Abstract
Flightless I (Fli I) is an evolutionarily conserved member of the gelsolin family, containing actin-binding and severing activity in vitro. The physiological function of Fli I during animal development remains largely undefined. In this study, we reveal a key role of the Caenorhabditis elegans Fli I homolog, fli-1, in specifying asymmetric cell division and in establishing anterior–posterior polarity in the zygote. The fli-1 gene also regulates the cytokinesis of somatic cells and the development of germline and interacts with the phosphoinositol-signaling pathway in the regulation of ovulation. The fli-1 reporter gene shows that the localization of FLI-1 coincides with actin-rich regions and that the actin cytoskeleton is impaired in many tissues in the fli-1 mutants. Furthermore, the function of fli-1 in C. elegans can be functionally substituted by the Drosophila Fli I. Our studies demonstrate that fli-1 plays an important role in regulating the actin-dependent events during C. elegans development.
THE actin microfilament cytoskeleton regulates multiple cellular processes, including cytokinesis, cell morphology, establishment of cell polarity, and cell motility. Actin-binding proteins, such as the members of the gelsolin family, have been shown to modulate the actin filament network by regulating the polymerization and depolymerization of actin (for review, see Sun et al. 1999). The actin filament severing and capping activity of gelsolin is regulated by Ca2+ and phosphoinositol 4,5-bisphosphate (PIP2) (for review, see Kwiatkowski 1999; Silacci et al. 2004). Despite their crucial role in regulating actin dynamics in tissue cultures, knockouts of gelsolin and several other related genes are viable in mice, indicating the existence of compensatory mechanisms in the regulation of actin cytoskeleton turnover and reorganization (Witke et al. 1995). Consequently, this functional redundancy of the actin-binding proteins complicates study of their physiological roles during animal development.
Flightless I (Fli I) is a unique member of the gelsolin family of proteins (Campbell et al. 1993). In addition to the gelsolin domain, Fli I contains 16 tandem leucine-rich repeats (LRR) at its N terminus. The LRR motif is known to be involved in protein–protein or protein–lipid interactions, suggesting a role for Fli I in linking the actin cytoskeleton with the signal transduction pathways (Claudianos and Campbell 1995; Campbell et al. 1997). Consistent with having the gelsolin domain, the human and Caenorhabditis elegans FLI-1 proteins bind to both actin monomers (G-actin) and actin filaments (F-actin) in vitro and possess F-actin-serving activity (Liu and Yin 1998; Goshima et al. 1999). Unlike other gelsolin family proteins, the actin-binding and severing activity of Fli I appears to be calcium independent (Goshima et al. 1999). Consistently, the residues essential for calcium binding in the gelsolin region of the other gelsolin family proteins are not conserved in Fli I (Goshima et al. 1999).
Analysis of the viable and lethal mutants in Drosophila suggests that Fli I may be involved in regulating the actin cytoskeleton reorganization (De Couet et al. 1995; Straub et al. 1996). Depletion of Fli I causes a defect in the cellularization of the syncytial blastoderm during early embryogenesis, a process with similarities to cytokinesis (Straub et al. 1996). This defect is associated with a disorganized cortical actin cytoskeleton in the embryo (Straub et al. 1996). However, flies lacking Fli I do not have defects in cytokinesis at other developmental stages, nor is Fli I needed for cell division of the germline (Straub et al. 1996). In mice, the Fli I knockout is embryonic lethal (Campbell et al. 2002). In humans, the Fli I locus is mapped to a region deleted in Smith–Magenis syndrome, a disorder that exhibits many developmental and behavioral abnormalities (Chen et al. 1995). Thus, elucidating the physiological function of Fli I during animal development will help us to understand the causes of the defects associated with Fli I loss of function in mammals.
Studies in C. elegans have revealed an essential role of the actin cytoskeleton in the establishment of cell polarity, asymmetric distribution of cell fate determinants, and morphogenesis (Strome and Wood 1983; Strome 1986). The establishment of cell polarity has been extensively characterized in the division of the one-cell-stage embryo (Albertson 1984; Gonczy et al. 1999). A polarized cytoplasmic flow, which involves the simultaneous movement of cortical cytoplasm away from and the interior cytoplasm toward the sperm pronucleus, occurs during the pronuclear stage (for review, see Cowan and Hyman 2004; Nance 2005). Correlated with the polarized cytoplasmic flow, some cytoplasmic components become asymmetrically distributed. For example, the germline-specific P granules are segregated exclusively in the posterior end of the P1 blastomere after first cell division (Hird et al. 1996; Kemphues and Strome 1997). Depolymerization of the actin microfilaments by cytochalasin D blocks polarized cytoplasmic flow, prevents P-granule segregation, and causes other losses in anterior/posterior (A/P) asymmetry (Hill and Strome 1988). Coordination of the dynamics of the actin cytoskeleton within different tissues also regulates other more complex biological processes during C. elegans development. For example, successful ovulation requires the orchestrated actions of the gonad sheath cell contracting and the spermatheca dilating (McCarter et al. 1997; Clandinin et al. 1998). Therefore, C. elegans offers a model by which to study comprehensively the physiological role of actin-binding proteins in the regulation of actin turnover and reorganization.
Here, we show that mutations in the C. elegans Fli I homolog, fli-1, cause defects in actin-based events, including cytokinesis, the establishment of cell polarity, asymmetric cell division, and ovulation. fli-1 is expressed in actin-rich regions. Abnormalities in the organization of actin filaments exist in fli-1 mutants. The function of fli-1 can be functionally substituted by the fly Fli I, suggesting a conserved role of Fli I in regulating the dynamics of the actin cytoskeleton during animal development.
MATERIALS AND METHODS
Strains and alleles:
The following mutant alleles were used in this study: LG I, lfe-2(sy326); LG III, fli-1(bp130), sma-3(e491), unc-32(e189), ruIs32(pie-1∷GFP∷H2B); LG IV, jcIs1(ajm-1∷gfp); LG V, pgl-1∷gfp, zuIs45(nmy-2∷gfp), wIs51(scm∷gfp), him-5(1490), bxIs14(pkd-2∷gfp); LG X, ipp-5(sy605). The location for qIs56(lim-7∷GFP) was not determined.
Identification, genetic mapping, and molecular cloning of fli-1:
fli-1(bp130) was identified in a screen to isolate mutants with altered numbers of seam cells in young adult animals. In brief, strains carrying the seam-cell-specific marker, scm∷gfp, were mutagenized by EMS and mutants with increased or reduced numbers of seam cells were cloned. From 6000 haploid genomes screened, 31 mutants were obtained. fli-1 mutants have reduced numbers of seam cells and other defects, including reduced brood size and defects in the development of germline and tail morphogenesis.
Three-factor mapping placed fli-1 between sma-3(-0.93) and unc-32(0.0) on linkage group III at an approximate chromosomal position of −0.1. From the + fli-1 +/sma-3 + unc-32 cross, 9 of 10 Sma non-Unc recombinants and 1 of 11 Unc non-Sma recombinants carried the fli-1 mutation.
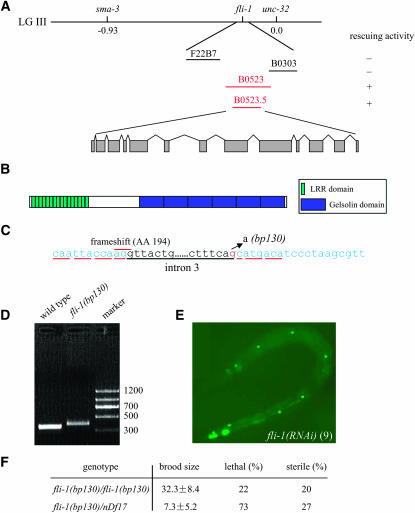
To identify the fli-1 gene in this region, cosmid DNA (provided by Alan Coulson at the Sanger Center, Cambridge, UK) was co-injected with the rol-6(su1006) dominant marker into fli-1/qC1; scm∷gfp animals. Heterozygous F1 Rol transformants were picked and stable transgenic lines were obtained. An individual F2 Rol animal was cloned and its progeny that did not segregate Dpy worms (qC1 animals are Dpy) were further analyzed for the number of seam cells and Emo phenotypes. We found that cosmid B0523 rescued the fli-1 mutant defects. The five candidate genes located in B0523 were PCR amplified for delimiting the rescuing activity and we found that B0523.5 gave rescue activity.
The fli-1(bp130) mutation was determined by sequencing the PCR products from the corresponding genomic sequence. To determine whether the bp130 mutation affects the fli-1 splicing, RT–PCR was performed using primers located in exon 3 (5′-atgcttccaccacagattcg-3′) and exon 5 (5′-ctgtcaattgattatggctc-3′) of fli-1 and the PCR products were sequenced.
