Abstract
FoxA transcription factors are critical regulators of gut development and function. FoxA proteins specify gut fate during early embryogenesis, drive gut differentiation and morphogenesis at later stages, and affect gut function to mediate nutritional responses. The level of FoxA is critical for these roles, yet we know relatively little about regulators for this family of proteins. To address this issue, we conducted a genetic screen for mutants that suppress a partial loss of pha-4, the sole FoxA factor of Caenorhabditis elegans. We identified 55 mutants using either chemical or insertional mutagenesis. Forty-two of these were informational suppressors that affected nonsense-mediated decay, while the remaining 13 were pha-4 suppressors. These 13 alleles defined at least six different loci. On the basis of mutational frequencies for C. elegans and the genetic dominance of four of the suppressors, we predict that many of the suppressors are either unusual loss-of-function mutations in negative regulators or rare gain-of-function mutations in positive regulators. We characterized one dominant suppressor molecularly and discovered the mutation alters a likely cis-regulatory region within pha-4 itself. A second suppressor defined a new locus, the predicted AAA+ helicase ruvb-1. These results indicate that our screen successfully found cis- or trans-acting regulators of pha-4.
DURING metazoan development, cells acquire specialized identities such as cell type, position, organ, or tissue. The developmental programs that establish these identities often depend on the activity of a subset of transcription factors called selector genes (Mann and Carroll 2002). Here we focus on the selector gene responsible for identity of cells of the foregut, which is the winged-helix transcription factor FoxA (Mango et al. 1994; Horner et al. 1998; Kalb et al. 1998).
FoxA, previously known as HNF3β (Kaestner et al. 2000), was identified as a protein enriched in liver nuclear extracts (Lai et al. 1990; Lai et al. 1991) and the gene defined by Drosophila fork head (Jurgens et al. 1984; Jurgens and Weigel 1988; Weigel et al. 1989). More recently, FoxA factors have been implicated in transcriptional control of breast cancer genes (Carroll et al. 2005; Laganiere et al. 2005), metabolic processes (Friedman and Kaestner 2006) and aging (Panowski et al. 2007). Loss of FoxA activity results in severe defects in foregut-derived organs such as liver and pancreas (Weigel et al. 1989; Ang and Rossant 1994; Mango et al. 1994; Weinstein et al. 1994; Dufort et al. 1998; reviewed in Zaret 2002; Lee et al. 2005). Conversely, ubiquitous expression of pha-4/FoxA in Caenorhabditis elegans is sufficient to induce ectopic foregut cells (Horner et al. 1998). These data reveal that appropriately regulated expression of pha-4/FoxA is critical for its normal functions.
Microarray studies have identified >300 potential targets of PHA-4 in C. elegans, and promoter analysis has revealed that many of these genes are direct PHA-4 targets (Gaudet and Mango 2002). The targets are expressed at varying times during the development, differentiation, or functioning of the pharynx. The appropriate timing of expression depends on combinatorial mechanisms among different cis-regulatory sites (Kalb et al. 2002; Ao et al. 2004; Gaudet et al. 2004; Vilimas et al. 2004; Raharjo and Gaudet 2007). In addition, the affinity of PHA-4 for its binding sites is an important contributor to temporal control (Gaudet and Mango 2002; Ao et al. 2004; Gaudet et al. 2004). FoxA proteins recognize the consensus TRTTKRY (R = A/G, K = T/G, and Y = T/C) in the cis-regulatory regions of gut-specific targets (Overdier et al. 1994; Kalb et al. 1998; Gaudet and Mango 2002). Target genes with high affinity sites are competent to fire early in embryogenesis, whereas genes with lower affinity sites are typically expressed late (Gaudet and Mango 2002; Gaudet et al. 2004). This regulatory configuration implies that the level and activity of PHA-4 are carefully controlled to achieve proper timing for pharyngeally expressed genes. This raises the question of what mechanisms control PHA-4 expression and activity. Here, we take advantage of our pha-4 alleles to identify mutants that can suppress a partial loss of pha-4 function. We chose a genetic approach over other possible schemes (e.g., yeast two-hybrid screen, co-immunoprecipitation) because this strategy could potentially identify proteins that do not physically touch PHA-4, such as components of signaling pathways, as well as those that do. Here we present the results and discuss the implications of this screen for regulators of PHA-4.
MATERIALS AND METHODS
Worm growth:
C. elegans strains were maintained as described in Brenner (1974). Wild type (WT) was N2 (Bristol). pha-4(ts) strains, SM190 smg-1(cc546ts)I; pha-4(zu225)V and SM568 smg-1(cc546ts)I; pha-4(q500) rol-9(sc148)V, KK822 par-1(zu310ts)V, SM279 smg-1(cc546ts)I five times outcrossed, SM141 or PGP is pxIs1(PHA-4∷GFP + rol-6) were described previously (Gaudet and Mango 2002; Alder et al. 2003; Kaltenbach et al. 2005; Updike and Mango 2006). Strains obtained from the CGC include TR1331 smg-1(r861)I, BC2511 dpy-18(e364); eT1III; unc-60(e677) dpy-11(e224) sDf35/eT1V, and CB4856 Hawaiian isolate. Strain TM2786 ruvb-1(tm2786) was obtained from the National Bioresource Project, Tokyo Women's Medical College, Tokyo (Gengyo-Ando and Mitani 2000).
Strains created for this study include the Mos1 mutagenesis strain SM948 smg-1(cc546ts)I; pha-4(zu225); oxIs30(Mos1 Transposase) X; oxEx229(Mos1 Transposon). SM948 was created from SM190 and EG2412, which carries the Mos1 transposon and transposase (Bessereau et al. 2001). SM1547 was used for single-nucleotide polymorphism (SNP) mapping and was generated by crossing SM190 10 times with Hawaiian CB4856, using pxEx259(sur-5∷GFP + unc-54∷GFP) to mark cross progeny. SM1153 was smg-1(cc546ts); pha-4(zu225) + (coelomocyte∷GFP). SM1261 was smg-1(cc546ts)I; pha-4(px63 zu225)/fog-2(q71) rol-9(sc148)V. SM1237 was dpy-11(e224) unc-42(e270) ruvb-1(px34)/evl-1(ar115) V. SM1238 was ruvb-1(px34)/unc-42(e270) sqt-3(sc63) V. SM1239 is ruvb-1(px34) sqt-3(sc63)/unc-42(e270) sqt-3(sc63) V. SM1262 was dpy-11(e224) unc-42(e270) ruvb-1(px34)/evl-1(ar115)V; pxEx210a(R05D5 + sur-5∷GFP). SM1263 was dpy-11(e224) unc-42(e270) ruvb-1(px34)/evl-1(ar115)V; pxEx210b(T10A5 + sur-5∷GFP). SM1276 was pxEx211(ruvb-1p∷GFP∷His2B + rol-6). SM1415 is smg-1(cc546ts)I; (px23)II; pha-4(zu225)V + (coelomocyte∷GFP). SM1261 was pha-4(px63 zu225)/fog-2(q71) rol-9(sc148). SM1592 was ruvb-1(tm2786)/unc-42(e270) sqt-3(sc63). Suppressor strains in the pha-4(ts) background are shown in Table 1.
TABLE 1.
List of non-NMD pha-4 suppressors
| Allele | Strain name | Back cross | Mutagen | Suppress lethalitya (%) | M−/− Z−/+ alive+ | M+/+ Z−/+ alivea (%) | M−/+ Z−/+ alive+ | Dominant/recessive | Map position | Locus |
|---|---|---|---|---|---|---|---|---|---|---|
| px8 | SM1150 | 4× | EMS | 7 | Yes | 0 | No | Rb | ND | |
| px11 | SM1114 | 4× | EMS | 58 | Yes | 0 | No | Rb | IIIM | |
| px12 | SM1591 | 5× | EMS | 29 | Yes | 0 | No | Rb | IVL | |
| px16 | SM1118 | 4× | EMS | 55 | Yes | 0 | No | Rb | II | |
| px17 | SM1569 | 4× | EMS | 12 | Yes | 0 | Yes | MatDc | IIIM | |
| px23 | SM1415 | 7× | EMS | 62 | Yes | 0 | No | Rb | IIR | |
| px28 | SM1430 | 4× | EMS | 18 | Yes | 0 | No | Rb | IIR | |
| px31 | SM1570 | 4× | EMS | 43 | Yes | 0 | No | Rb | ND | |
| px33 | SM1108 | 4× | EMS | 32 | Yes | 0 | Yes | MatDc | IIIM | |
| px34 | SM1149 | 6× | EMS | 86d | d | 0 | Yes | MatDc | V: +2.22 | ruvb-1 |
| px63 | SM1134 | 10× | Mos1 | 10d | d | 6 | Yes | ZygDe | V: +25.11 | pha-4 |
| px70 | SM1135 | 4× | Mos1 | 78 | Yes | 0 | No | Rb | ND | |
| px71 | SM1133 | 4× | Mos1 | 28 | Yes | 0 | No | Rb | ND |
None of these suppressors were components of the nonsense-mediated decay (NMD) pathway, according to rpl-7a mRNA analysis. None grew at 15° in combination with pha-4(ts), and none stained with an antibody that recognizes the carboxyl terminus of PHA-4. −, suppressor allele (note that it is unknown if a given allele is a loss of function). +, scored as “yes” if there were at least five viable progeny from ≤100 total. ND, no data.
Percentage viable at 20°, n = 100. pha-4(ts) is 0% viable at 20°.
Maternal absence effect, recessive.
Maternal dominant.
px34 and px63 are homozygous lethal, so analysis was performed on heterozygotes.
Zygotic dominant.
Isolation of pha-4(ts) suppressors:
pha-4(ts) strains were mutagenized with 50 mm ethylmethanesulfonate (EMS) for 4 hr at 24°. P0's were rinsed with M9 to remove EMS, and fourth larval stage (L4) worms were picked to new plates and incubated at 24°. F1 progeny were shifted to the intermediate restrictive temperature of 20° at the L3–L4. Viable F2 progeny were selected, and these strains were maintained at 20°, with no more than one F2 suppressor line isolated from one F1.
Mos1 transposon mutagenesis was performed with SM948. This strain carries pha-4(ts) and an extrachromosomal array with the Mos1 transposon (oxEx229) and an integrated HS∷Mos1 transposase (oxIs30) (Bessereau et al. 2001). Mutagenesis was performed as described in (Bessereau et al. 2001). To synchronize populations, several plates of SM948 worms were treated with bleach to isolate embryos. Embryos were grown to the adult stage to provide the P0 population. Synchronized P0 hermaphrodites were incubated at 33° for 1 hr to induce the transposase and then dispensed to several 10-cm plates at a density of ∼100 P0 animals per plate. F1 progeny were shifted to 20° at the L3–L4 stage. Viable F2 progeny were selected, and their progeny maintained at 20°, with no more than one F2 suppressor line isolated from each plate.
PCR amplification of a Mos1 sequence was used to detect an insertion event after the Mos1 extrachromosomal array was lost from the strain (Bessereau et al. 2001). No Mos1 insertion events were detected in the pha-4(ts) suppressor strains. A Mos1 insertion in unc-74 provided by Dan Williams and Erik Jorgensen was used as a positive control in these experiments and was always detected (Williams et al. 2005; D.W. and E.J.L., personal communication).
