Abstract
Family 18 of glycosyl hydrolases encompasses chitinases and so-called chi-lectins lacking enzymatic activity due to amino acid substitutions in their active site. Both types of proteins widely occur in mammals although these organisms lack endogenous chitin. Their physiological function(s) as well as evolutionary relationships are still largely enigmatic. An overview of all family members is presented and their relationships are described. Molecular phylogenetic analyses suggest that both active chitinases (chitotriosidase and AMCase) result from an early gene duplication event. Further duplication events, followed by mutations leading to loss of chitinase activity, allowed evolution of the chi-lectins. The homologous genes encoding chitinase(-like) proteins are clustered in two distinct loci that display a high degree of synteny among mammals. Despite the shared chromosomal location and high homology, individual genes have evolved independently. Orthologs are more closely related than paralogues, and calculated substitution rate ratios indicate that protein-coding sequences underwent purifying selection. Substantial gene specialization has occurred in time, allowing for tissue-specific expression of pH optimized chitinases and chi-lectins. Finally, several family 18 chitinase-like proteins are present only in certain lineages of mammals, exemplifying recent evolutionary events in the chitinase protein family.
CHITIN, the linear polymer of N-acetylglucosamine, is the second most abundant polysaccharide in nature and serves as an indispensable structural component in a variety of organisms, including fungi and arthropods (Tharanathan and Kittur 2003). On the basis of sequence homologies, chitinases fall into two groups: families 18 and 19 of glycosyl hydrolases (Henrissat 1991). Members of family 18 employ a substrate-assisted reaction mechanism (Terwisscha van Scheltinga et al. 1995; Van Aalten et al. 2001), whereas those of family 19 adopt a fold-and-reaction mechanism similar to that of lysozyme (Monzingo et al. 1996), suggesting that these families evolved independently to deal with chitin.
Early reports on chitinolytic activity in vertebrates (Jeuniaux 1961) were confirmed following investigations on Gaucher disease, the most common lysosomal storage disorder in humans caused by an inherited deficiency in glucocerebrosidase (Beutler and Grabowski 1995). In the plasma of symptomatic patients with Gaucher disease, activity toward the artificial substrate 4-methylumbelliferyl-chitotriose is elevated several hundredfold (Hollak et al. 1994). The responsible enzyme, named chitotriosidase, was shown to be a true chitinase, hydrolyzing natural chitin and showing high sequence homology to chitinases from lower organisms (Hollak et al. 1994; Boot et al. 1995; Renkema et al. 1995). Other members of the mammalian chitinase family have been discovered since, including a second chitinase which, given its acidic pH optimum, was named acidic mammalian chitinase (AMCase) (Boot et al. 2001). Since chitin is an important structural component of pathogens like fungi as well as a constituent of the mammalian diet, a dual function for mammalian chitinases in innate immunity and food digestion has been envisioned (Suzuki et al. 2002; Boot et al. 2005a). Indeed, for human chitotriosidase, an enzyme predominantly expressed by phagocytes, a fungistatic effect has been demonstrated recently (Van Eijk et al. 2005). Several studies have tried to link a common chitotriosidase deficiency (Boot et al. 1998) to susceptibility for infection by chitin-containing parasites (reviewed by Bussink et al. 2006). The physiological function of the second mammalian chitinase, AMCase, has recently attracted considerable attention due to a report linking the protein to the pathophysiology of asthma (Zhu et al. 2004).
In addition to active chitinases, highly homologous mammalian proteins lacking enzymatic activity due to substitution of active-site catalytic residues have been identified. Despite their lack of enzymatic activity, these proteins have retained active-site carbohydrate binding and hence have been named chi-lectins (Renkema et al. 1998; Houston et al. 2003; Bussink et al. 2006). Like the active chitinases, chi-lectins belong to family 18 of glycosyl hydrolases, consisting of a 39-kDa catalytic domain having a TIM-barrel structure, one of the most versatile folds in nature (Sun et al. 2001; Wierenga 2001; Fusetti et al. 2002, 2003; Houston et al. 2003). In contrast to both chitinases, chi-lectins lack the conserved additional chitin-binding domain (Boot et al. 1995, 2001; Renkema et al. 1997). Despite the detailed knowledge regarding structure, insight into the exact physiological function of the various chi-lectins is limited (reviewed by Bussink et al. 2006). Similar to chitotriosidase and AMCase, chi-lectins are secreted locally or into the circulation and a role in inflammatory conditions is suggested. For example, human cartilage GP39 (Hcgp39/YKL-40/CHI3L1), a protein expressed by chondrocytes and phagocytes, has been implicated in arthritis, tissue remodeling, fibrosis, and cancer (Hakala et al. 1993; Johansen et al. 1993; Verheijden et al. 1997; Recklies et al. 2002; reviewed by Johansen 2006). Similarly, the human chi-lectin YKL-39 (CHI3L2) and the murine Ym1 (Chi3L3/ECF-L) have been associated with the pathogenesis of arthritis (Hu et al. 1996; Tsuruha et al. 2002) and allergic airway inflammation, respectively (Chang et al. 2001; Ward et al. 2001; Homer et al. 2006).
The high-molecular-weight oviductins, consisting of the amino-terminal 39-kDa catalytic domain followed by a heavily glycosylated serine/threonine-rich domain, are secreted by nonciliated oviductal epithelial cells and have been shown to play a role in fertilization and early embryo development (reviewed by Buhi 2002).
To elucidate the evolution of members of family 18 of glycosyl hydrolases in mammals, we extensively surveyed available sequence information in terms of conserved protein features, molecular phylogeny, substitution rate ratios, and chromosomal distribution. This combined approach renders new insights into evolutionary origins, selective pressures, and genomic synteny. Moreover, our investigation has established the true orthologous relationships among mammalian chitinase(-like) proteins.
MATERIALS AND METHODS
Database searches:
DNA and protein sequences for all of the members of glycosyl hydrolase family 18 were obtained by BLAST searches (blastn and blastp and translated blasts) of public databases, mainly the NCBI and ENSEMBL databases (http://www.ncbi.nlm.nih.gov and http://www.ensembl.org/index.html). The sequences were checked against published sequences in literature, as well as genomic information. For mBclp2, only a partial sequence was available and the entire coding sequence was determined from the genomic sequence on the basis of consensus splice sites. All cDNA sequences obtained were checked against protein sequences and vice versa, by both BLAST searches and manual inspection.