Time-lapse recordings:
Time-lapse Nomarski images were performed as described previously (McCarter et al. 1997). In brief, worms were anesthetized in 0.1% tricaine and 0.01% tetramisole in M9 for 30 min, mounted on 2% agarose pads, and then observed under a Zeiss Axivert microscope with a ×100 Fluor objective (numerical aperture 1.3). Images were captured by CCD (AxioCam) and recorded every 10 sec for 60–100 min (Axiovision Re14.2).
RNA interference:
Single-stranded RNA was transcribed from the T7- and SP6-flanked PCR templates. The primers used for amplifying the fli-1 templates for synthesizing RNA are 5′-cactagatttaggtgacactatagacgaacaggtgcctgatgagctg-3′ and 5′-cactagtaatacgactcactatagccgacgccagcgattttcgac-3′. The double-stranded RNA was then injected into animals carrying the scm∷gfp reporter. Eggs laid by the injected animals between 4 and 48 hr were collected for further analysis. The average number of seam cells was 13.4 (ranging from 11 to 15, n = 20) in fli-1(RNAi) animals. Furthermore, 2 of 16 fli-1(RNAi) animals showed defects in tail morphogenesis and 6 of 16 fli-1(RNAi) animals showed the endomitotic oocyte (EMO) phenotype.
Construction of fli-1∷gfp reporter and fli-1∷Fli I cDNA:
The fli-1∷gfp reporter was constructed with a PCR-fusion-based approach. The fused PCR products were derived from two overlapping PCR DNA fragments. One contained the DNA derived from fosmid WRM0621aG11 (nt 20547–30147), which includes a 2-kb promoter region and the entire ORF of fli-1. Another one contained gfp and the unc-54 3′-UTR from pPD95.67 (the nuclear localization signal was not included in our reporters). The reporter DNA was co-injected with pRF4(rol-6). fli-1∷gfp rescued the fli-1 mutant defects (Table 2).
TABLE 2.
The phenotypes of fli-1(bp130) mutant animals carrying various transgenes
| Genotype | Brood size | No. of germ cellsa | EMOb | No. of seam cells |
|---|---|---|---|---|
| Wild type | 189.0 ± 32.2 (n = 5) | 320.4 ± 42.5 (n = 5) | 0 (n = 5) | 16.3 ± 0.4 (n = 32) |
| fli-1(bp130) | 13.0 ± 8.6 (n = 8) | 78.4 ± 34.2 (n = 12) | 6 (n = 8)c | 11.2 ± 0.5 (n = 83) |
| fli-1(bp130); fli-1∷gfp | ||||
| Line 1 | 32.0 ± 8.4 (n = 2) | 189.3 ± 10.1 (n = 3) | 1 (n = 10) | 14.5 ± 1.3 (n = 4) |
| Line 2 | 38.5 ± 4.9 (n = 4) | 192.5 ± 30.1 (n = 4) | 1 (n = 9) | 15.3 ± 0.75 (n = 6) |
| Line 3 | 68.3 ± 7.4 (n = 3) | 210.0 ± 62.3 (n = 4) | 0 (n = 12) | 15.7 ± 1.0 (n = 6) |
| Line 4 | 58.7 ± 6.3 (n = 3) | 86.5 ± 32.4 (n = 2) | 2 (n = 11) | 15.6 ± 1.3 (n = 8) |
| fli-1(bp130); scm∷fli-1∷gfp | ||||
| Line 1 | 18.2 ± 5.8 (n = 5) | 76.3 ± 8.9 (n = 3) | 8 (n = 8) | 14.7 ± 1.6 (n = 6) |
| Line 2 | 11.6 ± 6.0 (n = 3) | 89.2 ± 12.6 (n = 5) | 8 (n = 9) | 16.2 ± 0.6 (n = 7) |
| Line 3 | 12.6 ± 2.3 (n = 3) | 92.5 ± 10.6 (n = 2) | 7 (n = 7) | 13.8 ± 1.2 (n = 4) |
| fli-1(bp130); Fli I∷gfp | ||||
| Line 1 | 64.3 ± 10.6 (n = 3) | 168.3 ± 34.2 (n = 3) | 1 (n = 8) | 15.7 ± 2.3 (n = 7) |
| Line 2 | 74.5 ± 13.8 (n = 4) | 154.5 ± 20.2 (n = 4) | 0 (n = 9) | 16.2 ± 1.2 (n = 6) |
| Line 3 | 56.0 ± 5.7 (n = 2) | 192.0 ± 27.7 (n = 4) | 1 (n = 12) | 14.3 ± 1.5 (n = 9) |
The number of germ cells was counted in each gonad arm by DAPI staining.
The number of animals that displayed the EMO phenotype.
The other two mutants were sterile. The total number of animals examined are shown in parentheses.
The chimeric gene fli-1(promoter)∷Fli I cDNA was constructed in the following way: the full length of fly Fli I cDNA was subcloned into the pPD95.67 backbone that contains the fli-1 promoter (WRM0621aG11, nt 28146–30147) gfp and unc-54 3′-UTR. The construct was injected into fli-1/qC1 animals (Table 2).
Immunostaining:
A rapid one-step fixation/permeabilization/staining procedure was used for R-phalloidin (R-ph) visualization of F-actin as previously described by Strome (1986). In brief, the dissected animal parts, such as gonad arms, or the cut gravid adult hermaphrodites (for body muscle staining) were placed in 5 μl M9 on a polylysine-treated microscope slide. Samples were fixed (1.5% paraformaldehyde, 0.1% glutaraldehyde) and stained with R-phalloidin (0.33 μm in M9) for 20–30 min at room temperature, washed by PBS, and then observed. As for staining of F-actin in embryos, the eggs were collected from the bleached gravid adult hermaphrodites. Embryos were washed twice in M9 buffer and then were fixed and stained.
Dye-filling assays of the phasmid:
The worms were stained with 25 μg/ml 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate solution at room temperature for 2 hr and then destained for 1 hr. The stained animals were visualized under a fluorescence microscope using a rhodamine filter.
RESULTS
The fli-1 mutants display defects in the asymmetric cell division at post-embryonic stage:
Defects in the post-embryonic development of seam cells:
In wild-type animals, seam cells divide at each of the four larval stages, with one daughter cell fusing with the hypodermal syncytium, hyp7, and the other daughter cell maintaining seam cell fate for further division. At the L2 larval stage, seam cells H1, V1–V4, and V6 undergo an extra round of symmetric cell division with both daughter cells adopting seam cell fate, resulting in an increase in the number of seam cells from 10 at the hatching stage to 16 at the later L2 larval stage and onward (Figure 1, A and B) (Sulston and Horvitz 1977). To determine how the stage-specific seam cell division is specified, we performed genetic screens to identify mutants with altered numbers of seam cells at the adult stage. The bp130 mutation that caused a reduced number of seam cells was identified (Figure 1C). In the bp130 mutant adults, the average number of seam cells was 11.2 (n = 83, ranging from 6 to 14), compared to an average number of 16.3 (n = 32, ranging from 15 to 17) in wild-type animals. Subsequent genetic and molecular analysis indicated that bp130 encodes the C. elegans Fli I homolog, fli-1. fli-1(bp130) showed a maternal effect (Table 1) and only the fli-1(bp130) mutants derived from homozygous fli-1(bp130) were analyzed in the subsequent studies unless otherwise noted.
Figure 1.—
Defects in the post-embryonic development of seam cells in the fli-1 mutants. (A) The post-embryonic division pattern of a subset of seam cells. The proliferative S2 seam cell division is highlighted by the red box. Seam cell T undergoes a distinct division pattern at the L1 larval stage, with one daughter cell generating neuronal structures (highlighted by the red circle) and the other maintaining seam cell fate. The post-embryonic stages are indicated along the vertical axis, separated by larval stage. (B) Sixteen seam cells, visualized by the seam-cell-specific marker scm∷gfp, are evenly distributed along the anterior–posterior axis in a wild-type young adult. (C) In the fli-1 mutant animal shown, there were 11 seam cells, which were also unevenly distributed. (D) Defects in the asymmetric seam cell division in the fli-1 mutants. Both V1 daughter cells adopted seam cell fate, while both V4 daughter cells failed to adopt the seam cell fate (The V4 daughters remained unfused with hyp7 at the stage shown and thus retained a weak gfp signal.) (E) Two cells labeled by ajm-1∷gfp (arrows) did not express scm∷gfp in a fli-1 mutant animal. (F) Presence of two nuclei in one seam cell (arrows) in a fli-1 mutant animal, indicating a defect in the cytokinesis. (G) The phasmid structure, which takes up the dye in the dye-filling assay, is generated on each side of a wild-type animal (two socket neurons in each phasmid can be stained by the dye). (H) Failure of staining with dye in a fli-1 mutant side. (I and J) Asymmetric cell division of seam cell T in a wild-type L2 larva. The anterior daughter cell (arrow in I) maintains the hypodermal seam cell fate (expressing scm∷gfp) and the posterior daughter adopts a neuronal fate, which has a distinct nuclear morphology. (K and L) In a fli-1 mutant larva, both daughter cells of T had a hypodermal cell appearance and expressed scm∷gfp (arrows in K and L). (I and K) Nomarski micrograph. (J and L) Expression of scm∷gfp in the same animal shown in I and K, respectively.