Genetic analysis:
To test for suppression in heterozygous cross progeny, 10 SM1153 males were crossed to five to eight sup; pha-4(ts) hermaphrodites at 20°. GFP-positive F1 cross progeny were observed for all suppressors. SM1153 hermaphrodite controls had no viable F1 progeny. To determine whether suppressors were dominant, five GFP-positive F1 cross progeny were picked to new plates in L4 and examined for viable F2 progeny at 20°.
To test for dominant zygotic suppression, males were generated for each of the 13 pha-4 suppressor strains. Twenty suppressor males were crossed to 10 pha-4(ts) worms at 20°. Ten pha-4(ts) males were used for a control. After two dates of mating, P0 mothers were picked from the plates. Two days later, the percentage of progeny larger than the L1 stage were scored (n = 100 progeny).
To examine the gonad, pha-4(ts) animals were shifted to 20° at the L1 or the pretzel stage of embryogenesis. Worms were examined for gonad defects under the light microscope two days later.
RT–PCR of rpl-7a:
For total RNA extraction, 100 μl of frozen worm pellets were crushed and resuspended in Trizol Reagent (Gibco BRL, Grand Island, NY), followed by chloroform extraction, according to the manufacturer's specifications. RNA was precipitated with isopropanol and washed with 70% ethanol. Resuspended RNA was treated with DNase, extracted with phenol:chloroform and precipitated with ethanol. A FirstStrand reaction kit (Gibco BRL) was used to perform the reverse transcriptase reaction. This reaction was followed by phenol:chloroform extraction and ethanol precipitation. Amplification of rpl-7a from the cDNA was performed as described in (Mitrovich and Anderson 2000), and PCR products were analyzed on a 1% agarose gel.
Antibody staining:
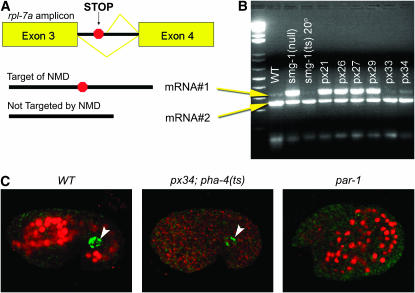
To detect possible nonsense-codon, readthrough suppression, embryos were stained as described previously with an antibody that recognizes the carboxyl terminus of PHA-4 (Horner et al. 1998; Kaltenbach et al. 2005). Embryos from pha-4 suppressing strains were stained with α-carboxyl PHA-4 at a 1:20 dilution and costained with α-P granule (OIC1D4) at 1:10 [received from the Developmental Studies Hybridoma Bank, University of Iowa (Beanan and Strome 1992)]. par-1 mutant embryos (Kemphues et al. 1988) were placed on the same slide as the suppressors as a positive control for PHA-4 staining. Their altered morphology allowed them to be distinguished from pha-4(ts) embryos.
To assess early pharyngeal development, embryos were stained with pharyngeal muscle antibody 3NB12 (Priess and Thomson 1987) and costained with α-LIN-26 antibody (Labouesse et al. 1996), as described in (Horner et al. 1998).
To count pharyngeal cells, embryos were stained with a 1:2000 dilution of affinity purified pan-PHA-4 antibody as described previously (Kaltenbach et al. 2005). Confocal sections were taken every 0.25 μm and the number of PHA-4 positive nuclei counted in 6–12 embryos for each strain.
Single nucleotide polymorphism mapping of suppressors:
SM1547, which is pha-4(ts) outcrossed 10 times in the Hawaiian CB4856 background, males were crossed to sup; pha-4(ts) hermaphrodites (Bristol background) at 20° (px17, px33, px34, and px63) or 24° (px8, px11, px12, px16, px23, px28, px31, px70, and px71). GFP fusions with sur-5 (Yochem et al. 1998) or unc-54 (Sha and Fire 2005) in SM1547 males were used to identify F1 cross progeny. Hermaphrodite F1 cross progeny were cloned to individual plates at 20°. Viable F2 progeny were picked to individual plates at 20°. Individual F2 animals that gave rise to viable F3 progeny at 20° were picked into 20 μl of worm lysis buffer with proteinase K and frozen for later analysis.
Worms in lysis buffer (50 mm KCl, 10 mm Tris pH 8.3, 2.5 mm MgCl2, 0.45% IGEPAL CA-630, 0.45% Tween 20, 0.01% gelatin, 60 μg/ml proteinase K) were incubated at 65° for 1 hr, followed by 15 min at 95°. This lysate was used as a template in PCR reactions to amplify SNPs spanning all chromosomes. After PCR, the appropriate restriction enzymes and buffers were added to detect SNPs between the Hawaiian and N2 isolates (Wicks et al. 2001; Davis et al. 2005). Digested amplicons were analyzed by 2% agarose gel electrophoresis.
SNP mapping data:
F2 segregation ratio of N2:N2/Hi:Hi is given for suppressors linked to chromosomes. Map positions and physical locations in this article correspond to WormBase release WS174.
px11: Chromosome III. snp_Y71H2B[2] at −12.05 cM 28:1:2. pkP3101 at −0.92 cM 29:1:1. snp_F56C9[1] at −0.75 cM 26:4:1. snp_Y39A1[9] at +4.13 cM 29:0:2.
px12: Chromosome IV. snp_Y41D4B[2] at −17.51 cM 6:1:0.
px16: Chromosome II. pkP2101 at −15.91 cM 45:6:1. pkP2103 at −6.21 cM 45:6:1. snp_T24B8[1] at +0.98 cM 43:6:2. pkP2116 at +15.77 cM 45:3:4. snp_Y39G8B[1] at +21.19 cM 45:3:4.
px17: Chromosome III. snp_Y71H2B[2] at −12.05 cM 142:210:28. Of the 28 Hi/Hi at −12.05 cM, only 1 was still Hi/Hi at pkP3076 at −0.85 cM, and it was N2/Hi by snp_F56C9[1] at −0.75 cM. snp_Y39A1[9] at +4.13 cM 145:209:11. Of the 11 Hi/Hi at +4.13 cM, only 2 were still Hi/Hi at pkP3105 at −0.59 cM, and both were N2/Hi by snp_F56C9[1] at −0.75 cM.
px23: Chromosome II. pkP2116 at +15.77 cM 89:4:0. Of the four N2/Hi at +15.77 cM, two were N2 homozygotes by pkP2118 at +20.71 cM. snp_Y39G8B[1] at +21.19 cM 92:0:0.
px28: Chromosome II. pkP2116 at +15.77 cM 6:1:0. snp_Y39G8B[1] at +21.19 cM 7:0:0
px33: Chromosome III. snp_F56C9[1] at −0.75 cM 5:10:0.
px34: Chromosome V. nP118 at −18.81 cM 3:6:1. snp_K12B6[7] at −0.18 cM 10:13:1. pkP5117 at +2.00 cM 10:12:1. snp_F46F3[1] at +3.25 cM 12:12:0. pkP5125 at +5.83 cM 9:15:1. Next, F2 arrested L3 worms were picked for finer mapping instead of suppressors. snp_K12B6[7] at −0.18 cM 12:9:0. pkP5117 at +2.00 cM 68:1:0. Remaining N2/Hi is N2/N2 by snp_F58E6[1] at +2.14 cM. pkP5064 at +2.51 cM 47:1:0. Remaining N2/Hi is still N2/Hi by snp_C12D8[2] at +2.40 cM, but is N2/N2 by snp_C08B6[1] at 2.28 cM. snp_F29F11[1] at +2.72 67:2:0. snp_F46F3[1] at +3.25 cM 18:3:0. Three-point mapping demonstrated that px34 mapped to the right of unc-42, which is at +2.16 cM. For finer mapping from the left, non-L3 arrested Unc recombinants were mapped from px34 animals marked with unc-42. From 14 non-L3 arrested Unc recombinants, 2 were still N2/N2 by pkP5119 at +2.20 cM, but N2/Hi by pkP5064 at +2.51 cM. For finer mapping from the right, non-L3 arrested Sqt recombinants were mapped from px34 animals marked with sqt-3. From 48 non-L3 arrested Sqt recombinants, 10 were still N2/N2 by snp_F29F11[1] at +2.72 cM, only 1 was N2/N2 by snp_C08B6[1] at 2.28 cM, but N2/Hi by pkP5119 at +2.20 cM.
px63: Chromosome V. pkP5117 at +2.00 cM 4:6:0.
Southern blot analysis:
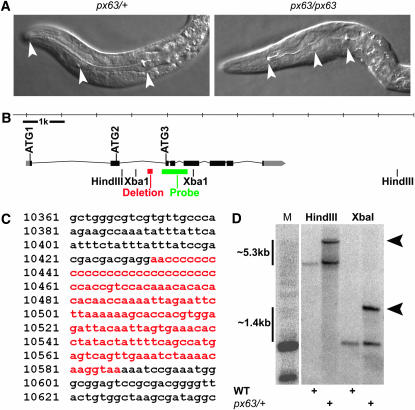
Genomic DNA was prepared from frozen 0.5-ml pellets of N2 or smg-1(cc546ts); pha-4(zu225)/pha-4(px63 zu225) worms by standard proteinase K lysis, phenol:chloroform extraction, and ethanol precipitation (Sulston and Hodgkin 1988). A total of 5 μg of genomic DNA from each strain was digested overnight with EcoRI, HindIII, XbaI, or HindIII and XbaI. Digested DNA was run in a 0.8% agarose gel, blotted, and cross linked to a nylon membrane. 32P-labeled PCR amplicons were used as probes and hybridized to the blot. Primers flanking HindIII internal amplicon: (left-CAGGTCCCCTGACAAG) and (right-GGTGTAGTCAATTCCTGGATAACTGC). Primers flanking XbaI internal amplicon: (left-CAGGTTCAGAGGTCAAACGGGAC) and (right-CGGTGGTGCCAGTGGTAAAAC). These probes placed the px63 rearrangement between XbaI sites in intron 2 and exon 5.
px23 and px28 complementation:
SM1415 (+coelomocyte∷GFP) males were crossed to SM1430 hermaphrodites at 20°. As complementation controls, SM1153(+coelomocyte∷GFP) males were crossed to SM1415 (−coelomocyte∷GFP) and SM1430 hermaphrodites at 20°. From each of the three crosses 10 L4 cross progeny (+coelomocyte∷GFP) were cloned to individual plates at 20°. As noncomplementation controls, 10 SM1415 and SM1430 L4 animals were cloned to individual plates at 20°. 10/10 plates from SM1415/SM1430 (+coelomocyte∷GFP) cross progeny had viable F2 progeny, as did 10/10 plates from SM1415 and SM1430. 0/10 SM1415/+ and 0/10 SM1430/+ cross progeny had viable F2 offspring.