Protein—alignment and features:
Signal peptides and chitin-binding domain-coding sequences were omitted; hence only sequence information corresponding with the 39-kDa catalytic domain of the active chitinases was used for the alignment. ClustalX was implemented for both protein and DNA alignments (Thompson et al. 1997), which were checked manually, and, where necessary, alignments were edited. ESPript 2.2 (Gouet et al. 1999) was used for visualization of the protein alignment (using RISLER similarity scoring) and automatic superimposition of secondary structures (Gouet et al. 2003) of human chitotriosidase on the basis of a published crystal structure (pdb accession code 1LQ0). The assigned helices, strands, and turns were checked manually against the published chitotriosidase structures.
Calculated isoelectric points were determined using the pI/MW tool on the ExPASy server (http://www.expasy.org/tools/pi_tool.html) (Gasteiger et al. 2003).
Synteny analysis:
Genomic mapping was performed manually by linking the coding sequences retrieved (see above) to genomic location according to both the NCBI and Ensemble databases.
Phylogenetic analysis:
Maximum-likelihood and parsimony analyses based on the cDNA alignment were performed by PHYLIP version 3.65 (available at http://evolution.genetics.washington.edu/phylip.html; Felsenstein 2006), making use of the DnaML (after rejection of the molecular clock hypothesis by performing likelihood-ratio testing) and DnaPars programs. Support values were generated on a thousand bootstrapped replicate data sets. For the likelihood analyses, rates were considered equal over sites. To avoid bias caused by order of the sequence input, the order was randomized in all analyses. The consensus tree and bootstrap support values were determined using the Consense program, implemented in the PHYLIP package. The cladogram was generated by aligning the nucleotide sequences coding for the 39-kDa domain with the aid of ClustalX. The input comprised 34 taxa (excluding monkey sequences and including the outgroup sequence), with 1127 characters in each taxon. Inclusion and exclusion of gaps resulted in identical branching and near identical bootstrap values. The input for supplemental Figure 2 at http://www.genetics.org/supplemental/ was extended with the two Xenopus tropicalis chitinase sequences.
Calculation of substitution rates:
Rate ratios of nonsynonymous-to-synonymous substitutions dN/dS (ω) were calculated by PAML, version 3.15 (Yang 1997), using a maximum-likelihood approach based on the consensus tree topology as determined by PHYLIP (see supplemental Figure 1 at http://www.genetics.org/supplemental/). The program Codeml was used to calculate ω-values for branches under the “free-ratio” model that allows a different ω for specified branches (model 2). Codeml was also used to calculate ω-values for sites (specific codons) under models that either exclude (M0 and M1a) or allow (M2a) positive selection. The different models were compared by performing likelihood-ratio testing.
RESULTS
Identification of mammalian chitinase protein family members:
Mining the literature and using NCBI or ENSEMBL BLAST searches led to the identification of 44 members of the chitinase protein family from 11 different mammalian species. Supplemental Table 1A at http://www.genetics.org/supplemental/ provides an overview of all sequences retrieved, including species specification, common protein aliases, existence of expression data, and NCBI accession numbers. Included are two chitinases from X. tropicalis and a chitinase from Caenorhabditis elegans that was used as an outgroup in the phylogenetic analyses. Supplemental Table 1B provides an overview of relevant genes identified in the mammalian genomes that are completely (Homo sapiens, Pan Troglodytes, and Mus musculus) or nearly completely sequenced (Rattus norvegicus and Bos taurus).
The true chitinase genes (coding for chitotriosidase and AMCase) are present in all mammalian species for which (near) complete genome data exist. The bovine chitotriosidase gene, mapped to chromosome 16, could be only partially aligned due to incompleteness of the genomic sequence.
The genes of the chi-lectins CHI3L1 (encoding Hcgp39 or GP39) and OVGP1 (oviductin) are also present in all analyzed species. Only in the case of rat oviductin could the complete coding sequence not be retrieved due to gaps in the available genomic sequence. The bovine genome contains a second GP39-like gene that is highly homologous (96% nucleotide identity), named BP40, that seems specific for the artiodactyls (even-toed ungulates or hoofed mammals).
Other chi-lectin genes are more specific for particular species. For example, the CHI3L2 (YKL-39) is present in the primate and cow genomes but not in the genomes of rodents. The opposite is the case for the Chi3l3 (Ym1), Chi3l4 (Ym2), and Bclp2 (brain chitinase-like protein 2) genes found only in rodent and not in primate genomes. Interestingly, BLAST searches on the rodent genomes revealed the presence of a previously unidentified paralogue, here referred to as BYm (basic Ym). BYm lacks the catalytic glutamic acid, suggesting that it is a chi-lectin and shows a high homology to Ym1 and Ym2. Expressed sequence tag data indicate that the gene is transcribed in murine olfactory epithelium and neonates. Previously, Jin et al. (1998) identified a cluster of four genes related to Ym1, naming the other three Ym2, Ym3, and Ym4. The newly identified BYm is not identical to any of these genes, suggesting that it is a novel enzymatically inactive member of the chitinase protein family. So far, there are no indications that the Ym3 gene is expressed. The Ym4 gene is not a complete gene since it lacks most upstream exons while those that are present contain a stop codon, suggesting that it is a pseudogene (Jin et al. 1998).
