Abstract
Nonstop mRNA decay, a specific mRNA surveillance pathway, rapidly degrades transcripts that lack in-frame stop codons. The cytoplasmic exosome, a complex of 3′–5′ exoribonucleases involved in RNA degradation and processing events, degrades nonstop transcripts. To further understand how nonstop mRNAs are recognized and degraded, we performed a genomewide screen for nonessential genes that are required for nonstop mRNA decay. We identified 16 genes that affect the expression of two different nonstop reporters. Most of these genes affected the stability of a nonstop mRNA reporter. Additionally, three mutations that affected nonstop gene expression without stabilizing nonstop mRNA levels implicated the proteasome. This finding not only suggested that the proteasome may degrade proteins encoded by nonstop mRNAs, but also supported previous observations that rapid decay of nonstop mRNAs cannot fully explain the lack of the encoded proteins. Further, we show that the proteasome and Ski7p affected expression of nonstop reporter genes independently of each other. In addition, our results implicate inositol 1,3,4,5,6-pentakisphosphate as an inhibitor of nonstop mRNA decay.
MESSENGER RNA turnover is an important process that many organisms use to control gene expression. In many instances, changes in mRNA decay rates modulate gene expression in response to a variety of signals. Short-lived mRNAs also allow rapid adjustments to steady-state RNA levels after up- or downregulation of transcription.
Studies using the budding yeast Saccharomyces cerevisiae as a model system have identified two general pathways that degrade mRNAs. These two pathways are conserved in most, if not all, eukaryotes. Normally, the major deadenylase, Ccr4p, gradually removes the poly(A) tail and initiates mRNA degradation (Shyu et al. 1991; Muhlrad and Parker 1992; Tucker et al. 2001). This triggers two deadenylation-dependent decay pathways. One pathway involves removal of the 5′-cap (decapping) by Dcp1p and Dcp2p (Decker and Parker 1993; Hsu and Stevens 1993; Muhlrad et al. 1995; Beelman et al. 1996; Dunckley and Parker 1999; Steiger et al. 2003). Decapping the transcript allows its degradation from the 5′-end by Xrn1p, a 5′–3′ exoribonuclease (Larimer et al. 1992; Hsu and Stevens 1993; Muhlrad and Parker 1994). In the second pathway, the transcript body is degraded from the 3′-end by a 3′–5′ exoribonuclease complex: the exosome (Muhlrad et al. 1995; Jacobs Anderson and Parker 1998). Although all mRNAs appear to be degraded by these two pathways, the rate at which individual steps occur can vary widely depending on the mRNA. However, it is currently not known which mechanisms target this basal degradation machinery preferentially to some mRNAs.
In addition to affecting the expression of normal cellular genes, mRNA turnover also is important as a quality control mechanism to maintain the overall fidelity of gene expression. Eukaryotes have evolved intricate mechanisms for gene expression. These intricacies introduce not only potential points of gene regulation, but also potential errors in the form of aberrant mRNAs. While many mechanisms exist to ensure high fidelity of gene expression, errors can occur that lead to aberrant mRNAs. Hence, specialized mRNA turnover pathways, termed mRNA surveillance, degrade these aberrant mRNAs. mRNA surveillance prevents accumulation of aberrant, dominant-negative, or truncated proteins that may cause harmful effects (Pulak and Anderson 1993).
Interestingly, the same enzymes degrade normal and aberrant transcripts. Transcripts containing premature stop codons, retained introns, or extended 3′-UTRs are all targets for the nonsense-mediated decay pathway (Zaret and Sherman 1984; He et al. 1993; Muhlrad and Parker 1994, 1999). Rapid degradation of nonsense transcripts bypasses deadenylation and instead triggers rapid decapping (Muhlrad and Parker 1994). Similarly, the exosome, independently of prior deadenylation, degrades transcripts that lack all in-frame termination codons, i.e., nonstop transcripts (Frischmeyer et al. 2002; van Hoof et al. 2002). Thus, understanding the molecular mechanisms that are responsible for the rapid decay of aberrant transcripts may provide insight into how the mRNA decay machinery targets some mRNAs preferentially.
Nonstop mRNAs arise in various ways. One source is premature polyadenylation due to inaccurate 3′-end processing events or cryptic polyadenylation signals within the coding region of the transcript (Mayer and Dieckmann 1991; Sparks and Dieckmann 1998; Frischmeyer et al. 2002). Mutations or errors in transcription that cause a change in the normal stop codon are other mechanisms that produce nonstop transcripts.
It is estimated that ∼30% of all human disease alleles generate a nonsense transcript (Frischmeyer and Dietz 1999). While alleles encoding nonstop transcripts have not been studied in similar detail, generation of a nonstop transcript can indeed result in disease. Mutation of the stop codon in the human adenine phosphoribosyltransferase (APRT) gene leads to 2,8-dihydroxyadenine urolithiasis (Taniguchi et al. 1998). Similarly, mutation in the normal stop codon of a G-protein-coupled receptor gene that regulates puberty (GPR54) causes hypogonadotrophic hypogonadism and sterility in affected individuals (Seminara et al. 2003). In both cases, the nonstop mutation leads to reduced levels of the nonstop mRNA and the encoded protein. Importantly, in hypogonadotrophic hypogonadism, overexpression of the nonstop GPR54 transcript can produce a functional protein. This observation suggests that partial inhibition of the nonstop mRNA decay machinery in these patients may prove to be beneficial.
In the current model for nonstop mRNA decay, the ribosome continues translation to the end of the poly(A) tail of nonstop transcripts (van Hoof et al. 2002). Upon reaching the end of the transcript, the ribosome stalls. This stalled ribosome is thought to be recognized by the C-terminal domain of the Ski7p. This hypothesis is based on the similarity of this domain to eEF1A and eRF3, which are known to interact with the ribosome during translation elongation and termination, respectively (Benard et al. 1999; van Hoof et al. 2002). Consistent with this hypothesis, this C-terminal domain is necessary for nonstop mRNA decay, but not for other exosome functions (van Hoof et al. 2002). In contrast, the N-terminal domain of Ski7p physically interacts not only with the exosome, but also with a complex of Ski2p, Ski3p, and Ski8p (Araki et al. 2001). This interaction is thought to recruit the exosome to the nonstop mRNA–ribosome complex, resulting in degradation of the nonstop mRNA (van Hoof et al. 2002).
Recently, it has been shown that proteins encoded by several nonstop reporters fail to accumulate, which cannot be fully explained by nonstop mRNA degradation (Inada and Aiba 2005; Ito-Harashima et al. 2007). This suggests that additional mechanisms exist by which nonstop mRNAs are downregulated. To address the possibility that there may be additional factors required for exosome-mediated nonstop mRNA degradation and to identify factors required for any other aspects of nonstop mRNA metabolism, we used a genomic screen in S. cerevisiae. Here, we show that, in addition to the Ski7p, Ski2p, Ski3p, Ski8p, and the exosome, there are indeed additional trans-acting factors that are required for the efficient recognition or degradation of nonstop mRNA transcripts. Additionally, we provide evidence that the proteasome degrades the translated nonstop protein, which may explain why the nonstop protein fails to accumulate.