Quantitation of nuclear PHA-4:
In situ antibody staining was performed as described in Horner et al. (1998). Single layer confocal images were taken using a Ziess LSM510. Intensity of PHA-4 staining was quantified in four nuclei for each of 10 1.5-fold embryos using ImageJ. The intensity of PHA-4, after background subtraction, was normalized to the intensity of PHA-4 in 10 par-1 mutant embryos on the same slide (Kaltenbach et al. 2005) [P ≤ 0.0072, t-test; significance of PHA-4 levels in suppressors from pha-4(ts) at 20°]. For this experiment, all embryos were in the smg-1(cc546ts)I; pha-4(zu225)V background, with the exception of px12 and px17, which were in smg-1(cc546ts)I; pha-4(q500) rol-9(sc148)V.
ruvb-1 expression and rescue:
DNA was prepared from cosmids R05D5 and T10A5 with QIAGEN (Valencia, CA) spin miniprep columns. A total of 30 ng/μl of T10A5 was injected with 2 ng/μl of a sur-5∷GFP reporter (Yochem et al. 1998) to rescue ruvb-1 lethality in SM1237. Of the four lines carrying T10A5 in SM1237, two lines produced Dpy Unc progeny that suppressed L3 lethality of ruvb-1 (60%, n = 5; and 33%, n = 9; rescue). A total of 11 ng/μl of R05D5 was injected with 2 ng/μl of sur-5∷GFP to rescue ruvb-1-associated lethality of SM1237 and SM1238. Of the five lines carrying R05D5 in SM1237, two lines produced Dpy Unc progeny that suppressed L3 lethality (33%, n = 15; and 69%, n = 16; rescue). Of the seven lines carrying R05D5 in SM1238, one produced non-Unc non-Dpy non-Rol progeny that suppressed L3 lethality (28% rescue at 25°, n = 36). A total of 0.2 ng/μl of a 3777-bp amplicon containing ruvb-1 (−677 to +3100 relative to the ATG) was injected with 2 ng of an amplicon containing the sur-5∷GFP reporter and 98 ng/μl herring sperm carrier DNA to rescue ruvb-1 lethality in SM1238, and three non-Unc non-Dpy non-Rol sur-5∷GFP positive transgenic animals suppressed L3 lethality.
The ruvb-1 promoter was amplified from R05D5, encompassing nucleotide positions −677 to −1 relative to the ruvb-1 start codon. This promoter fragment was engineered with 5′ SphI and 3′ KpnI restriction sites to facilitate cloning into the GFP∷HIS2B reporter vector pAP.10 (Gaudet and Mango 2002). GFP expression was examined on a Zeiss LSM510 confocal microscope.
Suppression of pha-4 using RNAi:
For pha-4(ts) suppression, HT115 bacteria expressing GFP, ruvb-1 or ruvb-2 double-stranded RNA were grown overnight in 60 μg/ml carbenicillin at 37°, spotted to unseeded plates containing 1 mm IPTG and 60 μg/ml carbenicillin and incubated at 37° overnight (Kamath et al. 2003). Five L4 stage pha-4(ts) or smg-1; ruvb-1 pha-4/+ pha-4 worms were picked to five HT115 (RNAi) or OP50 (control) plates and incubated at 20°. P0 animals were allowed to lay eggs for 1 day and then removed. Two days later, the percentage of animals older than L1 (as determined by size) were counted (∼40 F1 animals/plate).
For pha-4(RNAi) suppression, HT115 bacteria expressing GFP or pha-4 double-stranded RNA were grown overnight in 60 μg/ml carbenicillin at 37°. Three parts of a GFP dsRNA-expressing culture were mixed with one part pha-4 dsRNA-expressing culture to weaken the potency of pha-4(RNAi). The mixture was spotted to unseeded plates containing no IPTG or antibiotic and incubated at 37° overnight. Five L4 stage WT N2 or ruvb-1(px34)/unc-42(e270) sqt-3(sc63) worms were picked to six HT115 (RNAi) plates and incubated at 25°. P0 animals were allowed to lay eggs for 36 hr and then removed. Two days later the percentage of animals older than L1 were counted (∼100 F1 animals/plate). P-value = 5.9 × 10−5 for significance between N2 and ruvb-1 suppression (t-test).
RESULTS
Identification of suppressors of pha-4(ts) lethality:
To screen for pha-4 suppressors, we used two temperature-sensitive (ts) configurations of pha-4 that allowed us to regulate PHA-4 accumulation. pha-4(zu225) and pha-4(q500) each carry a premature stop codon predicted to truncate PHA-4 after the DNA binding domain and render pha-4 mRNA subject to degradation by the nonsense-mediated decay (NMD) pathway (Mango 2001; Gaudet and Mango 2002; Kaltenbach et al. 2005; Updike and Mango 2006). Our previous analyses revealed that animals homozygous for these alleles were viable provided the NMD pathway was also inactivated (Kaltenbach et al. 2005). We took advantage of a ts allele of an NMD component smg-1(cc546ts), which exhibits robust NMD activity at 15° but compromised activity at higher temperatures (Pulak and Anderson 1993; Mango 2001; Grimson et al. 2004; A. Fire, personal communication). At 15°, SMG-1 was active and pha-4 mRNA from either pha-4(zu225) or pha-4(q500) was degraded. We call this “restrictive temperature” because it results in Pha-4 lethality at L1 (Kaltenbach et al. 2005). At the “permissive temperature” of 24°, SMG-1 was compromised and pha-4 mRNA was stabilized. Truncated PHA-4 protein accumulated, and worms survived (Kaltenbach et al. 2005). At 20°, an intermediate level of truncated PHA-4 accrued, producing a phenotype less severe than the null phenotype, but lethal nonetheless (Kaltenbach et al. 2005). We define pha-4(ts) strains as those strains carrying one of the two pha-4 nonsense alleles and smg-1(cc546ts).
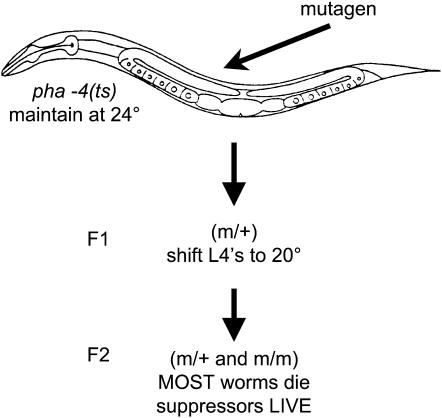
To identify pha-4 suppressors, we mutagenized pha-4(ts) worms at the permissive temperature of 24° and shifted the first generation progeny (F1) to 20° at the L4 (Figure 1). The second generation (F2), which could be homozygous or heterozygous for any induced mutation, was selected for suppression of pha-4 lethality at the intermediate temperature of 20°. We selected suppressors at the intermediate temperature because we were most interested in proteins within the pha-4 pathway (not bypass mutations), which would likely require some pha-4 activity. We predicted that this screen would find regulators or cofactors of pha-4, but not downstream targets. PHA-4 has many downstream targets (Gaudet and Mango 2002), and it is unlikely that mutation of a single target could restore viability to pha-4(ts) mutants. We will determine if this prediction holds true in the future, as the suppressors are analyzed at the molecular and mechanistic levels.
Figure 1.—
Strategy for pha-4 suppressor screen EMS or Mos1 transposon mutagenesis was performed on pha-4(ts) animals at 24°. F1 progeny were shifted to 20°. F2 progeny died in L1 at 20° unless a suppressing mutation had been introduced.
Twenty-six suppressors from 15,000 haploid genomes were obtained using the alkylating agent EMS as a mutagen (Tables 1 and 2). Twenty-nine more suppressors from 1,160,000 haploid genomes were obtained using the Mos1 transposon as a mutagen (Bessereau et al. 2001). Surprisingly, none of our transposon-induced alleles carried a Mos1 insertion, despite robust positive controls (see materials and methods). These alleles likely represent either spontaneous or “hit-and-run” mutations by Mos1 (Williams et al. 2005).
TABLE 2.
List of NMD suppressors
| Allele | Strain name | Mutagen | Growth at 15° |
|---|---|---|---|
| px9 | SM579 | EMS | Yes |
| px10 | SM580 | EMS | No |
| px13 | SM583 | EMS | Yes |
| px14 | SM584 | EMS | No |
| px15 | SM585 | EMS | No |
| px19 | SM600 | EMS | Yes |
| px20 | SM601 | EMS | Yes |
| px21 | SM604 | EMS | Yes |
| px22 | SM610 | EMS | Yes |
| px24 | SM612 | EMS | Yes |
| px25 | SM613 | EMS | No |
| px26 | SM621 | EMS | Yes |
| px27 | SM622 | EMS | Yes |
| px29 | SM624 | EMS | No |
| px30 | SM625 | EMS | No |
| px32 | SM635 | EMS | Yes |
| px52 | SM701 | Mos1 | Yes |
| px53 | SM702 | Mos1 | Yes |
| px54 | SM703 | Mos1 | Yes |
| px55 | SM704 | Mos1 | Yes |
| px56 | SM705 | Mos1 | Yes |
| px57 | SM706 | Mos1 | Yes |
| px58 | SM780 | Mos1 | Yes |
| px59 | SM781 | Mos1 | Yes |
| px60 | SM785 | Mos1 | Yes |
| px61 | SM786 | Mos1 | Yes |
| px62 | SM787 | Mos1 | Yes |
| px64 | SM827 | Mos1 | Yes |
| px65 | SM828 | Mos1 | Yes |
| px66 | SM866 | Mos1 | Yes |
| px67 | SM867 | Mos1 | Yes |
| px68 | SM876 | Mos1 | Yes |
| px69 | SM949 | Mos1 | No |
| px72 | SM956 | Mos1 | Yes |
| px73 | SM957 | Mos1 | Yes |
| px74 | SM958 | Mos1 | Yes |
| px75 | SM959 | Mos1 | Yes |
| px76 | SM961 | Mos1 | Yes |
| px77 | SM964 | Mos1 | No |
| px78 | SM965 | Mos1 | Yes |
| px79 | SM966 | Mos1 | No |
| px80 | SM967 | Mos1 | No |
NMD was established by rpl-7a RT–PCR.
We predicted we would isolate two classes of pha-4 suppressors. First, we expected informational suppressors, including mutations in NMD components. Second, we expected “true” pha-4 suppressors, including regulators of pha-4 expression or modifiers of PHA-4 activity. We performed three tests to distinguish informational suppressors from true pha-4 suppressors. First, we examined a natural target of NMD rpl-7a (Mitrovich and Anderson 2000). This gene produces two mRNAs by alternative splicing (Figure 2A). The larger isoform is created by alternative splicing of exon 3, which introduces a stop codon and makes this mRNA an NMD target. The smaller mRNA isoform, which lacks the entire third intron, is not targeted by NMD. RT–PCR of rpl-7a from WT animals amplifies primarily the smaller mRNA. In animals defective for NMD, RT–PCR also amplifies the larger transcript since this mRNA is now stable. rpl-7a RT–PCR of the suppressors revealed that 16/26 EMS and 26/29 transposon-mutagenized suppressors were components of NMD, leaving 13 potential regulators of PHA-4 activity or expression (Figure 2B; Tables 1 and 2).
