Abstract
Many plant species, including important crops like wheat, are polyploids that carry more than two sets of genetically related chromosomes capable of meiotic pairing. To safeguard a diploid-like behavior at meiosis, many polyploids evolved genetic loci that suppress incorrect pairing and recombination of homeologues. The Ph1 locus in wheat was proposed to ensure homologous pairing by controlling the specificity of centromere associations that precede chromosome pairing. Using wheat chromosomes that carry rye centromeres, we show that the centromere associations in early meiosis are not based on homology and that the Ph1 locus has no effect on such associations. Although centromeres indeed undergo a switch from nonhomologous to homologous associations in meiosis, this process is driven by the terminally initiated synapsis. The centromere has no effect on metaphase I chiasmate chromosome associations: homologs with identical or different centromeres, in the presence and absence of Ph1, pair the same. A FISH analysis of the behavior of centromeres and distal chromomeres in telocentric and bi-armed chromosomes demonstrates that it is not the centromeric, but rather the subtelomeric, regions that are involved in the correct partner recognition and selection.
POLYPLOIDY is widely acknowledged as a major mechanism of adaptation and speciation in plants. It is estimated that between 47 and 70% of angiosperms are polyploid (Ramsey and Schemske 1998). Most polyploid plant species, including important crops like wheat, are allopolyploids that arose after hybridization between related diploid progenitors. The polyploid condition confers some advantages such as heterosis or gene redundancy but implies disadvantages such as the propensity to produce aneuploid meiotic products that reduce fertility (Comai 2005). This effect on fertility is conditioned by the presence of more than two genetically related chromosome sets capable of meiotic pairing. Many polyploid species have evolved genetic regulatory systems that ensure a diploid-like behavior with efficient disjunction of homologous chromosomes at the first division (Jenczewski and Alix 2004). The best-studied example is common bread wheat, Triticum aestivum, an allohexaploid species (2n = 6x = 42) with three genomes, A, B, and D, from three related diploid species. In spite of the genetic synteny between homeologous chromosomes, bread wheat forms 21 bivalents at diakinesis and metaphase I (MI) of meiosis. Several loci have been identified that affect chromosome pairing in hexaploid wheat (reviewed by Sears 1976). The exclusive formation of homologous bivalents at MI is principally controlled by the Ph1 (pairing homeologous) locus on the long arm of chromosome 5B (Riley and Chapman 1958; Sears and Okamoto 1958). The Ph1 locus has been recently localized to a 2.5-Mb region containing a segment of subtelomeric heterochromatin inserted into a cluster of cdc-2 related genes (Griffiths et al. 2006). However, its mode of action remains to be elucidated.
Three major meiotic processes—chromosome pairing (i.e., an interaction of chromosomes that results in the alignment of homologs), synapsis [i.e., the formation of the proteinaceous synaptonemal complex (SC) structure between each homologous pair], and crossing over—are involved in the formation of bivalents. Homologous chromosomes previously distributed throughout the nucleus (Bass et al. 2000; Maestra et al. 2002) must approach and recognize each other to enter into intimate contact and form bivalents. How homologous chromosomes get into close physical proximity with each other to undergo interaction represents one of the least-understood mechanisms of the meiotic process (Roeder 1997; Zickler and Kleckner 1998; Page and Hawley 2003; Pawlowski et al. 2003; Pawlowski and Cande 2005). In most organisms, telomeres attach to the inner nuclear envelop and congregate to form the so-called meiotic bouquet (Bass et al. 2000; Niwa et al. 2000; Trelles-Sticken et al. 2000; Cowan et al. 2001; Scherthan 2001; Harper et al. 2004). This chromosome arrangement is thought to facilitate homologous recognition.
An ultrastructural analysis of spread silver-stained meiotic nuclei of hexaploid wheat by Holm (1986) revealed that, at the beginning of the zygotene stage, telomeres aggregate and chromosome pairing and SC formation is initiated distally. In nuclei at mid-zygotene, generally the longest SC segments were those joining the distal segments. The presence of only one pairing partner exchange in most SC multivalents formed in polyploid wheats is also in agreement with the initiation of pairing and synapsis in distal chromosome regions (Martínez et al. 2001a,b). Distal chromosome pairing initiation in wheat explains the failure of homologous synapsis after colchicine-induced inhibition of bouquet formation (Corredor and Naranjo 2007). Distal, but not proximal, regions of wheat chromosomes are also critical for MI chiasmate chromosome associations (Lukaszewski 1997; Jones et al. 2002). The commencement of pairing usually at distal sites and succeeded by numerous intercalary initiations has been observed in other plant species such as maize (Gillies 1975), Lilium (Holm 1977), rye (Gillies 1985), or Allium (Albini and Jones 1987).
Studies denoting polarization and association of centromeres in premeiotic cells postulated a possible role of this chromosome structure in meiotic pairing. Three-dimensional reconstruction of microsporocyte nuclei from electron micrographs of serial thin sections in Allium fistulosum, Lilium speciosum, Ornithogalum virens, wheat, rye (Secale cereale L.), and triticale (X Triticosecale Wittmack) showed polarization of centromeres opposite the telomeres but were not conclusive in establishing whether presynapstic centromere association was based on homology (Church and Moens 1976; Bennett et al. 1979; Church 1981; Delfosse and Church 1981). A fluorescence in situ hybridization (FISH) analysis of chromosome arrangement in hexaploid wheat showed that centromeres associate prior to meiosis, usually in pairs (Aragón-Alcaide et al. 1997; Martínez-Pérez et al. 1999, 2001). This suggested that the Ph1 locus suppresses homeologous pairing through the control of the specificity of centromere association. However, Maestra et al. (2002) reported that, in a majority of cells at premeiotic interphase and leptotene, two homologous chromosomes added to wheat occupied separated territories both in the presence and in the absence of the Ph1 locus. In early leptotene in wheat, centromeres associate in multimeric structures. On the basis of the formation of seven centromere structures in a small number of microsporocytes at leptotene, Martínez-Pérez et al. (2003) proposed that these clusters represent an important component of the chromosome-sorting mechanism. Each cluster would include three pairs of centromeres corresponding to chromosomes of the same homeologous group. After homologous recognition, clusters resolve into pairs of homologous centromeres under the control of Ph1. This hypothesis might seem feasible in the light of the chromosome dynamics at meiosis in yeast (Tsubouchi and Roeder 2005). Centromeres of yeast chromosomes associate in pairs that initially are nonhomologous and then undergo switching until all pairs are homologous. Unlike in yeast, however, the composition of individual centromere clusters in wheat has never been established because of the paucity of chromosome- or genome-specific DNA probes.