MATERIALS AND METHODS
Plasmids:
Plasmid pAV188 has been described previously (van Hoof et al. 2002). It contains a his3-nonstop reporter gene, a URA3 selectable marker, and a centromere. Plasmid pAV240 is identical to pAV188 except that it contains a LEU2 selectable marker gene instead of URA3. pAV240 was created by digesting pAV188 with BamHI and SacI to isolate the his3-nonstop reporter. The digested his3-nonstop reporter was cloned into the BamHI and SacI site of pRS415 (Sikorski and Hieter 1989). Plasmid pAV184 contains a Protein A-nonstop reporter gene under control of the GAL1 promoter and with a PGK1 3′-UTR. It was created by PCR amplifying the PGK1–nonstop 3′-UTR with oligonucleotides oRP1073 (cgacgggatccggtaaggaattgccaggtgtt) and oRP1074 (ggccagtgccaagctttaacg) from the PGK1-nonstop plasmid described by Frischmeyer et al. (2002). The resulting PCR product was digested with BamHI and HindIII and cloned into the BamHI and HindIII sites of pAV182. Plasmid pAV185 contains a Protein A (with a stop codon) reporter gene under the control of the GAL1 promoter with a PGK1 3′-UTR. It was created by the same method used to create plasmid pAV184 with the exception that wild-type PGK1 3′-UTR was used for PCR amplification. The pRS416 plasmid has been previously described (Sikorski and Hieter 1989). pAV182 was obtained from Rhett Michelson and Ted Weinert (University of Arizona). pAV182 is a derivative of pMOV with two Z domains of Protein A under the control of the GAL1 promoter (Lydall and Weinert 1997).
Transformation and mutant screen:
To identify additional trans-acting factors in nonstop mRNA metabolism, we obtained the yeast deletion collection from Open Biosystems and transformed each individual strain with pAV188. Transformation was carried out by a modified version of a previously described protocol (Gietz and Woods 2002). Briefly, cells were grown on a YPD plate and transferred to a 96-well plate containing 10 μg of carrier DNA and 0.5 μg of pAV188 in a total volume of 10 μl. Next, 150 μl of PLATE solution was added (40% PEG 3350, 0.1 m lithium acetate, 10 mm TRIS–HCl, pH 8.0, 1 mm EDTA) and the plate was vortexed and incubated at room temperature (1 hr to overnight). Cells were heat-shocked for 15 min at 42°, pelleted, resuspended in 10 μl water, spotted on SC–URA, and incubated for 5 days at 30° to select for transformants. Transformants were then replica plated onto SC–HIS and incubated for 3 days at 30° to identify genes that suppress the his3-nonstop phenotype. Most strains yielded URA+ transformants on the first attempt. For those strains where the first transformation failed, a second attempt to transform was made. Overall, 99% of strains were successfully transformed.
Rescreen by serial dilution:
Strains that suppressed the his3-nonstop phenotype were rescreened by individually retransforming these strains with the his3-nonstop reporter using a standard yeast transformation protocol. Single colonies were picked and restreaked onto SC–URA to be used for serial dilutions.
Serial dilutions were done by growing liquid cultures of transformants in SC–URA overnight at 30°. The following day, cells were diluted in SC–URA to a starting OD of 0.2. Cultures were grown until they reached an OD of 0.8. Cells were serially diluted in 96-well plates by a factor of 5 and spotted onto SC–HIS. These plates were then incubated for 3 days at 30° to qualitatively assay growth relative to wild-type and ski7Δ mutants. These experiments were repeated in triplicate.
Confirmation that the His+ phenotype is linked to the deletion:
To ensure that the suppression of his3-nonstop was indeed caused by the annotated deletion, we PCR amplified the disrupted gene from the knockout strain, using primers 500 nt on either side of the open reading frame (ORF) (primer sequences available upon request). The resulting PCR products were used to knock out the genes in BY4741. Although similar analysis on >30 strains indicated that the right gene had indeed been deleted, we identified two strains that were mislabeled in the collection obtained from Open Biosystems. The two knockouts that were mislabeled were identified by PCR amplifying and sequencing of the “molecular barcodes” included in the knockout cassettes. We also identified three cases in which the his3-nonstop suppression was not recreated, presumably because the phenotype of the knockout strain was caused by an unlinked mutation.
Stability of Protein A-nonstop mRNA:
To determine the half-life of the Protein A-nonstop mRNA reporter, each strain was transformed with pAV184. Transformants were grown overnight in 20 ml of SC–URA+2% galactose to induce expression of the Protein A-nonstop reporter. The following day, strains were diluted in 50 ml of SC–URA+2% galactose to a starting OD of 0.2 and grown to a final OD of 0.8. Cells were then pelleted and resuspended in 20 ml of SC–URA (no sugar). A 2-ml sample from each strain was taken and pelleted and stored immediately on dry ice. The remaining liquid culture was incubated on a shaker at 30°. A total of 2 ml of 40% dextrose was added to each strain and 2-ml samples were taken (as above) at the 1-, 2-, 3-, 4-, 6-, 8-, 10-, 15-, 30-, and 60-min time points. Next, RNA was isolated from each sample and Northern blot analysis was performed.
Stability of Protein A-nonstop protein:
To determine the half-life of the Protein A-nonstop protein, wild-type (yAV670) and proteasome-defective (pre9Δ, yAV720) yeast strains transformed with pAV184 were grown to midlog phase in media containing galactose. At this point, transcription and translation were terminated by replacement with media containing 4% glucose and 100 μg/ml cycloheximide, respectively. Aliquots were taken at the times indicated and total protein was isolated. Western blot analysis was performed with antibodies specific for Protein A (Sigma, St. Louis) and Pgk1p (Molecular Probes, Eugene, OR). Signals were detected by chemiluminescence (Amersham, Piscataway, NJ), scanned using a Phosphoimager (Amersham), and quantitated using ImageQuant software.
Creation of the ski7Δ pre9Δ and ipk1Δ ipk2Δ double mutant:
yAV987 (ski7Δ∷HygMX4) (Table 1) and yAV1052 (ipk1Δ∷HygMX4) were created as previously described (Goldstein and McCusker 1999). yAV720 (pre9Δ∷KanMX4) was mated with yAV987, and yAV1054 (ipk2Δ∷KanMX4) was mated with yAV1052. Haploid progeny spores were obtained by the hydrophobic spore isolation method essentially as described (Rockmill et al. 1991) and plated on YPD. Double-mutant strains were identified by replica plating on YPD+geneticin and YPD+hygromycin.
TABLE 1.
Strains used
| Strain | Genotype |
|---|---|
| BY4741 | MATa, his3Δ1, leu2Δ0, ura3Δ0, met15Δ0 |
| BY4742 | MATα, his3Δ1, leu2Δ0, ura3Δ0, lys2Δ0 |
| yAV760 | MATα, his3Δ1, leu2Δ0, ura3Δ0, met15Δ0, lys2Δ0, dcp2-7∷URA3, can1Δ∷MFA1-HIS3∷MFα1-LEU2 |
| yAV747 | MATa, his3Δ1, leu2Δ0, ura3Δ0, met15Δ0, lys2Δ0, dcp2-7∷URA3 |
| yAV987 | MATα, his3Δ1, leu2Δ0, ura3Δ0, lys2Δ0, ski7Δ∷HYG |
| yAV996 | MATα, his3Δ1, leu2Δ0, ura3Δ0, lys2Δ0, ski7Δ∷HYG, pre9∷NEO |
| yAV1052 | MATa, his3Δ1, leu2Δ0, ura3Δ0, met15Δ0, ipk1Δ∷HYG |
| yAV1053 | MATα, his3Δ1, leu2Δ0, ura3Δ0, met15Δ0, ipk1Δ∷HYG, ipk2∷NEO |
| yAV1054 | MATα, his3Δ1, leu2Δ0, ura3Δ0, lys2Δ0, ipk2∷NEO |
Creation of dcp2-7ts double mutants:
Although DCP2 is annotated as an essential gene (http://www.yeastgenome.org), this annotation is incorrect. This conclusion is based on our unpublished observation that the heterozygous diploid dcp2Δ strain 22958 (Open Biosystems) gives two wild-type and two very slow-growing dcp2Δ spores per tetrad. To introduce the dcp2-7ts temperature-sensitive allele into the same genetic background as the knockout collection, strain 22958 was transformed with pRP989 (Dunckley and Parker 2001). The resulting URA+ geneticin-sensitive strain was sporulated to yield yAV747 (Table 1). Strain yAV747 was crossed with Y3656 (Tong et al. 2001) to give yAV760.