TABLE 1.
fli-1(bp130) mutants display maternal effect
| Lethality
|
||||||
|---|---|---|---|---|---|---|
| Brood sized | No. of germ cellse | Sterility (%) | Embryo(%) | Larvae (%) | Nf | |
| fli-1(bp130)a | 32.3 ± 8.4 (n = 6) | 124.2 ± 32.2 (n = 9) | 19.5 | 11.7 | 9.9 | 194 |
| fli-1(bp130)b | 13.0 ± 8.6 (n = 8) | 78.4 ± 20.4 (n = 12) | 24.5 | 32.3 | 21.5 | 104 |
| Wild typec | 189.0 ± 32.2 (n = 5) | 320.4 ± 42.5 (n = 5) | 4.0 | 1.5 | 0.5 | 945 |
Six fli-1 mutant animals derived from m/+ hermaphrodites were analyzed for brood size, sterility, and lethality.
Eight fli-1 mutant animals derived from fli-1(bp130)/fli-1(bp130) mother were analyzed.
Five animals carrying the wIs51(scm∷gfp) transgene were analyzed.
Brood size refers to the eggs laid by the mother.
The number of germ cells was counted in each gonad arm by DAPI staining.
The number of progeny analyzed.
We further analyzed seam cell division in the fli-1 mutants and found that the asymmetric cell division of seam cells was defective, resulting in both daughter cells either fusing with hyp 7 or retaining seam cell fates (Figure 1D). Twenty-five percent and 67% of fli-1 mutant animal sides (n = 21, seam cells on each side of the animal develop independently, representing separate developmental processes) have one or more seam cells with both daughter cells retaining the seam cell fate or fusing with hyp 7, respectively. Other abnormalities in the seam cell development in fli-1 mutants included the transformation of the seam cell fate to other hypodermal cell types and failure to divide (Figure 1, E and F). In 15% of fli-1 mutant animal sides (n = 32), some daughter seam cells expressed the epidermal cell marker ajm-1∷gfp, but failed to express the seam-cell-specific marker, scm∷gfp (Figure 1E), indicating a change of the seam cell fate to other hypodermal cell fates. Moreover, in 9% of mutant animal sides (n = 32), two nuclei were present in one cell, indicating a defect in the cytokinesis of seam cells (Figure 1F). Thus, the development of seam cells shows multiple defects in fli-1 mutants.
We further characterized the development of seam cell T, which undergoes asymmetric cell division with the anterior daughter cell maintaining seam cell fate and the posterior daughter cell acquiring a neuronal fate, giving rise to a group of neuronal cells, called phasmid (Figure 1A). The phasmid can be detected by its ability to take up dye (Figure 1G) (Herman and Horvitz 1994). One or both of the two socket cells in the phasmid failed to take up dye in the dye-filling assay in 46 and 10% of the fli-1 mutant animal sides (n = 100), respectively (Figure 1H). Consistent with this, we found that both daughters of T, T.a and T.p, expressed the seam hypodermal fate in 33% of the animal sides (n = 12) (Figure 1, I–L). In summary, asymmetric cell division of seam cells is defective in the fli-1 mutants.
Defects in the development and division of distal tip cells:
The development of distal tip cells (DTCs), located at the tip of each gonad arm (Figure 2, A and B), was also analyzed in the fli-1 mutants. The DTC plays an essential role in the migration and extension of the gonad arm. In 12% of the fli-1 mutant animals (n = 43), two DTCs (labeled by a lag-2∷gfp reporter) were present in one gonad arm (Figure 2C). Consistently, two gonad branches were formed in those gonad arms containing two DTCs (Figure 2D). In 5% of the fli-1 mutant branches, two nuclei were found in one DTC, indicating a defect of the cytokinesis (data not shown). The morphology of DTC was also abnormal in the fli-1 mutants. In wild-type young adult animals, the processes of DTC extend and branch down the side of the germline (Figure 2E) (Finger et al. 2003). In 72% of the fli-1 mutant young adults (n = 45), the processes of the DTCs were much longer and disorganized (Figure 2F).
Figure 2.—
Defects in the development of somatic gonad in the fli-1 mutants (A and B) One DTC, labeled by lag-2∷gfp, is present on each gonad arm in a wild-type hermaphrodite. The tip of the gonad, expressing lag-2∷gfp, is indicated by an arrow (B). (C and D) In a fli-1 mutant animal, two DTCs were present on one side. Consistently, two gonad branches were present (arrows in D). (E) The processes of DTC (arrow) extend down the side of the germline and contain two longer branches at the outer edges in a wild-type hermaphrodite gonad. (F) The processes of DTC (arrow) were much longer and less organized in a fli-1 mutant. (G) Migration of the gonad arm in a wild-type hermaphrodite. The gonad grows out of the vulva position, then makes a turn from the ventral to the dorsal site, and then reorients and migrates back to the mid-body. The anterior gonad is shown in G and H. (H) Defects in the migration of the gonad in a fli-1 mutant hermaphrodite. The turning from the ventral to the dorsal site was defective in the mutant gonad arm shown (arrow).
The migration of the gonad was also defective in the fli-1 mutants. Normally, in wild-type animals, the gonad migrates away from the mid-body region and then makes a turn from the ventral to the dorsal site. Finally, it reorients and migrates back toward the mid-body (Figure 2G) (Finger et al. 2003). However, in the fli-1 mutants, 67% of the gonad branches (n = 102) migrated for only a short distance along the ventral body-wall muscles. Forty-five percent of the mutant gonad branches (n = 102) showed defects in the migration from the ventral to the dorsal site (Figure 2H), while 54% (n = 102) showed defects in the migration back to the mid-body (note that a single gonad arm could display multiple migration defects). Taken together, wild-type fli-1 is required for the DTC and gonadal development.
Reduction of the function of fli-1 causes defects in the establishment of the A/P polarity and cytokinesis in the first mitotic cell cycle:
To further determine the role of fli-1 in asymmetric cell division, we analyzed the first mitotic cell division in living fli-1 mutant embryos by time-lapse recording. In wild-type zygotes, the polarized cytoplasmic flow, shown by the movement of the cortical yolk droplets, occurs rapidly in the posterior region of the embryo (see supplemental Movie 1 at http://www.genetics.org/supplemental/) (Hird and White 1993; Hird et al. 1996). Correlated with the polarized flow, the maternal pronucleus migrates to meet the paternal pronucleus in the posterior hemisphere of the embryo, ∼70% of the length of the embryo (Figure 3A) (for review, see Cowan and Hyman 2004). After the pronuclear meeting, the first mitotic spindle forms and becomes displaced posteriorly, resulting in the production of a smaller P1 cell and a larger AB cell (Figure 3E; supplemental Movie 1). We found that reducing the function of fli-1 caused multiple defects in the division of the zygotes. The movement of the yolk droplets was greatly reduced in the fli-1 mutants (see supplemental Movie 2). The polarized localization of P granules in the posterior end of the embryo and the asymmetric meeting of the pronuclei were also severely impaired in the fli-1 mutant zygotes (Figure 3; Figure 4) (Hird et al. 1996; Kawasaki et al. 1998). In 30% of the fli-1 mutant embryos (n = 42), pgl-1∷gfp-labeled P granules were distributed in the whole P1 cell instead of being confined to the posterior end of the P1 cell (Figure 4, A–D). In ∼5% of these mutant embryos (n = 42), the P granules were distributed in both the AB and the P cells (Figure 4, E and F). We analyzed the first mitotic cell division in 12 normal-looking fli-1 mutant zygotes in detail. The maternal and paternal pronuclei were initially properly positioned and the pseudocleavage furrow was formed in most of the fli-1 mutants. The migration of the pronucleus, however, was abnormal in 4 of the 12 fli-1 zygotes (Figure 3, B–D). The paternal pronucleus migrated only a small distance and the pronuclei met near the ∼80–90% of the embryo in the posterior region in two mutant embryos (Figure 3B), while both pronuclei migrated and met in the center of the embryo in the other two (Figure 3C). Three of these four embryos with abnormal pronuclear meeting proceeded through the mitotic cycle without division and underwent endomitotic division. The rest of the eight fli-1 mutant embryos successfully progressed through the first division. However, five mutant embryos took 1 hr or more to complete division, compared to ∼20 min in wild-type embryos (Figure 3F).
