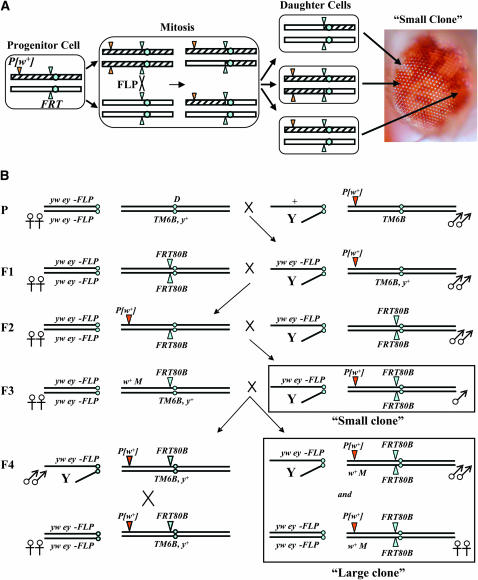
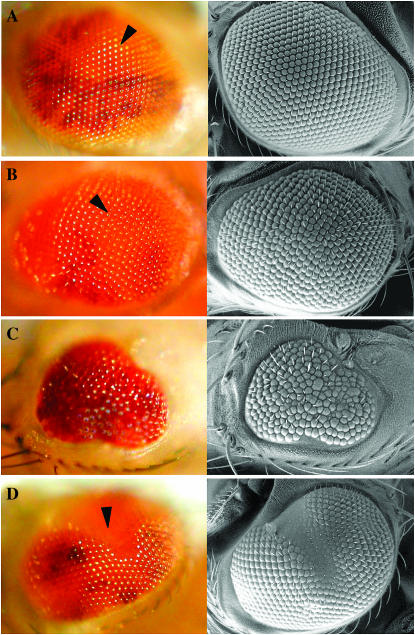
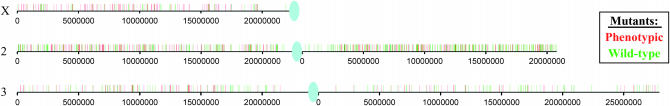
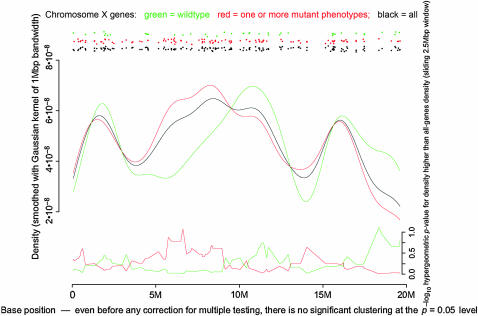
Gerald B Call
Gerald B Call
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,1,2,
John M Olson
John M Olson
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,1,
Jiong Chen
Jiong Chen
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,3,
Nikki Villarasa
Nikki Villarasa
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Kathy T Ngo
Kathy T Ngo
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Allison M Yabroff
Allison M Yabroff
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,4,
Shawn Cokus
Shawn Cokus
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Matteo Pellegrini
Matteo Pellegrini
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Elena Bibikova
Elena Bibikova
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Chris Bui
Chris Bui
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Albert Cespedes
Albert Cespedes
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Cheryl Chan
Cheryl Chan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Stacy Chan
Stacy Chan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Amrita K Cheema
Amrita K Cheema
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Akanksha Chhabra
Akanksha Chhabra
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Vida Chitsazzadeh
Vida Chitsazzadeh
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Minh-Tu Do
Minh-Tu Do
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Q Angela Fang
Q Angela Fang
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Andrew Folick
Andrew Folick
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Gelsey L Goodstein
Gelsey L Goodstein
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Cheng R Huang
Cheng R Huang
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Tony Hung
Tony Hung
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Eunha Kim
Eunha Kim
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
William Kim
William Kim
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Yulee Kim
Yulee Kim
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Emil Kohan
Emil Kohan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Edward Kuoy
Edward Kuoy
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Robert Kwak
Robert Kwak
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Eric Lee
Eric Lee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
JiEun Lee
JiEun Lee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Henry Lin
Henry Lin
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
H-C Angela Liu
H-C Angela Liu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Tatiana Moroz
Tatiana Moroz
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Tharani Prasad
Tharani Prasad
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sacha L Prashad
Sacha L Prashad
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Alexander N Patananan
Alexander N Patananan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Alma Rangel
Alma Rangel
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Desiree Rosselli
Desiree Rosselli
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sohrab Sidhu
Sohrab Sidhu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Daniel Sitz
Daniel Sitz
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Chelsea E Taber
Chelsea E Taber
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jingwen Tan
Jingwen Tan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Kasey Topp
Kasey Topp
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
PhuongThao Tran
PhuongThao Tran
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Quynh-Minh Tran
Quynh-Minh Tran
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Mary Unkovic
Mary Unkovic
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Maggie Wells
Maggie Wells
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jessica Wickland
Jessica Wickland
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Kevin Yackle
Kevin Yackle
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Amir Yavari
Amir Yavari
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jesse M Zaretsky
Jesse M Zaretsky
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Christopher M Allen
Christopher M Allen
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Latifat Alli
Latifat Alli
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Ju An
Ju An
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Abbas Anwar
Abbas Anwar
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sonia Arevalo
Sonia Arevalo
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Danny Ayoub
Danny Ayoub
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Shawn S Badal
Shawn S Badal
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Armonde Baghdanian
Armonde Baghdanian
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Arthur H Baghdanian
Arthur H Baghdanian
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sara A Baumann
Sara A Baumann
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Vivian N Becerra
Vivian N Becerra
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Hei J Chan
Hei J Chan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Aileen E Chang
Aileen E Chang
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Xibin A Cheng
Xibin A Cheng
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Mabel Chin
Mabel Chin
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Fleurette Chong
Fleurette Chong
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Carlyn Crisostomo
Carlyn Crisostomo