Figure 2.—
Identification of informational suppressors. (A) rpl-7a encodes two transcripts, the longer of which contains a premature nonsense codon and is degraded by NMD (Mitrovich and Anderson, 2000). (B) PCR amplification of rpl-7a cDNA from mutant strains identifies when NMD is compromised and the longer mRNA accumulates [smg-1(null) and suppressors px21, px26, px27, and px29]. WT strains exhibit normal NMD activity. NMD activity is partially restored in the remaining strains grown at 20° [smg-1(ts) and suppressors px33 and px34] (see also Kaltenbach et al. 2005). (C) pha-4 suppression is not caused by readthrough of carboxyl-terminal nonsense alleles. Red, α-PHA-4 carboxy-terminal antibody (Horner et al. 1998); green, P-granule costain (arrowhead) (Strome and Wood 1982). par-1 embryos have excess pharyngeal cells (Kemphues et al. 1988) and were used as an on-slide control for PHA-4 staining. The aberrant morphology of par-1 embryos allowed us to distinguish control embryos from sup; pha-4(ts) embryos.
As a second test that these 13 suppressors were not mutations in the NMD pathway or bypass mutations that no longer required PHA-4, we examined suppression at the more restrictive temperature of 15°. We reasoned that mutations that completely inactivated the NMD pathway would suppress pha-4(ts) at any temperature (Kaltenbach et al. 2005). For each suppressor, we shifted 10 worms at L4 from 20° to 15°, and searched for progeny that were viable past L1. None of the 13 suppressors were viable when grown at 15° (Table 1). These data suggest that either we have isolated cold-sensitive lethal alleles, which is unlikely, or none of the 13 suppressors can bypass the requirement for some pha-4 activity. These data also demonstrate that none of the 13 suppressors are null alleles of NMD components.
Third, we wanted to determine whether suppression could be explained by readthrough of the pha-4(zu225) or pha-4(q500) nonsense mutations. We used antibodies that recognize an epitope located in the carboxyl terminus of WT PHA-4, but are absent from the truncated proteins made by pha-4(zu225) or pha-4(q500) (Kaltenbach et al. 2005). We detected no staining with the PHA-4 carboxyl terminus antibody for any of the 13 pha-4 suppressors (Figure 2C). PHA-4 staining of par-1 mutant embryos (Kemphues et al. 1988) placed on the same slides served as a positive control for staining (Figure 2C). These results indicate that suppression of Pha-4 lethality cannot be explained by translational readthrough of pha-4(zu225) or pha-4(q500) nonsense mutations. We conclude that 13 of 55 pha-4 suppressors are not informational suppressors. We focus on the 13 pha-4 suppressors here; the NMD suppressors will be described elsewhere.
pha-4 suppressors increase PHA-4 levels:
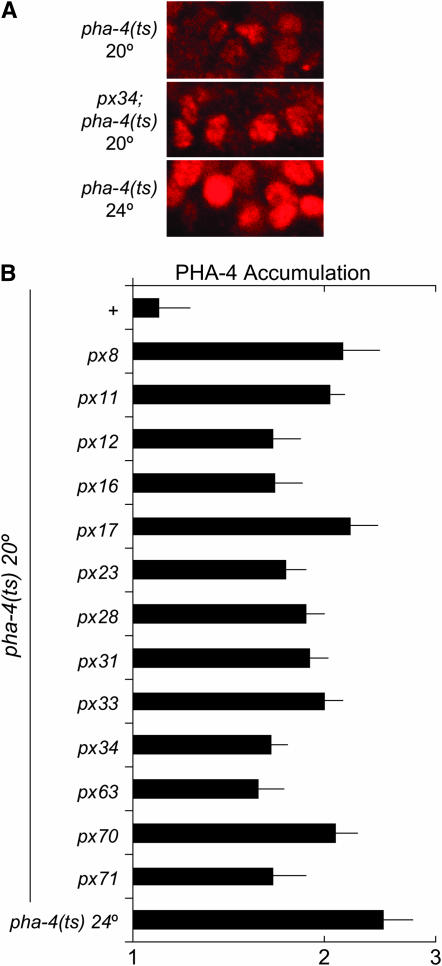
We wanted to investigate the effect of the suppressors on pha-4 expression. We compared levels of endogenous PHA-4 from pha-4(ts) and suppressed pha-4(ts) embryos at the restrictive temperature of 20°. We measured the nuclear accumulation of truncated PHA-4 protein in individual nuclei, using a pan-PHA-4 antibody, and compared it to PHA-4 in par-1 mutant embryos placed on the same slide (our positive control). Levels were expressed as the ratio between the query strain and par-1. In previous experiments, this approach was the most sensitive measure of PHA-4 accumulation (Kaltenbach et al. 2005). Our analysis revealed that PHA-4 accumulated to higher levels in sup; pha-4(ts) embryos compared to pha-4(ts) embryos (Figure 3). We observed a range of 1.6- to 2.2-fold higher expression, which resembled the level of PHA-4 in pha-4(ts) embryos at permissive temperature (24°). Surprisingly, however, we did not observe a strong correlation between PHA-4 levels and suppressor strength (Table 1), which may reflect regulation of PHA-4 activity as well as levels for suppression. For example, px8 embryos had high levels of PHA-4 but poor suppression, whereas px34 mutants had relatively lower levels of PHA-4 but strong suppression (Figure 3; Table 1). These data reveal that the 13 suppressors lead to a modest increase in PHA-4 accumulation at restrictive temperature. We note that this experiment did not address whether the increase in PHA-4 was direct or indirect.
Figure 3.—
PHA-4 restoration in sup; pha-4(ts) strains. (A) px34 increases endogenous PHA-4 levels in the pha-4(ts) background at 20°. pan-PHA-4 antibody (red) (Kaltenbach et al. 2005). (B) PHA-4 levels increase in pha-4(ts) strains at 20° in the presence of each of the pha-4 suppressors. PHA-4 was quantified in individual nuclei as described in materials and methods [P ≤ 0.0072 for suppressor strain compared to pha-4(ts) alone]. The ratio of PHA-4 in the suppressed strain vs. a par-1 control is shown.
Four dominant pha-4 suppressors:
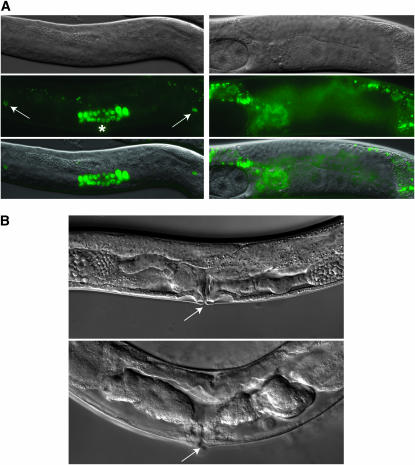
All 13 suppressors [i.e., sup; pha-4(ts) or sup/+;pha-4(ts)] had viable progeny when crossed with pha-4(ts) males at 20°, indicating that pha-4 suppression was either dominant (gain-of-function or haploinsufficient) or subject to a maternal-absence effect. To distinguish between these possibilities, heterozygous sup/+;pha-4(ts) hermaphrodites were tested for their ability to produce viable F2 progeny at 20°. Four heterozygous suppressors (px17, px33, px34, and px63) generated viable F2 progeny at 20°, indicating these four alleles were dominant (Table 1). Five of the remaining suppressors (px11, px16, px31, px70, and px71) had dead F2 progeny, indicating that suppression required both maternal copies of the suppressing gene be mutated (Table 1). F1 heterozygotes for the final four suppressors (px8, px12, px23, and px28) were sterile and gave no F2 progeny. The sterility of the heterozygotes may reflect poor feeding, due to altered pharyngeal development. Alternatively, gonad formation may require pha-4 activity directly during postembryonic development (Figure 4). First stage larvae do not express PHA-4∷GFP in their gonads (data not shown), and L1 animals with pha-4(null) mutations are born with a normal gonad primordium (Mango et al. 1994). However, we observed that as larval development progressed, PHA-4∷GFP (Alder et al. 2003) was activated in the developing gonad (Figure 4A; Kalb et al. 1998; Gaudet et al. 2004) and was required for its proper formation (Figure 4B; Ao et al. 2004). A shift of pha-4(ts) larvae to nonpermissive temperature (20°) at the L1 stage produced sterile adults (Figure 4B). A shift to nonpermissive temperature (15°) at the pretzel stage of embryogenesis generated larvae that arrested at the L2 or L3 stage, with appropriately undeveloped gonads (data not shown). These data indicate that pha-4 is required to form the gonad, and that px8, px12, px23, and px28 heterozygotes cannot rescue the gonad defects of pha-4(ts).
Figure 4.—
PHA-4 is required in the developing gonad. (A) PHA-4∷GFP is broadly expressed in the somatic gonad of a third stage larval worm (asterisk, left). Arrows denote the distal tip cells. PHA-4∷GFP is low or undetectable in the gonads of young adult animals (right) and older L4 larvae (not shown). Some epifluorescence derives from the intestine. (B) Somatic gonad development after 2 days of growth at 20° from the L1. Somatic gonad of a WT worm (top). A sterile pha-4(ts) adult with abnormal somatic gonad development (bottom). Arrowhead denotes the vulva.
To determine whether the suppressors could suppress pha-4 lethality when contributed paternally, 20 males from each suppressor [sup; pha-4(ts) or for px34 and px63, sup/+;pha-4(ts)] were crossed to 10 pha-4(ts) L4 hermaphrodites at the restrictive temperature of 20°. Only px63/+ males gave rise to viable F1 progeny, suggesting that suppression by px63 was contributed zygotically, whereas the remaining three dominant suppressors (px17, px33, and px34) required a maternal contribution. We note that zygotic suppression did not track with overall strength of suppression. px63 M+/− Z+/− gave one of the lowest suppression rates (10%), yet this allele could still suppress as M+/+ Z+/−. px34 M+/− Z+/−, on the other hand, was one of the strongest suppressors, yet M+/+ Z+/− did not suppress pha-4(ts). These behaviors indicate that suppression reflected maternal vs. zygotic contributions rather than overall strength of suppression.
Location of pha-4 suppressors:
We were able to assign a linkage group (LG) to 9 of the 13 suppressors using single nucleotide polymorphisms (SNPs). Mapping data from the 9 suppressors revealed that at least six different loci were involved.
LG II: px23 and px28 mapped to the same location on the far right arm of chromosome II (see materials and methods). When grown at the restrictive temperature of 20°, px23/px28; pha-4(ts) heterozygotes produced viable progeny, unlike px23/+; pha-4(ts) or px28/+; pha-4(ts) mothers [progeny scored from 10 px23/px28, px23/+ or px28/+ animals in the pha-4(ts) background]. Thus, px23 and px28 fail to complement each other for suppression of pha-4(ts) lethality and are likely alleles of the same gene. A third suppressor, px16, also mapped to chromosome II with a recessive segregation ratio, but was not linked to the far right arm, indicating it was distinct from px23 and px28. None of the three alleles had an obvious phenotype when the pha-4 and smg-1 mutations were removed (data not shown).