Telomeres and centromeres are involved in complex multimeric structures formed in presynaptic meiotic cells, in which previously separate chromosomes can interact. Functionally equivalent regions of different chromosomes, such as telomeres or centromeres, provide excellent starting points for homology recognition that avoids scanning of the entire genome, a complicated task in large genomes. However, because wheat chromosomes are bi-armed and very large—in mid-zygotene they reach an average length of 112 μm (Martínez et al. 2001b)—it is difficult to envisage how a mechanism of chromosome recognition operating at the centromeres may trigger the initiation of synapsis at the telomeres.
We have taken advantage of wheat chromosomes with centromere introgressions from rye to study centromere positioning in early meiotic cells and their effect on MI chiasmate chromosome associations, both in the presence and in the absence of Ph1. These centromere introgressions were produced by recurrent centric breakage-fusion events (Lukaszewski 1993; Zhang et al. 2001). Rye centromeres contain species-specific repeats that can be identified by FISH with probe pAWRC.1 when in a wheat background (Langridge et al. 1998; Franki 2001). We find that homologous centromeres are mainly separate in presynaptic stages and that the transition to homologous association is driven by synapsis. Modification of the centromere constitution in homozygous or heterozygous condition has no effect on MI chiasmate chromosome associations either in the wild-type or in the Ph1− mutant. On the other hand, using chromosome-specific markers, we verify that terminal and subterminal regions pair earlier than centromeres of bi-armed chromosomes. This excludes centromeres as components of the chromosome-sorting mechanism.
MATERIALS AND METHODS
Plant material:
The following hexaploid wheat (T. aestivum; 2n = 6x = 42; genome AABBDD) genotypes were used to analyze the role of the centromere in chromosome pairing: the wild-type (Ph1) and the ph1b (Ph1−) mutant lines in cv. Pavon 76 homozygous and heterozygous for chromosome 1B with the centromere from rye chromosome 1R (1Brc), homozygous and heterozygous for chromosome 2B with the centromere from rye chromosome 2R (2Brc), and a double monosomic 2Brc, 2R. The role of distal regions in homologous recognition was assessed in two lines with different conformation for the long arm (L) of rye chromosome 2R: the Robertsonian 2BS.2RL translocation line of cv. Pavon and the ditelocentric 2RL (Dt2RL) addition line of cv. Chinese Spring.
Analysis of chiasmate chromosome associations:
Anthers with pollen mother cells (PMCs) at MI were fixed in a 3:1 ethanol–acetic acid solution. Squashed preparations were C-banded as previously described (Giráldez et al. 1979) or subject to FISH with pAWRC.1 containing a rye-specific centromere repeat (Langridge et al. 1998; Franki 2001) for identification of marked chromosomes. Observations were under a Nikon Eclipse E400 microscope or Zeiss Axioscope 20. MI chiasmate chromosome associations were scored in plants grown in two environments and in three different growing seasons. As there were no significant differences between sites and seasons, all data were pooled. The overall levels of homologous and homeologous MI chromosome associations in Ph1+ and Ph1− plants was carried out by C-banding in samples of 100 PMCs for each line.
Fluorescence in situ hybridization, microscopy, and image processing:
Fixed anthers were digested in a pectolytic enzyme mixture, transferred to a clean slide, and spread according to Zhong et al. (1996). This procedure involves no mechanical pressure to spread the cells on the slide and the three-dimensional information is largely preserved. Preparations were pretreated as previously described (Maestra et al. 2002).
For the analysis of the behavior of centromeres and distal chromosome regions, the following repeat DNA probes were used: clone pAtT4 containing the Arabidopsis telomeric tandem repeat (Richards and Ausubel 1988), clone 6C6 containing a cereal-specific centromere repeat (Zhang et al. 2004), clone pAWRC.1 containing a rye-specific centromere repeat, clone pSc74 containing a rye-specific 480-bp tandem repeat (Bedbrook et al.1980; Cuadrado and Schwarzacher 1998), clone pSc119.2 containing a 120-bp tandem repeated sequence unit from rye that identifies wheat B-genome chromosomes (Bedbrook et al.1980; Mukai et al. 1993; Cuadrado and Jouve 1994), and sonicated or boiled rye genomic DNA (fragment size <2 kb). These clones were used in different combinations for in situ hybridization as previously described (Maestra et al. 2002; Corredor and Naranjo 2007). Concentrations of DNA probes in the different hybridization mixes were 5 ng/μl for pAtT4; 10 ng/μl for 6C6, pAWRC.1, pSc74, and pSc119.2; and 2.2 ng/μl for rye genomic DNA.
