochondrial genome: dynamic, yet functional. Trends Genet. 11: 228–235. [DOI] [PubMed] [Google Scholar]
- Fauron, C., B. Moore and M. Casper, 1995. b Maize as a model of higher plant plasticity. Plant Sci. 112: 11–32. [Google Scholar]
- Fauron, C., J. O. Allen, S. Clifton and K. J. Newton, 2004. Plant mitochondrial genomes, pp. 155–171 in Molecular Biology and Biotechnology of Plant Organelles, edited by H. Daniell and C. Chase. Kluwer Academic, Dordrecht, The Netherlands.
- Freudenstein, J. V., and M. W. Chase, 2001. Analysis of mitochondrial nad1b-c intron sequences in Orchidaceae: utility and coding of length-change characters. Syst. Bot. 26: 643–657. [Google Scholar]
- Giege, P., Z. Konthur, G. Walter and A. Brennicke, 1998. An ordered Arabidopsis thaliana mitochondrial cDNA library on high-density filters allows rapid systematic analysis of plant gene expression: a pilot study. Plant J. 15: 721–726. [DOI] [PubMed] [Google Scholar]
- Giege, P., M. Hoffmann, S. Binder and A. Brennicke, 2000. RNA degradation buffers asymmetries of transcription in Arabidopsis mitochondria. EMBO Rep. 1: 164–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., C. Abajian and P. Green, 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8: 195–202. [DOI] [PubMed] [Google Scholar]
- Gromiha, M. M., S. Ahmad and M. Suwa, 2004. Neural network-based prediction of transmembrane beta strand segments in outer membrane proteins. J. Comput. Chem. 25: 762–767. [DOI] [PubMed] [Google Scholar]
- Handa, H., 2003. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 31: 5907–5916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa, H., G. Bonnard and J. M. Grienenberger, 1996. The rapeseed mitochondrial gene encoding a homologue of the bacterial protein Ccl1 is divided into two independently transcribed reading frames. Mol. Gen. Genet. 252: 292–302. [DOI] [PubMed] [Google Scholar]
- Handa, H., K. Itani and H. Sato, 2002. Structural features and expression analysis of a linear mitochondrial plasmid in rapeseed (Brassica napus L.). Mol. Genet. Genomics 267: 797–805. [DOI] [PubMed] [Google Scholar]
- Hanson, M. R., and S. Bentolila, 2004. Interactions of mitochondrial and nuclear genes that affect male gametophyte development. Plant Cell 16(Suppl.): S154–S169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson, M. R., and O. Folkerts, 1992. Structure and function of the higher plant mitochondrial genome. Int. Rev. Cytol. 141: 129–172.
- Heazlewood, J. L., J. Whelan and A. H. Millar, 2003. The products of the mitochondrial orf25 and orfB genes are FO components in the plant F1FO ATP synthase. FEBS Lett. 540: 201–205. [DOI] [PubMed] [Google Scholar]
- Hermanns, J., and H. D. Osiewacz, 1992. The linear mitochondrial plasmid pAL2–1 of a long-lived Podospora anserina mutant is an invertron encoding a DNA and RNA polymerase. Curr. Genet. 22: 491–500. [DOI] [PubMed] [Google Scholar]
- Hirokawa, T., S. Boon-Chieng and S. Mitaku, 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–379. [DOI] [PubMed] [Google Scholar]
- Houchins, J. P., H. Ginsburg, M. Rohrbaugh, R. M. Dale, C. L. Schardl et al., 1986. DNA sequence analysis of a 5.27-kb direct repeat occurring adjacent to the regions of S-episome homology in maize mitochondria. EMBO J. 5: 2781–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, M. A., S. R. Payne and A. J. Bendich, 1996. Moving pictures and pulsed-field gel electrophoresis show only linear mitochondrial DNA molecules from yeasts with linear-mapping and circular-mapping mitochondrial genomes. Curr. Genet. 30: 3–11. [DOI] [PubMed] [Google Scholar]
- Kim, D. H., and B. D. Kim, 2006. The organization of mitochondrial atp6 gene region in male-fertile and CMS lines of pepper (Capsicum annuum L.). Curr. Genet. 49: 59–67. [DOI] [PubMed] [Google Scholar]
