Abstract
The previously reported negative regulatory activity of HIM-8 on the Sox protein EGL-13 is shared by the HIM-8-related ZIM proteins. Furthermore, mutation of HIM-8 can modulate the effects of substitution mutations in the DNA-binding domains of at least four other transcription factors, suggesting broad regulatory activity by HIM-8.
THE Caenorhabditis elegans him-8 gene was identified on the basis of its high-incidence-of-males (him) phenotype where hermaphrodites produce a high proportion of male progeny due to meiotic nondisjunction of the X chromosome (Hodgkin et al. 1979; Broverman and Meneely 1994). HIM-8 is a C2H2 zinc-finger protein. In meiotic cells, it binds at or near the X chromosome pairing center and associates with the nuclear periphery (Phillips et al. 2005). HIM-8 functions to ensure pairing and subsequent synapsis of X chromosomes (Phillips et al. 2005). We recently demonstrated that HIM-8 has a broader function; it also acts in nonmeiotic cells to negatively regulate the activity of the Sox transcription factor EGL-13 (Nelms and Hanna-Rose 2006), which is encoded on the X chromosome and is important for maintenance of uterine-seam cell fate (Hanna-Rose and Han 1999; Cinar et al. 2003). The connection of gonad (Cog) morphology defect and the functional egg-laying (Egl) defect that result from the lack of uterine-seam cell-fate maintenance in egl-13 mutants are suppressed by mutation of the zinc-finger region of HIM-8 (Nelms and Hanna-Rose 2006).
HIM-8 is encoded in an operon with three similar proteins, ZIM-1 (zinc finger in meiosis), ZIM-2, and ZIM-3. Each of the four proteins has one C2H2 zinc finger with atypical spacing in the intervening region between the two cysteines and the two histidines, followed by a second typical C2H2 zinc finger (Phillips et al. 2005; Phillips and Dernburg 2006).
The ZIM proteins bind to the region of the chromosome-pairing center of specific subsets of autosomes. ZIM-1 binds to chromosomes II and III, ZIM-2 to chromosome V, and ZIM-3 to chromosomes I and IV. ZIM proteins also localize to the nuclear periphery and promote autosome pairing, analogous to the function of HIM-8 on the X chromosome (Phillips and Dernburg 2006). In this study, we report on suppression of egl-13 mutant defects by mutations in zim genes. We also demonstrate that HIM-8 has broader regulatory activity on transcription factors other than EGL-13, acts in tissues other than the somatic gonad, and can affect genes that are encoded on an autosome.
Mutations in the zim genes suppress egl-13(ku207) phenotypes:
Because of the strong similarity between HIM-8 and the ZIM proteins in the zinc fingers (Phillips et al. 2005; Phillips and Dernburg 2006), which are critical for HIM-8 suppression activity (Nelms and Hanna-Rose 2006), we asked if suppression of egl-13 was specific to mutation of him-8 or might be shared more broadly in this family of proteins. We tested deletion alleles of zim-1, zim-2, and zim-3 (from the National Bioresource Project, Tokyo Women's Medical University) for the ability to suppress egl-13(ku207) phenotypes.
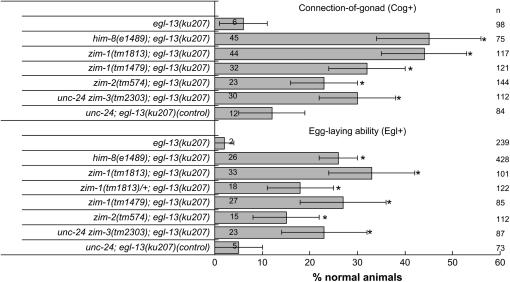
Mutations in the zim genes suppress the Cog morphology and the functional Egl defects of egl-13(ku207) (Figure 1). zim-1(tm1813) is as potent a suppressor as him-8(e1489), but suppression by zim-2(tm574) appears to be weaker (Figure 1). We previously demonstrated that him-8 mutations are semidominant suppressors due to haplo-insufficiency of the him-8 locus (Nelms and Hanna-Rose 2006). zim-1(tm1813) has a similar semidominant effect (Figure 1). As with suppression by him-8 (Nelms and Hanna-Rose 2006), suppression by zim-1(tm1813) is specific to non-null alleles of egl-13. zim-1(tm1813); egl-13(ku194 null) animals are not suppressed (0% Egl+, n = 70). Thus, these related C2H2 zinc-finger proteins share another function in addition to their meiotic function: each appears to negatively regulate EGL-13 activity.