Protein alignment and features:
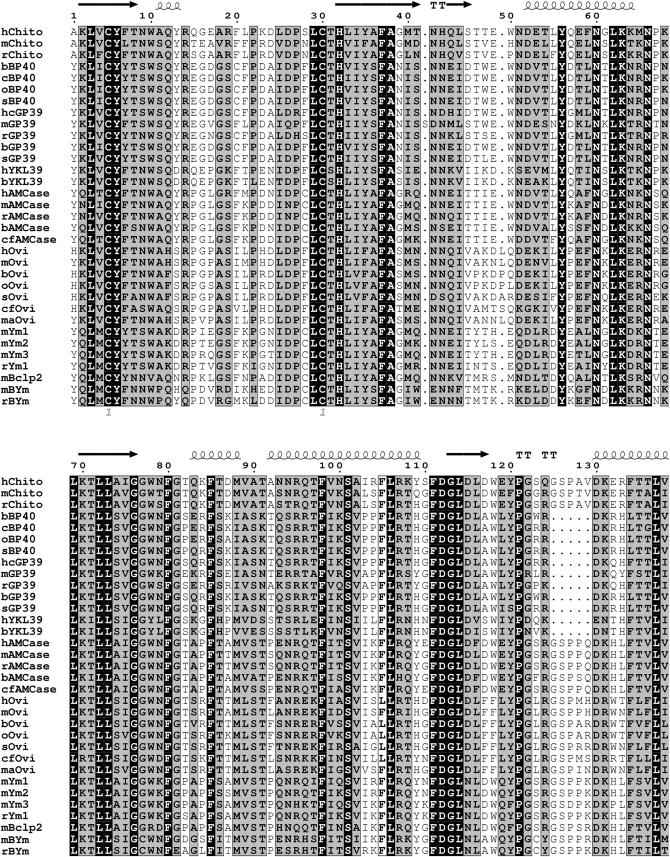
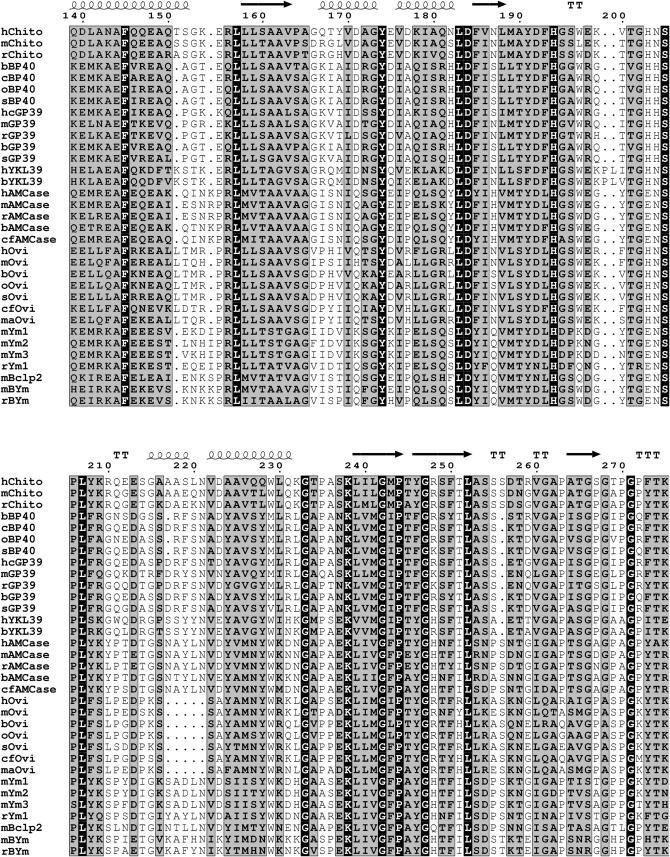
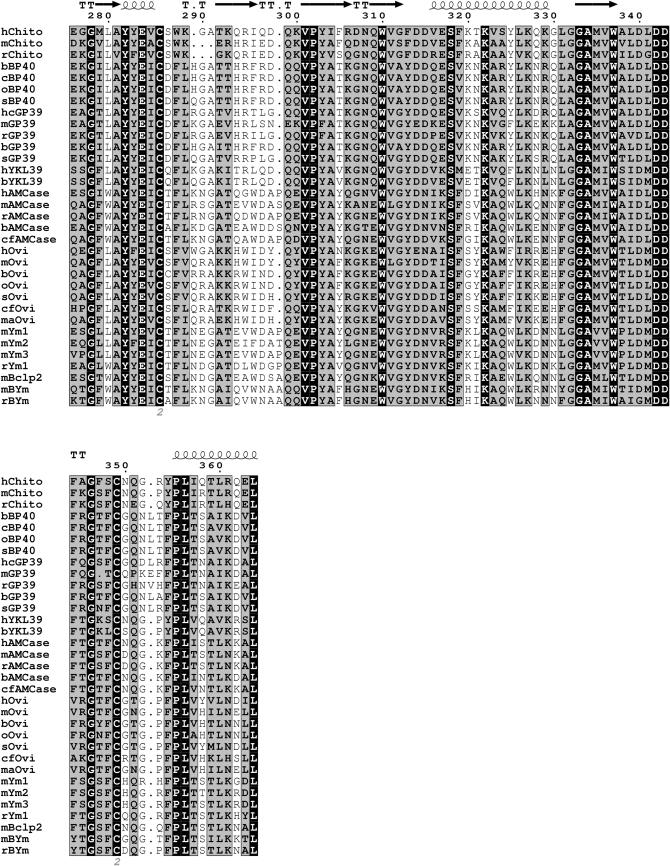
To investigate the overall homology and shared features of the members of the chitinase protein family listed in supplemental Table 1 at http://www.genetics.org/supplemental/, we aligned the 39-kDa TIM-barrel domain. Left out of this alignment were primate species other than humans (chimpanzee and macaque), given the extreme homology. Figure 1 shows the high overall homology in the mammalian TIM-barrel domains, as indicated by the shaded and solid backgrounds. The secondary structures of chitotriosidase, as observed in its crystal structure, generally correspond with regions of higher conservation, whereas loops separating two helices or strands correspond with gaps in the alignment. When secondary structures seen in crystal structures of two other family members, Hcgp39 and Ym1 (Houston et al. 2003 and Sun et al. 2001, respectively) are superimposed on the alignment, the outcome is similar (not shown).
Figure 1.—
Protein alignment of the mature 39-kDa catalytic domain of mammalian chitinase(-like) proteins. Completely conserved amino acids are indicated a with solid baskground and those with a high similarity score (according to RISLER) are in a shaded background. The secondary structures (strands, arrows; helices and turns, T's) of human chitotriosidase, as published, are superimposed on the alignment. Cysteines involved in disulfide bridging are indicated by the numbers below the alignment. Chito, chitotriosidase; hcGP39, human cartilage glycoprotein 39; Ovi, oviductin; h, H. sapiens; m, M. musculus; r, R. norvegicus; b, B. taurus; c, Capra hircus; o, Ovis aries; s, Sus scrofa; cf, Canis familiaris; ma, Mesocricetus auratus. For further details, see text.