To create yeast deletions strains that also contained a temperature-sensitive mutation in the decapping machinery, we mated the yeast deletion strains with strain yAV760. Haploid progeny spores were obtained by the hydrophobic spore isolation method essentially as described by Rockmill et al. (1991) and plated on CSM –Arg –Ura –His plus canavanine at 23° to select for MATa dcp2-7ts progeny. MATa dcp2-7ts progeny were then replica plated to YPD+geneticin to identify progeny that also contained the deletion of interest.
To determine whether the strains that we identified in our screen control exosome function, strains that were dcp2-7ts and deleted for an ORF of interest were grown in YPD overnight at 23°. The following day, cells were diluted to an OD of 0.2 and grown to an OD of 0.8. Cells were then serially diluted in 96-well plates by a factor of 5 and spotted onto YPD media plates and grown for 3 days at 23°, 30°, and 37°. These experiments were done in triplicate.
Other methods:
Western and Northern blotting were done according to standard methods. Western blots were probed with an antibody against Protein A (Sigma) or the loading control Pgk1p (molecular probes). Northern blots were probed for Protein A using oligo oAV72 (tctactttcggcgcctgagcatcattt) and for the 7S RNA subunit of the signal recognition particle using oRP100 (gtctagccgcgaggaagg).
RESULTS
A genomic screen identifies mutants that suppress a his3-nonstop mutation:
Nonstop mRNA decay is not an essential process in yeast (van Hoof et al. 2002). In addition, mutations that inactivate nonstop mRNA decay partially suppress a his3-nonstop allele. Specifically, van Hoof et al. (2002) showed that a wild-type strain containing a his3-nonstop allele is auxotrophic for histidine. However, a ski7Δ mutant containing the same his3-nonstop allele is no longer auxotrophic for histidine, presumably because the his3-nonstop mRNA is stabilized and produces enough His3p for histidine biosynthesis (van Hoof et al. 2002). With this knowledge, and to expand our understanding of nonstop mRNA decay, a genetic screen utilizing a deletion collection of almost 5000 nonessential open reading frames in S. cerevisiae was used to identify additional genes involved in the nonstop mRNA decay pathway. These strains contained null mutations in the URA3 and HIS3 genes and a complete deletion in a nonessential open reading frame. Each strain from the collection was individually transformed with a plasmid containing a selectable URA3 gene and a his3-nonstop reporter. Transformants were selected by growth in the absence of uracil and replica plated onto media lacking histidine. Potential genes involved in nonstop mRNA decay were identified on the basis of the ability of the mutant to grow on media lacking histidine. To eliminate false positives, we restreaked each strain on media lacking uracil and selected single colonies. These strains were placed in 96-well plates with fivefold serial dilutions and plated onto media lacking histidine or uracil. The transformation was then repeated using a high-efficiency transformation protocol to eliminate additional false positives. This screen yielded a number of mutants that reproducibly suppressed the his3-nonstop phenotype to varying extents. Here, we concentrated on the mutants that increased his3-nonstop expression approximately as much as a ski7Δ mutant. In addition to these, we isolated several mutants that showed small increases in growth, but grew significantly slower than the ski7Δ control (data not shown).
The his3-nonstop suppression phenotype is tightly linked to the deletion:
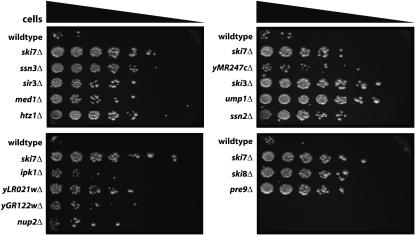
During creation and maintenance of the deletion collection, some strains may have been mislabeled or may have accumulated unlinked mutations (e.g., Hughes et al. 2000). Therefore, to ensure that the observed his3-nonstop suppression phenotype was indeed caused by the deletion, we recreated each of the newly identified mutants in the wild-type strain BY4741. As controls, we included the ski3Δ, ski7Δ, and ski8Δ mutants. Indeed, this analysis identified two strains that were mislabeled in the knockout collection and three strains where the original his3-nonstop suppression phenotype was not tightly linked to the deletion. Presumably, these latter three strains from the collection contain an unlinked mutation that suppressed the his3-nonstop phenotype. However, for 15 genes that were initially identified in our screen, the his3-nonstop suppression phenotype was indeed linked to their deletion mutants (Figure 1 and Table 2).
Figure 1.—
Isolation of deletion mutants of S. cerevisiae that suppress a his3-nonstop reporter. Cells containing a his3-nonstop reporter as the only source of His3p grow slowly on plates lacking histidine. The yeast deletion collection was transformed with a his3-nonstop reporter. To assay suppression of the his3-nonstop phenotype, each of the indicated strains was serially diluted and spotted on media lacking histidine.
TABLE 2.
Genes that suppress the nonstop phenotype
| Protein A-nonstop
|
|||||
|---|---|---|---|---|---|
| Gene | Function/putative function | his3-nonstop | Steady state | Stability | dcp synthetic lethal |
| SKI2 | Exosome-mediated mRNA degradation | ++ | ND | ND | ND |
| SKI3 | Exosome-mediated mRNA degradation | ++ | 2.0 | 2.7 | + |
| SKI7 | Exosome-mediated mRNA degradation | ++ | 2.7 | 2.5 | + |
| SKI8 | Exosome-mediated mRNA degradation | ++ | 2.1 | 2.5 | + |
| PRE9 | Proteasome-mediated protein degradation | ++ | 0.4 | ND | ND |
| UMP1 | Proteasome-mediated protein degradation | ++ | 0.4 | ND | ND |
| yMR247c | Proteasome-mediated protein degradation | +++ | 0.6 | ND | ND |
| SSN2 | Subunit of the RNA polymerase II mediator; interacts with mRNA deadenylase | ++ | 2.6 | 1.1 | − |
| SSN3 | Kinase of the RNA polymerase II mediator; interacts with mRNA deadenylase | ++ | 2.3 | 1.5 | − |
| MED1 | Subunit of the RNA polymerase II mediator | ++ | 1.5 | 1.2 | − |
| IPK1 | mRNA export, RNA editing, inositol phosphate kinase | ++ | 3.0 | 1.6 | − |
| HTZ1 | Variant of histone H2 | ++ | 2.0 | 1.1 | + |
| SIR3 | Gene silencing | ++ | 1.7 | 1.4 | ND |
| yGR122w | Unknown | ++ | 1.5 | ND | ND |
| NUP2 | Subunit of the nuclear pore complex | ++ | 2.1 | 0.9 | + |
| yLR021w | Unknown | ++ | 0.5 | 1.7 | + |
| Wild type | — | − | 1.0 | 0.9 | − |
His3-nonstop growth is indicated as −, indicating little or no growth; ++, indicating significant growth; and +++, indicating growth that was even more pronounced. dcp2-7 synthetic lethality is indicated with a + sign and − sign indicates no synthetic growth defect. ND, not determined.