Clones pAtT4, 6C6, pAWRC.1, pSc74, and pSc119.2 were labeled by nick translation with biotin-16-dUTP or digoxigenin-11-dUTP, and rye genomic DNA was random primed labeled with digoxigenin-11-dUTP. In the Dt2RL and 2BS.2RL plants, pAtT4 was labeled with both digoxigenin-11-dUTP and biotin-16-dUTP to produce an orange color. Two sequential rounds of hybridization were used for multicolor painting of chromosome 2RL with pAtT4, pAWRC.1, pSc199.2, and rye total genomic DNA probes in somatic cells. The digoxigenin-labeled probes were detected with 6–8 ng/μl of the FITC-conjugated antidigoxigenin antibody (Sigma, St. Louis) in 4B (0.5% blocking reagent in 4× SSC) and biotin-labeled probes with 10–15 ng/μl of the Cy3-conjugated avidine (Sigma) in 4B.
Microscopy and image processing have been previously described (Corredor and Naranjo 2007).
RESULTS
Chromosome pairs studied:
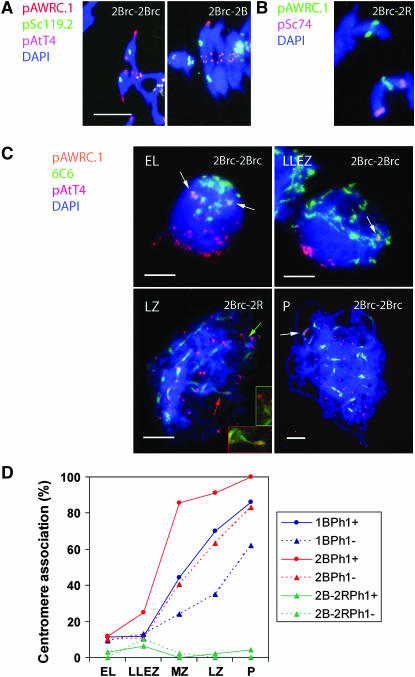
We have used wheat chromosomes 1B and 2B with the rye centromeres (rc) (1Brc and 2Brc, respectively) to produce lines that have modified the centromere constitution of one chromosome pair in both the presence and absence of the Ph1 locus. The three chromosome combinations that we studied were (i) chromosome pairs 1Brc–1Brc and 2Brc–2Brc (homologous chromosomes–homologous centromeres; Figure 1A; supplemental Figure 1 at http://www.genetics.org/supplemental/), (ii) chromosome pairs 1B–1Brc and 2B–2Brc (homologous chromosomes–homeologous centromeres; Figure 1A), and (iii) chromosome pair 2Brc–2R, the first from wheat and the second from rye (homeologous chromosomes–homologous centromeres; Figure 1B; supplemental Figure 1).
Figure 1.—
Rye-specific DNA sequences in centromeres of wheat and rye chromosomes and their arrangement at early meiosis. (A) Bivalents 2Brc–2Brc and 2B–2Brc at MI with rye centromeres labeled with probe pAWRC.1. Signals of the pSc119.2 probe identify B-genome chromosomes. (B) Rye centromere in univalents 2Brc and 2R at MI; chromosome 2R shows the location of the rye-specific pSc74 DNA repeat. (C) Nuclei at early leptotene (EL), leptotene–zygotene transition (LLEZ), late zygotene (LZ), and pachytene (P) of homozygotes 2Brc–2Brc and double monosomic 2Brc–2R. Homologous centromeres of rye (arrows) are separated in EL and LZ and associated in LLEZ and P, respectively. Wheat centromeres and telomeres were labeled with the 6C6 and pAtT4 DNA probes, respectively. (D) Frequency of associations of homologous rye centromeres in pairs 1Brc–1Brc, 2Brc–2Brc, and 2Brc–2R at EL, LLEZ, mid-zygotene (MZ), LZ, and P stages in Ph1+ and Ph1− wheat lines. Mean number of PMCs, n = 43 ± 3. Bars, 10 μm.
The wild-type and Ph1− mutant phenotypes:
The main phenotypic feature of Ph1− mutant wheat is the formation of multivalent configuration at MI, which is accompanied by a reduction in the number of chiasmate chromosome associations. We have verified the genotype of the lines studied by quantification of the overall levels of chromosome configurations at MI in samples of 100 PMCs. The results obtained appear in Table 1. As expected, in the mutant lines, the mean number of multivalents per cell increases and the mean number of bound arms decreases relative to wild-type lines.
TABLE 1.
Mean values of univalents, ring bivalents, open bivalents, trivalents, and other multivalents and bound arms per cell at metaphase I in samples of 100 PMCs of different wheat lines
| Wheat line | Univalents | Ring bivalents | Open bivalents | Trivalents | Multivalents | Bound arms per cell |
|---|---|---|---|---|---|---|
| Ph1+ | ||||||
| 1Brc–1Brc | 0.18 ± 0.06 | 18,61 ± 0.17 | 2.19 ± 0.16 | 0.00 | 0.04 ± 0.02 | 39.56 ± 0.19 |
| 1B–1Brc | 0.13 ± 0.05 | 19.44 ± 0.17 | 1.34 ± 0.16 | 0.01 ± 0.01 | 0.05 ± 0.02 | 40.42 ± 0.18 |
| 2Brc–2Brc | 0.00 | 20.06 ± 0.12 | 0.86 ± 0.11 | 0.00 | 0.04 ± 0.02 | 41.13 ± 0.05 |
| 2B–2Brc | 0.00 | 20.10 ± 0.11 | 0.88 ± 0.11 | 0.00 | 0.01 ± 0.01 | 41.12 ± 0.10 |
| 2Brc–2R | 2.10 ± 0.04a | 18.78 ± 0.12 | 1.13 ± 0.12 | 0.00 | 0.02 ± 0.01 | 38.77 ± 0.13 |
| Ph1− | ||||||
| 1Brc–1Brc | 1.36 ± 0.05 | 13.89 ± 0.22 | 5.05 ± 0.21 | 0.23 ± 0.05 | 0.52 ± 0.07 | 35.03 ± 0.26 |
| 1B–1Brc | 0.66 ± 0.13 | 15.81 ± 0.20 | 3.82 ± 0.18 | 0.03 ± 0.02 | 0.51 ± 0.08 | 37.15 ± 0.21 |
| 2Brc–2Brc | 0.67 ± 0.11 | 15.64 ± 0.21 | 4.19 ± 0.20 | 0.07 ± 0.02 | 0.36 ± 0.05 | 36.86 ± 0.23 |
| 2B–2Brc | 0.83 ± 0.12 | 15.66 ± 0.21 | 3.88 ± 0.19 | 0.05 ± 0.02 | 0.48 ± 0.05 | 37.03 ± 0.24 |
| 2Brc–2R | 2.71 ± 0.14b | 14.73 ± 0.20 | 3.54 ± 0.16 | 0.12 ± 0.04 | 0.59 ± 0.07 | 35.25 ± 0.21 |
2B univalent + 2R univalent = 2.00.