- Kubo, T., S. Nishizawa, A. Sugawara, N. Itchoda, A. Estiati et al., 2000. The complete nucleotide sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.) reveals a novel gene for tRNA(Cys)(GCA). Nucleic Acids Res. 28: 2571–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, R., and C. S. Levings, III, 1993. RNA editing of a chimeric maize mitochondrial gene transcript is sequence specific. Curr. Genet. 23: 154–159. [DOI] [PubMed] [Google Scholar]
- Kumar, R., L. Marechal-Drouard, K. Akama and I. Small, 1996. Striking differences in mitochondrial tRNA import between different plant species. Mol. Gen. Genet. 252: 404–411. [DOI] [PubMed] [Google Scholar]
- Kuzmin, E. V., and I. V. Levchenko, 1987. S1 plasmid from cms-S-maize mitochondria encodes a viral type DNA-polymerase. Nucleic Acids Res. 15: 6758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin, E. V., I. V. Levchenko and G. N. Zaitseva, 1988. S2 plasmid from cms-S-maize mitochondria potentially encodes a specific RNA polymerase. Nucleic Acids Res. 16: 4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughnan, J., and S. Gabay-Laughnan, 1983. Cytoplasmic male sterility in maize. Annu. Rev. Genet. 17: 27–48. [DOI] [PubMed] [Google Scholar]
- Leaver, C. J., P. G. Isaac, I. D. Small, J. Bailey-Serres, A. D. Liddell et al., 1988. Mitochondrial genome diversity and cytoplasmic male sterility in higher plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 319: 165–176. [Google Scholar]
- Levings, C. S., III, and D. R. Pring, 1976. Restriction endonuclease analysis of mitochondrial DNA from normal and Texas cytoplasmic male-sterile maize. Science 193: 158–160. [DOI] [PubMed] [Google Scholar]
- Levings, C. S., III, and R. R. Sederoff, 1983. Nucleotide sequence of the S-2 mitochondrial DNA from the S cytoplasm of maize. Proc. Natl. Acad. Sci. USA 80: 4055–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, A. A., C. P. Andre and V. Walbot, 1991. Analysis of a 120-kilobase mitochondrial chromosome in maize. Genetics 128: 417–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loessl, A., N. Adler, R. Horn, U. Frei and G. Wenzel, 1987. Chondriome-type characterization of potato: mt alpha, beta, gamma, delta, epsilon and novel plastid-mitochondrial configurations in somatic hybrids. Theor. Appl. Genet. 99: 1–10. [Google Scholar]
- Lonsdale, D. M., T. P. Hodge and C. M. Fauron, 1984. The physical map and organisation of the mitochondrial genome from the fertile cytoplasm of maize. Nucleic Acids Res. 12: 9249–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe, T. M., and S. R. Eddy, 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, B., and M. R. Hanson, 1999. A single homogenous form of ATP6 protein accumulates in petunia mitochondria despite the presence of differentially edited atp6 transcripts. Plant Cell 6: 1955–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, S. R., R. F. Pohlman, J. Vieira, A. G. Smith and J. Messing, 1985. The nucleotide sequence of a mitochondrial replicon form maize. Gene 38: 131–138. [DOI] [PubMed] [Google Scholar]
- Lupold, D. S., A. G. Caoile and D. B. Stern, 1999. The maize mitochondrial cox2 gene has five promoters in two genomic regions, including a complex promoter consisting of seven overlapping units. J. Biol. Chem. 274: 3897–3903. [DOI] [PubMed] [Google Scholar]
- Makaroff, C. A., I. J. Apel and J. D. Palmer, 1989. The atp6 coding region has been disrupted and a novel reading frame generated in the mitochondrial genome of cytoplasmic male-sterile radish. J. Biol. Chem. 264: 11706–11713. [PubMed] [Google Scholar]
- Marienfeld, J. R., and K. J. Newton, 1994. The maize NCS2 abnormal growth mutant has a chimeric nad4-nad7 gene and is associated with reduced complex I function. Genetics 138: 855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marienfeld, J., M. Unseld, P. Brandt and A. Brennicke, 1996. Genomic recombination of the mitochondrial atp6 gene in Arabidopsis thaliana at the protein processing site creates two different presequences. DNA Res. 3: 287–290. [DOI] [PubMed] [Google Scholar]
- Meyer, L. J., 2004. ORF analysis and tissue-specific differential gene expression in maize mitochondria. M. S. Thesis, University of Missouri, Columbia, MO.
