Abstract
The assembly of cytochrome c oxidase (CcO) in yeast mitochondria is shown to be dependent on a new assembly factor designated Coa1 that associates with the mitochondrial inner membrane. Translation of the mitochondrial-encoded subunits of CcO occurs normally in coa1Δ cells, but these subunits fail to accumulate. The respiratory defect in coa1Δ cells is suppressed by high-copy MSS51, MDJ1 and COX10. Mss51 functions in Cox1 translation and elongation, whereas Cox10 participates in the biosynthesis of heme a, a key cofactor of CcO. Respiration in coa1Δ and shy1Δ cells is enhanced when Mss51 and Cox10 are coexpressed. Shy1 has been implicated in formation of the heme a3-CuB site in Cox1. The interaction between Coa1 and Cox1, and the physical and genetic interactions between Coa1 and Mss51, Shy1 and Cox14 suggest that Coa1 coordinates the transition of newly synthesized Cox1 from the Mss51:Cox14 complex to the heme a cofactor insertion involving Shy1. coa1Δ cells also display a mitochondrial copper defect suggesting that Coa1 may have a direct link to copper metallation of CcO.
Keywords: copper, cytochrome c oxidase, heme a, mitochondria, Shy1
Introduction
Cytochrome c oxidase (CcO) is the terminal enzyme of the energy-transducing respiratory chain in mitochondria of eukaryotes, coupling the reduction of molecular oxygen to proton translocation across the inner membrane (IM), which drives ATP synthesis. Eukaryotic CcO consists of 12–13 subunits, with three mitochondrial encoded subunits (Cox1–Cox3) forming the core enzyme. The remaining subunits are encoded by the nuclear genome and consist of small polypeptides that surround the catalytic core (Tsukihara et al, 1995). The core enzyme contains three copper atoms and two heme a, a cofactor specific to CcO (Tsukihara et al, 1995). The enzyme is embedded within the IM with a portion of the molecule protruding into the intermembrane space (IMS) and a separate portion extending into the matrix compartment.
Assembly of CcO requires a myriad of steps including the assembly of subunits translated on cytoplasmic and mitochondrial ribosomes, modification of protoheme to heme a and the subsequent insertion of this moiety, along with that of copper, zinc and magnesium ions, into the enzyme complex (Carr and Winge, 2003; Khalimonchuk and Rodel, 2005). Two separate copper centers CuA and CuB are functionally important in both Cox2 and Cox1, respectively. The assembly process of CcO requires over 30 accessory proteins (Tzagoloff and Dieckmann, 1990; Barrientos et al, 2002a) and appears to follow a sequential order of subassembly intermediates in mammalian tissues (Nijtmans et al, 1998; Williams et al, 2004).
Translation of mitochondrial-encoded subunits occurs within the matrix on ribosomes associated with both the inner boundary membrane and cristae membrane (Vogel et al, 2006). Transcripts for COX1–COX3 are recruited to the IM in yeast by a series of IM-associated translational activators (Sanchirico et al, 1998). These activators bind sequences in the 5′ untranslated regions of the mRNA and promote cotranslational membrane insertion (Naithani et al, 2003). Translation of the COX1 transcript is activated by Pet309 (Manthey and McEwen, 1995), the COX2 transcript by Pet111 (Green-Willms et al, 2001) and the COX3 transcript by Pet54, Pet122 and Pet494 (McMullin and Fox, 1993). In addition to Pet309, Mss51 is required for COX1 translational initiation, and has an additional role in translational elongation (Siep et al, 2000; Perez-Martinez et al, 2003; Barrientos et al, 2004). Mss51 also binds newly synthesized Cox1 and collaborates with Cox14 in a post-translational step in CcO assembly (Barrientos et al, 2004). Cox1–Cox3 are inserted into the IM by translocases, one of which is Oxa1 (He and Fox, 1997; Hell et al, 2001).