Many his3-nonstop suppressing mutations also increase the abundance of a Protein A-nonstop mRNA:
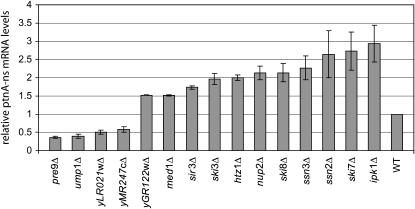
There are at least two explanations for the suppression of the his3-nonstop phenotype in the mutants that we identified. One possibility is that the phenotype is specific to the HIS3 gene. For example, any mutation that dramatically increases transcription from the HIS3 promoter could result in histidine autotrophy. A second possibility is that the phenotype that we observed may be due to a general defect in the nonstop mRNA degradation pathway. In this latter case, expression of all nonstop mRNAs in the cell would be increased in the mutant. To distinguish between these two possibilities, we analyzed the effect of the mutations on a second nonstop reporter. This Protein A-nonstop reporter shares no sequence homology with the his3-nonstop gene. Specifically, the his3-nonstop reporter contains the HIS3 promoter, HIS3 coding region, and HIS3 3′-UTR, while the Protein A-nonstop reporter contains the GAL1 promoter, Protein A-coding region, and the PGK1 3′-UTR. After transforming each one of our deletion mutants with the Protein A-nonstop reporter, RNA was isolated and Protein A-nonstop mRNA levels were analyzed relative to wild type and ski7Δ controls (Figure 2). This experiment showed that deletion of 11 of the 15 mutants tested also increased the abundance of our Protein A-nonstop mRNA (Figure 2 and data not shown). Importantly, these data showed that suppression in these 11 strains was not specific to the his3-nonstop reporter, but was most likely due to a general defect in the nonstop mRNA decay pathway.
Figure 2.—
Many of the genes that suppress the his3-nonstop phenotype also increase mRNA levels of a second nonstop mRNA reporter. Strains isolated as suppressors of his3-nonstop were transformed with a plasmid containing a Protein A-nonstop reporter. The steady-state level of Protein A-nonstop mRNA was assayed by Northern blot. Shown is the average and the spread of two independent experiments after correcting for loading difference using a probe for the RNA subunit of the signal recognition particle (7S RNA). The mRNA level is expressed as a value relative to that found in an isogenic wild-type strain.
Mutations implicating the proteasome increase Protein A-nonstop protein, but without a corresponding increase in mRNA levels:
The above analysis indicated that four of the deletion mutant strains increased his3-nonstop expression but not Protein A-nonstop mRNA abundance. Intriguingly, three of these four genes are implicated in proteasome function, which suggests that the proteasome may normally degrade the his3-nonstop protein. Pre9p is a proteasome β-type subunit. It is the only nonessential component of the 20S proteasome and thus the only 20S proteasome subunit in the deletion collection (Emori et al. 1991; Giaever et al. 2002). Ump1p is a short-lived chaperone that is involved in the maturation of the proteasome (Ramos et al. 1998). YMR247C encodes a putative ubiquitin-conjugating enzyme that copurifies with the 19S regulatory subunit of the proteasome (Verma et al. 2000; Braun et al. 2007). One possible reason that deletions in the PRE9, UMP1, and YMR247C genes suppressed the his3-nonstop phenotype is that the protein encoded by the his3-nonstop mRNA is normally rapidly degraded by the proteasome. In this instance, suppression of the his3-nonstop phenotype would not be due to increased stability of the mRNA, but to increased stability of the protein.
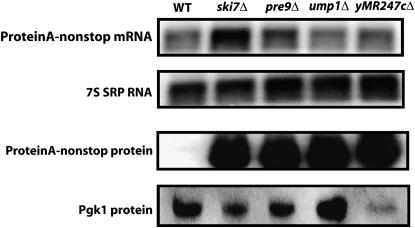
To further characterize the effects of the PRE9, UMP1, and YMR247C genes on protein levels encoded from nonstop reporter transcripts, we examined the levels of Protein A-nonstop protein. Interestingly, far more Protein A-nonstop protein was present in extracts made from the pre9Δ, ump1Δ, and ymr247cΔ mutants compared to extracts from wild-type cells (Figure 3). This occurred despite the relatively low nonstop mRNA levels in these mutants. Therefore, mutations affecting proteasome function most likely suppressed the his3-nonstop phenotype because they stabilize the His3-nonstop protein.
Figure 3.—
Mutations implicating the proteasome increase the steady-state protein level of a nonstop reporter, without increasing the steady-state mRNA level. (Top) Total RNA was isolated from yeast strains, and Protein A-nonstop mRNA levels were analyzed by Northern blot. The RNA subunit of the signal recognition particle (7S RNA) served as a loading control. (Bottom) Protein was isolated from yeast strains expressing Protein A-nonstop protein and analyzed by Western blot. Pgk1p served as a loading control.
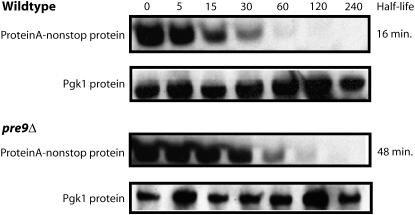
From the above analysis, it is not entirely clear when the proteasome degrades the protein encoded by Protein A-nonstop mRNA. One possibility is that the proteasome degrades the protein cotranslationally, while a second possibility is that the encoded protein is degraded after synthesis is completed. To distinguish between these two possibilities, we measured the degradation rate of fully synthesized Protein A encoded by a nonstop mRNA in wild-type and pre9 cells. Cells were grown in the presence of galactose, and then glucose and cycloheximide were added to prevent further synthesis of Protein A. Analysis of the levels of Protein A at various times after this treatment showed that decay from the steady-state pool was relatively rapid in wild-type cells, but approximately threefold slower in pre9 cells (Figure 4). Therefore, the pre9 deletion affects the decay of Protein A out of the steady-state pool. We conclude that Pre9p, and presumably Ump1p and yMR247Cp, affect the post-translational degradation of the protein encoded by the Protein A-nonstop reporter.
Figure 4.—
The pre9 deletion increases the stability of the protein encoded by the Protein A-nonstop reporter. Wild-type and pre9Δ strains were transformed with a Protein A-nonstop reporter under the control of a galactose-inducible promoter. Expression of the reporter was repressed by the addition of glucose and cycloheximide. The stability of the Protein A-nonstop protein was determined by Western blot with antibodies specific to Protein A. Pgk1p served as a loading control.