The cysteine residues involved in disulfide-bond formation, known to be essential for correct folding and stability of the TIM-barrel, are completely conserved (see Figure 1). Interestingly, both the AMCases and Ym proteins have two additional conserved cysteine residues, potentially allowing formation of a third disulfide bond at positions 28 and 371 (Boot et al. 2001; Sun et al. 2001). The latter cysteine is located just outside the 39-kDa domain and is therefore not depicted in Figure 1.
The calculated isoelectric points (pl) of the chitinase(-like) proteins are also strongly conserved, as shown in Table 1. Both mouse and rat BYm have a basic pI. All AMCases have an acidic pI, being neutral in an acidic environment, in agreement with the observed expression in the gastrointestinal tract (Suzuki et al. 2002; Boot et al. 2005a). The oviductins, thought to be ubiquitously expressed in the slightly basic oviduct (Hugentobler et al. 2004), have basic pI's. The largest variation in pI exists among chitotriosidases (see Table 1). Taken together, the data on overall homology, conserved cysteines, and isoelectric point indicate interspecies retention of protein structure among orthologs.
TABLE 1.
Theoretically determined isoelectric points
| Chito | AMCase | GP39/BP40 | YKL39 | Oviductin | Ym1/2/3 and Bclp2 | BYm | |
|---|---|---|---|---|---|---|---|
| H. sapiens | 6.52 | 5.42 | 8.65 | 7.24 | 9.17 | — | — |
| P. troglodytes | 7.10 | — | 8.65 | 7.24 | 9.17 | — | — |
| Macaca mulatta | 7.76 | 4.90 | 8.19 | 6.83 | 9.17 | — | — |
| M. musculus | 5.94 | 5.14 | 8.65 | — | 8.49 | 5.16, 5.45, 5.62, 5.16 | 8.92 |
| R. norvegicus | 8.49 | 5.02 | 8.98 | — | — | 5.67 | 8.82 |
| B. taurus | — | 5.12 | 8.65, 8.80 | 8.89 | 9.41 | — | — |
| Capra hircus | — | — | 8.93 | — | — | — | — |
| Ovis aries | — | — | 8.66 | — | 9.44 | — | — |
| Sus scrofa | — | — | 9.14, 8.92 | — | 9.22 | — | — |
| Canis familiaris | — | 4.68 | — | — | 9.33 | — | — |
| Mesocricetus auratus | — | — | — | — | 8.71 | — | — |
Phylogenetic analyses:
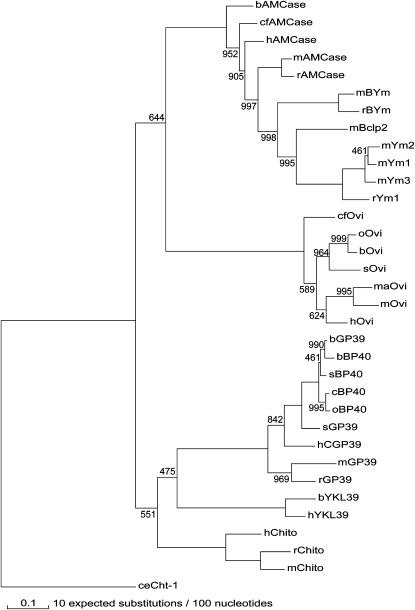
The maximum-likelihood tree shown in Figure 2, generated as described in materials and methods, is supported by high bootstrap support values. Importantly, a consensus maximum-parsimony analysis resulted in identical branching (see supplemental Figure 1 at http://www.genetics.org/supplemental/). The tree reveals clustering of all orthologs that are grouped in either the AMCase or the chitotriosidase clade. Both clades contain chi-lectins next to the chitinases, allowing a discrimination of AMCase-lectins and chitotriosidase-lectins. The chitotriosidase clade contains YKL39, GP39, and BP40 homologs. The latter two show complete conservation of the putative N-linked glycosylation sequence NIS near the N terminus (see Figure 1). The AMCase clade contains all oviductins and the rodent Ym proteins.
Figure 2.—
Maximum-likelihood tree of mammalian chitinase(-like) genes. The number on every node indicates the support bootstrap value for the likelihood analyses of a thousand replicate data sets. Only values other than 1000 are shown. The tree was rerooted by an outgroup (ceCht-1, Caenorhabditis elegans chitinase-1). Abbreviations as in Figure 1. For further details, see text.
Substitution rate ratios:
The phylogenetic tree provides information on evolutionary relationships of chitinase(-like) protein-coding genes. However, it does not provide information on selective forces. Since the AMCases have evolved to function in an acid environment, it might be hypothesized that there have been episodes of positive selection after the initial gene duplication to allow for rapid adaptation, in analogy to mammalian stomach lysozymes (Messier and Stewart 1997; Yang 1998). In addition, the repeated occurrence of the chi-lectin (loss of enzymatic function) mutations suggests site-specific positive selection. To evaluate more closely such selective forces, substitution rate ratios of nonsynonymous vs. synonymous mutations (dN/dS, ω) have been calculated with PAML. Values <1 indicate the occurrence of purifying (negative) selection, i.e., elimination of mutations that would result in a change in protein composition, whereas values >1 indicate a selective pressure to maintain the changes in the protein (positive or adaptive selection).
ω-Values were first calculated using the branch model to assess selection within clades. As a positive control, a data set consisting of seven primate lysozyme sequences was used, in which the occurrence of positive selection had previously been determined (Messier and Stewart 1997; Yang 1998). Although the results obtained by Yang (1998) could be exactly reproduced, applying various similar models to the chitinase family data set did not reveal episodes of adaptive selection. For the most parameter-rich model used, in which, in addition to a single ω for each ortholog, every ancestral branch was allowed a different ω, all ω-values were found to be substantially <1, thus giving no direct support for the occurrence of positive selection. Table 2 shows free in-group ω-values for all orthologs using this model. Values for ancestral branches ranged from 0.11 to 0.44 in all models.
TABLE 2.