Heme a biogenesis involves farnesylation and oxidation of protoheme catalyzed by Cox10 and Cox15. Although eukaryotic heme a insertion is not well understood, heme a is likely inserted in Cox1 before its association with other subunits (Williams et al, 2004). In the absence of Cox2, Cox1 accumulates in Rhodobacter sphaeroides with a single heme a moiety in the heme a site, leaving the a3 site unpopulated (Bratton et al, 2000). Heme a3 formation likely occurs concurrent with Cox11-mediated CuB site formation in Cox1. Shy1 appears to be important for heme a3 insertion or stabilization of the intermediate before Cox2 binding, because R. sphaeroides lacking SHY1 have a CcO assembly defect in heme a3 and CuB (Smith et al, 2005). Shy1 is not absolutely required for CcO assembly in yeast, since shy1Δ cells retain 10–15% residual CcO activity. Mutations in SURF1, the human ortholog of SHY1, cause a CcO deficiency resulting in Leigh syndrome (Zhu et al, 1998).
Copper insertion is required in the maturation of both Cox1 and Cox2. A number of accessory factors are important in the copper metallation of CcO within the IMS. The source of copper for the formation of the CuA and CuB cofactor sites in CcO derives from a copper pool within the mitochondrial matrix. This pool of copper is present in a ligand complex (CuL) that is conserved in eukaryotic cells (Cobine et al, 2006a). Targeting of heterologous copper-binding proteins to the matrix attenuates this CuL complex, resulting in an impairment of CcO assembly. This predicts the presence of mitochondrial IM proteins for the transport of the CuL complex (Cobine et al, 2006b).
A group of mitochondrial proteins of unknown function were designated as the Found in Mitochondrial Proteome (FMP) family in a yeast proteomic study (Sickmann et al, 2003). In an attempt to identify proteins involved in the translocation of the CuL complex to mitochondria, we screened a collection of FMP gene deletion strains for those exhibiting respiratory defects and abnormal mitochondrial copper concentrations. In the present study, we identify one FMP protein (Yil157c/Fmp35) that when deleted from cells, results in low mitochondrial copper levels and attenuated CcO activity. This IM protein, renamed Coa1 for Cytochrome Oxidase Assembly 1, is shown presently to be important for Cox1 assembly. Deletion of COA1 or SHY1 leads to very similar phenotypes that can be suppressed by MSS51 and COX10, arguing that Coa1 and Shy1 function at a closely related step in Cox1 assembly. Genetic and physical interaction of Coa1 with Mss51, Cox14 and Shy1 suggest that Coa1 is important for coordinating Cox1 translation and cofactor assembly in this subunit.
Results
coa1Δ cells are respiratory deficient and have low level of mitochondrial copper
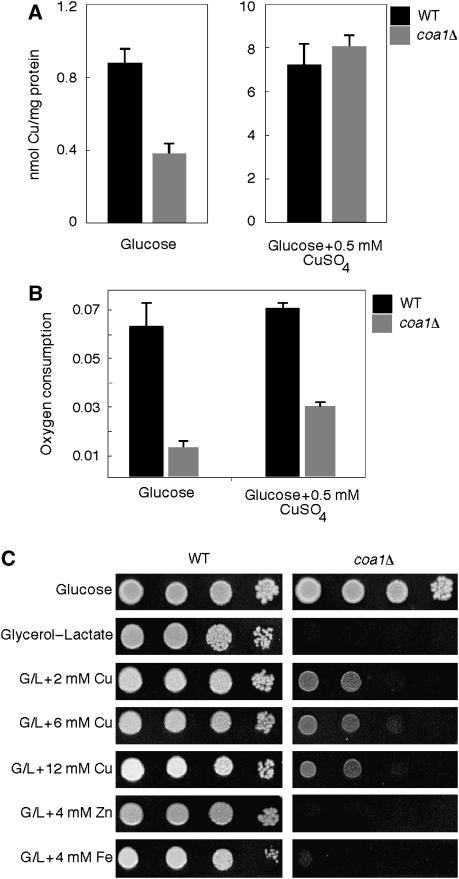