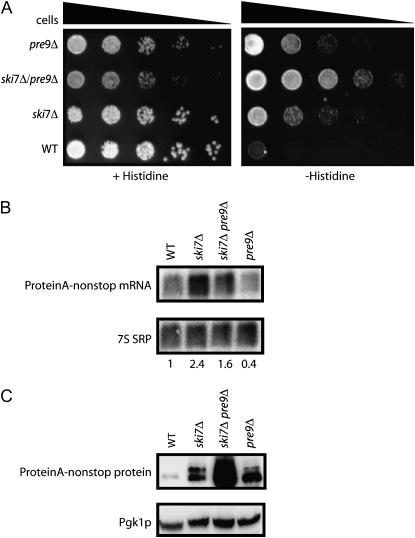
If ski7Δ suppressed the his3-nonstop phenotype by inactivating the cytoplasmic exosome and stabilizing the nonstop mRNA, and pre9Δ suppressed his3-nonstop phenotype by inactivating the proteasome and stabilizing the nonstop protein, a pre9Δ ski7Δ double mutant might exhibit an additive effect on the suppression phenotype. This was indeed the case since the ski7Δ pre9Δ double mutant grew better in the absence of histidine than either the ski7Δ or pre9Δ single mutants when all contained the his3-nonstop reporter (Figure 5A). We also noted that the ski7Δ pre9Δ double mutant grew slower in the presence of histidine than either single mutant. This genetic interaction was consistent with the hypothesis that SKI7 and PRE9 genes may act in pathways that are functionally related. In addition, Northern and Western blot analysis of the ski7Δ pre9Δ double mutant showed that while Protein A-nonstop mRNA levels are not significantly increased, it accumulated more Protein A-nonstop protein than either single mutant alone (Figure 5, B and C). We conclude that ski7Δ and pre9Δ suppressed the his3-nonstop phenotype by independent mechanisms (see discussion).
Figure 5.—
The effects of ski7Δ and pre9Δ on nonstop gene expression are additive. (A) Yeast strains containing mutations in the exosome (ski7Δ), proteasome (pre9Δ), or the exosome and proteasome (ski7Δ, pre9Δ) were transformed with a his3-nonstop reporter, serially diluted, and spotted on media without histidine to assay for suppression of the his3-nonstop phenotype. The same strains were transformed with a Protein A-nonstop reporter and analyzed by Northern blotting (B) and Western blotting (C).
Many his3-nonstop suppressing mutations decrease the rate of Protein A-nonstop mRNA decay:
While our screen was designed to isolate mutants that stabilize the his3-nonstop mRNA, other types of mutants might also suppress the his3-nonstop phenotype. For example, mutants that increase transcription from the HIS3 promoter could also result in higher levels of his3-nonstop mRNA and thus in increased growth in the absence of histidine. This latter possibility is especially relevant for three of the mutants identified in our screen that have previously been implicated in transcription: ssn2, ssn3, and med1. These genes have been implicated in both transcriptional activation and repression of a variety of genes, suggesting that they might increase his3-nonstop expression by increasing the strength of the HIS3 promoter (Kelleher et al. 1990; Flanagan et al. 1991; Thompson et al. 1993). On the other hand, Ssn2p and Ssn3p interact with the CCR4/NOT complex, which is the major cytoplasmic deadenylation complex, suggesting that they might have a direct role in mRNA decay (Liu et al. 2001; Tucker et al. 2001; Huh et al. 2003).
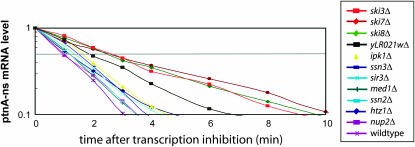
We directly measured Protein A-nonstop mRNA decay rates to distinguish between increased transcription and increased mRNA stability in all mutants that did not implicate proteasome function. In this experiment, mRNA was isolated at various time points after transcription of the Protein A-nonstop reporter was repressed by the addition of glucose and the half-life of the transcript was determined by Northern blot analysis. These measurements were repeated three or four times and the average is plotted in Figure 6. As expected, the decay rate of Protein A-nonstop mRNA was about threefold slower in ski3Δ, ski7Δ, and ski8Δ strains than in wild-type cells. Interestingly, the eight other mutants isolated in our genomic screen also increased the stability of the Protein A-nonstop mRNA, although to a lesser extend than the ski deletions. The largest increase in Protein A-nonstop half-life (approximately twofold) was observed for the deletion of yLR021w, an uncharacterized open reading frame of unknown function. These data are consistent with the hypothesis that the deletions that we identified indeed increased the half-life of nonstop reporter mRNAs rather than increased transcription.
Figure 6.—
Mutants that increase steady-state levels of the Protein A-nonstop mRNA also increase stability of the mRNA. Each of the indicated strains was transformed with a plasmid directing expression of the Protein A-nonstop reporter from a GAL1 promoter. mRNA stability was assayed by inhibiting transcription from the GAL1 promoter with the addition of glucose and isolating RNA at the indicated time points. The mRNA remaining at various points after transcription inhibition was quantitated. Plotted is the average mRNA remaining from three independent experiments for each mutant and from four independent experiments for the wild type.
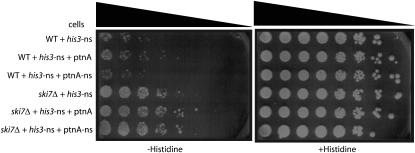
One possible explanation for the increased stability of the Protein A-nonstop reporter in the mediator mutants is that these mutants increase transcription from the nonstop reporter and that this increased transcription saturates the nonstop mRNA decay machinery, in turn leading to an increased mRNA stability. To determine whether or not we could saturate the nonstop mRNA decay pathway, we introduced the Protein A-nonstop reporter, which is expressed from the strong GAL1 promoter into a strain that also contains a his3-nonstop reporter expressed from the HIS3 promoter. As a control, we used a plasmid that encodes a normal Protein A mRNA from the GAL1 promoter. The production of Protein A-nonstop mRNA did not affect the growth phenotype of the his3-nonstop mutation (Figure 7). We conclude that the nonstop mRNA decay pathway is not easily saturated. Therefore, these data indicate that the increased expression of his3-nonstop and Protein A-nonstop in mediator mutants is unlikely to result from the saturation of the nonstop mRNA decay machinery.
Figure 7.—
Overexpression of Protein A-nonstop does not saturate the nonstop mRNA decay pathway. To determine if the nonstop mRNA decay pathway could easily be saturated, wild-type and ski7Δ strains were transformed with plasmids expressing his3-nonstop from the HIS3 promoter and either Protein A-nonstop expressed from the GAL1 promoter or the indicated control plasmids. Strains were serially diluted and spotted on media containing 2% galactose as the sole carbon source and either lacking or containing histidine.
The accumulation of inositol 1,3,4,5,6-pentakisphosphate suppresses his3-nonstop:
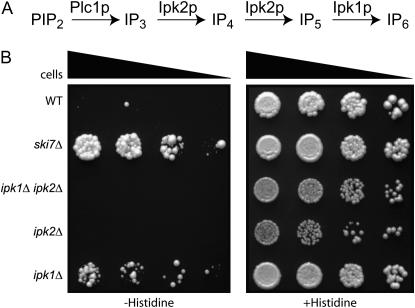
One of the deletions that suppressed the phenotype of his3-nonstop was ipk1Δ. The IPK1 gene encodes the enzyme inositol 1,3,4,5,6-pentakisphosphate 2-kinase that produces inositol 1,2,3,4,5,6-hexakisphosphate (IP6) from inositol 1,3,4,5,6-pentakisphosphate (IP5) (York et al. 1999). IP6 has known roles in regulating RNA metabolism. Yeast mutants lacking IP6 accumulate poly(A)+ RNAs in the nucleus and have defects in tRNA modification (York et al. 1999; Macbeth et al. 2005). To investigate whether the role of Ipk1p in his3-nonstop suppression was related to these previously known functions of Ipk1p, we analyzed other mutants lacking IP6. IP6 production is a four-step metabolic pathway (Figure 8A) that also requires phospholipase C (Plc1p) and Ipk2p. Strikingly, plc1Δ and ipk2Δ mutations were not identified in our genomic screen and, upon direct testing, had no effect on his3-nonstop expression. In contrast, all other known defects of ipk1Δ mutants are shared with ipk2Δ and plc1Δ. Thus, the effect of ipk1Δ on his3-nonstop expression is not due to a lack of IP6.