Figure 1.—
Mutation of the zim genes suppresses the Cog and Egl defects of egl-13(ku207). The histogram illustrates the Cog+ and Egl+ percentage of egl-13(ku207) and suppressor; egl-13(ku207) double mutants. The original zim-3(tm2303) chromosome was associated with linked mutations causing lethality (Phillips and Dernburg 2006). Thus, we used an unc-24(e138) zim-3(tm2303) recombinant chromosome, which causes less lethality (Phillips and Dernburg 2006), to assay suppression by zim-3 and tested unc-24(e138) as a control. Cog and Egl phenotypes were scored as reported (Hanna-Rose and Han 1999). The him-8(e1489) data were published previously (Nelms and Hanna-Rose 2006) and shown here for comparison. Note that wild-type animals have 100% normal gonad morphology and egg-laying ability. For Figures 1–3, basic principles of proportional analysis were applied as previously described to obtain 95% confidence intervals shown as error bars (Nelms and Hanna-Rose 2006). Actual percentage is cited within each bar, and sample sizes (n) are indicated at the right in Figures 1–3. Asterisks indicate values for double mutants in Figures 1–3 that are significantly different from the relevant single mutants (P < 0.005, Fisher's exact test).
The high degree of protein sequence similarity among HIM-8, ZIM-1, ZIM-2, and ZIM-3 might suggest some degree of redundancy. However, the proteins play distinct roles in meiosis (Phillips and Dernburg 2006) and mutation of each gene results in suppression (Figure 1), indicating that the genes are not fully redundant. Partial redundancy in function is possible but is difficult to test due to the proximity of the genes in an operon.
Because genes in an operon share transcriptional regulatory sequences (Blumenthal and Gleason 2003), we considered the possibility that the mutations in the upstream zim genes act as suppressors due to indirect negative effects on expression of him-8, the last gene in the operon. If this were the case, the strong zim-1(tm1813) suppressor would be predicted to reduce him-8 activity significantly, similar to the him-8(e1489) mutant, and to cause a Him phenotype as a result. In contrast, none of the zim mutants cause a highly penetrant Him phenotype (Phillips and Dernburg 2006). We cannot rule out the scenario that disruption of him-8 expression could be making a contribution to the suppression phenomenon, but additional direct effects of the zim mutations seem most likely.