Figure 3.—
Defects in the first mitotic cell division in the fli-1 mutants. (A) The pronucleus meets at ∼70% egg length in the posterior in a wild-type zygote. Anterior is to the left in all panels. (B and C) Defects in the polarized meeting of the pronucleus in the fli-1 zygotes. The pronucleus meets at ∼80% (B) or 50% (C) of the zygotes in the posterior. (D) Schematic of the position where pronuclei meet in eight fli-1 mutant zygotes analyzed. (E) Time-lapse DIC images of the first cleavage of a wild-type zygote. The pronuclear, pronuclear meeting, fusion and centration, formation of the spindle, formation of cleavage furrow, and the two-cell stages are shown. The time when the event occurred is shown in minutes and seconds. The first mitotic division required ∼20 min to complete. Bar, 10 μm. (F) Time-lapse DIC images of the first cleavage of a fli-1 mutant embryo. The pronucleus meets in the center of the embryo. The mutant embryo shown required much more time to complete division. (G and H) Equal amount of DNA, labeled by H2B∷GFP, is segregated into the AB and P cells in a wild-type embryo. (I and J) Unequal distribution of DNA into two daughters of a fli-1 mutant embryo. More H2B∷GFP containing DNA content was segregated into the posterior P1 cell. (K and L) The second mitotic division in a wild-type embryo. The division of the AB cell completes prior to the initiation of the cleavage of the P1 cell. (M and N) Both AB and P1 cells appeared to be at the same cell division stage in a fli-1 mutant embryo. The arrangement of the AB and P1 cells was also abnormal.
Figure 4.—
Defects in the asymmetric segregation of P granules in the fli-1 mutants. (A and B) Distribution of P granules, labeled with PGL-1∷GFP, in a wild-type embryo after the first mitotic cell division. P granules are confined in the posterior end of the P1 cell. (A) Nomarski micrograph. (B) Fluorescence micrograph of the same embryo. (C–F) Abnormal segregation of P granules in the fli-1(bp130) mutant embryos. P granules are localized in the whole P1 cell (C) or are localized in both AB and P1 cells (E). The localization of P granules in the P1 cell in F is marked with arrow, which is not at the same focus plane as the ones in the anterior AB cell. (C and E) Nomarski micrograph. (D and F) Fluorescence micrograph of the same embryos shown in C and E, respectively.
Mutations in fli-1 cause other defects in the first mitotic cell division. For example, segregation of chromosomes, visualized by the histone 2B (H2B∷GFP) marker, showed abnormality at anaphase in 9% of fli-1 mutant embryos (n = 23) (Figure 3, G–J). In addition to the defects in the first mitotic cell division, mutations in fli-1 also affected subsequent cell divisions. For example, in wild-type embryos, the AB cell completes division prior to the initiation of the cleavage of the P1 cell (Figure 3, K and L). In the fli-1 mutants, however, the temporal order of the division of the AB and P cells could be disrupted, such that P1 cell division occurred before that of the AB cell in 13% of mutant embryos (n = 23) (Figure 3, M and N).
To determine whether the defects in early cell division in fli-1 mutants were the indirect consequence of defects in eggshell production, we examined the DAPI absorption of the embryos (Kawasaki et al. 2004). fli-1(bp130) and wild-type embryos were released from the uterus of mothers into water containing DAPI (final concentration 10 μg/ml). We found that all seven fli-1(bp130) one-cell-stage embryos excluded DAPI. Thus, fli-1 mutant embryos produced functional eggshells.
In summary, mutations in fli-1 cause multiple defects in the first mitotic cell division, including defects in the polarized cytoplasmic flow; in the establishment of the A/P asymmetry of P granules; in the migration of the pronuclei; in the formation of the cleavage furrow; and in the chromosomal segregation.
The fli-1 mutants display defects in other developmental processes:
The fli-1 mutants also showed defects in morphogenesis, cell migration, and axon guidance. Of 20 fli-1 mutant embryos, 4 displayed bulges at dorsal surfaces, a phenotype that is similar to that of hmp-1 and other mutants causing the humpback phenotype (Figure 5, A and B) (Costa et al. 1998). Also instead of having a gradually tapered tail spike, the fli-1 mutant tail was malformed and contained a bulge (100%, n = 13) (Figure 5, C and D). The development of male-specific structures was also severely deformed in the fli-1 mutants. For example, the cuticular fan structure was small and the male-specific sensory rays were missing in all the male sides examined (n = 6) (Figure 5, E and F). Thus, fli-1 plays an important role in morphogenesis and/or in cell fate determination.
Figure 5.—
Mutations in fli-1 cause other developmental defects. (A and B) Compared to a wild-type embryo (A), a fli-1 mutant embryo exhibits a bulge on the body surface (arrow in B). (C) The wild-type hermaphrodite has a tapered tail spike (arrow). (D) The tail morphology is malformed in a fli-1 mutant animal (arrow). (E) Nine pairs of rays, embedded in the cuticule fan-like structure, are present in a wild-type male tail. (F) The fli-1 mutant male tail is grossly abnormal. In the mutant animal shown, most of the rays are missing. Other male-specific structures, including the spicule (arrow), are also abnormal. (G) AVM (arrow), labeled by mec-7∷gfp, positions anteriorly to the ALMs (arrowhead) in a wild-type animal. (H) In fli-1 mutants, AVM positions posteriorly to the ALMs. (I) PKD-2∷GFP marks the axons of B-type neurons of all the rays. R1B has a distinct pathfinding route (arrow). (J) Defects in the axon pathfinding of R1B in a fli-1 mutant male. The R1B axons failed to make a turn to the ventral side and continued to migrate toward the anterior body region (arrow).
The migration of Q cell descendants and axon guidance was also defective in the fli-1 mutants. In wild-type animals, the mec-7∷gfp-labeled touch neuron AVM, a descendant of the QR neuroblast, migrates to the anterior and positions anteriorly to the touch neuron ALMs (Figure 5G) (Ch'ng et al. 2003). In 8% of the fli-1 mutant sides (n = 52), the migratory distance of AVM was shorter and positioned posteriorly to ALMs (Figure 5H). In 11% of the fli-1 mutant sides (n = 23), AVM was missing, indicating a defect in the development of QR. The axon guidance of various neuronal types, including DD and VD motor neurons, the PVM touch neuron, and the male ray neurons, was defective in fli-1 mutants. For example, in 33% of the fli-1 mutants (n = 16), the axon trajectory followed by ray 1 B-type neuron (R1B), labeled by pkd-2∷gfp, failed to make the turn around the body, but continued to migrate to the anterior (Figure 5, I and J) (Jia and Emmons 2006). Therefore, fli-1 plays essential roles in controlling the migration of cells and neuronal axons.
Mutations in fli-1 cause defects in germ cell development:
Having established the important role of fli-1 in the development of somatic cells, we next investigated the role of fli-1 in the development of the germline. During germline development, germ cells undergo mitotic cell division at the distal tip of the gonad and enter meiosis as they move proximally (for review, see Crittenden et al. 2003). In the wild-type gonad, the meiotic germ cells are nearly round in shape and are evenly distributed at the germ cell plasma membrane, forming a single layer of nuclei around the nucleus-free center (the rachis) (Figure 6, A and C). The number of germ cells was greatly reduced from an average of 320 in wild-type animals (n = 5) to an average of 78 in the fli-1 mutants (n = 12). Also, in the fli-1 mutants the shapes of the germ cell were irregular and the size of the germ cell varied (Figure 6B). The alignment of the germ cells in the gonad was also disorganized and the rachis was misshaped in 12 of 19 fli-1 mutant gonad arms examined (Figure 6, B and D). Thus, fli-1 is required for the development of germ cells.
Figure 6.—
Defects in germline development in the fli-1 mutants. (A) In the meiotic region of a wild-type hermaphrodite, the germ cells are round in shape (arrow) and evenly aligned at the germ cell plasma membrane. (A and B) The upper surface of the gonad. (B) The morphology of the germ cell is irregular, the size of the germ cell varies, and the germ cells are disorganized in a fli-1 mutant (arrows). (C) In a wild-type hermaphrodite, the syncytial germ cells are arranged around a central cytoplasm core, termed the rachis (arrow). The gonad was stained with R-phalloidin and the mid-focal plane of the gonad is shown in C and D. (D) The rachis in fli-1 mutants is smaller (arrow) and the alignment of the germ cells in the rachis is disorganized in a fli-1 mutant animal (arrowheads).