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sanjit Datta
Sanjit Datta
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
†,
Angela Delosreyes
Angela Delosreyes
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Francie Diep
Francie Diep
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Preethika Ekanayake
Preethika Ekanayake
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Mark Engeln
Mark Engeln
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Elizabeth Evers
Elizabeth Evers
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Farzin Farshidi
Farzin Farshidi
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Katrina Fischer
Katrina Fischer
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Arlene J Formanes
Arlene J Formanes
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jun Gong
Jun Gong
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Riju Gupta
Riju Gupta
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Blake E Haas
Blake E Haas
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Vicky Hahm
Vicky Hahm
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Michael Hsieh
Michael Hsieh
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
James Z Hui
James Z Hui
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Mei L Iao
Mei L Iao
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sophia D Jin
Sophia D Jin
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Angela Y Kim
Angela Y Kim
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Lydia S-H Kim
Lydia S-H Kim
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Megan King
Megan King
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
‡,
Chloe Knudsen-Robbins
Chloe Knudsen-Robbins
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
David Kohanchi
David Kohanchi
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Bogdana Kovshilovskaya
Bogdana Kovshilovskaya
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Amy Ku
Amy Ku
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Raymond W Kung
Raymond W Kung
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Mark E L Landig
Mark E L Landig
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Stephanie S Latterman
Stephanie S Latterman
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Stephanie S Lauw
Stephanie S Lauw
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Daniel S Lee
Daniel S Lee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Joann S Lee
Joann S Lee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Kai C Lei
Kai C Lei
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Lesley L Leung
Lesley L Leung
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Renata Lerner
Renata Lerner
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jian-ya Lin
Jian-ya Lin
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Kathleen Lin
Kathleen Lin
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Bryon C Lim
Bryon C Lim
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Crystal P Y Lui
Crystal P Y Lui
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Tiffany Q Liu
Tiffany Q Liu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Vincent Luong
Vincent Luong
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jacob Makshanoff
Jacob Makshanoff
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
An-Chi Mei
An-Chi Mei
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Miguel Meza
Miguel Meza
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Yara A Mikhaeil
Yara A Mikhaeil
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Majid Moarefi
Majid Moarefi
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Long H Nguyen
Long H Nguyen
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Shekhar S Pai
Shekhar S Pai
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Manish Pandya
Manish Pandya
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Aadit R Patel
Aadit R Patel
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Paul D Picard
Paul D Picard
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
§,
Michael M Safaee
Michael M Safaee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Carol Salame
Carol Salame
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Christian Sanchez
Christian Sanchez
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Nina Sanchez
Nina Sanchez
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Christina C Seifert
Christina C Seifert
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Abhishek Shah
Abhishek Shah
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Oganes H Shilgevorkyan
Oganes H Shilgevorkyan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Inderroop Singh
Inderroop Singh
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Vanessa Soma
Vanessa Soma
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Junia J Song
Junia J Song
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Neetika Srivastava
Neetika Srivastava
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jennifer L StaAna
Jennifer L StaAna
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Christie Sun
Christie Sun
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Diane Tan
Diane Tan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Alison S Teruya
Alison S Teruya
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Robyn Tikia
Robyn Tikia
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Trinh Tran
Trinh Tran
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Emily G Travis
Emily G Travis
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jennifer D Trinh
Jennifer D Trinh
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Diane Vo
Diane Vo