Mutation of him-8 suppresses a non-null mutation in the DNA-binding domain of POP-1:
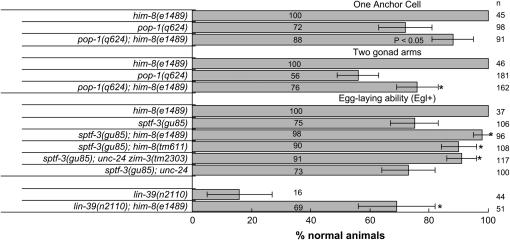
We next investigated the question of whether mutation of him-8 might have effects on genes other than the egl-13 gene. We first tested for suppression of a mutation in a transcription factor related to EGL-13. POP-1 is the C. elegans TCF-1/LEF-1 family protein (Lin et al. 1995; Korswagen 2002), which are downstream effectors of canonical wnt signal transduction pathways (Brantjes et al. 2002). TCF-1/LEF-1 proteins bind to DNA via an HMG box motif related to the Sox (Sry-related HMG box) domain of EGL-13 (Laudet et al. 1993). POP-1 is encoded on chromosome I and functions in multiple tissues (Lin et al. 1995; Herman 2001; Siegfried and Kimble 2002). pop-1(q624) causes a substitution of an isoleucine for asparagine 224 within the HMG box (Siegfried and Kimble 2002). Asparagine 224 is conserved in the TCF/LEF proteins but is not a conserved residue of the HMG box motif in general (Laudet et al. 1993). Thus, q624 is likely to have an adverse effect on DNA binding or target-site specificity but is unlikely to be a null, consistent with the incompletely penetrant phenotypes, including extra anchor cells, missing gonad arms, and lethality of first larval stage animals (Siegfried and Kimble 2002). Mutation of him-8 suppresses the gonad arm and anchor cell defects of pop-1(q624) animals (Figure 2). We conclude that mutation of him-8 can increase the effective activity of at least one other mutant transcription factor other than EGL-13 in the somatic gonad. Furthermore, mutation of him-8 can affect a gene encoded on an autosome. Interestingly, him-8(e1489) has no effect on the penetrance of larval lethality caused by pop-1(q624). pop-1(q624) mutants are 61% (n = 319) L1 lethal and pop-1(q624); him-8(e1489) double mutants are 62% (n = 719) L1 lethal.
Figure 2.—
Mutation of him-8 suppresses non-null mutations in the DNA-binding domains of POP-1, SPTF-3, and LIN-39. The histogram illustrates the percentage of normal phenotypes. The percentage of normal pop-1(q624) animals reported here agrees closely with previously published data (Siegfried and Kimble 2002). We also scored these phenotypes in pop-1(q624); syIs50[cdh-3∷GFP] and pop-1(q624);e1489; syIs50[cdh-3∷GFP] animals and obtained similar results (data not shown). cdh-3∷GFP is strongly expressed in the anchor cell, allowing easier scoring of the double-anchor cell defect. Note that 100% of wild-type animals are Egl+ and have one anchor cell and two gonad arms.
Mutation of him-8 suppresses non-null mutations in an Sp1-related zinc-finger protein and a Hox domain transcription factor:
SPTF-3, which is encoded on chromosome I, is a transcription factor with three C2H2 zinc fingers related to Sp1 (S. F. Sleiman and H. M. Chamberlin, unpublished results). sptf-3(gu85) is a missense mutation of an invariant phenylalanine in zinc finger 2. sptf-3(gu85) causes an incompletely penetrant Egl defect that is suppressed by him-8(e1489) and him-8(tm611), as well as by zim-3(tm2303) (Figure 2). sptf-3(tm607), which is a deletion allele and a putative null that disrupts the first two zinc fingers, causes embryonic lethality that is not suppressed by him-8(e1489). Neither sptf-3(tm607)/hT2[qIs48 GFP+] nor sptf-3(tm607)/hT2[qIs48 GFP+]; him-8(e1489) animals have viable sptf-3(tm607) (GFP-negative) progeny.
LIN-39, which is encoded on chromosome III, is a Hox family DNA-binding factor (Clark et al. 1993). lin-39(n2110) is a missense mutation, E179K, in the Hox domain that results in a highly penetrant Egl defect (Clark et al. 1993). lin-39(n2110) is efficiently suppressed by him-8(e1489) (Figure 2). However, the Egl phenotype of lin-39(n1880), a putative null allele that introduces a stop at codon 100 prior to the Hox domain (Clark et al. 1993), is not suppressed by him-8(e1489). Both lin-39(n1880) and lin-39(n1880); him-8(e1489) are 0% Egl+. We conclude that mutation of him-8 can increase the effective activity of compromised transcription factors with DNA-binding domains other than an HMG box but cannot compensate for the loss of these factors.