Mutations in fli-1 cause ovulation defects:
The fli-1 mutants displayed a defect in ovulation. In wild-type hermaphrodites, oocytes are aligned along the proximal–distal axis (Figure 7A). During ovulation, the proximal oocytes are engulfed by the spermatheca and pulled into the oviduct (Figure 7C) (McCarter et al. 1997, 1999). We analyzed 11 single ovulation processes in the fli-1 mutants. In 5 of them, the proximal oocyte was not engulfed by the spermatheca and the trapped oocyte in the distal side underwent multi-rounds of DNA replication, causing an EMO phenotype (Figure 7B) (Iwasaki et al. 1996). In 3 of the ovulation processes analyzed in the fli-1 mutants, oocytes were spliced during the dilation of the spermatheca. These went on to be fertilized, developing into small embryos in two of the cases and undergoing endomitosis in the third case (Figure 7D). The egg-laying apparatus also appeared to be defective in the fli-1 mutants, as the fertilized embryos were often trapped in the uterus.
Figure 7.—
Mutations in fli-1 cause defects in ovulation. (A and B) Anterior gonad. (A) DAPI staining in the proximal gonad in a wild-type hermaphrodite. Chromosomes of the oocytes in the proximal gonad are condensed in diakinesis (arrow). The spermatheca contains characteristic small, compact nuclei (arrowhead). (B) DAPI staining showed endomitotic oocyte nuclei in a fli-1 mutant animal (arrows). (C) The oocyte (arrow) is engulfed by the spermatheca and located in the uterus in a wild-type hermaphrodite. (D) In the fli-1 mutant shown, the oocyte was spliced into two pieces (arrows) during ovulation. One part was engulfed in the spermatheca and the small part was left behind. (E) Suppression of the ovulation defect in the fli-1 mutants by mutations in ipp-5 and lfe-2. The alleles used were fli-1(bp130), ipp-5(sy605), and lfe-2(sy326). The number of animals analyzed are the following: fli-1, 8 and 83 (for brood size and number of seam cells, respectively); ipp-5, 15 and 12; fli-1; ipp-5, 7 and 18; lfe-2, 18 and 17; and lfe-2; fli-1, 12 and 12.
Ovulation requires the coordinated action of the contraction of the gonad sheath cells and the dilation of the spermatheca (McCarter et al. 1997). In wild-type animals, the contraction of the sheath cells and spermatheca is vigorous and increases in rate and intensity during ovulation (see supplemental Movie 3 at http://www.genetics.org/supplemental/). However, in the fli-1 mutants, we found that the contraction of the sheath cells and the spermatheca was weak and infrequent (see supplemental Movie 4). The number and organization of the sheath and spermathecal cells appeared to be normal in the fli-1 mutants (data not shown), suggesting that other defects, such as the interaction between the oocyte and the spermatheca, may cause the failure of ovulation in the fli-1 mutant animals.
The ovulation defect in the fli-1 mutants can be partially rescued by mutations in lfe-2 and ipp-5 in the phosphoinositol signaling pathway:
The LIN-3 (expressed in the oocyte)/LET-23 (expressed in the spermatheca and sheath cells) EGF signaling pathway has been shown to play an important role in controlling the contraction of sheath cells and the dilation of spermatheca through the regulation of the cellular level of inositol 1,4,5-trisphosphate (IP3) (Clandinin et al. 1998; Yin et al. 2004). Loss of function of lfe-2 (encoding the IP3 kinase) or ipp-5 (encoding type I inositol polyphosphate 5-phosphatase) bypasses the defect in the interaction between the oocyte and the spermatheca (Clandinin et al. 1998; Bui and Sternberg 2002). We found that loss of function of ipp-5 and lfe-2 partially rescued the defects in the contraction of the sheath cells and the dilation of the spermatheca in the fli-1 mutants. Consistently, the brood size of fli-1 mutants was dramatically increased (P < 0.05) in fli-1; ipp-5 and lfe-2; fli-1 double mutants (Figure 7E). Other mutant phenotypes in the fli-1 mutants, including defects in seam cell division and abnormal tail morphology, however, were not rescued by mutations in lfe-2 and ipp-5 (Figure 7E and data not shown). Thus, the defect in the ovulation in the fli-1 mutants is probably in part due to the defective signaling between the oocyte and the spermatheca.
fli-1 encodes the C. elegans Flightless I homolog:
Genetic mapping placed bp130 between sma-3 and unc-32 on chromosome III. Cosmids from this region were used for transformation rescue experiments. We found that cosmid B0523 rescued the mutant phenotypes in the fli-1 mutants in all four stable transgenic lines analyzed. Further analysis demonstrated that the PCR product containing the single predicted gene, B0523.5, had rescuing activity (Figure 8A). Furthermore, B0523.5 (RNAi) exhibited phenotypes similar to those observed in the fli-1(bp130) mutants (for details see materials and methods and Figure 8E), indicating that the defects are in fact due to loss of function.
Figure 8.—
fli-1 encodes the C. elegans Flightless I homolog. (A) Cosmid B0523 and PCR product containing the single predicted gene, B0523.5, had the rescuing activity. (B) fli-1 encodes the fly Flightless I homolog; FLI-1 contains 16 repeats of leucine-rich motif and six folds of the gelsolin domain. (C) fli-1(bp130) contains a G-to-A mutation at the 3′ splicing site of the third intron of fli-1. The reading frame is underlined in red. Failure of splicing of intron 3 caused a frameshift at amino acid 194. (D) The RT–PCR products covering exon 3 to exon 5 of fli-1 in wild-type and fli-1(bp130) mutants. Failure of removing intron 3 led to the increased size of the product from 355 bp in wild-type animals to 398 bp in fli-1(bp130) mutants. (E) fli-1(RNAi) also showed a reduced number of seam cells. In the animal side shown, only nine seam cells were present. (F) fli-1(bp130)/nDf17 caused more severe defects in lethality and sterility than fli-1(bp130) homozygotes did, indicating that fli-1(bp130) is a partial loss-of-function allele. nDf17 is a deletion in chromosome III from −1.50 to 2.11. Twenty-five percent of lethal progeny derived from fli-1(bp130)/nDf17 should be nDf17 homozygotes, which are embryonic lethal. The progeny derived from six fli-1(bp130) homozyogtes (n = 194) and 11 fli-1(bp130)/nDf17 (n = 80) were analyzed. Both fli-1(bp130) and fli-1(bp130)/nDf17 were derived from m/+ hermaphrodites.
B0523.5 encodes the C. elegans Fli I homolog (Campbell et al. 1993). The N terminus of FLI-1 consists of 16 tandem repeats of a 23-amino-acid (aa) leucine-rich motif and the C terminus consists of six copies of a 125-aa residue gelsolin-related repeating unit (Figure 8B).
By sequencing, we found that an invariant residue located at the 3′ splicing site of the third intron is mutated from G to A in the fli-1(bp130) mutants (Figure 8C). RT–PCR was performed using primers located in exon 3 and exon 5 of fli-1. We found that intron 3 was present in fli-1(bp130) mRNA (Figure 8D), which resulted in a frameshift at amino acid 194.
fli-1(bp130) does not appear to be null, as the mutant defects in the fli-1(bp130)/nDf17 animals were more severe than the ones in the homozygous fli-1(bp130) mutant (Figure 8F). The non-null nature of fli-1(130) could be due to the fact that truncated FLI-1(bp130), which contains the C-terminal eight copies of LRR repeats, retains some function or could be due to the presence of other alternative spliced forms of fli-1(bp130) that were not detected in our analysis.
fli-1 functions cell autonomously in determining the seam cell division:
To further examine the role of fli-1 in controlling seam cell division, we expressed fli-1 using a seam-cell-specific promoter. The defects of seam cells in fli-1(bp130), including the reduced number of seam cells, were rescued in the animals carrying a scm∷fli-1 transgene (Table 2). This indicates that fli-1 acts cell autonomously in regulating the development of seam cells. This also suggests that the pleiotropic defects observed in fli-1 mutants cannot be solely attributed to the defects in the development of germline or early embryos.
fli-1 can be functionally substituted by the fly Fli I:
C. elegans FLI-1 displays 49% identity to Drosophila Fli I at the protein sequence level. We determined whether Drosophila Fli I can substitute for the function of fli-1 in C. elegans. The fly Fli I cDNA was expressed under the control of the fli-1 promoter. This transgene was functional in rescuing the defects in the fli-1(bp130) mutants, including the number of seam cells, EMO, and reduced brood size (Table 2), indicating that the function of Fli I is evolutionarily conserved.
fli-1 is wildly expressed and localized in actin-rich regions:
To determine the expression pattern of fli-1, a fli-1 green fluorescence reporter was constructed, including the 2-kb promoter region and the full-length genomic DNA of fli-1. This reporter was functional in rescuing the defects in the fli-1(bp130) mutants (Table 2). The reporter was widely expressed and the expression level was dynamic, varying among tissues and developmental stages. The GFP was localized mainly in the cytoplasm. The expression of fli-1∷gfp was first detected during embryogenesis before the apparent morphogenesis occurred (Figure 9, A and B). In the comma-stage embryo, expression of fli-1∷gfp was detected in muscle cells (Figure 9, C and D). The expression of fli-1∷gfp was not observed before the 32-cell-stage embryos (n > 40 embryos examined). The early developmental defects in fli-1 mutants suggest that maternally produced FLI-1 is present in the early embryo. Such maternal expression cannot be recapitulated by the reporter construct due to the germline silencing effect.