LG III: The alleles px11, px17, and px33 mapped to the center of chromosome III. Both px17 and px33 were maternally dominant, whereas px11 was recessive. The dominant attributes of px17 and px33 prevented further complementation analysis. These three genes may comprise an allelic series for one locus or represent individual genes located near each other. None of the three genes had an obvious phenotype when the pha-4 and smg-1 mutations were removed (data not shown).
LG IV: px12 was the only suppressor that mapped to the left end of chromosome IV. px12 exhibited a recessive segregation ratio, where homozygous but not heterozygous px12 animals gave rise to viable F3 progeny. px12 did not reveal an obvious phenotype when the pha-4 and smg-1 mutations were removed (data not shown).
LG V: SNP mapping was used to position px63 to the right arm of chromosome V. Homozygous px63 pha-4(ts) animals from balanced smg-1; pha-4(px63 zu225)/fog-2 rol-9 heterozygotes arrested as late-stage embryos or L1 larvae with a pharynx unattached (Pun) phenotype (Figure 5A).
Figure 5.—
px63 is a complex rearrangement in pha-4. (A) In px63 homozygotes, the pharynx is differentiated, but unattached (arrowheads). (B) Overview of the pha-4 locus, the 156-bp deletion in the second intron (red), and the probe used for Southern blotting (green). (C) 156-bp deletion (red) in intron 2. Sequence numbers correspond to the Z92633 GeneBank entry. (D) Southern blot comparing WT and px63/+ genomic DNA. A probe internal to the XbaI sites revealed an insertion of at least 5.3 kb (arrowheads).
px34 mapped to the center of chromosome V. Suppression was vigorous, as 90% of progeny from smg-1; ruvb-1(px34) pha-4/+ pha-4 mothers survived at 20°, compared to 0% in the absence of px34 (Table 3). Our data suggest that in addition to being dominant, px34 suppression is contributed maternally since >90% of progeny from a px34 heterozygous mother survived. Intriguingly, homozygous px34 animals arrested as larvae even in the absence of smg-1 or pha-4, suggesting the px34 mutation disrupts an essential gene (data not shown).
TABLE 3.
ruvb-1(px34) suppression of pha-4
| Genotype of parent | Viable > L1 (%) | SD (%) | Replicates |
|---|---|---|---|
| pha-4(ts) 20° | 0.0 | 0.0 | 5 |
| smg-1; ruvb-1 pha-4/+ pha-4 20° | 90.0 | 4.3 | 5 |
| GFP(RNAi); pha-4(ts) 20° | 0.9 | 2.1 | 5 |
| ruvb-1(RNAi) pha-4(ts) 20° | 48.8 | 9.0 | 5 |
| ruvb-2(RNAi) pha-4(ts) 20° | 47.6 | 4.8 | 5 |
| pha-4(RNAi)a | 14.7 | 6.5 | 6 |
| ruvb-1(px34)/+; pha-4(RNAi)a | 56.4 | 14.0 | 6 |
SD, standard deviation.
1:4 ratio of pha-4(RNAi) to GFP(RNAi) for weak pha-4(RNAi).
Additional alleles:
To date, we have not assigned a linkage group for px8, px31, px70, or px71. Our difficulty may reflect where these genes are located. To map suppression, the mutant alleles smg-1(cc546ts) and pha-4(zu225) from C. elegans Bristol were introduced into the Hawaiian CB4856 strain. The resulting Bristol/Hawaiian chimera was crossed with Hawaiian C. elegans 10 times to remove as much Bristol DNA as possible. Nevertheless, SNPs at I: −6 and I: −1, which are near the smg-1 locus on chromosome I, still harbored DNA from the Bristol isolate (data not shown). Thus, suppressors that map to the central region of chromosome I or to the far right end of chromosome V (near pha-4), would elude SNP mapping using our pha-4(ts) Hawaiian mapping strain.
px63 is an allele of pha-4:
The location of px63 on the right arm of chromosome V, near pha-4 itself, suggested that px63 might be an allele of pha-4. As a first test of this idea, we attempted to exchange the pha-4(zu225) allele of smg-1; pha-4(zu225) px63/pha-4(zu225) with pha-4(q500), another pha-4 nonsense allele targeted by NMD. We observed that the pha-4(zu225) allele could not be exchanged with pha-4(q500) rol-9 (n = 0/96 non-Rol F2 offpsring). This result suggested that either suppression was specific for the pha-4(zu225) allele or px63 was very closely linked to the pha-4 locus.
To determine whether px63 was an allele of pha-4, we sequenced the pha-4 locus from smg-1(cc546ts); pha-4(px63 zu225)/pha-4(zu225) animals (Figure 5C). We verified that the zu225 stop codon had not reverted to enable suppression, and we also found a 156-bp deletion in the second intron of pha-4 (Figure 5, B and C). In addition to the deletion, Southern blotting revealed an insertion of ≥5.3 kb between the 3′ end of intron 2 and the middle of exon 5 (Figure 5, B and D). Taken together, these results suggest that px63 is an allele of pha-4 that contains a complex rearrangement in the 3′ end of intron 2 and possibly other regions as well.
The pha-4 gene produces three transcripts that initiate at exons 1, 2, and 3 and that generate three in-frame proteins (Figure 5B) (Azzaria et al. 1996). The rearrangement associated with px63 affected the second intron, which could possibly function as a promoter for the third, most abundant mRNA (Azzaria et al. 1996). We predict that the rearrangement associated with pha-4(px63) either disrupts a negative regulatory element or introduces a positive element within the third pha-4 promoter. The identification of a pha-4 cis-acting mutation validated our suppressor screen as an effective method for finding regulators of pha-4.
px34 is an allele of the predicted AAA+ helicase ruvb-1:
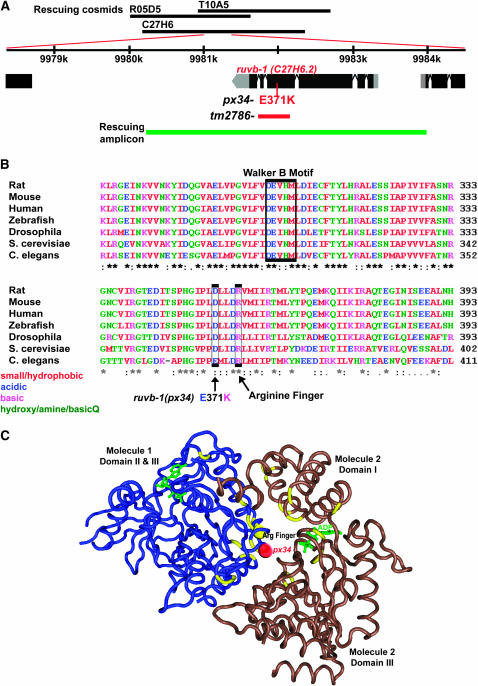
px34 was positioned to within 220 kb on chromosome V (V: 9906715 … 10124252) using standard SNP mapping (Figure 6A) (Wicks et al. 2001). Each of 10 cosmids spanning the 220-kb region were injected into dpy-11 unc-42 px34/evl-1 hermaphrodites and scored for rescue of lethality. Of the 10 cosmids, R05D5 and T10A5 could each rescue px34, implicating one of the five genes common to both of these cosmids (see materials and methods). One of these five genes, the predicted helicase ruvb-1 (C27H6.2) was associated with a larval arrest phenotype, suggesting ruvb-1 was a good candidate for px34 (Piano et al. 2002; Simmer et al. 2003).
Figure 6.—
Identification of px34 within the ruvb-1 locus. (A) px34 was mapped to the center of LG V. Rescuing cosmids R05D5 and T10A5 overlap ruvb-1. A PCR fragment (green) containing ruvb-1 was used to rescue the larval arrest phenotype. (B) Alignment of ruvb-1 with its orthologs. The E371K mutation associated with px34 (arrow) is located just upstream of the Arginine finger. (C) The E371K mutation is positioned at the predicted interface between RuvB molecules. The position of the C. elegans ruvb-1 E371K has been superimposed on the human RuvbL1 structure adapted from Matias et al. (2006).
Two lines of evidence revealed that px34 was an allele of ruvb-1. First, a 3777-bp fragment, which contained the ruvb-1 locus with 677 bp of 5′ upstream sequences, could rescue the lethality associated with px34 larvae (Figure 6A; materials and methods). Second, we detected a C-to-T transition within ruvb-1 that was predicted to change the conserved glutamate at position 371 to a lysine (Figure 6B). This mutation was located after the conserved Walker A and Walker B motifs required for ATP binding of the helicase and four residues before a conserved Arginine finger motif thought to regulate ATP hydrolysis and DNA binding (Putnam et al. 2001; Ohnishi et al. 2005; Matias et al. 2006). These data demonstrate that px34 is an allele of ruvb-1.
Four experiments suggested that ruvb-1(px34) was a loss-of-function mutation. First, RNAi against ruvb-1 could suppress the lethality of pha-4(ts) animals reared at 20°, demonstrating that reduced ruvb-1 was responsible for pha-4 suppression (Table 3). Similarly, RNAi against ruvb-2 could suppress pha-4(ts) lethality (Table 3). In other organisms, orthologs of RUVB-1 and RUVB-2 bind each other and function as a hexameric helicase (Ikura et al. 2000; Shen et al. 2000; Wood et al. 2000; Fuchs et al. 2001; Krogan et al. 2003; Doyon et al. 2004; Mizuguchi et al. 2004; Jin et al. 2005; Bakshi et al. 2006). Second, some ruvb-1(RNAi) animals arrested as larvae and resembled ruvb-1(px34) homozygotes (Simmer et al. 2003; Piano et al. 2002). Third, when ruvb-1(px34)/unc-42(e270) sqt-3(sc63) males were crossed to sDf35/+ heterozygotes, 24% (17/71) of the progeny arrested as larvae, with a phenotype resembling that of ruvb-1(px34). sDf35 removes the ruvb-1 region (Mckim et al. 1988). Fourth, we observed an identical larval arrest for a second ruvb-1 allele, tm2786, which contained a deletion in exons 4 and 5 (Figure 6A). A total of 23.5% of progeny from ruvb-1(tm2786)/unc-42(e270) sqt-3(sc63) mothers arrested, indicating a zygotic larval arrest phenotype (n = 251). These data indicate that the ruvb-1 E371K mutation causes a loss-of-function in ruvb-1 (see also discussion).