Mutation of him-8 has tissue-specific effects on a non-null mutation in EGL-38:
EGL-38, which is encoded on chromosome IV, is a C. elegans Pax 2/5/8 family protein (Chamberlin et al. 1997). Reduction-of-function mutations in egl-38 have defects in uv1 cell-fate specification, vulval morphology, egg laying, hindgut development (including hindgut expression of a lin-48∷GFP reporter), and male tail morphology (Chamberlin et al. 1997; Johnson et al. 2001; Zhang et al. 2005; Rajakumar and Chamberlin 2007). Different mutations within the EGL-38 DNA-binding domain can have specific effects on one or more of the phenotypes, but do not necessarily affect each phenotype equally (Zhang et al. 2005). egl-38(gu22) causes a substitution in the DNA-binding domain of isoleucine for methionine 29, which is conserved but not invariant within the Pax 2/5/8 family of proteins (Zhang et al. 2005).
A lin-48∷GFP reporter is expressed in four hindgut cells in wild-type animals but expression is absent from one or more hindgut cells in egl-38(gu22) mutants (Johnson et al. 2001; Zhang et al. 2005). Although egl-38(gu22) mutants lack efficient lin-48∷GFP expression, they have largely normal vulval morphology and egg-laying function (Zhang et al. 2005). The proportion of the population of egl-38(gu22) mutants that completely lack lin-48∷GFP hindgut expression is eliminated in egl-38(gu22) him-8(e1489) double mutants (Figure 3) and the intensity of expression relative to the egl-38 mutants with expression is increased as well (Figure 4). Thus, him-8(e1489) is a potent suppressor of egl-38(gu22) in the hindgut. him-8(e1489) also completely suppresses the Mab (male abnormal development) defect of egl-38(gu22) (Figure 3). Surprisingly, him-8(e1489) had an opposite effect on the vulval morphology and the related egg-laying defects of egl-38(gu22). him-8(e1489) dramatically exacerbates these defects (Figure 3).
Figure 3.—
him-8(e1489) suppresses the male tail defect and defective hindgut expression of a lin-48∷GFP reporter but exacerbates the vulval development and egg-laying defects caused by egl-38(gu22). The histogram illustrates the percentage of animals that express lin-48::GFP in at least one of the four hindgut cells with normal male tail morphology, normal vulval morphology, or normal egg-laying activity (Egl+). Phenotypes were assayed as reported (Chamberlin et al. 1997; Johnson et al. 2001; Zhang et al. 2005; Rajakumar and Chamberlin 2007).
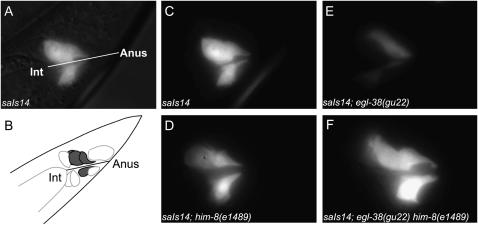
Figure 4.—
him-8(e1489) increases the intensity of lin-48∷GFP expression in the hindgut of an egl-38(gu22) mutant. (A) Fluorescent image of the hindgut region of an saIs14[lin-48∷GFP] hermaphrodite overlaid on the corresponding DIC image. (B) lin-48∷GFP expression is visible in four hindgut cells that flank the rectum. (C) Expression is consistently detected in all four cells at similar intensities in the transgenic parent strain. (D) The intensity of lin-48∷GFP expression in him-8(e1489); saIs14[lin-48∷GFP] animals is not detectably different from that of the parent strain. (E) Thirty-one percent of egl-38(gu22); unc-119(e2498); saIs14[lin-48∷GFP] animals lack all hindgut expression of lin-48∷GFP (Figure 3), and the intensity of lin-48∷GFP expression is weaker than that of the parental strain in all animals that have expression. (F) In addition to an increase in the percentage of the population that expresses the reporter faithfully (Figure 3), all suppressed egl-38(gu22) him-8(e1489); saIs14[lin-48∷GFP] animals have an increase in the intensity of lin-48∷GFP expression relative to the parental strain. Image exposure conditions were identical for A and C–F.