Figure 8.—
IP5 affects the expression of his3-nonstop. (A) Ipk1p catalyzes the conversion of IP5 to IP6 and acts downstream of Plc1p and Ipk2p. (B) ipk2Δ and ipk1Δ, ipk2Δ do not suppress the his3-nonstop phenotype. ipk1Δ, ipk2Δ, and ipk1Δ ipk2Δ deletion strains were transformed with a his3-nonstop reporter. To assay suppression of the his3-nonstop phenotype, each of the indicated strains was serially diluted and spotted on media lacking or containing histidine.
A second possible mechanism by which ipk1Δ affects his3-nonstop expression is that the mutant accumulates IP5, which may inhibit nonstop mRNA decay (York et al. 1999). This hypothesis predicts that Ipk1p can no longer affect his3-nonstop expression if IP5 accumulation is prevented by a mutation in IPK2. A third possibility is that Ipk1p is a bifunctional protein with completely separate roles in IP6 production and his3-nonstop suppression. Under this hypothesis, we predict that Ipk1p can still affect his3-nonstop expression in an ipk2 strain. To distinguish between these latter two possibilities, we tested suppression of the his3-nonstop phenotype in an ipk1Δ ipk2Δ double mutant (Figure 8). We observed that the double mutant does not suppress the his3-nonstop phenotype. Therefore, these results suggest that the suppression phenotype that we observed in an ipk1Δ strain is a result of the accumulation of IP5 and not a second, unrelated function of Ipk1p. To our knowledge, this is the first result that implicates IP5 as a regulatory molecule.
Some mutations affect nonstop mRNA decay without affecting other cytoplasmic exosome functions:
Mutations such as a ski7Δ that inactivate the cytoplasmic exosome stabilize nonstop mRNAs (van Hoof et al. 2002). Thus, at least two classes of mutants might be expected in our screen. One class would have defects in cytoplasmic exosome function (e.g., ski7Δ), while a second class of mutants might have specific defects in the recognition of nonstop mRNAs (e.g., ski7ΔC; van Hoof et al. 2002). Mutations disrupting cytoplasmic exosome function exhibit synthetic lethality with decapping defects, while nonstop mRNA recognition mutants would not be expected to exhibit a genetic interaction with decapping defects (Johnson and Kolodner 1995; Jacobs Anderson and Parker 1998; van Hoof et al. 2002). We therefore tested whether the new mutants that we identified were synthetically lethal with a decapping defect.
Each of the mutants was crossed with a temperature-sensitive decapping mutant (dcp2-7ts), and double mutants were isolated. At 37°, the dcp2-7ts allele inactivates the decapping enzyme and thereby inactivates the 5′–3′ mRNA degradation pathway. Importantly, dcp2-7ts strains are still able to grow at 37°, because the alternative 3′–5′ decay pathway is intact and sufficient for viability. However, when the dcp2-7ts allele is combined with a mutation inactivating the 3′–5′ decay pathway, both pathways are nonfunctional at 37°, and therefore such a strain cannot grow at 37°. Thus, an inability to grow at the nonpermissive temperature (37°) would suggest that the gene is required for general exosome-mediated decay of mRNAs. Strikingly, most of the genes that we identified in our genetic screen did not have growth defects at the nonpermissive temperature when combined with the decapping mutant (data not shown). This observation suggests that these genes are not required for general cytoplasmic exosome activity.
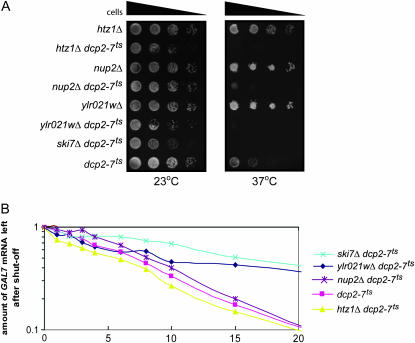
Although most of the genes tested did not show a synthetic lethal interaction with dcp2-7ts, three deletions did significantly reduce growth of the dcp2-7ts strain and thus may disrupt the activity of the exosome (i.e., nup2Δ, htz1Δ, and ylr021wΔ). As shown in Figure 9, at the nonpermissive temperature, these cells were either synthetically sick or lethal when combined with the decapping mutation. One explanation for these findings is that their function is not limited to nonstop mRNA decay, but that they have a more general function in cytoplasmic exosome function. To more directly determine whether NUP2, HTZ1, and YLR021W genes function in exosome-mediated decay of all mRNAs, we assayed their effects on stability of the GAL7 and GAL10 mRNAs. We grew the dcp2-7ts double mutants in YEP+galactose to induce expression of the GAL7 and GAL10 mRNAs. We then incubated the cells at 37° for 1 hr to inactivate the decapping enzyme and then added glucose to shut off transcription of the GAL7 and GAL10 mRNAs. Under these conditions, cytoplasmic mRNA decay is solely carried out by the exosome (Jacobs Anderson and Parker 1998). As expected, the GAL7 and GAL10 mRNAs were stabilized in the ski7Δ dcp2-7 double mutant, when compared to the dcp2-7 single mutant (Figure 9B). The nup2Δ, htz1Δ, and ylr021wΔ mutants did not have this same effect, suggesting that these three genes were not required for exosome-mediated decay of the GAL7 and GAL10 mRNAs. However, the ylr021wΔ mutant appeared to have a minor defect in that the GAL7 mRNA decayed with biphasic kinetics. Approximately half of the GAL7 mRNA decayed with normal kinetics (half-life of 8 min), while the other half was stabilized (half-life >20 min). While the significance of this biphasic decay is not understood, we conclude that these three genes are not required for all functions of the cytoplasmic exosome. Overall, analysis of dcp2-7ts double mutants suggests that we have identified a number of proteins that are required for exosome-mediated decay of his3-nonstop mRNA and Protein A-nonstop mRNA, but not for all exosome-mediated mRNA decay.
Figure 9.—
Mutations in Htz1p, Nup2p, and yLR021wp are synthetic lethal with decapping defects, but do not block exosome-mediated decay of normal mRNAs. Mutants that block cytoplasmic exosome function are synthetically lethal with defects in decapping. The majority of the genes isolated in our screen do not show synthetic lethality (not shown). (A) The htz1Δ, nup2Δ, and ylr021wΔ genes are synthetically lethal with a temperature-sensitive mutation that blocks decapping (dcp2-7), consistent with the possibility that they may have a more general effect on cytoplasmic exosome function. The indicated strains were serially diluted and spotted on YPD plates that were then incubated at 23° or 37°. At 23°, the decapping enzyme is functional and thus all strains grew. At 37°, the decapping enzyme is inactivated and defects in the cytoplasmic exosome affect growth. (B) To test whether the synthetic lethality shown in A was caused by a general block of cytoplasmic exosome function, the stability of the GAL7 mRNA was measured. The stability of the GAL7 mRNA was analyzed by growing cells at 25° in media containing galactose. The decapping enzyme was then inactivating by incubating the cells for 1 hr at 37°. Following the addition of glucose, total RNA was isolated from each strain at the time points indicated on the x-axis. The stability of the GAL7 mRNA was analyzed by Northern blotting. The 7S RNA was used as a loading control. Each time point represents the average of two experiments.