Figure 9.—
fli-1 is wildly expressed. (A and B) The onset of the expression of fli-1∷gfp was detected at the embryonic stage before obvious morphogenesis. (C and D) Expression of fli-1∷gfp in a comma-stage embryo. Strong expression was evident in muscle cells. (E–J) Expression of fli-1∷gfp in pharyngeal muscles (E), vulva muscles (F), spermatheca (G), male proctodeum muscles (H), distal tip cell (arrow, I), and axons (arrow, J). In the spermatheca, strong expression was seen in the valve connected to the uterus (arrows). (J–L) Colocalization of fli-1∷gfp and actin filaments in body-wall muscles. Both fli-1∷gfp and phalloidin-labeled actin filaments were enriched at the dense bodies (arrow). FLI-1∷GFP (K), actin filaments (L), merged picture (M).
At post-embryonic stages, expression of fli-1∷gfp was detected in many tissues, including the pharyngeal muscle, rectum muscle, vulva muscle, proctodeum muscle in males, and somatic gonad tissues (including spermatheca and distal tip cell) (Figure 9, E–I, and data not shown). The fli-1 expression in the spermatheca was unevenly distributed with strong expression in the distal and proximal part of spermatheca (Figure 9G). fli-1 was also expressed within neuronal migratory structures such as axons (Figure 9J). In the body-wall muscle, fli-1∷gfp was localized in a distinct striated pattern, with accumulation of FLI-1∷GFP at dense bodies, which is similar to the Z-disc in the muscle cells in other organisms (Figure 9K). The expression pattern of fli-1 in body-wall muscle cells is reminiscent of the distribution of the actin filaments. We stained F-actin in body-wall muscle cells with rhodamine-labeled phalloidin and found that it colocalized with FLI-1∷GFP (Figure 9, K–M). In summary, the expression pattern of fli-1 appears to be coincident with actin-based structures.
Organization of F-actin microfilaments is impaired in the fli-1 mutants:
We next examined the actin cytoskeleton in the fli-1 mutant animals at various stages and tissues. In the wild-type pronuclear one-cell embryo, F-actin is uniformly distributed in the cortex with the dispersed localization of actin foci (Strome 1986; Hill and Strome 1988). The actin filaments are enriched in the cortical surfaces where the cleavage furrow forms in multi-cell embryos (Figure 10, A–D) (Strome 1986). In three of five fli-1 mutant zygotes analyzed, the actin foci were reduced in number and increased in size. In 2 of 12 fli-1 mutant multi-cell embryos, F-actin was unevenly distributed along the membrane and it diffused in the peripheral cytoplasm membranes (Figure 10, E–H).
Figure 10.—
The actin filaments are disorganized in the fli-1 mutants. (A–C) The distribution of F-actin in a wild-type pronuclear-stage embryo. Actin is uniformly distributed and distinct actin foci are dispersed through the cortex. R-ph staining seen at the mid-focal plane (B) and on the top surface (C). (B) Actin filaments are enriched at the cleavage furrow structures in a wild-type four-cell stage embryo. (E–G) The distribution of F-actin in a fli-1 mutant pronuclear embryo. The actin foci were reduced in number and increased in size (arrow in G). (F) Mid-focal plane. (G) Top surface. (H) The actin filaments in the cortical surface of the embryo were not evenly distributed and reached varying depths in peripheral cytoplasm (arrow) in a fli-1 mutant multi-cell embryo. (I and K) In a wild-type animal, the actin filaments form a meshwork in the sheath cells (I) and are densely packed and aligned circumferentially around the spermathecal cell (K). (J and L) The distribution of F-actin was irregular and disorganized in the sheath cells (J) and in the spermathecal cells (L) in a fli-1 mutant. (M) Distribution of F-actin in body-wall muscles. The actin filaments form an organized myofibril lattice and are enriched in dense bodies (arrow). (N) The muscle thin fibers were abnormal in appearance in fli-1 mutants. The actin filaments were disorganized and appeared to concentrate in many places along the fibers (arrow). (O) A honeycomb arrangement of the actin network in the wild-type germline. The F-actin evenly surrounds the germ cell nuclei. (P) Distribution of F-actin in the fli-1 mutant germ cells. F-actin appeared to accumulate around the nuclei in some parts (arrow). (Q and R) NMY-2∷GFP displayed the same expression pattern as actin filaments in both wild-type germline (Q) and fli-1 mutant germline (R).
In gonad tissues, the actin filaments form cortical meshwork structures in the sheath cells and run roughly circumferentially in spermathecal cells, with strong accumulation at the valve from the spermatheca to the uterus (Figure 10, I and K) (Strome 1986; McCarter et al. 1997). In the fli-1 mutants, normal arrays of the actin filaments appeared to be formed in the sheath and spermathecal cells. However, the organization of the actin filaments was irregular and disorganized (Figure 10, J and L). In body-wall muscle cells, the fli-1 mutants had a variably disorganized myofilament lattice; the actin filaments were disorganized and unevenly distributed with more actin-rich dense body appearance structures (Figure 10, M and N).
During the wild-type germline development, the actin filaments are distributed along the membranes that surround germ nuclei, forming a honeycomb arrangement (Figure 10O) (Strome 1986). In the fli-1(bp130) mutants, the actin filaments were unevenly distributed along the membrane and appeared to diffuse in the periphery of the membrane (Figure 10P). Furthermore, actin foci were observed surrounding the germ nuclei. nmy-2, encoding the nonmuscle myosin II, displays the same expression pattern as actin filaments in the wild-type germline (Guo and Kemphues 1996; Piekny et al. 2003). Consistently, NMY-2∷GFP showed abnormal accumulation as actin filaments in the fli-1 mutant germline (Figure 10, Q and R). Taken together, wild-type fli-1 plays an essential role in organization of the actin filaments during C. elegans development.
DISCUSSION
Defects in the establishment of A/P polarity and asymmetric cell division in the fli-1 mutants:
We show here that fli-1 regulates the establishment of cell polarity in the one-cell-stage embryo, including polarized cytoplasmic flow, asymmetric meeting of pronuclei, and asymmetric segregation of P granules, while at the post-embryonic stage, fli-1 regulates asymmetric cell division.
How is fli-1 involved in specifying the distinct cell fate of daughters? Mutations in fli-1 can lead to disruption of the asymmetric distribution of the key cell fate determinants, such as P granules. The defects in the spindle position (data not shown) and chromosome segregation observed in the fli-1 mutant embryos indicate that fli-1 may also play a role in regulating the spindle alignment and function, which is crucial for asymmetric segregation of the cell fate determinants. Alternatively, cells in fli-1 mutants could be defective in responding to extrinsic cues that are involved in specifying the distinct cell fate.
The failure of cytokinesis in the one-cell-stage embryo and the presence of two nuclei in seam cells and DTCs also indicate that fli-1 is essential for cell division. In the fly, the cellularization of the syncytial blastoderm is defective (Straub et al. 1996). The cytoplasmic contraction waves and nuclear migration in the syncytium, however, are normal in Fli I mutants. Fly Fli I is also not needed for the cytokinesis at the late developmental stage and is not required for the development of the oocyte (Straub et al. 1996). The less severe defects in the fly could be due to the redundancy of the Fli I with other proteins, such as with other gelsolin family members, in controlling the dynamics of the actin network. The development of the muscle structure is defective in both C. elegans and the fly. In fli-1 mutants, the muscle fibers are irregular and malformed. Viable Fli I mutant flies show abnormal formation of the indirect flight muscle fibers, which display severely disrupted Z-discs (Miklos and De Couet 1990), suggesting a conserved role of Fli I in regulation of the development of muscle cells.