2B univalent + 2R univalent = 1.89.
Presynaptic centromere association is nonhomologous:
To determine the nature of the early centromere associations in the presence and absence of the Ph1 locus, we analyzed by FISH the physical distribution of homologous rye centromere pairs relative to wheat centromeres, which were hybridized with probe 6C6 in plants with 1Brc–1Brc, 2Brc–2Brc, and 2Brc–2R in meiocytes from leptotene to pachytene (Figure 1C). In leptotene, telomeres migrate to form a tight cluster and centromeres appear as compact structures. Chromatin undergoes a conformational change that results in chromosome elongation (Mikhailova et al. 1998; Maestra et al. 2002), which is apparent in centromere signals at the leptotene–zygotene transition. As synapsis progresses in mid-zygotene, the telomere bouquet disintegrates (Holm 1986). Late zygotene and pachytene are postbouquet stages, with high levels of synapsis or with complete synapsis (Holm 1986; Mikhailova et al. 1998), and differ by the degree of chromatin condensation.
In leptotene of the Ph1+ lines, rye centromere pairs 1Brc–1Brc and 2Brc–2Brc were physically separated and included in different clusters (Figure 1C) in 88% of meiocytes (Figure 1D). The labeled centromeres were considered to be physically associated in the remainder meiocytes (12%), which showed only one FISH signal or two signals in the same cluster and were separated by <1 μm. The latter was an infrequent event. The frequency of associations of these rye centromeres increased with the progression of meiosis, reaching 100% for 2Brc–2Brc at pachytene. A low frequency of homologous centromere associations in leptotene and a gradual increase through the zygotene–pachytene suggests that these associations are a result of the synaptonemal complex expansion and not some presynaptic event or process. In Ph1−, the arrangement of labeled centromeres of the 1Brc–1Brc and 2Brc–2Brc pairs at leptotene was similar to that in Ph1+. The level of associations in pairs increased in the course of prophase I but the frequencies observed in zygotene and pachytene were lower than the corresponding frequencies in the wild type. We interpret this as being in agreement with a delay in the initiation and development of synapsis in the absence of a functional Ph1 allele, known to exist in wheat (Mikhailova et al. 1998; Maestra et al. 2002). Homologous centromeres in the 2Brc–2R pair were separated in 97 and 100% of Ph1+ and Ph1− cells at leptotene, respectively, and their association level did not increase in the course of prophase I. These observations support the conclusion that, in wheat, centromere clustering in early meiosis is not based on homology and therefore cannot promote recognition of homologous chromosomes. The transition from nonhomologous to homologous centromere associations in meiotic prophase is driven by synapsis; because synapsis begins at the ends of homologs, eventually homologous centromeres associate.
Centromere heterozygosity does not affect pairing and recombination:
If synapsis expansion brings homologous centromeres together, centromere identity cannot influence the selection of the correct partner for pairing and recombination. We have tested this hypothesis by studying the level of chiasmate chromosome associations at MI for chromosomes with rye centromeres. Each pair can form a ring bivalent or an open bivalent or fail to pair (Figure 1, A and B). We have quantified the frequency of arms bound for each chromosome pair in the presence and absence of the Ph1 locus (Table 2).
TABLE 2.
MI frequencies of bound arms per chromosome for chromosome pairs with replaced centromeres
| Chromosome pair | Ph1+ | Ph1− |
|---|---|---|
| 1Brc–1Brc | 1.89 (241) | 1.76 (650)* |
| 1B–1Brc | 1.92 (192) | 1.85 (287)* |
| 2Brc–2Brc | 1.89 (183) | 1.82 (174) |
| 2B–2Brc | 1.88 (243) | 1.75 (230) |
| 2Brc–2R | 0.0 (387) | 0.07 (189) |
Numbers of microsporocytes analyzed are in parentheses. *χ2 = 7.13, P < 0.01.
Homozygosity or heterozygosity for the centromere had no effect on the frequencies of the MI chiasmate associations of labeled chromosome in Ph1+. Pairs 1Brc–1Brc and 1B–1Brc and pairs 2Brc–2Brc and 2B–2Brc showed almost equal numbers of bound arms. In the Ph1− lines, pairs 2Brc–2Brc and 2B–2Brc behaved in the same way but the 1Brc–1Brc homozygote showed a decrease in the mean number of arms bound per pair relative to the 1B–1Brc heterozygote. This result is explained by reduced overall chiasmate bonds in the former (Table 1). The Ph1 locus is known to suppress pairing between standard chromosomes 2B and 2R (Naranjo et al. 1987); in Ph1−, there is practically no chiasmate association of the short arms and some chiasmate association of the long arms (Naranjo and Fernández-Rueda 1996). This did not change when rye 2R and wheat 2B had identical rye centromeres. Homologous centromeres did not make homeologous chromosomes pair and recombine, even in Ph1−. Therefore, the centromere does not determine whether or not two homeologous chromosomes can pair.