DISCUSSION
The nonstop mRNA decay pathway identifies and degrades aberrant transcripts that may encode proteins with the potential to cause deleterious effects in the cell. Therefore nonstop mRNA decay is a part of the cell's mRNA surveillance mechanisms that maintain the overall fidelity of gene expression. To better understand how nonstop mRNAs are removed from eukaryotic cells, we took a genetic approach in S. cerevisiae. Using the yeast deletion mutant collection, we identified known and unknown nonessential genes involved in nonstop mRNA metabolism. One measure of the completeness of this screen is that we identified all four nonessential genes known to be required for nonstop mRNA decay (i.e., SKI2, SKI3, SKI7, and SKI8). Importantly, most of the newly identified genes do not appear to be required for general cytoplasmic exosome functions and suggest that nonstop mRNA metabolism is more complex than previously known, involving roles in diverse functions like proteolysis and phosphoinositide signaling.
The one possible exception to the observation that the newly identified genes do not affect other exosome functions may be yLR021w. Unlike most of the newly identified genes, ylr021wΔ is synthetically lethal with decapping defects and has a small effect on the degradation of GAL7 mRNA. ylr021wΔ also had the largest effect on Protein A-nonstop mRNA stability and thus may encode a regulator of the cytoplasmic exosome. The function of yLR021w is completely uncharacterized; the protein encoded by yLR021w is not similar to any protein with a known function.
Our results suggest a role for genes encoding functions for the eukaryotic proteasome in nonstop mRNA surveillance. The proteasome was implicated three times in our genetic screen, which strongly suggests that it may be involved in degrading the his3-nonstop protein. The PRE9 gene encodes a subunit of the 20S proteasome and is the only nonessential proteasome gene (Emori et al. 1991; Giaever et al. 2002). The UMP1 gene encodes a chaperone required for 20S proteasome assembly (Ramos et al. 1998). The YMR247C ORF encodes a protein that copurifies with the proteasome and has very recently been identified as a RING domain containing ubiquitin-conjugating enzyme (Verma et al. 2000; Braun et al. 2007). Deletions in these genes do not cause increased abundance of nonstop transcripts but instead cause increased levels of the nonstop protein product. Consistent with our conclusion that the his3-nonstop protein is normally degraded by the proteasome, Ito-Harashima et al. 2007 very recently published that addition of eight or more lysine residues could target His3p to proteasome-mediated degradation and that this proteolysis contributes to the reduced expression of nonstop reporter genes.
In Eubacteria, the signal that identifies a transcript as nonstop is thought to arise from the stalled ribosome (Keiler et al. 1996; Ueda et al. 2002). When this occurs, an RNP composed of tmRNA and SmpB recognizes the stalled ribosome and this recognition adds a C-terminal peptide tag to the protein encoded by the nonstop mRNA (Keiler et al. 1996; Karzai et al. 1999; Hallier et al. 2004). The addition of this tag targets the protein encoded by the nonstop mRNA for rapid proteolysis. Our identification of three mutants that implicate the proteasome in the degradation of the his3-nonstop and Protein A-nonstop proteins suggests the possibility that eukaryotes may also actively recognize and degrade proteins encoded by nonstop mRNAs. There are several ways in which the proteins encoded by the his3-nonstop and Protein A-nonstop mRNAs might be targeted to the proteasome. Analogous to the prokaryotic system, the stalled ribosome at the end of an mRNA could target the encoded protein for degradation. However, two lines of evidence do not support this idea. First, Ski7p most likely plays a central role in recognizing the stalled ribosome, and analysis of a ski7Δ pre9Δ double mutant clearly shows that even in the absence of the Ski7p, the his3-nonstop and Protein A-nonstop proteins are still targeted to the proteasome (Figure 5C). Second, direct measurement of Protein A-nonstop protein stability indicates that Pre9p acts post-translationally, rather than cotranslationally (Figure 4). Future experiments are required to understand the physiological significance of the proteasome in degrading proteins encoded by nonstop mRNAs.
Another unexpected finding in our genetic screen is the identification of the IPK1 gene, which is involved in cellular signaling and nuclear transport. The IPK1 gene encodes the enzyme inositol 1,3,4,5,6-pentakisphosphate 2-kinase that produces IP6 from IP5. (York et al. 1999). Analysis of other mutants in the IP6 pathway implicated IP5 as an inhibitor of his3-nonstop expression. Most importantly, when IP5 production in the ipk1Δ strain was inhibited by also deleting IPK2, the his3-nonstop suppression was reversed. To our knowledge, this is the only known role of IP5. In contrast, IP6 has been implicated in several aspects of RNA metabolism: ipk1, ipk2, and plc1 mutants accumulate polyadenylated RNA in the nucleus, and thus IP6 may have a role in nuclear export of poly(A)+ mRNAs (York et al. 1999). Interestingly, exosome mutants also accumulate polyadenylated RNA in the nucleus, suggesting the possibility that both IP5 and IP6 regulate diverse exosome functions. In addition, IP6 is an important component of adenine deaminases that act on mRNA and tRNA (ADARs and ADATs, respectively), and mutants lacking IP6 have defects in tRNA modification (Macbeth et al. 2005). Overall, these results suggest that phosphoinositides might regulate diverse aspects of RNA processing and degradation.
Acknowledgments
We thank Roy Parker for his encouragement, helpful discussions, and suggestions during this work and Tom Vida for his critical reading of the manuscript. This work was funded by the National Institutes of Health (GM069900) and a PEW Scholarship in the Biomedical Sciences to A.V. and by an American Society for Microbiology Robert D. Watkins Fellowship to M.A.W.