The role of FLI-1 in controlling the dynamics of the actin network:
The fli-1 mutant phenotypes argue that fli-1 plays an important role in controlling the dynamic arrangement of the actin filaments or in generation of the force of the actomyosin structure. First, the defects in fli-1 mutant embryos resemble some of the effects caused by cytochalasin-D-induced actin depolymerization (Strome and Wood 1983; Hill and Strome 1988). Mutations in two actin-associated proteins, NMY-2, the nonmuscle myosin II, and MLC-4, the nonmuscle myosin II regulatory light chain, also result in similar defects in the polarized cytoplasm flow, establishment of the A/P polarity, and cytokinesis in the first mitotic cell division (Guo and Kemphues 1996; Shelton et al. 1999). Mutations in several genes that are involved in regulating actomyosin have also been reported to influence the timing of cytokinesis. For example, in mel-11 (which encodes myosin phosphatase) mutants, the furrow ingression completed approximately twice as fast as in wild type, while mutations in let-502, which encodes Rho-binding kinase, had slower cytokinesis (Piekny and Mains 2002). Multinucleate single embryos were observed in mlc-4 and let-502 mutants due to cytokinesis failures (Shelton et al. 1999; Piekny and Mains 2002). Second, FLI-1 is localized at the sites where dynamic actin rearrangement appears to occur, such as in the distal tip cells, spermatheca, and vulval muscle cells. Moreover, FLI-1 is colocalized with actin bundles in the body-wall muscles. Third, the distribution of the actin filaments is impaired in the fli-1 mutants. In general, the actin filaments appear to be disorganized and also accumulate in some parts in the fli-1 mutants. Fourth, FLI-1 can directly interact with the G- and F-actin and contains the actin-severing activity (Goshima et al. 1999). The abnormality of chromosome segregation in fli-1 mutants suggests that fli-1 may also regulate some microtubule-dependent events, such as spindle alignment and function. In mammalian cells, Fli I is also localized to centrosomes and accumulated at the mid-body (Davy et al. 2001). Alternatively, the defects in chromosome segregation in fli-1 mutants could be an indirect effect.
How does FLI-1 modulate the actin network? FLI-1 could function as a structural component, holding the actin filaments together. FLI-1 might also be involved in the delivery of actin to or in the stabilization of the actin network. Furthermore, FLI-1 could have a regulatory role in establishing the actin cytoskeleton. The LRR repeats of FLI-1 have been shown to directly bind to Ras in vitro (Goshima et al. 1999), raising the possibility that FLI-1 integrates the Ras-signaling pathway with the actin filaments.
Ovulation defect in the fli-1 mutants:
The fli-1 mutants show a great reduction in the frequency and intensity of the contraction of the sheath cells and the dilation of the spermatheca during ovulation. The contraction of the sheath cells and dilation of the spermatheca is triggered by LIN-3/LET-23 EGF signaling, which most likely stimulates hydrolysis of PIP2 into IP3 (Bui and Sternberg 2002; Yin et al. 2004). In other systems, IP3 has been shown to excite intracellular calcium release channels and cause a transient increase in intracellular calcium (Berridge 1993). Mutations in ipp-5 and lfe-2 suppress the fli-1 ovulation defect, strongly arguing in favor of a defect in signaling between the oocyte and the spermatheca in fli-1 mutants. However, it is unlikely that the increase in intracellular calcium concentration directly regulates the activity of FLI-1, as the actin-binding activity of FLI-1 is calcium independent (Goshima et al. 1999). In mammalian cells, the EGF receptor signaling pathway drives cytoskeleton rearrangement and cell protrusion by regulating the activity of gelsolin. The F-actin-severing activity of gelsolin is regulated by PIP2 (Janmey and Stossel 1987; Janmey et al. 1992; Yu et al. 1992). EGF signaling activates phospholipase C-γ (PLCγ), which hydrolysis PIP2, and subsequently leads to the dissociation of gelsolin from the plasma membrane (Chou et al. 2002). Thus, it is possible that the activity of FLI-1 in modulating the actin cytoskeleton in the spermatheca is regulated by PIP2, which could be dissociated from FLI-1 in lfe-2 and ipp-5 mutants. Alternatively, the defects in ovulation could be due to the oocyte, which secretes EGF, being defective in fli-1 mutants. Further studies could help us to elucidate how the EGF-signaling pathway regulates the activity of fli-1 in controlling the dynamics of the actin network during animal development.
In summary, our studies reveal a key role of fli-1 in establishing anterior–posterior polarity, specifying asymmetric cell division, interacting with the phosphoinositol signaling pathway in the regulation of ovulation, and other actin-dependent events. These findings have significant implications for our understanding of the causes of the defects associated with Fli I loss of function in mammals and also provide insight into the physiological functions of gelsolin family members during animal development, especially in early embryogenesis and muscle development.
Acknowledgments
We thank Xiaochen Wang, Andrea Christoforou, and members in our laboratory for their helpful comments on the manuscript. We also thank Bob Goldstein for the pgl-1∷gfp strain. Some strains used in this work were received from the Caenorhabditis Genetics Center, which is supported by a grant from the National Institutes of Health. This work was supported by National High Technology Projects 863 (2005AA210910).
References
- Albertson, D. G., 1984. Formation of the first cleavage spindle in nematode embryos. Dev. Biol. 101: 61–72. [DOI] [PubMed] [Google Scholar]
- Berridge, M. J., 1993. Inositol triphosphate and calcium signaling. Nature 361: 315–325. [DOI] [PubMed] [Google Scholar]
- Bui, Y. K., and P. W. Sternberg, 2002. Caenorhabditis elegans inositol 5-phosphatase homolog negatively regulates inositol 1,4,5-triphosphate signaling in ovulation. Mol. Biol. Cell 13: 1641–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, H. D., T. Schimansky, C. Claudianos, N. Ozsarac, A. B. Kasprzak et al., 1993. The Drosophila melanogaster flightless-I gene involved in gastrulation and muscle degeneration encodes gelsolin-like and leucine-rich repeat domains and is conserved in Caenorhabditis elegans and humans. Proc. Natl. Acad. Sci. USA 90: 11386–11390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, H. D., S. Fountain, I. G. Young, C. Claudianos, J. D. Hoheisel et al., 1997. Genomic structure, evolution and expression of human FLII, a gelsolin and leucine-rich-repeat family member: overlap with LLGL. Genomics 42: 46–54. [DOI] [PubMed] [Google Scholar]
- Campbell, H. D., S. Fountain, I. S. McLennan, L. A. Berven, M. F. Crouch et al., 2002. Fliih, a gelsolin-related cytoskeletal regulator essential for early mammalian embryonic development. Mol. Cell. Biol. 22: 3518–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. S., P. H. Gunaratne, J. D. Hoheisel, I. G. Young, G. L. Miklos et al., 1995. The human homologue of the Drosophila melanogaster flightless-I gene (fliI) maps within the Smith-Magenis microdeletion critical region in 17p11.2. Am. J. Hum. Genet. 56: 175–182. [PMC free article] [PubMed] [Google Scholar]
- Ch'ng, Q., L. Williams, Y. S. Lie, M. Sym, J. Whangbo et al., 2003. Identification of genes that regulate a left-right asymmetric neuronal migration in Caenorhabditis elegans. Genetics 164: 1355–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou, J., D. B. Stolz, N. A. Burke, S. C. Watkins and A. Wells, 2002. Distribution of gelsolin and phosphoinositol 4,5-bisphosphate in lamellipodia during EGF-induced motility. Int. J. Biochem. Cell Biol. 34: 776–790. [DOI] [PubMed] [Google Scholar]
- Clandinin, T. R., J. A. DeModena and P. W. Sternberg, 1998. Inositol triphosphate mediates a ras-independent response to LET-23. Cell 92: 523–533. [DOI] [PubMed] [Google Scholar]
- Claudianos, C., and H. D. Campbell, 1995. The novel flightless-I gene brings together two gene families, actin binding proteins related to gelsolin and leucine-rich-repeat proteins involved in ras signal transduction. Mol. Biol. Evol. 12: 405–414. [DOI] [PubMed] [Google Scholar]