Restoration of pharyngeal cells by ruvb-1(px34):
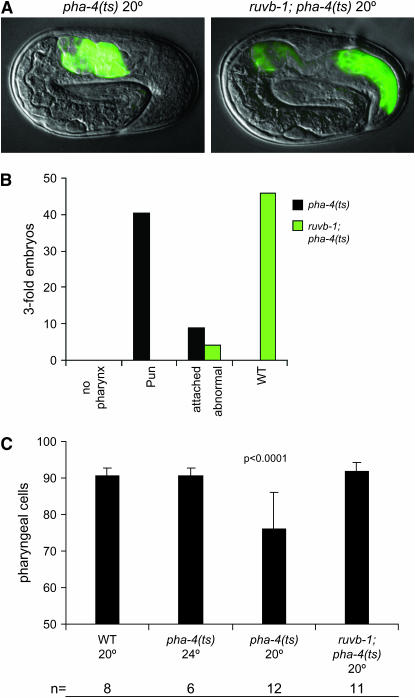
PHA-4 is required during embryogenesis to form the pharynx (Mango et al. 1994; Kiefer et al. 2007), suggesting that the pha-4 suppressors function embryonically to restore pharynx development. To explore this idea, we examined one suppressor [ruvb-1(px34)] in greater detail. At 20°, 82% (n = 50) of pha-4(ts) embryos had an unattached (Pun) pharynx, compared to 0% (n = 50) for embryos from a smg-1; ruvb-1 pha-4/+ pha-4 mother (Figure 7, A and B). We reasoned that the pharyngeal phenotype associated with pha-4(ts) could reflect either fewer pharyngeal cells or defective pharyngeal morphology (Mango 2007). To distinguish between these possibilities, embryos were stained with a pan-α-PHA-4 antibody, and pharyngeal cells counted at the 1.5-fold stage of embryogenesis. The number of pharyngeal cells was reduced for pha-4(ts) worms incubated at 20° (76 ± 10.21, n = 12) compared to 24° (90.5 ± 2.35, n = 6) (Figure 7C). In embryos from smg-1; ruvb-1 pha-4/+ pha-4 mothers, the number of pharyngeal cells at 20° was restored to the WT level (91.8 ± 2.40, n = 11). These data indicate that suppression of Pha-4 lethality by ruvb-1(px34) is embryonic and affects accumulation of PHA-4. We note that we did not observe ectopic expression of the ectodermal factor LIN-26 (Labouesse et al. 1996) within presumptive pharyngeal cells in either pha-4(ts) or smg-1; ruvb-1 pha-4/+ pha-4 embryos (data not shown). This result differs from pha-4 null embryos, which express ectopic LIN-26 within cells that would normally become pharyngeal (Horner et al. 1998; Kiefer et al. 2007).
Figure 7.—
ruvb-1(px34) suppression of pha-4(ts) restores pharyngeal cells. (A) Suppression of the pha-4(ts) Pun phenotype by ruvb-1(px34). Green, 3NB12 antibody, which stains pharynx muscle (Priess and Thomson 1987). (B) Severity of pha-4(ts) pharyngeal defects is suppressed by ruvb-1(px34) (n = 50 threefold embryos for each strain; Pun, pharynx unattached). (C) Reduction of pharyngeal cell number (PHA-4+ cells within the head) in pha-4(ts) at 20° is suppressed by ruvb-1(px34).
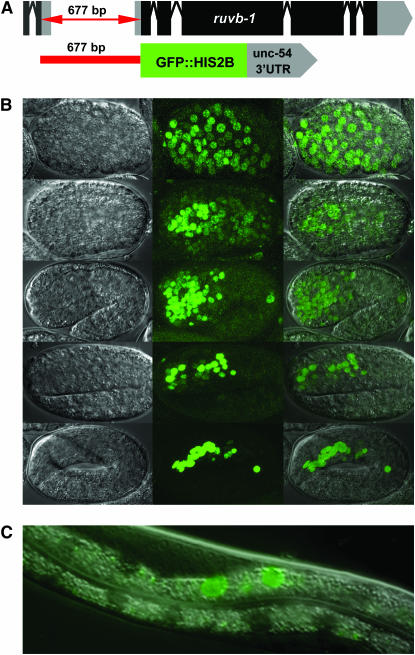
ruvb-1 is expressed in the pharynx during embryogenesis:
To monitor zygotic expression of ruvb-1, we designed a transcriptional, nuclear GFP reporter driven by 677 bp of ruvb-1 promoter (Figure 8A). This construct included all bases between the ruvb-1 ATG and the upstream gene C27H6.3, which corresponds to the sequences used in the ruvb-1 rescuing construct. We observed expression in most cells of the early embryo, but as development progressed to the twofold stage, expression became restricted to pharyngeal cells (Figure 8B). We also observed GFP in two intestinal cells near the center of larval stage worms, which could reflect artifactual intestinal expression (Figure 8C) (Reece-Hoyes et al. 2007). The expression of ruvb-1 in pharyngeal cells supports its role as a regulator of pharyngeal development.
Figure 8.—
ruvb-1 expression. (A) Design of ruvb-1 transcriptional GFP reporter. (B) ruvb-1 transcriptional GFP reporter is expressed in most cells of the early embryo, but becomes restricted to pharyngeal cells during late embryogenesis. (C) ruvb-1 transcriptional GFP reporter is frequently expressed in two intestinal cells during larval development.
DISCUSSION
Selector genes specify regional, tissue, organ, and cell identity in processes that require the precise temporal and spatial expression of target genes. In this study, we used a forward genetic screen to identify potential regulators or cofactors for the selector gene pha-4/FoxA. We isolated 55 mutants, of which 13 were true pha-4 suppressors and the remainder were likely NMD components. The 13 suppressors represented at least six different loci and included rare dominant alleles, as well as recessive alleles that exhibited a maternal-absence effect. Previous studies have identified genes that functioned upstream of pha-4 in either the ABa or EMS cell lineage (Mango et al. 1994). The screen described here complements these previous approaches since it likely required activation of pha-4-dependent processes in both ABa- and EMS-derived cells.
The recessive suppressors px23 and px28 failed to complement each other and likely define the same gene. Supporting this conclusion, both map to the same area on the far right arm of chromosome II. px16 also maps to chromosome II, but shows no preference for the right arm of the chromosome. A potential allelic series mapped to the center of chromosome III and consisted of px11, px17, and px33. px12 was the only suppressor to map to the far left arm of chromosome IV. We were unable to assign a linkage group to px8, px31, px70, or px71, which may reflect the SNP mapping strain. With the exception of ruvb-1(px34) and pha-4(px63), we did not observe obvious phenotypes other than suppression of pha-4(ts) lethality.
PHA-4 is first expressed at the 28–50 cell stage of embryogenesis (Horner et al. 1998). Given this early expression, we were not surprised to uncover a maternal contribution for the nine recessive suppressors and three of the four dominant suppressors (px17, px33, and px34). Genes affecting the activity or expression of PHA-4 might need to be present during early embryogenesis, which could be accomplished by a maternal contribution. The only suppressor that clearly did not have a maternal requirement was px63, which was a mutation in the pha-4 locus. pha-4 loss-of-function mutations are also zygotic (Mango et al. 1994).
The px63 mutation was a complex rearrangement in a likely regulatory region of pha-4. There are three transcriptional initiation sites for pha-4, which produce three mRNAs and three PHA-4 proteins (Azzaria et al. 1996). The location of the px63 rearrangement is predicted to affect the third and strongest initiation site, but could also affect the two upstream promoters. We note that all sequence conservation between FoxA proteins and PHA-4 is located within the third open reading frame (Azzaria et al. 1996). Thus, the transcript and protein derived from the third promoter, and affected by px63, appear to be critical for PHA-4 function.
px63 was generated by Mos1 transposon mutagenesis (Bessereau et al. 2001), yet we were unable to detect Mos1 sequences within pha-4. Mos1-mediated mutagenesis can create deletions where the transposon inserts into a region of DNA and then excises, leaving a nucleotide footprint (Williams et al. 2005). It is also possible that repair of a double-strand break created by Mos1 transposase or insertion of a non-Mos1 substrate could be responsible for creating a complex rearrangement. We note that the strain we used harbored an integrated copy of the transposase but an extrachromosomal array of the transposase substrate (i.e., the transposon). Transposase in worms, including worms lacking the extrachromosomal array, might target genomic DNA at low frequency, and our selection scheme would have enabled these rare events to be detected.
The frequencies at which we obtained suppressors by EMS mutagenesis suggest that many of our suppressors represent unusual loss-of-function mutations or rare gain-of-function mutations. Typically, null mutations are generated at a frequency of 1/2000 haploid genomes (Jorgensen and Mango 2002). If a typical loss-of-function mutation were able to suppress pha-4(ts) lethality, we would have obtained approximately seven alleles/gene from EMS mutagenesis of 15,000 haploid genomes. One explanation for our low frequency is that our screen selected for viability. If regulators of pha-4 are essential, we would not be able to isolate null alleles. This phenomenon may explain the ratio of NMD mutations vs. pha-4 suppressors in EMS vs. Mos1-mediated mutagenesis. Only 3 of 29 suppressors (∼10%) from the Mos1 transposon mutagenesis were pha-4 suppressors, compared to 10 of 26 (∼40%) from the EMS mutagenesis (P < 0.02, Fisher's exact test). Unlike EMS mutagenesis, which typically creates point mutations, Mos1 transposon creates insertions or deletions, which often have stronger loss-of-function phenotypes (Williams et al. 2005). For example, smg-4 was identified at a low frequency by traditional genetic screens using EMS (Hodgkin et al. 1989), and smg-4(r1169) is a large deletion (Aronoff et al. 2001). An alternative explanation for the high frequency of NMD alleles by Mos mutagenesis is that Mos1, but not EMS, could inactivate pairs of redundant, linked NMD genes, such as those found in operons.
Role of RuvB proteins in development:
A second dominant suppressor was associated with the predicted helicase ruvb-1. RUVB-1 belongs to the AAA+ ATPase class of helicases represented by RuvB in bacteria and RUVBL1 in mammals (e.g., TIP49a, TAP54α, Pontin52) (Kanemaki et al. 1997; Bauer et al. 1998; Ikura et al. 2000). We isolated a single allele of ruvb-1, px34, which carried a lysine substitution for glutamate at position 371; we obtained no alleles of ruvb-2. A comparison of C. elegans ruvb-1 with the crystal structure for human RuvbL1 (Matias et al. 2006) suggests that the px34 mutation lies at the interface between RUVB molecules within the hexameric complex (see Figure 6C). Intriguingly, multiple dominant-negative alleles of RuvB have been generated in bacteria, and these map to the interface between two RuvB molecules in the hexameric ring (Iwasaki et al. 2000; Putnam et al. 2001). These observations suggest that the lysine substitution of RUVB-1 may disrupt the formation or stability of the hexameric ring, despite the presence of WT RUVB-1 and RUVB-2 proteins. Thus, this mutation may be a weak dominant negative and not a simple loss of function. Consistent with this hypothesis, we observed dominant suppression of pha-4 lethality by ruvb-1(px34).
The role of RuvB in bacteria is DNA repair and recombination, where RuvB is required to translocate the Holliday junction along DNA (West 1996). Eukaryotes posses a RuvB homolog but not RuvA or RuvC, raising the question of what RuvB proteins do in higher organisms. Expression of ruvb-1 orthologs in Drosophila, Xenopus, zebrafish, and mice suggests a possible role in gut development. Strong expression of Pontin/ruvb-1 and Reptin/ruvb-2 is found in the anterior and posterior midgut of Drosophila (Bauer et al. 2000). In Xenopus, xPontin/ruvb-1 is expressed in different gastrointestinal organs (Etard et al. 2000). Zebrafish express higher levels of zReptin/ruvb-2 in the heart, brachial arches, liver, exocrine pancreas, and intestine (Rottbauer et al. 2002). In mice, robust expression of mPontin/ruvb-1 and mReptin/ruvb-2 is found in the gut and pancreas (Chauvet et al. 2005). The pharyngeal expression pattern we observed in C. elegans, together with the expression of RuvB genes in Drosophila, Xenopus, zebrafish, and mice, suggest an evolutionarily conserved role for RuvB in gut development.