Substitution rates dN, dS, and their ratio ω (dN/dS) for each orthologous group
| Chito | AMCase | GP39/BP40 | YKL39 | Oviductin | Ym1/2/3 and Bclp2 | BYm | |
|---|---|---|---|---|---|---|---|
| dN | 0.11 | 0.11 | 0.17 | 0.17 | 0.20 | 0.04 | 0.11 |
| dS | 0.48 | 0.61 | 0.60 | 0.83 | 0.82 | 0.10 | 0.29 |
| ω (dN/dS) | 0.23 | 0.18 | 0.28 | 0.21 | 0.25 | 0.37 | 0.38 |
It is conceivable that the chitinase(-like) proteins contain constrained amino acid sites subjected to purifying selection with ω close to zero as well as sites that could be subjected to positive selection. A large number of constrained sites would mask a signal of positive selection when the ω-values are averaged over all sites. ω-Values were therefore also calculated using the so-called site model. Swanson et al. (2001) had to employ a similar approach to identify a few amino acid positions in oviductins that were subjected to positive selection. An analysis using a data set consisting only of oviductin sequences confirmed the reported findings by Swanson et al. (2001) but expansion of the data set with other family members did not identify amino acid positions in other chitinase(-like) proteins subjected to positive selection.
Genomic synteny of chitotriosidase and AMCase loci in humans and rodents:
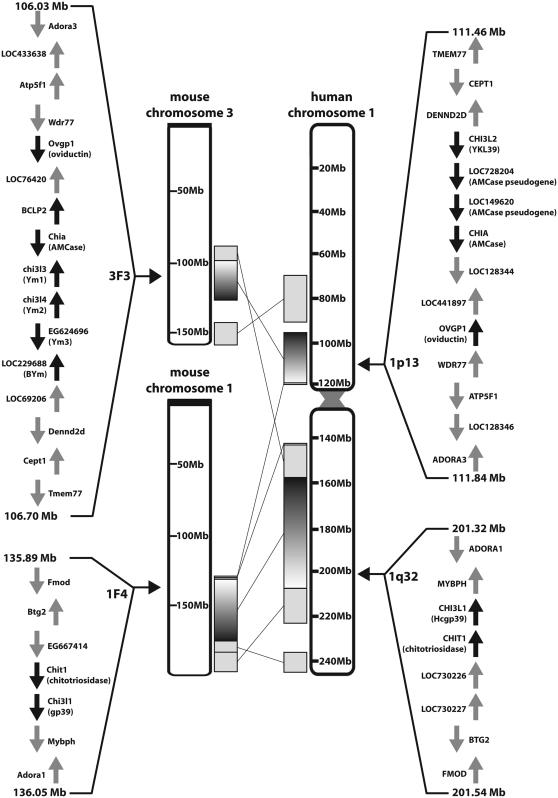
The chromosomal location of all chitinase(-like) protein-coding genes in mice and humans has been mapped. The chitotriosidase locus (1q32 in humans and 1F4 in mice) encoding chitotriosidase and GP39 and flanked by the genes coding for adenosine A1 receptor (ADORA1) and fibromodulin is syntenic (Boot et al. 2005a). In Figure 3 the synteny analysis is extended to the AMCase loci, corresponding to locus 1p13 in humans and 3F3 in mice. Again, the human and murine regions are flanked by the same genes, in this case coding for adenosine A3 receptor and transmembrane protein 77. The presence of an ADORA paralogue on chitotriosidase and AMCase loci suggests that the genes encoding the two active chitinases result from a large-scale duplication.
Figure 3.—
Synteny of loci encoding chitinase(-like) proteins between mice and humans. Schematic overview of the synteny of mouse locus 1F4 with human 1q32 and mouse locus 3F3 with human 1p13. The orientation and position of the genes are indicated with arrows. The genes of members of the chitinase protein family are depicted by solid arrows, whereas the other genes in the loci are depicted by shaded arrows.
The AMCase locus reveals major differences between mice and humans. First, additional open reading frames exist in the mouse genome encoding Ym1, Ym2, Bclp2, and BYm (encoded by Chi3l3, Chi3l4, BCLP2, and LOC229688, respectively), whereas these genes are absent from human chromosome 1p13. The opposite holds true for Chi3L2 encoding YKL39. Second, additional AMCase-like pseudogenes, LOC728204 and LOC149620, are present on human chromosome 1. Finally, the orientation of many genes in the human and mouse AMCase loci differs (see Figure 3), suggesting the occurrence of multiple and diverse recombination events.
It is of interest to note that the chi-lectin gene YKL39 is part of the AMCase locus in humans and cows, but, on the basis of both phylogenetic analyses and protein features, YKL39 likely results from a gene duplication event in the chitotriosidase locus. Apparently, in the case of the YKL39 gene, an additional rearrangement occurred.
DISCUSSION
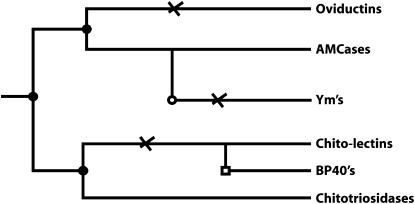
Our investigation rendered new insights into the evolutionary relationships of mammalian chitinase(-like) proteins. Both phylogenetic analyses and genomic synteny point to the same evolution of mammalian family 18 chitinase proteins (Figure 4). First, a gene duplication event allowed the specialization of two active chitinases, chitotriosidase and AMCase. Duplications of both chitotriosidase and AMCase genes, followed by loss-of-enzymatic-function mutations, led to the subsequent evolution of chi-lectins.
Figure 4.—
Overview of the evolution of chitinase(-like) genes. •, the “ancestral” gene duplications; ○, rodent-specific gene duplication; □, artiodactyle-specific gene duplication; a cross indicates the loss of catalytic activity mutations. “Chito-lectins” are chi-lectins evolved from the chitotriosidase gene (duplication).