Distal regions pair earlier than centromeres:
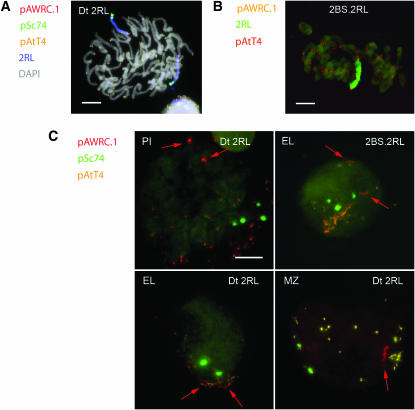
We assessed the role of terminal regions on homologous recognition relative to centromeres by studying the arrangement of centromeric, subtelomeric, and telomeric markers of rye chromosome arm 2RL in the presynaptic and synaptic stages of meiosis in two different lines of wheat: one with a Robertsonian translocation 2BS.2RL and another with a ditelosomic addition 2RL (Dt2RL). In the Robertsonian translocation, the 2RL arm is fused to 2BS and the centromere is located in the central chromosome region. In the telocentric 2RL, the centromere is located at one telomere. Multicolor FISH labeling of 2RL highlights the positions of the centromere, the telomeres, a terminal heterochromatic knob, and another knob that is subterminal (Figure 2, A and B). These two knobs can be recognized by a difference in signal size.
Figure 2.—
The centromere and the terminal and subterminal knobs of chromosome 2RL and their arrangement in premeiotic interphase and in early meiosis. (A) The centromere and knobs of telocentric 2RL in a mitotic prometaphase after two rounds of FISH. (B) The arms 2RL and their rye centromeres in the ring bivalent formed by the 2BS.2RL translocation pair at MI after two-color FISH. (C) PMCs at the premeiotic interphase (PI), early leptotene (EL), and mid-zygotene (M) in the Dt2RL and 2BS.2RL lines showing the arrangement of the rye centromeres (red arrows), knobs (green), and telomeres (orange). Centromeres are separated in PI and ELs and associated in MZ. In the EL of Dt2RL, rye centromeres are incorporated at the telomere pole. Distal knobs are separated in PI and associated in the three other PMCs while subdistal knobs are associated only in the EL and MZ PMCs of Dt2RL. Bars, 10 μm.
In the prebouquet stage, in both chromosomes 2RL and 2BS.2RL, the terminal and subterminal knobs were associated in <8% of PMCs (Figure 3). Telomere convergence increased the frequency of these associations. At the leptotene–zygotene transition, the terminal knobs on 2RL were associated in 65 and 68% of PMCs in the Dt2RL and 2BS.2RL lines, while the subterminal knobs were associated in 37 and 27% of the meiocytes, respectively. These differences are consistent with the progression of synapsis from the telomere toward the center of the chromosome. At late zygotene, the levels of associations of the terminal and subterminal knobs were >92% for both chromosomes. In summary, the terminal and subterminal regions of the 2RL arm in the telocentric and the translocation chromosomes behaved the same. The centromeres, on the other hand, behaved in a similar fashion only in the presynaptic stages when they were located at the centromere pole of the nucleus and physically separated in 85% of cells. At early zygotene, because of the telomere dominance in bouquet formation (Maestra et al. 2002), the centromeres of the telocentrics were able to enter the bouquet and were paired in 59% of PMCs; centromeres of the bi-armed translocated chromosomes remained stationary and separated (93%) at the centromere pole of the nucleus. The difference in the level of physical association of the centromeres of the two types of chromosomes remained the same through mid-zygotene and decreased at late zygotene. Physical association of the centromeres of telocentrics was at a level comparable to that of the distal knob.
Figure 3.—
The frequencies (%) of association of centromeres (Cen), and the terminal (Tm) and subterminal (Sm) knobs at the premeiotic interphase and in early meiosis in the Dt2RL and 2BS.2RL wheat lines. PI, premeiotic interphase. EL, early leptotene, LLEZ, late leptotene–early zygotene. MZ, mid-zygotene. LZ, late zygotene. Mean number of PMCs, n = 52 ± 6.
DISCUSSION
Our results on the behavior of labeled centromeres in early meiotic stages are unequivocal in showing that centromeres do not take part in the homologous recognition. Both the centromeres of chromosomes 1Rrc and 2Brc and the centromeres of the translocated 2BS.2RL and telocentric 2RL are separated in leptotene. This arrangement was also observed for the centromeres of chromosomes 5R and 5RL in wheat–rye additions (Corredor and Naranjo 2007). All chromosomes studied also show that the transition from nonhomologous to homologous centromere associations is affected once synapsis has been triggered at the telomeres. Exceptions are centromeres of telocentric chromosomes, such as those of chromosomes 2RL studied here and of 5RL (Corredor and Naranjo 2007). These centromeres abandon the centromere pole of the nucleus during leptotene, dragged along by their telomeric sequence with which they are capped, and incorporate into the bouquet, behaving like any distal chromosome regions. On the other hand, homologous centromeres of nonhomologous chromosomes, such as the 2Brc–2R pair here, that remain unpaired at MI, probably by not undergoing synapsis, remain separated during the entire prophase I. Thus, progression of synapsis in zygotene from the telomere toward the centromere is the main force that brings homologous centromere regions together.