It has been proposed that the paralogs ruvb-1/Pontin/Tip49 and ruvb-2/Reptin/Tip48 have antagonistic roles for wingless signaling in Drosophila and zebrafish (Bauer et al. 2000; Rottbauer et al. 2002). In contrast, Xenopus Xpontin and Xreptin appear synergistic (Etard et al. 2005). Our results resemble those of Xenopus, since both ruvb-1 and ruvb-2 negatively regulate pha-4 and pharyngeal development (Table 3). Biochemical assays suggest that RUVB-1 and RUVB-2 are present together in multiprotein complexes in other organisms (Ikura et al. 2000; Shen et al. 2000; Wood et al. 2000; Fuchs et al. 2001; Krogan et al. 2003; Doyon et al. 2004; Mizuguchi et al. 2004; Jin et al. 2005; Bakshi et al. 2006). Direct interaction between these paralogs has been reported in multiple systems, most likely in the form of a dodecameric RUVB-1/2 ring (Puri et al. 2007). This configuration may help explain why ruvb-1 and ruvb-2 have synergistic roles for Xenopus and C. elegans development (Gohshi et al. 1999; Kanemaki et al. 1999; Bauer et al. 2000; Ikura et al. 2000; Wood et al. 2000; Walhout et al. 2002; Puri et al. 2007).
Biochemical studies have identified RuvB orthologs as members of several multiprotein complexes including those that control DNA repair (RuvABC complex, TIP60/SRCAP, and URI-1), transcription [c-Myc, TIP60/SRCAP, and Polycomb repressive complex 1 (PRC1)], and ribosome biogenesis (snoRNP assembly complex) (Ikura et al. 2000; Shen et al. 2000; Fuchs et al. 2001; Jonsson et al. 2001; Frank et al. 2003; Krogan et al. 2003; Doyon et al. 2004; Kobor et al. 2004; Jonsson et al. 2004; Taubert et al. 2004). How might these interactions explain a role for ruvb-1/2 during pharyngeal development? Vertebrate FoxA binds chromatin and modulates compaction in vitro, indicating FoxA factors function by modifying the chromatin environment (Cirillo et al. 2002). Previous work from our lab has shown that homologs of the TIP60/SRCAP histone acetyltransferase and nucleosome-remodeling complex cooperate with pha-4 to achieve the proper timing and expression of pharyngeal genes (Updike and Mango 2006). Therefore, one possibility is that ruvb-1/2 modulates the activity of the TIP60/SRCAP complex. PRC1 is another attractive candidate, however C. elegans lacks an obvious correlate to this complex. We favor the notion that ruvb-1 and ruvb-2 modulate pha-4 activity through interactions with the chromatin environment.
Acknowledgments
We thank Dan Williams, Wayne Davis, Marc Hammarlund, and Erik Jorgensen for sharing unpublished information; Jeb Gaudet and numerous rotation students for help with the genetics; and Michel Labouesse and the National Bioresource Project for reagents. Some strains used in this work were obtained by the Caenorhabditis Genetics Center. This work was supported by T32 HD007491 to D.L.U. S.E.M. was supported by National Institutes of Health R01 GM056264, the Huntsman Cancer Institute, and the Department of Oncological Sciences.
References
- Alder, M. N., S. Dames, J. Gaudet and S. E. Mango, 2003. Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA 9: 25–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, S. L., and J. Rossant, 1994. HNF-3 beta is essential for node and notochord formation in mouse development. Cell 78: 561–574. [DOI] [PubMed] [Google Scholar]
- Ao, W., J. Gaudet, W. J. Kent, S. Muttumu and S. E. Mango, 2004. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science 305: 1743–1746. [DOI] [PubMed] [Google Scholar]
- Aronoff, R., R. Baran and J. Hodgkin, 2001. Molecular identification of smg-4, required for mRNA surveillance in C. elegans. Gene 268: 153–164. [DOI] [PubMed] [Google Scholar]
- Azzaria, M., B. Goszczynski, M. A. Chung, J. M. Kalb and J. D. McGhee, 1996. A fork head/HNF-3 homolog expressed in the pharynx and intestine of the Caenorhabditis elegans embryo. Dev. Biol. 178: 289–303. [DOI] [PubMed] [Google Scholar]
- Bakshi, R., A. K. Mehta, R. Sharma, S. Maiti, S. Pasha et al., 2006. Characterization of a human SWI2/SNF2 like protein hINO80: demonstration of catalytic and DNA binding activity. Biochem. Biophys. Res. Commun. 339: 313–320. [DOI] [PubMed] [Google Scholar]
- Bauer, A., S. Chauvet, O. Huber, F. Usseglio, U. Rothbacher et al., 2000. Pontin52 and reptin52 function as antagonistic regulators of beta-catenin signalling activity. EMBO J 19: 6121–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, A., O. Huber and R. Kemler, 1998. Pontin52, an interaction partner of beta-catenin, binds to the TATA box binding protein. Proc. Natl. Acad. Sci. USA 95: 14787–14792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beanan, M. J., and S. Strome, 1992. Characterization of a germ-line proliferation mutation in C. elegans. Development 116: 755–766. [DOI] [PubMed] [Google Scholar]
- Bessereau, J. L., A. Wright, D. C. Williams, K. Schuske, M. W. Davis et al., 2001. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature 413: 70–74. [DOI] [PubMed] [Google Scholar]
- Brenner, S., 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer et al., 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122: 33–43. [DOI] [PubMed] [Google Scholar]
- Chauvet, S., F. Usseglio, D. Aragnol and J. Pradel, 2005. Analysis of paralogous pontin and reptin gene expression during mouse development. Dev. Genes Evol. 215: 575–579. [DOI] [PubMed] [Google Scholar]
- Cirillo, L. A., F. R. Lin, I. Cuesta, D. Friedman, M. Jarnik et al., 2002. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell 9: 279–289. [DOI] [PubMed] [Google Scholar]
- Davis, M. W., M. Hammarlund, T. Harrach, P. Hullett, S. Olsen et al., 2005. Rapid single nucleotide polymorphism mapping in C. elegans. BMC Genomics 6: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyon, Y., W. Selleck, W. S. Lane, S. Tan and J. Cote, 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol. Cell Biol. 24: 1884–1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dufort, D., L. Schwartz, K. Harpal and J. Rossant, 1998. The transcription factor HNF3beta is required in visceral endoderm for normal primitive streak morphogenesis. Development 125: 3015–3025. [DOI] [PubMed] [Google Scholar]
- Etard, C., D. Gradl, M. Kunz, M. Eilers and D. Wedlich, 2005. Pontin and Reptin regulate cell proliferation in early Xenopus embryos in collaboration with c-Myc and Miz-1. Mech. Dev. 122: 545–556. [DOI] [PubMed] [Google Scholar]
- Etard, C., D. Wedlich, A. Bauer, O. Huber and M. Kuhl, 2000. Expression of Xenopus homologs of the beta-catenin binding protein pontin52. Mech. Dev. 94: 219–222. [DOI] [PubMed] [Google Scholar]
- Frank, S. R., T. Parisi, S. Taubert, P. Fernandez, M. Fuchs et al., 2003. MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4: 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman, J. R., and K. H. Kaestner, 2006. The Foxa family of transcription factors in development and metabolism. Cell. Mol. Life Sci. 63: 2317–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, M., J. Gerber, R. Drapkin, S. Sif, T. Ikura et al., 2001. The p400 complex is an essential E1A transformation target. Cell 106: 297–307. [DOI] [PubMed] [Google Scholar]
- Gaudet, J., and S. E. Mango, 2002. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science 295: 821–825. [DOI] [PubMed] [Google Scholar]
- Gaudet, J., S. Muttumu, M. Horner and S. E. Mango, 2004. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol. 2: e352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gengyo-Ando, K., and S. Mitani, 2000. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 269: 64–69. [DOI] [PubMed] [Google Scholar]
- Gohshi, T., M. Shimada, S. Kawahire, N. Imai, T. Ichimura et al., 1999. Molecular cloning of mouse p47, a second group mammalian RuvB DNA helicase-like protein: homology with those from human and Saccharomyces cerevisiae. J. Biochem. 125: 939–946. [DOI] [PubMed] [Google Scholar]
- Grimson, A., S. O'Connor, C. L. Newman and P. Anderson, 2004. SMG-1 is a phosphatidylinositol kinase-related protein kinase required for nonsense-mediated mRNA Decay in Caenorhabditis elegans. Mol. Cell. Biol. 24: 7483–7490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin, J., A. Papp, R. Pulak, V. Ambros and P. Anderson, 1989. A new kind of informational suppression in the nematode Caenorhabditis elegans. Genetics 123: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner, M. A., S. Quintin, M. E. Domeier, J. Kimble, M. Labouesse et al., 1998. pha-4, an HNF-3 homolog, specifies pharyngeal organ identity in Caenorhabditis elegans. Genes Dev. 12: 1947–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura, T., V. V. Ogryzko, M. Grigoriev, R. Groisman, J. Wang et al., 2000. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 102: 463–473. [DOI] [PubMed] [Google Scholar]
- Iwasaki, H., Y. W. Han, T. Okamoto, T. Ohnishi, M. Yoshikawa, et al., 2000. Mutational analysis of the functional motifs of RuvB, an AAA+ class helicase and motor protein for holliday junction branch migration. Mol. Microbiol. 36: 528–538. [DOI] [PubMed] [Google Scholar]
- Jin, J., Y. Cai, T. Yao, A. J. Gottschalk, L. Florens et al., 2005. A mammalian chromatin remodeling complex with similarities to the yeast INO80 complex. J. Biol. Chem. 280: 41207–41212. [DOI] [PubMed] [Google Scholar]
- Jonsson, Z. O., S. K. Dhar, G. J. Narlikar, R. Auty, N. Wagle et al., 2001. Rvb1p and Rvb2p are essential components of a chromatin remodeling complex that regulates transcription of over 5% of yeast genes. J. Biol. Chem. 276: 16279–16288. [DOI] [PubMed] [Google Scholar]
- Jonsson, Z. O., S. Jha, J. A. Wohlschlegel and A. Dutta, 2004. Rvb1p/Rvb2p recruit Arp5p and assemble a functional Ino80 chromatin remodeling complex. Mol. Cell. 16: 465–477. [DOI] [PubMed] [Google Scholar]
- Jorgensen, E. M., and S. E. Mango, 2002. The art and design of genetic screens: caenorhabditis elegans. Nat. Rev. Genet. 3: 356–369. [DOI] [PubMed] [Google Scholar]
- Jurgens, G., and D. Weigel, 1988. Terminal versus segmental development in the Drosophila embryo: the role of the homeotic gene fork head. Rouxs Arch. Dev. Biol. 197: 345–354. [DOI] [PubMed] [Google Scholar]
- Jurgens, G., E. Wieschaus, C. Nusslein-Volhard and H. Kluding, 1984. Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II: Zygotic loci on the third chromosome. Rouxs Arch. Dev. Biol. 193: 283–295. [DOI] [PubMed] [Google Scholar]