The duplication of the active chitinase most likely has an ancient origin as Xenopus already has two active chitinases, one of which, like AMCase, is expressed in the stomach (Fujimoto et al. 2002). Indeed, the Xenopus AMCase homolog clusters with the mammalian AMCases (see supplemental Figure 2 at http://www.genetics.org/supplemental/). This suggests that the gene duplication allowing evolution of chitotriosidases and AMCases occurred very early in tetrapod evolution in the wake of the development of the acidic stomach. The evolution of the various mammalian chi-lectins is most likely a more recent event. The existence of a molluscan chi-lectin remarkably homologous to Hcgp39 has been reported (Badariotti et al. 2006). However, the molluscan sequence does not clade within the GP39 group (results not shown), suggesting that it is a product of an independent gene duplication and loss-of-function mutation. BLAST searches on the genome of the nematode C. elegans reveal several genes likely to encode chi-lectins, yet phylogenetic analyses again show none of them to group within the tree (results not shown). Recently, the occurrence in several plant species of chi-lectins homologous to chitinases has also been reported (van Damme et al. 2006). Mutations resulting in loss of chitinolytic activity have apparently occurred independently in a variety of lineages.
It is theoretically conceivable that the various types of chi-lectins in mammalian species have interdependently evolved by concerted evolution, a process driven by unequal crossover and gene conversion (reviewed in Nei and Rooney 2005). The lack of conservation of gene orientation within the AMCase locus (Figure 3) indeed suggests that recombination has occurred. However, phylogenetic analyses show that orthologs are far more closely related than paralogues, which does not substantiate concerted evolution as an important contributing mechanism underlying the diversification of chi-lectins. Instead, on the basis of observed relationships and selective pressures, the evolution of this gene family is in accordance with a form of multigene family evolution now referred to as “birth-and-death evolution under strong purifying selection” (reviewed in Nei and Rooney 2005).
Our study revealed the existence in mice of a previously unidentified chi-lectin, referred to as a hypothetical protein (BYm). Despite the fact that BYm belongs to the group of AMCase- (or acidic-) lectins, it displays a basic isoelectric point, suggesting substantial specialization. The absence of the gene in animals other than rodents points to a relatively recent gene duplication, nicely illustrating the remarkable ongoing evolution of chitinase(-like) proteins in mammals. Likewise, the occurrence of GP40 seems restricted to artiodactyls. GP40 has been found to be present in dry mammary secretions at times when extensive tissue remodeling occurs (Srivastava et al. 2006); hence its presence may reflect differences in mammary function between artiodactyls and other mammals.
Structural features among orthologs of the chitinase protein family are extremely well conserved among mammals, yet there are differences in expression among species. A striking example in this respect is chitotriosidase in humans and rodents (Boot et al. 2005a). The marked conservation of structural features of the catalytic domain of chitinases may be imposed by severe restrictions in changes compatible with preservation of catalytic function. Mammalian chitinases appear strongly subjected to negative (purifying) selection, substantially more than the functionally similar lysozymes. This is nicely illustrated by the fact that comparison of substitution rate ratios between lysozyme from Rhesus macaque and from hominoids point strongly toward positive selection whereas such comparisons for chitotriosidase and AMCase result in ω-values far <1. A large number of constrained sites in both chitinases should mask any signal of positive selection when the ω-values are averaged over all sites. Site-specific models for ω calculation did not give values >1, although positive selection in the diversification of the two chitinases and their respective chi-lectins clearly occurred. To detect a very strong indicator of positive selection (ω > 1), it is necessary to look not only at a specific site but also at a specific time interval (Sharp 1997; Zhang et al. 2005).
The high homology among members of the chitinase(-like) protein family may cause confusion when comparing genes of different species (Boot et al. 2005b; Raes et al. 2005; Reese et al. 2007). There are considerable differences between species regarding presence or absence of particular chi-lectins. One example in this respect is Ym1. This protein has been extensively studied since it is secreted by alternatively activated macrophages in mice under a variety of inflammatory conditions (Chang et al. 2001; Welch et al. 2002; Zhao et al. 2005; Iwashita et al. 2006; Reese et al. 2007). The implications for human pathology, however, may be considered limited since no true human ortholog of Ym1 exists.
In conclusion, our investigation of family 18 glycosyl hydrolases has revealed that active chitinases and chi-lectins are widespread and conserved in the mammalian kingdom. An ancient gene duplication first allowed the specialization of two active chitinases, chitotriosidase and AMCase, and subsequent gene duplications followed by loss-of-enzymatic-function mutations, have led to the evolution of a broad spectrum of chi-lectins in mammals.
Acknowledgments
We gratefully acknowledge Roy Erkens of the University of Utrecht, The Netherlands, for his skillful technical assistance and useful discussions relating to the phylogenetic analyses.
References
- Badariotti, F., M. Kypriotou, C. Lelong, M. P. Dubos, E. Renard et al., 2006. The phylogenetically conserved molluscan chitinase-like protein 1 (Cg-Clp1), homologue of human HC-gp39, stimulates proliferation and regulates synthesis of extracellular matrix components of mammalian chondrocytes. J. Biol. Chem. 281: 29583–29596. [DOI] [PubMed] [Google Scholar]
- Beutler, E., and G. A. Grabowski, 1995. Gaucher disease, pp. 2641–2670 in The Metabolic and Molecular Bases of Inherited Disease, Ed. 7, edited by C. R. Scriver, A. L. Beaudet, W. S. Sly and D. Valle. McGraw-Hill, New York.
- Boot, R. G., G. H. Renkema, A. Strijland, A. J. van Zonneveld and J. M. Aerts, 1995. Cloning of a cDNA encoding chitotriosidase, a human chitinase produced by macrophages. J. Biol. Chem. 270: 26252–26256. [DOI] [PubMed] [Google Scholar]
- Boot, R. G., G. H. Renkema, M. Verhoek, A. Strijland, J. Bliek et al., 1998. The human chitotriosidase gene. Nature of inherited enzyme deficiency. J. Biol. Chem. 273: 25680–25685. [DOI] [PubMed] [Google Scholar]
- Boot, R. G., E. F. Blommaart, E. Swart, K. Ghauharali-van der Vlugt, N. Bijl et al., 2001. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J. Biol. Chem. 276: 6770–6778. [DOI] [PubMed] [Google Scholar]
- Boot, R. G., A. P. Bussink, M. Verhoek, P. A. de Boer, A. F. Moorman et al., 2005. a Marked differences in tissue-specific expression of chitinases in mouse and man. J. Histochem. Cytochem. 53: 1283–1292. [DOI] [PubMed] [Google Scholar]
- Boot, R. G., A. P. Bussink and J. M. Aerts, 2005. b Human acidic mammalian chitinase erroneously known as eosinophil chemotactic cytokine is not the ortholog of mouse YM1. J. Immunol. 175: 2041–2042. [DOI] [PubMed] [Google Scholar]
- Buhi, W. C., 2002. Characterization and biological roles of oviduct-specific, oestrogen-dependent glycoprotein. Reproduction 123: 355–362. [DOI] [PubMed] [Google Scholar]
- Bussink, A. P., M. van Eijk, G. H. Renkema, J. M. Aerts and R. G. Boot, 2006. The biology of the Gaucher cell: the cradle of human chitinases. Int. Rev. Cytol. 252: 71–128. [DOI] [PubMed] [Google Scholar]
- Chang, N. C., S. I. Hung, K. Y. Hwa, I. Kato, J. E. Chen et al., 2001. A macrophage protein, Ym1, transiently expressed during inflammation is a novel mammalian lectin. J. Biol. Chem. 276: 17497–17506. [DOI] [PubMed] [Google Scholar]
- Felsenstein, J., 2006. PHYLIP (Phylogeny Inference Package), Version 3.65. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle.