An argument could be raised that, unlike the three-dimensional study of Martínez-Pérez et al. (1999, 2001, 2003), the chromosome spreading technique used in this study could disrupt the presynaptic association of homologous centromeres and hence provide misleading results. It needs, therefore, to be pointed out that both technical approaches—the three-dimensional confocal microscopy on preparations from anther sections (Martínez-Pérez et al. 1999, 2001, 2003) and the analyses of spread nuclei (Maestra et al. 2002; Corredor and Naranjo 2007)—reveal the same associations of centromeres in pairs in premeiotic interphase and the formation of more complex structures in early leptotene. Thus, spreading preserves the overall presynaptic centromere arrangement detected in intact nuclei.
As deduced from the MI chromosome association results, centromere constitution does not affect the ability of chromosomes to recombine. Homologous chromosomes carrying the same or different centromeres show the same frequencies of chiasmate associations at MI, and homeologous chromosomes rarely pair although they possess homologous centromeres. In wheat, genetic mapping of the physical attributes of chromosomes and deletion mapping of genetic markers has shown that crossing over was concentrated in the terminal segments of the chromosome arms and was practically absent from the proximal halves of the arms (Jones et al. 2002 and references therein). In fungi, mammals, and plants, but not in Drosophila or Caenorhabditis elegans, chromosome pairing is largely dependent on the initiation and progression of recombination (reviewed in Pawlowski and Cande 2005; Zickler 2006). The absence of any effect of centromeres in wheat on the presence or absence of chiasmate bonds at MI is in agreement with this link between pairing and recombination, as well as with the observation that pairing progresses from the chromosome ends to the center of the chromosome.
What is the role of centromere association in leptotene? During this stage, chromosomes undergo profound changes in the chromatin conformation and its spatial arrangement. Wheat chromosomes multiply their length fivefold in leptotene relative to premeiotic interphase (Mikhailova et al. 1998). Because the size of the nucleus remains the same at the leptotene–zygotene transition or is even reduced (Maestra et al. 2002), chromosomes span the entire nucleus. Meanwhile, telomeres move to converge in a small region of the nuclear periphery opposite the centromeres. Both the chromosome elongation movement and telomere migration are concurrent and affect all chromosomes. Chromatin conformational changes may generate random chromosome movement, which could interfere with the oriented telomere migration. Centromere association may act to stabilize the centromere pole to reduce the degree of disorder introduced by chromosome elongation and to maintain a reference point for the telomere migration.
The data presented here do not support in any way the statement that the Ph1 locus in wheat controls bivalent pairing through the centromeres. This is consistent with an earlier observation that, in a chromosome formed by a fusion of two homeologous arms at the centromere, intrachromosomal homeologous pairing takes place only in the absence of the Ph1 locus but is suppressed in its presence (Dvorak and Lukaszewski 2000). On the other hand, long and perfectly homologous segments of an asymmetrical isochromosome do not pair in MI probably because telomere clustering does not juxtapose homologous segments of these arms (Lukaszewski 1997). The formation of multivalents at metaphase I in the absence of Ph1 is preceded by different synaptonemal complex dynamics. Although both wild-type and mutant wheats form synaptonemal complex multivalents, these are transformed into bivalents at the end of the zygotene in the wild type but not in the mutant (Holm and Wang 1988; Martínez et al. 2001a,b). Failure of the pairing correction mechanism allows for both homologous and homeologous chromosomes to form chiasmata and for multivalents to become evident in MI. Failure of the multivalent correction mechanism in the mutant is accompanied by, and might be dependent on, a delay of synapsis. Dubcovsky et al. (1995) reported that recombination between homeologous chromosome segments is dramatically reduced by the presence of Ph1 even when they are introgressed in intercalary positions of homologs. Taking into account that in most organisms the initiation of recombination interacts with chromosome pairing, it is not possible to conclude whether the suppression of recombination induced by Ph1 is a consequence of the SC multivalent correction or, by contrast, is the trigger of the pairing correction mechanism. A realistic explanation of how the homology of synapsed chromosomes is scrutinized in the presence of the Ph1 locus needs more data but we do not see any evidence that the control is exercised through the centromere.
Acknowledgments
We thank B. Friebe, P. Landgridge, and A. Cuadrado for kindly supplying clones 6C6, pAWRC.1, and pSc119.2, respectively. This work has been supported by grant 2003-04 from New Del Amo Program from the University of California-Universidad Complutense de Madrid, by grant BFU2004-02261 from Dirección General de Investigación, Ministerio de Educación y Ciencia of Spain, and by grant PR27/05-13984 from Banco Santander-Universidad Complutense de Madrid.
References
- Albini, S. M., and G. H. Jones, 1987. Synaptonemal complex spreading in Allium cepa and A. Fistulosum. I. The initiation and sequence of pairing. Chromosoma 95: 324–338. [Google Scholar]
- Aragón-Alcaide, L., S. Reader, A. Beven, P. Shaw, T. Miller et al., 1997. Association of homologous chromosomes during floral development. Curr. Biol. 7: 905–908. [DOI] [PubMed] [Google Scholar]
- Bass, H. W., O. Riera-Lizarazu, E. V. Ananiev, S. J. Bordoli, H. W. Rines et al., 2000. Evidence for the coincident initiation of homolog pairing and synapsis during the telomere-clustering (bouquet) stage of meiotic prophase. J. Cell Sci. 113: 1033–1042. [DOI] [PubMed] [Google Scholar]
- Bedbrook, J. R., J. Jones, M. O'Dell, R. Thompson and R. B. Flavell, 1980. A molecular description of telomeric heterochromatin in Secale species. Cell 19: 545–560. [DOI] [PubMed] [Google Scholar]
- Bennett, M. D., J. B. Smith, S. Simpson and B. Wells, 1979. Intranuclear fibrillar material in cereal pollen mother cells. Chromosoma 71: 289–332. [Google Scholar]
- Church, K., 1981. The architecture of and chromosome movements within the premeiotic interphase nucleus, pp. 83–102 in Mitosis/Citokinesis, edited by A. M. Zimmerman and A. Forer. Academic Press, San Diego.