- Mulligan, R. M., P. Leon and V. Walbot, 1991. Transcriptional and posttranscriptional regulation of maize mitochondrial gene expression. Mol. Cell. Biol. 11: 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, K. J., 1994. Procedures for isolating mitochondria and mitochondrial DNA and RNA, pp. 549–556 in The Maize Handbook, edited by M. Freeling and V. Walbot. Springer-Verlag, New York.
- Newton, K. J., B. Winberg, K. Yamato, S. Lupold and D. B. Stern, 1995. Evidence for a novel mitochondrial promoter preceding the cox2 gene of perennial teosintes. EMBO J. 14: 585–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton, K. J., J. M. Mariano, C. M. Gibson, E. Kuzmin and S. Gabay-Laughnan, 1996. Involvement of S2 episomal sequences in the generation of NCS4 deletion mutation in maize mitochondria. Dev. Genet. 19: 277–286. [DOI] [PubMed] [Google Scholar]
- Newton, K. J., S. Gabay-Laughnan and R. DePaepe, 2004. Mitochondrial mutations in plants, pp. 121–142 in Plant Mitochondria: From Genome to Function, edited by D. Day, A. H. Miller and J. Whelan. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Notsu, Y., S. Masood, T. Nishikawa, N. Kubo, G. Akiduki et al., 2002. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genomics 268: 434–445. [DOI] [PubMed] [Google Scholar]
- Oda, K., K. Yamato, E. Ohta, Y. Nakamura, M. Takemura et al., 1992. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA: a primitive form of plant mitochondrial genome. J. Mol. Biol. 223: 1–7. [DOI] [PubMed] [Google Scholar]
- Oeser, B., and P. Tudzynski, 1989. The linear mitochondrial plasmid pC1K1 of the phytopathogenic fungus Claviceps purpurea may code for a DNA polymerase and an RNA polymerase. Mol. Gen. Genet. 217: 132–140. [DOI] [PubMed] [Google Scholar]
- Ogihara, Y., Y. Yamazaki, K. Murai, A. Kanno, T. Terachi et al., 2005. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 33: 6235–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paillard, M., R. R. Sederoff and C. S. Levings, III, 1985. Nucleotide sequence of the S-1 mitochondrial DNA from the S cytoplasm of maize. EMBO J. 4: 1125–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, J. D., 1995. Miropeats: graphical DNA sequence comparisons. Comput. Appl. Biosci. 11: 615–619. [DOI] [PubMed] [Google Scholar]
- Perrotta, G., J. M. Grienenberger and J. M. Gualberto, 2002. Plant mitochondrial rps2 genes code for proteins with a C-terminal extension that is processed. Plant Mol. Biol. 50: 523–533. [DOI] [PubMed] [Google Scholar]
- Pring, D. R., and C. S. I. Levings, 1978. Heterogeneity of maize cytoplasmic genomes among male-sterile cytoplasms. Genetics 89: 121–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads, D. M., C. I. Kaspi, C. S. Levings, III and J. N. Siedow, 1994. N,N′-dicyclohexylcarbodiimide cross-linking suggests a central core of helices II in oligomers of URF13, the pore-forming T-toxin receptor of cms-T maize mitochondria. Proc. Natl. Acad. Sci. USA 91: 8253–8257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, K., J. Parkhill, J. Crook, T. Horsnell, P. Rice et al., 2000. Artemis: sequence visualization and annotation. Bioinformatics 16: 944–945. [DOI] [PubMed] [Google Scholar]
- Satoh, M., T. Kubo, S. Nishizawa, A. Estiati, N. Itchoda et al., 2004. The cytoplasmic male-sterile type and normal type mitochondrial genomes of sugar beet share the same complement of genes of known function but differ in the content of expressed ORFs. Mol. Genet. Genomics 272: 247–256. [DOI] [PubMed] [Google Scholar]
- Schardl, C. L., D. M. Lonsdale, D. R. Pring and K. R. Rose, 1984. Linearization of maize mitochondrial chromosomes by recombination with linear episomes. Nature 310: 292–296. [Google Scholar]
- Schardl, C. L., D. R. Pring and D. M. Lonsdale, 1985. Mitochondrial DNA rearrangements associated with fertile revertants of S-type male-sterile maize. Cell 43: 361–368. [DOI] [PubMed] [Google Scholar]