- Fujimoto, W., K. Kimura and T. Iwanaga, 2002. Cellular expression of the gut chitinase in the stomach of frogs Xenopus laevis and Rana catesbeiana. Biomed. Res. 23: 91–99. [Google Scholar]
- Fusetti, F., H. von Moeller, D. Houston, H. J. Rozeboom, B. W. Dijkstra et al., 2002. Structure of human chitotriosidase. Implications for specific inhibitor design and function of mammalian chitinase-like lectins. J. Biol. Chem. 277: 25537–25544. [DOI] [PubMed] [Google Scholar]
- Fusetti, F., T. Pijning, K. H. Kalk, E. Bos and B. W. Dijkstra, 2003. Crystal structure and carbohydrate-binding properties of the human cartilage glycoprotein-39. J. Biol. Chem. 278: 37753–37760. [DOI] [PubMed] [Google Scholar]
- Gasteiger, E., A. Gattiker, C. Hoogland, I. Ivanyi, R. D. Appel et al., 2003. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31: 3784–3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouet, P., E. Courcelle, D. I. Stuart and F. Metoz, 1999. ESPript: multiple sequence alignments in PostScript. Bioinformatics 15: 305–308. [DOI] [PubMed] [Google Scholar]
- Gouet, P., X. Robert and E. Courcelle, 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 31: 3320–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hakala, B. E., C. White and A. D. Recklies, 1993. Human cartilage gp-39, a major secretory product of articular chondrocytes and synovial cells, is a mammalian member of a chitinase protein family. J. Biol. Chem. 268: 25803–25810. [PubMed] [Google Scholar]
- Henrissat, B., 1991. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollak, C. E., S. van Weely, M. H. van Oers and J. M. Aerts, 1994. Marked elevation of plasma chitotriosidase activity: a novel hallmark of Gaucher disease. J. Clin. Invest. 93: 1288–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer, R. J., Z. Zhu, L. Cohn, C. G. Lee, W. I. White et al., 2006. Differential expression of chitinases identify subsets of murine airway epithelial cells in allergic inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 291: 502–511. [DOI] [PubMed] [Google Scholar]
- Houston, D. R., A. D. Recklies, J. C. Krupa and D. M. van Aalten, 2003. Structure and ligand-induced conformational change of the 39-kDa glycoprotein from human articular chondrocytes. J. Biol. Chem. 278: 30206–30212. [DOI] [PubMed] [Google Scholar]
- Hu, B., K. Trinh, W. F. Figueira and P. A. Price, 1996. Isolation and sequence of a novel human chondrocyte protein related to mammalian members of the chitinase protein family. J. Biol. Chem. 271: 19415–19420. [DOI] [PubMed] [Google Scholar]
- Hugentobler, S., D. G. Morris, M. T. Kane and J. M. Serena, 2004. In situ oviduct and uterine pH in cattle. Theriogenology 61: 1419–1427. [DOI] [PubMed] [Google Scholar]
- Iwashita, H., S. Morita, Y. Sagiya and A. Nakanishi, 2006. Role of eosinophil chemotactic factor by T lymphocytes on airway hyperresponsiveness in a murine model of allergic asthma. Am. J. Respir. Cell Mol. Biol. 35: 103–109. [DOI] [PubMed] [Google Scholar]
- Jeuniaux, C., 1961. Chitinase: an addition to the list of hydrolases in the digestive tract of vertebrates. Nature 192: 135–136. [DOI] [PubMed] [Google Scholar]
- Jin, H. M., N. G. Copeland, D. J. Gilbert, N. A. Jenkins, R. B. Kirkpatrick et al., 1998. Genetic characterization of the murine Ym1 gene and identification of a cluster of highly homologous genes. Genomics 54: 316–322. [DOI] [PubMed] [Google Scholar]
- Johansen, J. S., 2006. Studies on serum YKL-40 as a biomarker in diseases with inflammation, tissue remodelling, fibroses and cancer. Dan. Med. Bull. 53: 172–209. [PubMed] [Google Scholar]
- Johansen, J. S., H.S. Jensen and P. A. Price, 1993. A new biochemical marker for joint injury: analysis of YKL-40 in serum and synovial fluid. Br. J. Rheumatol. 32: 949–955. [DOI] [PubMed] [Google Scholar]
- Messier, W., and C. B. Stewart, 1997. Episodic adaptive evolution of primate lysozymes. Nature 385: 151–154. [DOI] [PubMed] [Google Scholar]
- Monzingo, A. F., E. M. Marcotte, P. J. Hart and J. D. Robertus, 1996. Chitinases, chitosanases, and lysozymes can be divided into procaryotic and eucaryotic families sharing a conserved core. Nat. Struct. Biol. 3: 133–140. [DOI] [PubMed] [Google Scholar]
- Nei, M., and A. P. Rooney, 2005. Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39: 121–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raes, G., R. van den Bergh, P. de Baetselier, G. H. Ghassabeh, C. Scotton et al., 2005. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J. Immunol. 174: 834–845. [DOI] [PubMed] [Google Scholar]
- Recklies, A. D., C. White and H. Ling, 2002. The chitinase 3-like protein human cartilage glycoprotein 39 (HC-gp39) stimulates proliferation of human connective-tissue cells and activates both extracellular signal-regulated kinase- and protein kinase B-mediated signalling pathways. Biochem. J. 365: 119–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese, T. A., H. E. Liang, A. M. Tager, A. D. Luster, N. van Rooijen et al., 2007. Chitin induces accumulation in tissue of innate immune cells associated with allergy. Nature 447: 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkema, G. H., R. G. Boot, A. O. Muijsers, W. E. Donker-Koopman and J. M. Aerts, 1995. Purification and characterization of human chitotriosidase, a novel member of the chitinase family of proteins. J. Biol. Chem. 270: 2198–2202. [DOI] [PubMed] [Google Scholar]