The opposite effects on egl-38(gu22) in the hindgut and the male tail vs. the vulva could be due to different effects of him-8 in each tissue or, more likely, due to the complicated tissue-specific activities of egl-38 (Zhang et al. 2005). For example, mutation of him-8 could act to increase the activity of egl-38 in both tissues, but increased activity of the EGL-38(M29I) mutant in the vulva would have a different and deleterious effect as opposed to the positive effect of increased activity of EGL-38(M29I) in the hindgut. In general, it is clear that, although mutation of him-8 can suppress a range of mutations in a range of tissues, the phenotypic suppression effect is not universal, and phenotypic enhancement is possible.
Models for HIM-8/ZIM suppression:
HIM-8 has more global effects on gene activity than might be expected from its primarily X-specific activity in meiosis. Although we have not tested the ZIM proteins as widely as HIM-8, they behave similarly to HIM-8 where examined (in modulation of EGL-13 and SPTF-3). How might these proteins be mediating their effects? Our previous results implicated the DNA-binding domain of HIM-8 in suppression activity. At the same time, we showed that increasing the level of mutant EGL-13 protein could mimic suppression (Nelms and Hanna-Rose 2006). Thus, a possible scenario for suppression is that eliminating or reducing HIM-8/ZIM protein activity results in upregulation of EGL-13, and increased levels of mutant protein can increase the level of function.
This model predicts that mutant him-8 might suppress a wide range of mutations that would benefit from increased gene dose. lin-15(n765ts) is an X-linked, temperature-sensitive, hypomorphic mutation, which can be almost completely suppressed by elevated X chromosome expression caused by mutations in the dosage compensation genes dpy-21 and dpy-26 (Meneely and Wood 1987; Hsu and Meyer 1994). However, him-8(e1489) cannot suppress lin-15(n765ts). lin-15(n765) mutants are 100% (n = 18) Multivulva (Muv) at 20°, and him-8(e1489); lin-15(n765) animals are still highly Muv (97%, n = 34). Moreover, if mutation of him-8 suppresses egl-13(ku207) simply by increasing egl-13 gene expression, mutations in dosage compensation genes, which are responsible for reducing expression from the X chromosome in hermaphrodites (Meyer 2000), might be expected to suppress egl-13(ku207) as well. However, we detected no suppression by the dosage compensation mutants dpy-28(y1) or dpy-21(e428) (B. L. Nelms and W. Hanna-Rose, unpublished results). Thus, we have accumulated no evidence in support of an upregulation of gene transcription model.
An alternative model is that the suppression is uniquely specific to missense mutations affecting the DNA-binding domain of transcription factors (and perhaps other proteins that interact with the chromosome). This model is consistent with the failure of him-8(e1489) to suppress egl-13, sptf-3, or lin-39 null mutants or to suppress non-null mutations in genes that do not encode transcription factors (below).
In addition to failing to suppress lin-15(n765) (above), him-8(e1489) does not suppress missense mutations in several other genes that encode proteins without DNA-binding activity. First, him-8(e1489) does not suppress egl-26(n481), a missense mutation (S275F) in a putative acyltransferase (Estes et al. 2007). egl-26(n481) is 35% (n = 206) Egl+, and egl-26(n481); him-8(e1489) is 35% (n = 83) Egl+.
Second, him-8(e1489) cannot suppress sur-6(ku123), a missense mutation (C302Y) in a regulatory subunit of protein phosphatase 2A (Sieburth et al. 1999). sur-6(ku123) suppresses the Muv phenotype of animals harboring one copy of the ras gain-of-function transgene kuIs14. kuIs14/+ animals are 68% (n = 71) Muv whereas sur-6(ku123); kuIs14/+ animals are 51% (n = 77) Muv. However, the presence of him-8(e1489) has no effect on sur-6(ku123) activity. sur-6(ku123); kuIs14/+; him-8(e1489) are indistinguishable from sur-6(ku123); kuIs14/+ at 51% (n = 85) Muv.