- Schnable, P. S., and R. P. Wise, 1998. The molecular basis of cytoplasmic male sterility and fertility restoration. Trends Plant Sci. 3: 175–180. [Google Scholar]
- Schwartz, S., Z. Zhang, K. A. Frazer, A. Smit, C. Riemer et al., 2000. PipMaker: a web server for aligning two genomic DNA sequences. Genome Res. 10: 577–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., L. Elnitski, M. Li, M. Weirauch, C. Riemer et al., 2003. MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 31: 3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, I., R. Suffolk and C. J. Leaver, 1989. Evolution of plant mitochondrial genomes via substoichiometric intermediates. Cell 58: 69–76. [DOI] [PubMed] [Google Scholar]
- Smith, A. G., and D. R. Pring, 1987. Nucleotide sequence and molecular characterization of a maize mitochondrial plasmid-like DNA. Curr. Genet. 12: 617–623. [DOI] [PubMed] [Google Scholar]
- Staden, R., 1996. The Staden sequence analysis package. Mol. Biotechnol. 5: 233–241. [DOI] [PubMed] [Google Scholar]
- Stern, D. B., and D. M. Lonsdale, 1982. Mitochondrial and chloroplast genomes of maize have a 12-kilobase DNA sequence in common. Nature 299: 698–702. [DOI] [PubMed] [Google Scholar]
- Tatusov, R. L., D. A. Natale, I. V. Garkavtsev, T. A. Tatusova, U. T. Shankavaram et al., 2001. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29: 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, X., J. Zheng, S. Hu and J. Yu, 2006. The rice mitochondrial genomes and their variations. Plant Physiol. 140: 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahleithner, J. A., J. L. MacFarlane and D. R. Wolstenholme, 1990. A sequence encoding a maturase-related protein in a group II intron of a plant mitochondrial nad1 gene. Proc. Natl. Acad. Sci. USA 87: 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Y. Zou, X. Li, Q. Zhang, L. Chen et al., 2006. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18: 676–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, B. L., R. S. Anderson and A. J. Bendich, 1981. The mitochondrial genome is large and variable in a family of plants (Cucurbitaceae). Cell 25: 793–803. [DOI] [PubMed] [Google Scholar]
- Weiner, J. H., P. T. Bilous, G. M. Shaw, S. P. Lubitz, L. Frost et al., 1998. A novel and ubiquitous system for membrane targeting and secretion of cofactor-containing proteins. Cell 93: 93–101. [DOI] [PubMed] [Google Scholar]
- Weissinger, A. K., D. H. Timothy, C. S. Levings, III, W. W. L. Hu and M. M. Goodman, 1982. Unique plasmid-like mitochondrial DNAs from indigenous maize races of Latin America. Proc. Natl. Acad. Sci. USA 79: 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissinger, A. K., D. H. Timothy, C. S. Levings, III and M. M. Goodman, 1983. Patterns of mitochondrial DNA variation in indigenous maize races of Latin America. Genetics 104: 365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendl, M. C., S. Dear, D. Hodgson and L. Hillier, 1998. Automated sequence preprocessing in a large-scale sequencing environment. Genome Res. 8: 975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise, R. P., A. E. Fliss, D. R. Pring and B. G. Gengenbach, 1987. Urf-13-T of T cytoplasm maize mitochondria encodes a 13 KD polypeptide. Plant Mol. Biol. 9: 121–126. [DOI] [PubMed] [Google Scholar]
- Xiao, H., F. Zhang and Y. Zheng, 2006. The 5′ stem-loop and its role in mRNA stability in maize S cytoplasmic male sterility. Plant J. 47: 864–872. [DOI] [PubMed] [Google Scholar]
- Yamamoto, M. P., T. Kubo and T. Mikami, 2005. The 5′-leader sequence of sugar beet mitochondrial atp6 encodes a novel polypeptide that is characteristic of Owen cytoplasmic male sterility. Mol. Genet. Genomics 273: 342–349. [DOI] [PubMed] [Google Scholar]
- Zabala, G., S. Gabay-Laughnan and J. R. Laughnan, 1997. The nuclear gene Rf3 affects the expression of the mitochondrial chimeric sequence R implicated in S-type male sterility in maize. Genetics 147: 847–860. [DOI] [PMC free article] [PubMed] [Google Scholar]