- Church, K., and P. Moens, 1976. Centromere behaviour during interphase and meiotic prophase in Allium fistulosum from 3-D, E.M. reconstruction. Chromosoma 56: 249–263. [Google Scholar]
- Comai, L., 2005. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6: 836–846. [DOI] [PubMed] [Google Scholar]
- Corredor, E., and T. Naranjo, 2007. Effect of colchicine and telocentric chromosome conformation on centromere and telomere dynamics at meiotic prophase I in wheat-rye additions. Chromosome Res. 15: 231–245. [DOI] [PubMed] [Google Scholar]
- Cowan, C. R., P. M. Carlton and W. Z. Cande, 2001. The polar arrangement of telomeres in interphase and meiosis: Rabl organization and the bouquet. Plant Physiol. 125: 532–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuadrado, A., and N. Jouve, 1994. Mapping and organization of highly-repeated DNA sequences by means of simultaneous and sequential FISH and C-banding in 6×-triticale. Chromosome Res. 2: 331–338. [DOI] [PubMed] [Google Scholar]
- Cuadrado, A., and T. Schwarzacher, 1998. The chromosomal organization of simple sequence repeats in wheat and rye genomes. Chromosoma 107: 587–594. [DOI] [PubMed] [Google Scholar]
- Delfosse, F., and K. Church, 1981. Presynaptic chromosome behaviour in Lillium. I. Centromere orientation and movement during premeiotic interphase in Lillium speciosum cv. Rosemede. Chromosoma 81: 701–716. [Google Scholar]
- Dubcovsky, J., M. C. Luo and J. Dvorak, 1995. Differentiation between homoeologous chromosomes 1A of wheat and 1Am of Triticum monococcum and its recognition by the wheat Ph1 locus. Proc. Natl. Acad. Sci. USA 92: 6645–6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak, J., and A. J. Lukaszewski, 2000. Centromere association is an unlikely mechanism by which the wheat Ph1 locus regulates metaphase I chromosome pairing between homoeologous chromosomes. Chromosoma 109: 410–414. [DOI] [PubMed] [Google Scholar]
- Franki, M. G., 2001. Identification of Bilby, a diverged centromeric Ty1-copia retrotransposon family from cereal rye (Secale cereale L.). Genome 44: 266–274. [DOI] [PubMed] [Google Scholar]
- Gillies, C. B., 1975. An ultrastructural analysis of chromosome pairing in maize. C. R. Trav. Lab. Carlsberg 40: 135–161. [Google Scholar]
- Gillies, C. B., 1985. An electron microscopical study of synaptonemal complex formation at zygotene in rye. Chromosoma 92: 165–175. [Google Scholar]
- Giráldez, R., M. C. Cermeño and J. Orellana, 1979. Comparison of C-banding pattern in the chromosomes of inbred lines and open pollinated varieties of rye, Secale cereale L. Z. Planzenzücht. 83: 40–48. [Google Scholar]
- Griffiths, S., R. Sharp, T. N. Foote, I. Bertin, M. Wanous et al., 2006. Molecular characterization of Ph1 as a major chromosome pairing locus in polyploid wheat. Nature 439: 749–752. [DOI] [PubMed] [Google Scholar]
- Harper, L., I. Golubovskaya and W. Z. Cande, 2004. A bouquet of chromosomes. J. Cell Sci. 117: 4025–4032. [DOI] [PubMed] [Google Scholar]
- Holm, P. B., 1977. Three-dimensional reconstruction of chromosome pairing during the zygotene stage of meiosis in Lilium longiflorum (Thunb.). Carlsberg Res. Commun. 42: 103–151. [Google Scholar]
- Holm, P. B., 1986. Chromosome pairing and chiasma formation in allohexaploid wheat: Triticum aestivum analyzed by spreading of meiotic nuclei. Carlsberg Res. Commun. 51: 239–294. [Google Scholar]
- Holm, P. B., and X. Wang, 1988. The effect of chromosome 5B on synapsis and chiasma formation in wheat Triticum aestivum cv. Chinese Spring. Carlsberg Res. Commun. 53: 191–208. [Google Scholar]
- Jenczewski, E., and K. Alix, 2004. From diploids to allopolyploids: the emergence of efficient pairing control genes in plants. Crit. Rev. Plant Sci. 23: 21–45. [Google Scholar]
- Jones, L. E., K. Rybka and A. J. Lukaszewski, 2002. The effect of a deficiency and a deletion on recombination in chromosome 1BL in wheat. Theor. Appl. Genet. 104: 1204–1208. [DOI] [PubMed] [Google Scholar]
- Langridge, P., P. Guo, M. Franki and U. Langridge, 1998. Isolation of λ and YAC clones from defined regions of the rye genome. Mol. Gen. Genet. 257: 568–575. [DOI] [PubMed] [Google Scholar]
- Lukaszewski, A. J., 1993. Reconstruction in wheat of complete chromosomes 1B and 1R from the 1RS.1BL translocation of ‘Kavkaz’ origin. Genome 36: 821–824. [DOI] [PubMed] [Google Scholar]
- Lukaszewski, A. J., 1997. The development and meiotic behavior of asymmetrical isochromosomes in wheat. Genetics 145: 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maestra, B., J. H. De Jong, K. Shepherd and T. Naranjo, 2002. Chromosome arrangement and behaviour of two rye telosomes at the onset of meiosis in disomic wheat-5RL addition lines with and without the Ph1 locus. Chromosome Res. 10: 655–667. [DOI] [PubMed] [Google Scholar]
- Martínez, M., T. Naranjo, C. Cuadrado and C. Romero, 2001. a The synaptic behaviour of Triticum turgidum with variable doses of the Ph1 locus. Theor. Appl. Genet. 102: 751–758. [Google Scholar]