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Thomas Walsh
Thomas Walsh
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Regan S Wong
Regan S Wong
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Katherine Wu
Katherine Wu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Ya-Whey Wu
Ya-Whey Wu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Nkau X V Yang
Nkau X V Yang
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Michael Yeranosian
Michael Yeranosian
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
James S Yu
James S Yu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jennifer J Zhou
Jennifer J Zhou
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Ran X Zhu
Ran X Zhu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Anna Abrams
Anna Abrams
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Amanda Abramson
Amanda Abramson
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Latiffe Amado
Latiffe Amado
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jenny Anderson
Jenny Anderson
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Keenan Bashour
Keenan Bashour
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Elsa Beyer
Elsa Beyer
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Allen Bookatz
Allen Bookatz
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sarah Brewer
Sarah Brewer
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Natalie Buu
Natalie Buu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Stephanie Calvillo
Stephanie Calvillo
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Joseph Cao
Joseph Cao
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Amy Chan
Amy Chan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jenny Chan
Jenny Chan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Aileen Chang
Aileen Chang
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Daniel Chang
Daniel Chang
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Yuli Chang,
YiBing Chen
YiBing Chen
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Joo Choi
Joo Choi
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jeyling Chou
Jeyling Chou
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Peter Dang
Peter Dang
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sumit Datta
Sumit Datta
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Ardy Davarifar
Ardy Davarifar
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Artemis Deravanesian
Artemis Deravanesian
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Poonam Desai
Poonam Desai
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jordan Fabrikant
Jordan Fabrikant
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Shahbaz Farnad
Shahbaz Farnad
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Katherine Fu
Katherine Fu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Eddie Garcia
Eddie Garcia
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Nick Garrone
Nick Garrone
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Srpouhi Gasparyan
Srpouhi Gasparyan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Phyllis Gayda
Phyllis Gayda
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sherrylene Go
Sherrylene Go
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Chad Goffstein
Chad Goffstein
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Courtney Gonzalez
Courtney Gonzalez
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Mariam Guirguis
Mariam Guirguis
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Ryan Hassid
Ryan Hassid
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Brenda Hermogeno
Brenda Hermogeno
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Julie Hong
Julie Hong
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Aria Hong
Aria Hong
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Lindsay Hovestreydt
Lindsay Hovestreydt
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Charles Hu
Charles Hu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Devon Huff
Devon Huff
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Farid Jamshidian
Farid Jamshidian
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
James Jen
James Jen
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Katrin Kahen
Katrin Kahen
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Linda Kao
Linda Kao
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Melissa Kelley
Melissa Kelley
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Thomas Kho
Thomas Kho
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Yein Kim
Yein Kim
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sarah Kim
Sarah Kim
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Brian Kirkpatrick
Brian Kirkpatrick
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Adam Langenbacher
Adam Langenbacher
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Santino Laxamana
Santino Laxamana
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Janet Lee
Janet Lee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Chris Lee
Chris Lee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
So-Youn Lee
So-Youn Lee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
ToHang S Lee
ToHang S Lee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Toni Lee
Toni Lee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Gemma Lewis
Gemma Lewis
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sheila Lezcano
Sheila Lezcano
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Peter Lin
Peter Lin
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Thanh Luu
Thanh Luu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Julie Luu