Mitochondria isolated from 54 FMP deletion strains were screened by ICP/OES analysis for abnormal mitochondrial copper levels. One strain, coa1Δ (yil157cΔ), showed reduced mitochondrial copper (Figure 1A). Copper levels were 43% of WT in gradient-purified mitochondria, yet total cellular copper was unaffected. This defect was copper specific as zinc, iron and manganese levels were not altered in coa1Δ cells. Mitochondria isolated from the deletion cells cultured in 0.5 mM CuSO4 contained the same elevated levels of copper as WT cells (Figure 1A). As copper is required for CcO function, we quantified oxygen consumption of coa1Δ cells. This deletion strain consumed less oxygen than the WT and the addition of 1 mM CuSO4 to the growth medium partially enhanced respiration (Figure 1B). Consistent with the reduced oxygen consumption of coa1Δ cells, the mutant cells were compromised for growth on non-fermentable medium containing glycerol/lactate (Figure 1C). The deletion of COA1 in the BY or W303 backgrounds resulted in an inability to grow on minimal glycerol/lactate medium. However, weak growth could be observed for W303 coa1Δ cells on rich glycerol/lactate medium, whereas BY4743 coa1Δ cells failed to grow (data not shown and Figure 1C). The addition of copper to rich glycerol/lactate medium enhanced the growth of coa1Δ cells (Figure 1C). The partial rescue in growth by copper salts was specific for copper, as high concentrations of iron or zinc salts failed to yield cell growth (Figure 1C).
Figure 1.
coa1Δ cells are respiratory deficient and have low mitochondrial copper. (A) Copper levels were assessed by ICP-OES in Nycodenz-purified mitochondria of coa1Δ cells and the isogenic WT BY4743 grown in YPD. (B) coa1Δ cells and the WT BY4743 were grown in YPD liquid media at 30°C with 1 mM CuSO4, where indicated and oxygen consumption (% O2/sec/OD600) was measured. (C) BY4743 and coa1Δ cells were grown in YPD then serially diluted and spotted on YP 2% glycerol–2% lactate (G/L) or 2% glucose plates. CuSO4, FeCl2 and ZnSO4 were added to the plate media at specified concentrations. Plates were kept at 30°C for 5 days.
coa1Δ cells have a specific defect in CcO assembly that is partially rescued by copper
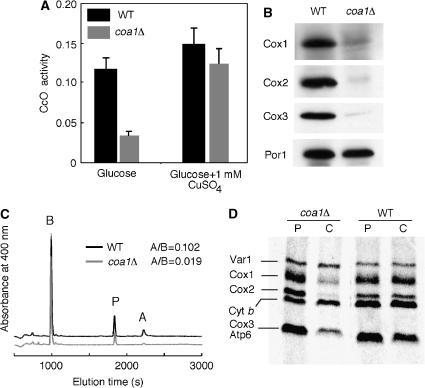
Mitochondria purified from coa1Δ cells exhibit low CcO activity (30% of WT) that can be partially restored by addition of copper to the growth medium (Figure 2A). Complex II and complex III activities were unaffected by the deletion of COA1 (data not shown). Cells lacking COA1 had reduced steady-state levels of CcO subunits Cox1–3 (Figure 2B), and the level of heme a in mitochondria from coa1Δ cells was only 20% of WT (Figure 2C), consistent with the low CcO activity. Heme b levels were comparable in mitochondria from coa1Δ or WT cells (Figure 2C), in accordance with the normal complex III activity.
Figure 2.