Third, him-8(e1489) cannot suppress unc-37(e262). unc-37(e262) is a missense mutation (H539Y) in a WD repeat motif of UNC-37, a Groucho-like corepressor protein (Pflugrad et al. 1997). All unc-37 animals and all unc-37(e262); him-8(e1489) animals abnormally coil instead of backing in response to a head touch (n > 60). Although UNC-37 Groucho is involved in transcriptional regulation, it does not bind directly to the DNA (Pickles et al. 2002). Our results suggest that mutation of the zinc fingers of HIM-8 specifically modulates the activity of DNA-binding transcription factors with compromised DNA-binding activity.
A DNA-binding-domain-specific model suggests that suppression occurs not due to direct changes in the suppressed gene, but rather due to altered activity of the protein product on its transcriptional targets. For example, disruption of HIM-8 might alter chromosome structure, allowing greater access to DNA targets, or otherwise enhance the in vivo DNA-binding activity of the compromised proteins. As most of the transcriptional targets responsible for the phenotypes assayed in this study are not known, it is possible that the difference between phenotypes that are suppressed and those that are enhanced could be the relative sensitivity of the target to the mutant protein or its genomic location. Our experiments leave open the possibility of direct interactions between the suppressed protein products and the suppressors as well. However, if protein–protein interactions between HIM-8 and transcription factors play a role in the mechanism, the transcription factors do not appear to alter HIM-8 meiotic activity since our double-mutant strains have Him phenotypes similar to that of him-8 single mutants. Future experiments will be required to better clarify the underlying mechanism for this unique function of HIM-8.
Acknowledgments
We thank Abby Dernburg and Meera Sundaram for strains and Shohei Mitani at the National Bioresource Project (Tokyo, Japan) for the tm deletion alleles. We thank members of our laboratories and the Malone lab for discussions and critical reading of our manuscript. This research was supported by award no. 0131287 to W.H.R. from the National Science Foundation and GM62336 to H.M.C. from the National Institutes of Health (NIH). Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources.
References
- Blumenthal, T., and K. S. Gleason, 2003. Caenorhabditis elegans operons: form and function. Nat. Rev. Genet. 4: 112–120. [DOI] [PubMed] [Google Scholar]
- Brantjes, H., N. Barker, J. van Es and H. Clevers, 2002. TCF: Lady Justice casting the final verdict on the outcome of Wnt signalling. Biol. Chem. 383: 255–261. [DOI] [PubMed] [Google Scholar]
- Broverman, S. A., and P. M. Meneely, 1994. Meiotic mutants that cause a polar decrease in recombination on the X chromosome in Caenorhabditis elegans. Genetics 136: 119–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlin, H. M., R. E. Palmer, A. P. Newman, P. W. Sternberg, D. L. Baillie et al., 1997. The PAX gene egl-38 mediates development patterning in Caenorhabditis elegans. Development 124: 3919–3928. [DOI] [PubMed] [Google Scholar]
- Cinar, H. N., K. L. Richards, K. S. Oommen and A. P. Newman, 2003. The EGL-13 SOX domain transcription factor affects the uterine π cell lineages in Caenorhabditis elegans. Genetics 165: 1623–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, S. G., A. D. Chisholm and H. R. Horvitz, 1993. Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell 74: 43–55. [DOI] [PubMed] [Google Scholar]
- Estes, K. E., R. Kalamegham and W. Hanna-Rose, 2007. Membrane localization of the NlpC/P60 family protein EGL-26 correlates with regulation of vulval cell morphogenesis in C. elegans. Dev. Biol. 308: 196–205. [DOI] [PubMed] [Google Scholar]