- Martínez, M., N. Cuñado, N. Carcelen and C. Romero, 2001. b The Ph1 and Ph2 loci play different roles in the synaptic behaviour of hexaploid wheat Triticum aestivum. Theor. Appl. Genet. 103: 398–405. [Google Scholar]
- Martínez-Pérez, E., P. Shaw, S. Reader, L. Aragón-Alcaide, T. E. Miller et al., 1999. Homologous chromosome pairing in wheat. J. Cell Sci. 112: 1761–1769. [DOI] [PubMed] [Google Scholar]
- Martínez-Pérez, E., P. Shaw and G. Moore, 2001. The Ph1 locus is needed to ensure specific somatic and meiotic centromere association. Nature 411: 204–207. [DOI] [PubMed] [Google Scholar]
- Martínez-Pérez, E., P. Shaw, L. Aragón-Alcaide and G. Moore, 2003. Chromosomes form into seven groups in hexaploid and tetraploid wheat as a prelude of meiosis. Plant J. 36: 21–29. [DOI] [PubMed] [Google Scholar]
- Mikhailova, E. I., T. Naranjo, K. Shepherd, J. Wennekes-Van Eden, C. Heyting et al., 1998. The effect of the wheat Ph1 locus on chromatin organisation and meiotic chromosome pairing analysed by genome painting. Chromosoma 107: 339–350. [DOI] [PubMed] [Google Scholar]
- Mukai, Y., Y. Nakahara and M. Yamamoto, 1993. Simultaneous discrimination of the three genomes in hexaploid wheat by multicolor fluorescence in situ hybridization using total genomic and highly repeated DNA probes. Genome 36: 489–494. [DOI] [PubMed] [Google Scholar]
- Naranjo, T., and P. Fernández-Rueda, 1996. Pairing and recombination between individual chromosomes of wheat and rye in hybrids carrying the ph1b mutation. Theor. Appl. Genet. 93: 242–248. [DOI] [PubMed] [Google Scholar]
- Naranjo, T., A. Roca, P. G. Goicoechea and R. Giráldez, 1987. Arm homoeology of wheat and rye chromosomes. Genome 29: 873–882. [Google Scholar]
- Niwa, O, M. Simanuki and F. Miki, 2000. Telomere-led bouquet formation facilitates homologous chromosome pairing and restricts ectopic interactions in fission yeast meiosis. EMBO J. 19: 3831–3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, S. L., and R. S. Hawley, 2003. Chromosome choreography: the meiotic ballet. Science 301: 785–789. [DOI] [PubMed] [Google Scholar]
- Pawlowski, W. P., and W. Z. Cande, 2005. Coordinating the events of the meiotic prophase. Trends Cell Biol. 15: 664–681. [DOI] [PubMed] [Google Scholar]
- Pawlowski, W. P., I. N. Golubovskaya and W. Z. Cande, 2003. Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 15: 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey, J., and D. W. Schemske, 1998. Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29: 467–501. [Google Scholar]
- Richards, E. J., and S. M. Ausubel, 1988. Isolation of a higher eukaryothic telomere sequence from Arabidopsis thaliana. Cell 53: 127–136. [DOI] [PubMed] [Google Scholar]
- Riley, R., and V. Chapman, 1958. Genetic control of the cytologically diploid behaviour of hexaploid wheat. Nature 182: 713–715. [Google Scholar]
- Roeder, G. S., 1997. Meiotic chromosomes: it takes two to tango. Genes Dev. 11: 2600–2621. [DOI] [PubMed] [Google Scholar]
- Scherthan, H., 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell Biol. 6: 975–987. [DOI] [PubMed] [Google Scholar]
- Sears, E. R., 1976. Genetic control of chromosome pairing in wheat. Annu. Rev. Genet. 10: 31–51. [DOI] [PubMed] [Google Scholar]
- Sears, E. R., and M. Okamoto, 1958. Intergenomic chromosome relationships in hexaploid wheat. Proc. Int. Congr. Genet. 10th 2: 258–259. [Google Scholar]
- Trelles-Sticken, E., M. E. Dresser and H. Scherthan, 2000. Meiotic telomere protein Ndj1p is required for meiosis-specific telomere distribution, bouquet formation and efficient homologue pairing. J. Cell Biol. 151: 95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi, T., and S. Roeder, 2005. A synaptonemal complex protein promotes homology-independent centromere coupling. Science 308: 870–873. [DOI] [PubMed] [Google Scholar]
- Zhang, P., B. Friebe, A. J. Lukaszewski and B. S. Gill, 2001. The centromere structure in Robertsonian wheat-rye translocation chromosomes indicates that centric breakage-fusion can occur at different positions within the primary constriction. Chromosoma 110: 335–344. [DOI] [PubMed] [Google Scholar]
- Zhang, P., W. Li, J. Fellers, B. Friebe and B. S. Gill, 2004. BAC-FISH in wheat identifies chromosome landmarks consisting of different types of transposable elements. Chromosoma 112: 288–299. [DOI] [PubMed] [Google Scholar]
- Zhong, X., P. F. Fransz, J. Wennekes-Van Eden, P. Zabel and A. Van Kammen et al., 1996. High-resolution mapping of pachytene chromosomes and extended DNA fibres by fluorescent in situ hybridisation. Plant Mol. Biol. Rep. 14: 232–242. [Google Scholar]
- Zickler, D., 2006. From early homologous recognition to synaptonemal complex formation. Chromosoma 115: 158–174. [DOI] [PubMed] [Google Scholar]
- Zickler, D., and N. Kleckner, 1998. The leptotene-zygotene transition of meiosis. Annu. Rev. Genet. 32: 619–697. [DOI] [PubMed] [Google Scholar]