Deletion of COA1 results in a CcO-specific defect. (A) coa1Δ cells and the isogenic WT BY4743 were grown in YPD with or without 1 mM CuSO4. Purified mitochondria were assayed for CcO activity (ΔOD550/min/10 μg protein). The data represent the average of three independent experiments. (B) Immunoblot of mitochondria (30 μg protein) from coa1Δ cells or from BY4743 for Cox1–Cox3. Porin is shown as a loading control. (C) Heme was extracted from mitochondria (2 mg protein) purified from coa1Δ or BY4743 cells and separated by reverse-phase HPLC. The area under the curve of the peaks of heme b (B) and heme a (A) were calculated and used to determine a heme a to heme b ratio as the levels of heme b were unaffected by the deletion of COA1. Protoporphyrin peaks (P) were found to vary as reported previously (Barros and Tzagoloff, 2002). (D) In vivo labeling of mitochondrial translation products. coa1Δ and BY4743 cells were pulsed for 15 min with 35S-labeled methionine at which point 20 mM of cold methionine was added. The reaction was either stopped (P) or allowed to chase (C) for 90 min at 30°C. The samples were run on 12% SDS–PAGE, the gel was dried and exposed to autoradiographic film.
Since Coa1 was reported to interact with Atp14 in a global two-hybrid interaction study (Ito et al, 2001), we assessed the level of complex V in coa1Δ cells. The migration and levels of monomeric and dimeric complex V on Blue native PAGE (BN-PAGE) were comparable to WT (data not shown), making it unlikely that Coa1 is important for complex V assembly.
To investigate whether Coa1 has a role in the translation of mitochondrial-encoded subunits of CcO, [35S]-Met incorporation into mitochondrial translation products was monitored in vivo. coa1Δ cells exhibit a labeling profile of mitochondrial-encoded proteins similar to WT cells after the 15-min pulse (Figure 2D). During the 90-min chase, the levels of Cox1–Cox3 diminished markedly in coa1Δ cells, whereas Var1 and Cyt b remained stable as in the WT cells. This selective degradation of Cox1–Cox3 is consistent with an assembly defect in CcO. Taken together, these data show that coa1Δ cells have a post-translational CcO-specific defect.
Coa1 is associated with the IM
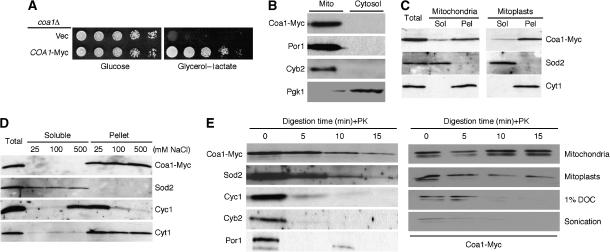
Glycerol/lactate growth was fully restored in coa1Δ cells when transformed with 3′ Myc-tagged COA1 on a YCp vector under the control of the MET25 promoter and CYC1 terminator (Figure 3A). Mitochondria isolated from coa1Δ COA1-Myc cells showed a prominent Coa1-Myc band at the expected mass of 22 kDa (Figure 3B). The protein is retained in mitoplasts and is not extracted by carbonate buffer (Figure 3C), low or high salt concentrations and sonication (Figure 3D), suggesting a tight association with either the IM or outer membrane. Digestion of Coa1-Myc by proteinase K in mitoplasts was slow compared with OM or IMS proteins and was most similar to the degradation pattern of the matrix protein Sod2 (Figure 3E). Coa1-Myc was degraded if the mitoplasts were initially solubilized with 1% deoxycholate (DOC) or disrupted by sonication, excluding the possibility that Coa1 is somehow resistant to degradation by proteinase K (Figure 3E). The hydropathy profile of Coa1 reveals a central, conserved hydrophobic stretch whose length is sufficient to form a transmembrane domain. Overall, these data strongly suggest that Coa1 spans the IM.
Figure 3.