Julie Luu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Will Marrs
Will Marrs
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Erin Marsh
Erin Marsh
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jamie Marshall
Jamie Marshall
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sarah Min
Sarah Min
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Tanya Minasian
Tanya Minasian
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Helena Minye
Helena Minye
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Amit Misra
Amit Misra
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Miles Morimoto
Miles Morimoto
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Yasaman Moshfegh
Yasaman Moshfegh
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jessica Murray
Jessica Murray
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Kha Nguyen
Kha Nguyen
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Cynthia Nguyen
Cynthia Nguyen
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Ernesto Nodado II
Ernesto Nodado II
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Amanda O'Donahue
Amanda O'Donahue
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Ndidi Onugha
Ndidi Onugha
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Nneka Orjiakor
Nneka Orjiakor
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Bhavin Padhiar
Bhavin Padhiar
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Eric Paul
Eric Paul
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Mara Pavel-Dinu
Mara Pavel-Dinu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Alex Pavlenko
Alex Pavlenko
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Edwin Paz
Edwin Paz
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sarah Phaklides
Sarah Phaklides
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Lephong Pham
Lephong Pham
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Preethi Poulose
Preethi Poulose
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Russell Powell
Russell Powell
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Aya Pusic
Aya Pusic
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Divi Ramola
Divi Ramola
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Kirsten Regalia
Kirsten Regalia
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Meghann Ribbens
Meghann Ribbens
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Bassel Rifai
Bassel Rifai
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Manyak Saakyan
Manyak Saakyan
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Pamela Saarikoski
Pamela Saarikoski
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Miriam Segura
Miriam Segura
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Farnaz Shadpour
Farnaz Shadpour
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Aram Shemmassian
Aram Shemmassian
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Ramnik Singh
Ramnik Singh
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Vivek Singh
Vivek Singh
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Emily Skinner
Emily Skinner
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Daniel Solomin
Daniel Solomin
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Kosha Soneji
Kosha Soneji
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Kristin Spivey
Kristin Spivey
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Erika Stageberg
Erika Stageberg
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Marina Stavchanskiy
Marina Stavchanskiy
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Leena Tekchandani
Leena Tekchandani
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Leo Thai
Leo Thai
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Jayantha Thiyanaratnam
Jayantha Thiyanaratnam
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Maurine Tong
Maurine Tong
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Aneet Toor
Aneet Toor
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Steve Tovar
Steve Tovar
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Kelly Trangsrud
Kelly Trangsrud
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Wah-Yung Tsang
Wah-Yung Tsang
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Marc Uemura
Marc Uemura
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Emily Vollmer
Emily Vollmer
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Emily Weiss
Emily Weiss
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Damien Wood
Damien Wood
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Joy Wu
Joy Wu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Sophia Wu
Sophia Wu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Winston Wu
Winston Wu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Qing Xu
Qing Xu
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Yuki Yamauchi
Yuki Yamauchi
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Will Yarosh
Will Yarosh
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Laura Yee
Laura Yee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
George Yen
George Yen
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,
Utpal Banerjee
Utpal Banerjee
*Department of Molecular, Cell, and Developmental Biology, University of California, Los Angeles, California 90095, †Palos Verdes Peninsula High School, Rolling Hills Estates, California 90274, ‡Culver City Independent Study School, Culver City, California 90230, §Loyola High School, Los Angeles, California 90006, **Molecular Biology Institute, University of California, Los Angeles, California 90095 and ††Department of Biological Chemistry, University of California, Los Angeles, California 90095
*,**,††,5