- Hanna-Rose, W., and M. Han, 1999. COG-2, a sox domain protein necessary for establishing a functional vulval-uterine connection in Caenorhabditis elegans. Development 126: 169–179. [DOI] [PubMed] [Google Scholar]
- Herman, M., 2001. C. elegans POP-1/TCF functions in a canonical Wnt pathway that controls cell migration and in a noncanonical Wnt pathway that controls cell polarity. Development 128: 581–590. [DOI] [PubMed] [Google Scholar]
- Hodgkin, J., H. R. Horvitz and S. Brenner, 1979. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics 91: 67–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, D. R., and B. J. Meyer, 1994. The dpy-30 gene encodes an essential component of the Caenorhabditis elegans dosage compensation machinery. Genetics 137: 999–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A. D., D. Fitzsimmons, J. Hagman and H. M. Chamberlin, 2001. EGL-38 Pax regulates the ovo-related gene lin-48 during Caenorhabditis elegans organ development. Development 128: 2857–2865. [DOI] [PubMed] [Google Scholar]
- Korswagen, H. C., 2002. Canonical and non-canonical Wnt signaling pathways in Caenorhabditis elegans: variations on a common signaling theme. BioEssays 24: 801–810. [DOI] [PubMed] [Google Scholar]
- Laudet, V., D. Stehelin and H. Clevers, 1993. Ancestry and diversity of the HMG box superfamily. Nucleic Acids Res. 21: 2493–2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, R. L., S. Thompson and J. R. Priess, 1995. pop-1 encodes an HMG box protein required for the specification of a mesoderm precursor in early C. elegans embryos. Cell 83: 599–609. [DOI] [PubMed] [Google Scholar]
- Meneely, P. M., and W. B. Wood, 1987. Genetic analysis of X-chromosome dosage compensation in Caenorhabditis elegans. Genetics 117: 25–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer, B. J., 2000. Sex in the wormcounting and compensating X-chromosome dose. Trends Genet. 16: 247–253. [DOI] [PubMed] [Google Scholar]
- Nelms, B. L., and W. Hanna-Rose, 2006. C. elegans HIM-8 functions outside of meiosis to antagonize EGL-13 Sox protein function. Dev. Biol. 293: 392–402. [DOI] [PubMed] [Google Scholar]
- Pflugrad, A., J. Y. Meir, T. M. Barnes and D. M. Miller, III, 1997. The Groucho-like transcription factor UNC-37 functions with the neural specificity gene unc-4 to govern motor neuron identity in C. elegans. Development 124: 1699–1709. [DOI] [PubMed] [Google Scholar]
- Phillips, C. M., and A. F. Dernburg, 2006. A family of zinc-finger proteins is required for chromosome-specific pairing and synapsis during meiosis in C. elegans. Dev. Cell 11: 817–829. [DOI] [PubMed] [Google Scholar]
- Phillips, C. M., C. Wong, N. Bhalla, P. M. Carlton, P. Weiser et al., 2005. HIM-8 binds to the X chromosome pairing center and mediates chromosome-specific meiotic synapsis. Cell 123: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles, L. M., S. M. Roe, E. J. Hemingway, S. Stifani and L. H. Pearl, 2002. Crystal structure of the C-terminal WD40 repeat domain of the human Groucho/TLE1 transcriptional corepressor. Structure 10: 751–761. [DOI] [PubMed] [Google Scholar]
- Rajakumar, V., and H. M. Chamberlin, 2007. The Pax2/5/8 gene egl-38 coordinates organogenesis of the C. elegans egg-laying system. Dev. Biol. 301: 240–253. [DOI] [PubMed] [Google Scholar]
- Sieburth, D. S., M. Sundaram, R. M. Howard and M. Han, 1999. A PP2A regulatory subunit positively regulates Ras-mediated signaling during Caenorhabditis elegans vulval induction. Genes Dev. 13: 2562–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegfried, K. R., and J. Kimble, 2002. POP-1 controls axis formation during early gonadogenesis in C. elegans. Development 129: 443–453. [DOI] [PubMed] [Google Scholar]
- Zhang, G., S. F. Sleiman, R. J. Tseng, V. Rajakumar, X. Wang et al., 2005. Alteration of the DNA binding domain disrupts distinct functions of the C. elegans Pax protein EGL-38. Mech. Dev. 122: 887–899. [DOI] [PubMed] [Google Scholar]