Mitochondrial localization of Coa1. (A) coa1Δ cells expressing a low-copy version of COA1-Myc were serially diluted on SC 2% glycerol–2% lactate or 2% glucose plates (control) and compared to the same strain containing an empty vector. (B) Purified mitochondria and the cytosolic fraction from the coa1Δ+COA1-Myc strain were isolated and probed by immunoblotting for c-Myc and for the mitochondrial markers porin (Por1) and cytochrome b2 (Cyb2), and the cytosolic marker phosphoglycerate kinase 1 (Pgk1). (C) Membrane association was assessed by carbonate extraction of Coa1-Myc from whole mitochondria or mitoplasts purified from the coa1Δ+COA1-Myc strain. The soluble and pellet fractions were assayed by immunoblotting with the c-Myc antibody and for the soluble matrix protein superoxide dismutase 2 (Sod2) or the IM protein cytochrome c1 (Cyt1). (D) The soluble and pellet fractions of Coa1-Myc mitoplasts were separated by centrifugation after sonication in buffer containing 25, 100 or 500 mM NaCl. The fractions were assayed by immunoblotting for c-Myc, Sod2 (soluble matrix protein), Cyt1 (integral IM protein) and Cyc1 (IM associated protein). (E) Mitoplasts were treated with proteinase K (PK) over a time course and analyzed by immunoblotting. As a control, whole mitochondria, mitoplasts, mitoplasts solubilized in 1% deoxycholate (DOC) and sonicated mitoplasts were treated with PK (right panel). Note: OM fragments remain associated with mitoplasts, hence the presence of some porin (Por1) in mitoplasts.
MSS51, COX10 and MDJ1 are high copy suppressors of the respiratory defect of coa1Δ cells
To address the role of Coa1 in CcO maturation, we screened for high-copy suppressors of the respiratory growth defect of coa1Δ cells. Cells transformed with a high-copy DNA bank prepared from coa1Δ cells were screened for growth on glycerol/lactate medium. From about 200 000 colonies replicated on glycerol/lactate medium, 51 colonies showed significant growth after 5–7 days of incubation at 30°C. The growth of most of these colonies was shown to be vector dependent, based on the loss of respiratory growth when the URA3-based vector was shed by 5-fluoroorotic acid selection. Sequencing showed that the plasmids contained different fragments of genomic DNA that encompassed one of three genes. MSS51 was present in 21 plasmids, COX10 in five and MDJ1 in six plasmids. MSS51 was a stronger suppressor relative to MDJ1 and COX10 (Figure 4A).
Figure 4.
High-copy suppressors of the respiratory defect of coa1Δ cells. (A) coa1Δ cells transformed with high-copy vectors isolated from the suppressor screen were grown in SC-2% raffinose, serially diluted and spotted on SC 2% glycerol–2% lactate or 2% glucose (control) with selection. The plates were incubated at 30°C for 2 days (glucose) or 7 days (glycerol-lactate). (B) The same cells as in panel A were spotted on rich media (YP) supplemented with 50 μM FeCl2, containing 2% glycerol–2% lactate (YPLG) or 2% glucose (YPD), and either the copper-specific chelator BCS or CuSO4. The plates were incubated at 30°C for 2 days (YPD) or 6 days (YPLG).
Due to the low mitochondrial copper in coa1Δ cells (Figure 1A), we assessed the efficiency of the high-copy suppressors upon modulation of the copper concentration in the growth medium. At 6 mM copper, the suppression of the respiratory defect of coa1Δ by Mdj1 or Cox10 is weakly enhanced (Figure 4B), but this may arise from the general copper suppression occurring in coa1Δ cells as shown in Figure 1C. When copper was limited by the chelator bathocuproine sulfonate (BCS), the growth of coa1Δ cells covered with COA1 on a low-copy plasmid was slightly attenuated compared with the YPLG medium; however, respiratory growth conferred by the high-copy MSS51 was more strongly attenuated (Figure 4B). Thus, the MSS51 suppression of coa1Δ cells appears linked to copper status.
Mdj1 is a DnaJ protein in the mitochondrial matrix that activates the ATPase activity of mtHsp70. Overexpression of mtHsp70 did not rescue the respiratory deficiency of coa1Δ cells, nor did it improve the suppression observed with MDJ1 when the two genes were overexpressed together (data not shown).