- Kaestner, K. H., W. Knochel and D. E. Martinez, 2000. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 14: 142–146. [PubMed] [Google Scholar]
- Kalb, J. M., L. Beaster-Jones, A. P. Fernandez, P. G. Okkema, B. Goszczynski et al., 2002. Interference between the PHA-4 and PEB-1 transcription factors in formation of the Caenorhabditis elegans pharynx. J. Mol. Biol. 320: 697–704. [DOI] [PubMed] [Google Scholar]
- Kalb, J. M., K. K. Lau, B. Goszczynski, T. Fukushige, D. Moons et al., 1998. pha-4 is Ce-fkh-1, a fork head/HNF-3alpha,beta,gamma homolog that functions in organogenesis of the C. elegans pharynx. Development 125: 2171–2180. [DOI] [PubMed] [Google Scholar]
- Kaltenbach, L. S., D. L. Updike and S. E. Mango, 2005. Contribution of the amino and carboxyl termini for PHA-4/FoxA function in Caenorhabditis elegans. Dev. Dyn. 234: 346–354. [DOI] [PubMed] [Google Scholar]
- Kamath, R. S., A. G. Fraser, Y. Dong, G. Poulin, R. Durbin et al., 2003. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421: 231–237. [DOI] [PubMed] [Google Scholar]
- Kanemaki, M., Y. Kurokawa, T. Matsu-ura, Y. Makino, A. Masani et al., 1999. TIP49b, a new RuvB-like DNA helicase, is included in a complex together with another RuvB-like DNA helicase, TIP49a. J. Biol. Chem. 274: 22437–22444. [DOI] [PubMed] [Google Scholar]
- Kanemaki, M., Y. Makino, T. Yoshida, T. Kishimoto, A. Koga et al., 1997. Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem. Biophys. Res. Commun. 235: 64–68. [DOI] [PubMed] [Google Scholar]
- Kemphues, K. J., J. R. Priess, D. G. Morton and N. S. Cheng, 1988. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell 52: 311–320. [DOI] [PubMed] [Google Scholar]
- Kiefer, J. C., P. A. Smith and S. E. Mango, 2007. PHA-4/FoxA cooperates with TAM-1/TRIM to regulate cell fate restriction in the C. elegans foregut. Dev. Biol. 303: 611–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobor, M. S., S. Venkatasubrahmanyam, M. D. Meneghini, J. W. Gin, J. L. Jennings et al., 2004. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2: E131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N. J., M. C. Keogh, N. Datta, C. Sawa, O. W. Ryan et al., 2003. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell 12: 1565–1576. [DOI] [PubMed] [Google Scholar]
- Labouesse, M., E. Hartwieg and H. R. Horvitz, 1996. The Caenorhabditis elegans LIN-26 protein is required to specify and/or maintain all non-neuronal ectodermal cell fates. Development 122: 2579–2588. [DOI] [PubMed] [Google Scholar]
- Laganiere, J., G. Deblois, C. Lefebvre, A. R. Bataille, F. Robert et al., 2005. From the Cover: Location analysis of estrogen receptor alpha target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc. Natl. Acad. Sci. USA 102: 11651–11656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, E., V. R. Prezioso, E. Smith, O. Litvin, R. H. Costa et al., 1990. HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 4: 1427–1436. [DOI] [PubMed] [Google Scholar]
- Lai, E., V. R. Prezioso, W. F. Tao, W. S. Chen and J. E. Darnell, Jr., 1991. Hepatocyte nuclear factor 3 alpha belongs to a gene family in mammals that is homologous to the Drosophila homeotic gene fork head. Genes Dev. 5: 416–427. [DOI] [PubMed] [Google Scholar]
- Lee, C. S., J. R. Friedman, J. T. Fulmer and K. H. Kaestner, 2005. The initiation of liver development is dependent on Foxa transcription factors. Nature 435: 944–947. [DOI] [PubMed] [Google Scholar]
- Mango, S. E., 2001. Stop making nonSense: the C. elegans smg genes. Trends Genet. 17: 646–653. [DOI] [PubMed] [Google Scholar]
- Mango, S. E., 2007. The C. elegans pharynx: a model for organogenesis, in Wormbook, edited by The C. elegans Research Community. Wormbook (http://www.wormbook.org). [DOI] [PMC free article] [PubMed]
- Mango, S. E., E. J. Lambie and J. Kimble, 1994. The pha-4 gene is required to generate the pharyngeal primordium of Caenorhabditis elegans. Development 120: 3019–3031. [DOI] [PubMed] [Google Scholar]
- Mann, R. S., and S. B. Carroll, 2002. Molecular mechanisms of selector gene function and evolution. Curr. Opin. Genet. Dev. 12: 592–600. [DOI] [PubMed] [Google Scholar]
- Matias, P. M., S. Gorynia, P. Donner and M. A. Carrondo, 2006. Crystal structure of the human AAA+ protein RuvBL1. J. Biol. Chem. 281: 38918–38929. [DOI] [PubMed] [Google Scholar]
- McKim, K. S., M. F. Heschl, R. E. Rosenbluth and D. L. Baillie, 1988. Genetic organization of the unc-60 region in Caenorhabditis elegans. Genetics 118: 49–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitrovich, Q. M., and P. Anderson, 2000. Unproductively spliced ribosomal protein mRNAs are natural targets of mRNA surveillance in C. elegans. Genes Dev. 14: 2173–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi, G., X. Shen, J. Landry, W. H. Wu, S. Sen et al., 2004. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348. [DOI] [PubMed] [Google Scholar]
- Ohnishi, T., T. Hishida, Y. Harada, H. Iwasaki and H. Shinagawa, 2005. Structure-function analysis of the three domains of RuvB DNA motor protein. J. Biol. Chem. 280: 30504–30510. [DOI] [PubMed] [Google Scholar]
- Overdier, D. G., A. Porcella and R. H. Costa, 1994. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol. Cell. Biol. 14: 2755–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panowski, S. H., S. Wolff, H. Aguilaniu, J. Durieux and A. Dillin, 2007. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature 447: 550–555. [DOI] [PubMed] [Google Scholar]
- Piano, F., A. J. Schetter, D. G. Morton, K. C. Gunsalus, V. Reinke et al., 2002. Gene clustering based on RNAi phenotypes of ovary-enriched genes in C. elegans. Curr. Biol. 12: 1959–1964. [DOI] [PubMed] [Google Scholar]
- Priess, J. R., and J. N. Thomson, 1987. Cellular interactions in early C. elegans embryos. Cell 48: 241–250. [DOI] [PubMed] [Google Scholar]
- Pulak, R., and P. Anderson, 1993. mRNA surveillance by the Caenorhabditis elegans smg genes. Genes Dev. 7: 1885–1897. [DOI] [PubMed] [Google Scholar]
- Puri, T., P. Wendler, B. Sigala, H. Saibil and I. R. Tsaneva, 2007. Dodecameric structure and ATPase activity of the human TIP48/TIP49 Complex. J. Mol. Biol. 366: 179–192. [DOI] [PubMed] [Google Scholar]
- Putnam, C. D., S. B. Clancy, H. Tsuruta, S. Gonzalez, J. G. Wetmur et al., 2001. Structure and mechanism of the RuvB Holliday junction branch migration motor. J. Mol. Biol. 311: 297–310. [DOI] [PubMed] [Google Scholar]
- Raharjo, I., and J. Gaudet, 2007. Gland-specific expression of C. elegans hlh-6 requires the combinatorial action of three distinct promoter elements. Dev. Biol. 302: 295–308. [DOI] [PubMed] [Google Scholar]
- Reece-Hoyes, J. S., J. Shingles, D. Dupuy, C. A. Grove, A. J. Walhout et al., 2007. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics 8: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottbauer, W., A. J. Saurin, H. Lickert, X. Shen, C. G. Burns et al., 2002. Reptin and pontin antagonistically regulate heart growth in zebrafish embryos. Cell 111: 661–672. [DOI] [PubMed] [Google Scholar]
- Sha, K., and A. Fire, 2005. Imprinting capacity of gamete lineages in Caenorhabditis elegans. Genetics 170: 1633–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, X., G. Mizuguchi, A. Hamiche and C. Wu, 2000. A chromatin remodelling complex involved in transcription and DNA processing. Nature 406: 541–544. [DOI] [PubMed] [Google Scholar]
- Simmer, F., C. Moorman, A. M. van der Linden, E. Kuijk, P. V. van den Berghe et al., 2003. Genome-wide RNAi of C. elegans using the hypersensitive rrf-3 strain reveals novel gene functions. PLoS Biol. 1: e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome, S., and W. B. Wood, 1982. Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 79: 1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston, J. E., and J. Hodgkin, 1988. Methods: In the Nematode C. elegans. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Taubert, S., C. Gorrini, S. R. Frank, T. Parisi, M. Fuchs et al., 2004. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol. Cell. Biol. 24: 4546–4556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike, D. L., and S. E. Mango, 2006. Temporal regulation of foregut development by HTZ-1/H2A.Z and PHA-4/FoxA. PLoS Genet 2: e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilimas, T., A. Abraham and P. G. Okkema, 2004. An early pharyngeal muscle enhancer from the Caenorhabditis elegans ceh-22 gene is targeted by the Forkhead factor PHA-4. Dev. Biol. 266: 388–398. [DOI] [PubMed] [Google Scholar]
- Walhout, A. J., J. Reboul, O. Shtanko, N. Bertin, P. Vaglio et al., 2002. Integrating interactome, phenome, and transcriptome mapping data for the C. elegans germline. Curr. Biol. 12: 1952–1958. [DOI] [PubMed] [Google Scholar]
- Weigel, D., G. Jurgens, F. Kuttner, E. Seifert and H. Jackle, 1989. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell 57: 645–658. [DOI] [PubMed] [Google Scholar]
- Weinstein, D. C., A. Ruiz i Altaba, W. S. Chen, P. Hoodless, V. R. Prezioso et al., 1994. The winged-helix transcription factor HNF-3 beta is required for notochord development in the mouse embryo. Cell 78: 575–588. [DOI] [PubMed] [Google Scholar]
- West, S. C., 1996. DNA helicases: new breeds of translocating motors and molecular pumps. Cell 86: 177–180. [DOI] [PubMed] [Google Scholar]
- Wicks, S. R., R. T. Yeh, W. R. Gish, R. H. Waterston and R. H. Plasterk, 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Williams, D. C., T. Boulin, A. F. Ruaud, E. M. Jorgensen and J. L. Bessereau, 2005. Characterization of Mos1-mediated mutagenesis in Caenorhabditis elegans: a method for the rapid identification of mutated genes. Genetics 169: 1779–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, M. A., S. B. McMahon and M. D. Cole, 2000. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell 5: 321–330. [DOI] [PubMed] [Google Scholar]
- Yochem, J., T. Gu and M. Han, 1998. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics 149: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret, K. S., 2002. Regulatory phases of early liver development: paradigms of organogenesis. Nat. Rev. Genet. 3: 499–512. [DOI] [PubMed] [Google Scholar]