- Renkema, G. H., R. G. Boot, A. Strijland, W. E. Donker-Koopman, M. van den Berg et al., 1997. Synthesis, sorting, and processing into distinct isoforms of human macrophage chitotriosidase. Eur. J. Biochem. 244: 279–285. [DOI] [PubMed] [Google Scholar]
- Renkema, G. H., R. G. Boot, F. L. Au, W. E. Donker-Koopman, A. Strijland et al., 1998. Chitotriosidase, a chitinase, and the 39-kDa human cartilage glycoprotein, a chitin-binding lectin, are homologues of family 18 glycosyl hydrolases secreted by human macrophages. Eur. J. Biochem. 251: 504–509. [DOI] [PubMed] [Google Scholar]
- Sharp, P. M., 1997. In search of molecular Darwinism. Nature 385: 111–112. [DOI] [PubMed] [Google Scholar]
- Srivastava, D. B., A. S. Ethayathulla, J. Kumar, N. Singh, S. Sharma et al., 2006. Crystal structure of a secretory signalling glycoprotein from sheep at 2.0A resolution. J. Struct. Biol. 156: 505–516. [DOI] [PubMed] [Google Scholar]
- Sun, Y. J., N. C. Chang, S. I. Hung, A. C. Chang, C. C. Chou et al., 2001. The crystal structure of a novel mammalian lectin, Ym1, suggests a saccharide binding site. J. Biol. Chem. 276: 17507–17514. [DOI] [PubMed] [Google Scholar]
- Suzuki, M., W. Fujimoto, M. Goto, M. Morimatsu, B. Syuto et al., 2002. Cellular expression of gut chitinase mRNA in the gastrointestinal tract of mice and chickens. J. Histochem. Cytochem. 50: 1081–1089. [DOI] [PubMed] [Google Scholar]
- Swanson, W. J., Z. Yang, M. F. Wolfner and C. F. Aquadro, 2001. Positive Darwinian selection drives the evoluion of several female reproductive proteins in mammals. Proc. Natl. Sci. USA 98: 2509–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwisscha van Scheltinga, A. C., S. Armand, K. H. Kalk, A. Isogai, B. Henrissat et al., 1995. Stereochemistry of chitin hydrolysis by a plant chitinase/lysozyme and X-ray structure of a complex with allosamidin: evidence for substrate assisted catalysis. Biochemistry 34: 15619–15623. [DOI] [PubMed] [Google Scholar]
- Tharanathan, R. N., and F. S. Kittur, 2003. Chitin: the undisputed biomolecule of great potential. Crit. Rev. Food Sci. Nutr. 43: 61–87. [DOI] [PubMed] [Google Scholar]
- Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin and D. G. Higgins, 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruha, J., K. Masuko-Hongo, T. Kato, M. Sakata, H. Nakamura et al., 2002. Autoimmunity against YKL-39, a human cartilage derived protein, in patients with osteoarthritis. J. Rheumatol. 29: 1459–1466. [PubMed] [Google Scholar]
- van Aalten, D. M., D. Komander, B. Synstad, S. Gaseidnes, M. G. Peter et al., 2001. Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc. Natl. Acad. Sci. USA 98: 8979–8984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Damme, E. J., R. Culerrier, A. Barre, R. Alvarez, P. Rouge et al., 2006. A novel family of lectins evolutionarily related to class V chitinases: an example of neofunctionalization in legumes. Plant Physiol. 144: 662–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijk, M., C. P. van Roomen, G. H. Renkema, A. P. Bussink, L. Andrews et al., 2005. Characterization of human phagocyte-derived chitotriosidase, a component of innate immunity. Int. Immunol. 17: 1505–1512. [DOI] [PubMed] [Google Scholar]
- Verheijden, G. F., A. W. Rijnders, E. Bos, C. J. Coenen-de Roo, C. J. van Staveren et al., 1997. Human cartilage glycoprotein-39 as a candidate autoantigen in rheumatoid arthritis. Arthritis Rheum. 40: 1115–1125. [DOI] [PubMed] [Google Scholar]
- Ward, J. M., M. Yoon, M. R. Anver, D. C. Haines, G. Kudo et al., 2001. Hyalinosis and Ym1/Ym2 gene expression in the stomach and respiratory tract of 129S4/SvJae and wild-type and CYP1A2-null B6, 129 mice. Am. J. Pathol. 158: 323–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, J. S., L. Escoubet-Lozach, D. B. Sykes, K. Liddiard, D. R. Greaves et al., 2002. TH2 cytokines and allergic challenge induce Ym1 expression in macrophages by a STAT6-dependent mechanism. J. Biol. Chem. 277: 42821–42829. [DOI] [PubMed] [Google Scholar]
- Wierenga, R. K., 2001. The TIM-barrel fold: a versatile framework for efficient enzymes. FEBS Lett. 492: 193–198. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1997. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 13: 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., 1998. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol. Biol. Evol. 15: 568–573. [DOI] [PubMed] [Google Scholar]
- Zhang, J., R. Nielsen and Z. Yang, 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol. Biol. Evol. 22: 2472–2479. [DOI] [PubMed] [Google Scholar]
- Zhao, J., H. Zhu, C. H. Wong, K. Y. Leung and W. S. Wong, 2005. Increased lungkine and chitinase levels in allergic airway inflammation: a proteomics approach. Proteomics 5: 2799–2807. [DOI] [PubMed] [Google Scholar]
- Zhu, Z., T. Zheng, R. J. Homer, Y. K. Kim, N. Y. Chen et al., 2004. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science 304: 1678–1682. [DOI] [PubMed] [Google Scholar]