Cox10 is the farnesyl transferase in heme a biogenesis, an obligate cofactor bound to Cox1. The suppression by COX10 is dependent on the catalytic function of the Cox10 farnesyl transferase, as a mutant allele that lacked catalytic activity (R212A, R216A) failed to act as a suppressor (data not shown).
MSS51 restores the CcO activity in coa1Δ cells without enhancing Cox1 translation
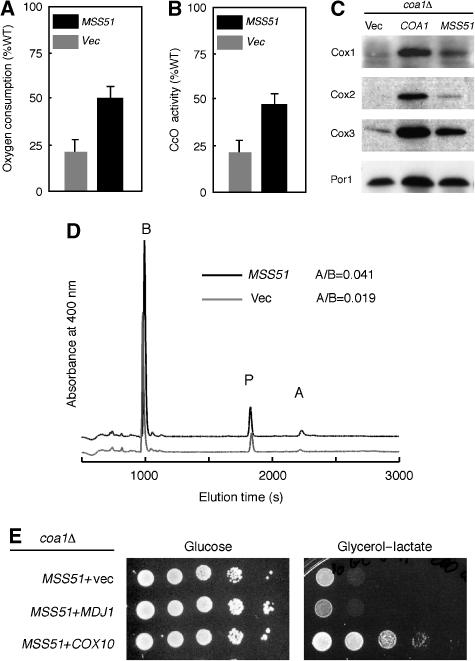
Mss51 is a multi-function protein acting as a translational activator of COX1 along with Pet309, as a translational elongation factor for COX1 and, in addition, binds newly synthesized Cox1 (Siep et al, 2000; Perez-Martinez et al, 2003; Barrientos et al, 2004). MSS51 is a known suppressor of the CcO deficiency in shy1Δ cells, by reversing the Cox1 translation defect (Barrientos et al, 2002b). As mentioned, Cox1 translation was normal in coa1Δ cells (Figure 2D). Overexpression of Mss51 in coa1Δ cells did not increase COX1 translation in an in vivo mitochondrial translation study during the pulse phase of the reaction (data not shown). Both oxygen consumption measured on intact coa1Δ cells overexpressing Mss51 and CcO activity of mitochondria purified from this strain increased to about 50% of WT (Figure 5A and B). Steady-state levels of Cox1–3 (Figure 5C) and heme a levels (Figure 5D) were also elevated in coa1Δ cells overexpressing Mss51. These data show that overexpression of Mss51 partially reverses the low CcO activity in coa1Δ cells presumably through a post-translational action of binding to Cox1.
Figure 5.
Restoration of CcO by high-copy MSS51 in coa1Δ cells. (A) coa1Δ cells with high-copy MSS51 or an empty vector (vec) were grown in SC-2% raffinose and assayed for oxygen consumption. (B) Mitochondria were purified from these cells and assayed for CcO activity. (C) Immunoblot of mitochondria from coa1Δ cells containing an empty vector (vec), a low-copy vector expressing COA1 or a high-copy vector expressing MSS51. (D) Heme profile from mitochondria purified from coa1Δ containing high-copy MSS51 or an empty vector. A/B represents the ratio of the area under the curve for the peaks of heme b (B) and heme a (A). (E) coa1Δ cells expressing high-copy MSS51 and containing an empty vector (vec) or high-copy vectors with MDJ1 or COX10 were grown in SC-2% raffinose, serially diluted and spotted on SC 2% glycerol–2% lactate or 2% glucose with selection. The plates were incubated at 30°C for 2 days (glucose) or 7 days (glycerol-lactate).
Since both Mss51 and Cox10 are related to Cox1 synthesis/maturation, we overexpressed these two proteins together in coa1Δ cells. Although COX10 and MDJ1 are only weak suppressors of coa1Δ cells, COX10, but not MDJ1, had a strong synergistic effect in combination with MSS51 (Figure 5E). The strong synergism between MSS51 and COX10 suggests that the two jointly correct a defect in coa1Δ cells relating to Cox1 through separate steps.