- Costa, M., W. Raich, C. Agbunag, B. Leung, J. Hardin et al., 1998. A putative catenin-cadherin system mediates morphogenesis of the Caenorhabditis elegans embryo. J. Cell Biol. 141: 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan, C.R., and A. A. Hyman, 2004. Asymmetric cell division in C. elegans: cortical polarity and spindle positioning. Annu. Rev. Cell Dev. Biol. 20: 427–453. [DOI] [PubMed] [Google Scholar]
- Crittenden, S. L., C. R. Eckmann, L. Wang, D. S. Bernstein, M. Wickens et al., 2003. Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philos. Trans. R. Soc. Lond B Biol. Sci. 358: 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy, D. A., H. D. Campbell, S. Fountain, D. de Jong and M. F. Crouch, 2001. The flightless I protein colocalizes with actin- and microtubule-based structures in motile Swiss 3T3 fibroblasts: evidence for the involvement of PI 3-kinase and Ras-related small GTPases. J. Cell Sci. 114(Pt. 3): 549–562. [DOI] [PubMed] [Google Scholar]
- de Couet, H. G., K. S. Fong, A. G. Weeds, P. J. McLaughlin and G. L. Miklos, 1995. Molecular and mutational analysis of a gelsolin-family member encoded by the flightless I gene of Drosophila melanogaster. Genetics 141: 1049–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger, F. P., K. R. Kopish and J. G. White, 2003. A role for septins in cellular and axonal migration in C. elegans. Dev. Biol. 261: 220–234. [DOI] [PubMed] [Google Scholar]
- Gonczy, P., H. Schnabel, T. Kaletta, A. D. Amores, T. Hyman et al., 1999. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J. Cell Biol. 144: 927–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, M., K. I. Kariya, Y. Yamawaki-Kataoka, T. Okada, M. Shibatohge et al., 1999. Characterization of a novel ras-binding protein Ce-FLI-1 comprising leucine-rich repeats and gelsolin-like domains. Biochem. Biophys. Res. Commun. 257: 111–116. [DOI] [PubMed] [Google Scholar]
- Guo, S., and K. J. Kemphues, 1996. A non-muscle myosin required for embryonic polarity in Caenorhabditis elegans. Nature 382: 455–458. [DOI] [PubMed] [Google Scholar]
- Herman, M. A., and H. R. Horvitz, 1994. The Caenorhabditis elegans gene lin-44 controls the polarity of asymmetric cell divisions. Development 120: 1035–1047. [DOI] [PubMed] [Google Scholar]
- Hill, D. P., and S. Strome, 1988. An analysis of the role of microfilaments in the establishment and maintenance of asymmetry in Caenorhabditis elegans zygotes. Dev. Biol. 125: 75–84. [DOI] [PubMed] [Google Scholar]
- Hird, S. N., and J. G. White, 1993. Cortical and cytoplasmic flow polarity in early embryonic cells of Caenorhabditis elegans. J. Cell Biol. 121: 1343–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird, S. N., J. E. Paulsen and S. Strome, 1996. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development 122: 1303–1312. [DOI] [PubMed] [Google Scholar]
- Iwasaki, K., J. McCarter, R. Francis and T. Schedl, 1996. emo-1, a Caenorhabditis elegans Sec61pg homologue, is required for oocyte development and ovulation. J. Cell Biol. 134: 699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey, P. A., and T. P. Stossel, 1987. Modulation of gelsolin function by phosphatidylinositol 4,5-bisphosphate. Nature 325: 362–364. [DOI] [PubMed] [Google Scholar]
- Janmey, P. A., J. Lamb, P. G. Allen and P. T. Matsudaira, 1992. Phosphoinositide-binding peptides derived from the sequences of gelsolin and villin. J. Biol. Chem. 267: 11818–11823. [PubMed] [Google Scholar]
- Jia, L., and S. W. Emmons, 2006. Genes that control ray sensory neuron axon development in the Caenorhabditis elegans male. Genetics 173: 1241–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, I., Y. H. Shim, J. Kirchner, J. Kaminker, W. B. Wood et al., 1998. PGL-1, a predicted RNA-binding component of germ granules, is essential for fertility in C. elegans. Cell 94: 635–645. [DOI] [PubMed] [Google Scholar]
- Kawasaki, I., A. Amiri, Y. Fan, N. Meyer, S Dunkelbarger et al., 2004. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics 167: 645–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemphues, K. J., and S. Strome, 1997. Fertilization and establishment of polarity in the embryo, pp. 335–360 in C. elegans II, edited by D. L. Riddle, T. Blumenthal, B. J. Meyer and J. R. Priess. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Kwiatkowski, D. J., 1999. Functions of gelsolin: motility, signaling, apoptosis, cancer. Curr. Opin. Cell Biol. 11: 103–108. [DOI] [PubMed] [Google Scholar]
- Liu, Y. T., and H. L. Yin, 1998. Identification of the binding partners for flightless I, a novel protein bridging the leucine-rich repeat and the gelsolin superfamilies. J. Biol. Chem. 273: 7920–7927. [DOI] [PubMed] [Google Scholar]
- McCarter, J., B. Bartlett, T. Dang and T. Schedl, 1997. Somagerm cell interactions in Caenorhabditis elegans: multiple events of hermaphrodite germline development require the somatic sheath and spermathecal lineages. Dev. Biol. 181: 121–143. [DOI] [PubMed] [Google Scholar]
- McCarter, J., B. Bartlett, T. Dang and T. Schedl, 1999. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev. Biol. 205: 111–128. [DOI] [PubMed] [Google Scholar]
- Miklos, G. L., and H. G. de Couet, 1990. The mutations previously designated as flightless-I3, flightless-O2 and standby are members of the W-2 lethal complementation group at the base of the X-chromosome of Drosophila melanogaster. J. Neurogenet. 6: 133–151. [DOI] [PubMed] [Google Scholar]
- Nance, J., 2005. PAR proteins and the establishment of cell polarity during C. elegans development. BioEssays 27: 126–135. [DOI] [PubMed] [Google Scholar]
- Piekny, A. J., and P. E. Mains, 2002. Rho-binding kinase (LET-502) and myosin phosphatase (MEL-11) regulate cytokinesis in the early Caenorhabditis elegans embryo. J. Cell Sci. 115: 2271–2282. [DOI] [PubMed] [Google Scholar]
- Piekny, A. J., J. L. Johnson, G. D. Cham and P. E. Mains, 2003. The Caenorhabditis elegans nonmuscle myosin genes nmy-1 and nmy-2 function as redundant components of the let-502/Rho-binding kinase and mel-11/myosin phosphatase pathway during embryonic morphogenesis. Development 130: 5695–5704. [DOI] [PubMed] [Google Scholar]
- Shelton, C. A., J. C. Carter, G. C. Ellis and B. Bowerman, 1999. The nonmuscle myosin regulatory light chain gene mlc-4 is required for cytokinesis, anterior-posterior polarity, and body morphology during Caenorhabditis elegans embryogenesis. J. Cell Biol. 146: 439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silacci, P., L. Mazzolaib, C. Gaucia, N. Stergiopulosa, H. L. Yinc et al., 2004. Gelsolin superfamily proteins: key regulators of cellular functions. Cell. Mol. Life Sci. 61: 2614–2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub, K. L., M. C. Stella and M. Leptin, 1996. The gelsolin-related flightless I protein is required for actin distribution during cellularisation in Drosophila. J. Cell Sci. 109: 263–270. [DOI] [PubMed] [Google Scholar]
- Strome, S., 1986. Fluorescence visualization of the distribution of microfilaments in gonads and early embryos of the nematode C. elegans. J. Cell Biol. 103: 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome, S., and W. B. Wood, 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35: 15–25. [DOI] [PubMed] [Google Scholar]
- Sulston, J. E., and H. R. Horvitz, 1977. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56: 110–156. [DOI] [PubMed] [Google Scholar]
- Sun, H. Q., M. Yamamoto, M. Mejillano and H. L. Yin, 1999. Gelsolin, a multifunctional actin regulatory protein. J. Biol. Chem. 274: 33179–33182. [DOI] [PubMed] [Google Scholar]
- Witke, W., A. H. Sharpe, J. H. Hartwig, T. Azuma, T. P. Stossel et al., 1995. Hemostatic, inflammatory and fibroblast responses are blunted in mice lacking gelsolin. Cell 81: 41–51. [DOI] [PubMed] [Google Scholar]
- Yin, X., N. J. Gower, H. A. Baylis and K. Strange, 2004. Inositol 1,4,5-trisphosphate signaling regulates rhythmic contractile activity of myoepithelial sheath cells in Caenorhabditis elegans. Mol. Biol. Cell 15: 3938–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, F. X., H. Q. Sun, P. A. Janmey and H. L. Yin, 1992. Identification of a polyphosphoinositide-binding sequence in an actin monomer-binding domain of gelsolin. J. Biol. Chem. 267: 14616–14621. [PubMed] [Google Scholar]