References
- Araki, Y., S. Takahashi, T. Kobayashi, H. Kajiho, S. Hoshino et al., 2001. Ski7p G protein interacts with the exosome and the Ski complex for 3′- to 5′- mRNA decay in yeast. EMBO J. 20: 4684–4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield et al., 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382: 642–646. [DOI] [PubMed] [Google Scholar]
- Benard, L., K. Carroll, R. C. Valle, D. C. Masison and R. B. Wickner, 1999. The ski7 antiviral protein is an EF1-alpha homolog that blocks expression of non-Poly(A) mRNA in Saccharomyces cerevisiae. J. Virol. 73: 2893–2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, M. A., P. J. Costa, E. M. Crisucci and K. M. Arndt, 2007. Identification of Rkr1, a nuclear RING domain protein with functional connections to chromatin modification in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 2800–2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C. J., and R. Parker, 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7: 1632–1643. [DOI] [PubMed] [Google Scholar]
- Dunckley, T., and R. Parker, 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18: 5411–5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunckley, T., and R. Parker, 2001. Yeast mRNA decapping enzyme. Methods Enzymol. 342: 226–233. [DOI] [PubMed] [Google Scholar]
- Emori, Y., T. Tsukahara, H. Kawasaki, S. Ishiura, H. Sugita et al., 1991. Molecular cloning and functional analysis of three subunits of yeast proteasome. Mol. Cell. Biol. 11: 344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan, P. M., R. J. Kelleher, III, M. H. Sayre, H. Tschochner and R. D. Kornberg, 1991. A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350: 436–438. [DOI] [PubMed] [Google Scholar]
- Frischmeyer, P. A., and H. C. Dietz, 1999. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 8: 1893–1900. [DOI] [PubMed] [Google Scholar]
- Frischmeyer, P. A., A. van Hoof, K. O'Donnell, A. L. Guerrerio, R. Parker et al., 2002. An mRNA surveillance mechanism that eliminates transcripts lacking termination codons. Science 295: 2258–2261. [DOI] [PubMed] [Google Scholar]
- Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles et al., 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387–391. [DOI] [PubMed] [Google Scholar]
- Gietz, R. D., and R. A. Woods, 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350: 87–96. [DOI] [PubMed] [Google Scholar]
- Goldstein, A. L., and J. H. McCusker, 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15: 1541–1553. [DOI] [PubMed] [Google Scholar]
- Hallier, M., N. Ivanova, A. Rametti, M. Pavlov, M. Ehrenberg et al., 2004. Pre-binding of small protein B to a stalled ribosome triggers trans-translation. J. Biol. Chem. 279: 25978–25985. [DOI] [PubMed] [Google Scholar]
- He, F., S. W. Peltz, J. L. Donahue, M. Rosbash and A. Jacobson, 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc. Natl. Acad. Sci. USA 90: 7034–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu, C. L., and A. Stevens, 1993. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 13: 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, T. R., C. J. Roberts, H. Dai, A. R. Jones, M. R. Meyer et al., 2000. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat. Genet. 25: 333–337. [DOI] [PubMed] [Google Scholar]
- Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson et al., 2003. Global analysis of protein localization in budding yeast. Nature 425: 686–691. [DOI] [PubMed] [Google Scholar]
- Inada, T., and H. Aiba, 2005. Translation of aberrant mRNAs lacking a termination codon or with a shortened 3′-UTR is repressed after initiation in yeast. EMBO J. 24: 1584–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito-Harashima, S., K. Kuroha, T. Tatematsu and T. Inada, 2007. Tanslation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21: 519–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs Anderson, J. S., and R. Parker, 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17: 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A. W., and R. D. Kolodner, 1995. Synthetic lethality of sep1 (xrn1) ski2 and sep1 (xrn1) ski3 mutants of Saccharomyces cerevisiae is independent of killer virus and suggests a general role for these genes in translation control. Mol. Cell. Biol. 15: 2719–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai, A. W., M. M. Susskind and R. T. Sauer, 1999. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA). EMBO J. 18: 3793–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler, K. C., P. R. Waller and R. T. Sauer, 1996. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271: 990–993. [DOI] [PubMed] [Google Scholar]
- Kelleher, R. J., III, P. M. Flanagan and R. D. Kornberg, 1990. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell 61: 1209–1215. [DOI] [PubMed] [Google Scholar]
- Larimer, F. W., C. L. Hsu, M. K. Maupin and A. Stevens, 1992. Characterization of the XRN1 gene encoding a 5′→3′ exoribonuclease: sequence data and analysis of disparate protein and mRNA levels of gene-disrupted yeast cells. Gene 120: 51–57. [DOI] [PubMed] [Google Scholar]
- Liu, H. Y., Y. C. Chiang, J. Pan, J. Chen, C. Salvadore et al., 2001. Characterization of CAF4 and CAF16 reveals a functional connection between the CCR4-NOT complex and a subset of SRB proteins of the RNA polymerase II holoenzyme. J. Biol. Chem. 276: 7541–7548. [DOI] [PubMed] [Google Scholar]
- Lydall, D., and T. Weinert, 1997. G2/M checkpoint genes of Saccharomyces cerevisiae: further evidence for roles in DNA replication and/or repair. Mol. Gen. Genet. 256: 638–651. [DOI] [PubMed] [Google Scholar]
- Macbeth, M. R., H. L. Schubert, A. P. Vandemark, A. T. Lingam, C. P. Hill et al., 2005. Inositol hexakisphosphate is bound in the ADAR2 core and required for RNA editing. Science 309: 1534–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, S. A., and C. L. Dieckmann, 1991. Yeast CBP1 mRNA 3′ end formation is regulated during the induction of mitochondrial function. Mol. Cell. Biol. 11: 813–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., and R. Parker, 1992. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 6: 2100–2111. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., and R. Parker, 1994. Premature translational termination triggers mRNA decapping. Nature 370: 578–581. [DOI] [PubMed] [Google Scholar]
- Muhlrad, D., and R. Parker, 1999. Recognition of yeast mRNAs as “nonsense containing” leads to both inhibition of mRNA translation and mRNA degradation: implications for the control of mRNA decapping. Mol. Biol. Cell 10: 3971–3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., C. J. Decker and R. Parker, 1995. Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol. 15: 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulak, R., and P. Anderson, 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7: 1885–1897. [DOI] [PubMed] [Google Scholar]
- Ramos, P. C., J. Hockendorff, E. S. Johnson, A. Varshavsky and R. J. Dohmen, 1998. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell 92: 489–499. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., E. J. Lambie and G. S. Roeder, 1991. Spore enrichment. Methods Enzymol. 194: 146–149. [DOI] [PubMed] [Google Scholar]
- Seminara, S. B., S. Messager, E. E. Chatzidaki, R. R. Thresher, J. S. Acierno, Jr. et al., 2003. The GPR54 gene as a regulator of puberty. N. Engl. J. Med. 349: 1614–1627. [DOI] [PubMed] [Google Scholar]
- Shyu, A. B., J. G. Belasco and M. E. Greenberg, 1991. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 5: 221–231. [DOI] [PubMed] [Google Scholar]
- Sikorski, R. S., and P. Hieter, 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks, K. A., and C. L. Dieckmann, 1998. Regulation of poly(A) site choice of several yeast mRNAs. Nucleic Acids Res. 26: 4676–4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiger, M., A. Carr-Schmid, D. C. Schwartz, M. Kiledjian and R. Parker, 2003. Analysis of recombinant yeast decapping enzyme. RNA 9: 231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi, A., M. Hakoda, H. Yamanaka, C. Terai, K. Hikiji et al., 1998. A germline mutation abolishing the original stop codon of the human adenine phosphoribosyltransferase (APRT) gene leads to complete loss of the enzyme protein. Hum. Genet. 102: 197–202. [DOI] [PubMed] [Google Scholar]
- Thompson, C. M., A. J. Koleske, D. M. Chao and R. A. Young, 1993. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell 73: 1361–1375. [DOI] [PubMed] [Google Scholar]
- Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader et al., 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368. [DOI] [PubMed] [Google Scholar]
- Tucker, M., M. A. Valencia-Sanchez, R. R. Staples, J. Chen, C. L. Denis et al., 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386. [DOI] [PubMed] [Google Scholar]
- Ueda, K., Y. Yamamoto, K. Ogawa, T. Abo, H. Inokuchi et al., 2002. Bacterial SsrA system plays a role in coping with unwanted translational readthrough caused by suppressor tRNAs. Genes Cells 7: 509–519. [DOI] [PubMed] [Google Scholar]
- van Hoof, A., P. A. Frischmeyer, H. C. Dietz and R. Parker, 2002. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 295: 2262–2264. [DOI] [PubMed] [Google Scholar]
- Verma, R., S. Chen, R. Feldman, D. Schieltz, J. Yates et al., 2000. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol. Biol. Cell 11: 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, J. D., A. R. Odom, R. Murphy, E. B. Ives and S. R. Wente, 1999. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science 285: 96–100. [DOI] [PubMed] [Google Scholar]
- Zaret, K. S., and F. Sherman, 1984. Mutationally altered 3′ ends of yeast CYC1 mRNA affect transcript stability and translational efficiency. J. Mol. Biol. 177: 107–135. [DOI] [PubMed] [Google Scholar]