Shy1 and Coa1 function at a related step of CcO assembly
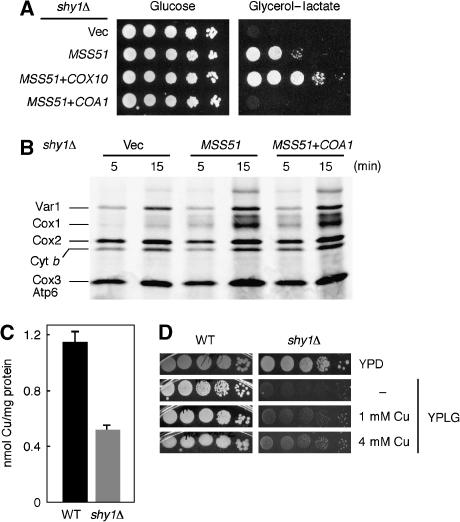
Given the additive effect of Mss51 and Cox10 coexpression in coa1Δ cells, we tested whether COX10 also had a synergistic effect with MSS51 in shy1Δ cells. The combination of MSS51 and COX10 clearly enhances respiratory growth in shy1Δ (Figure 6A). However, this phenotype is Mss51 dependent, as overexpression of COX10 by itself could not rescue the respiratory growth defect of shy1Δ cells (data not shown). While COX10 enhanced the stimulatory effect of MSS51 in shy1Δ cells, overexpression of COA1 abrogated it (Figure 6A). Overexpression of Coa1 in shy1Δ cells by itself did not diminish the residual CcO activity, as assessed by oxygen consumption and CcO activity measurements (data not shown). Therefore, the negative effect of Coa1 seen in shy1Δ cells overexpressing Mss51 is likely due to an impairment of Mss51 function. The level of expression of Coa1 had no effect on the steady-state level of Mss51 (data not shown). To determine whether overexpression of Coa1 attenuated the translational stimulation by Mss51 in shy1Δ cells, we evaluated the incorporation of [35S]-Met into mitochondrial translation products. The enhanced translation of Cox1 seen in shy1Δ cells overexpressing Mss51 was unaffected by the overexpression of Coa1 (Figure 6B), making it likely that Coa1 exerts its negative effect at a post-translational step. Similarly, we assessed whether Shy1 could have a negative effect on the Mss51 suppression of coa1Δ cells. Overexpression of Shy1 neither rescued the respiratory growth defect of coa1Δ cells, nor affected the suppression of this defect by high levels of Mss51 (Supplementary Figure 1).
Figure 6.
High-copy suppression of shy1Δ cells and low mitochondrial copper levels. (A) shy1Δ cells expressing high-copy MSS51 and an empty vector (vec) or high-copy vectors containing COX10 or COA1 were grown in SC-2% raffinose, serially diluted and spotted on SC 2% glycerol–2% lactate or 2% glucose with selection. The plates were incubated as in Figure 5E. (B) In vivo labeling of mitochondrial translation products of shy1Δ cells containing an empty vector, high-copy MSS51 in combination or not with high-copy COA1. The labeling was stopped after 5 and 15 min of pulse. (C) Copper levels in Nycodenz-purified mitochondria of shy1Δ cells or the isogenic WT BY4743 grown in YP-2% raffinose were measured by ICP-OES. The data are the average of three independent experiments. (D) shy1Δ cells or WT cells were grown in SC-2% raffinose, serially diluted and spotted on YP 2% glucose (YPD) or YP 2% glycerol–2% lactate (YPLG) supplemented with 1 or 4 mM CuSO4, as indicated.
The similarities in residual CcO activity and in MSS51/COX10 suppression of shy1Δ and coa1Δ strains led us to investigate whether shy1Δ cells resembles coa1Δ strains in having a mitochondrial copper defect. shy1Δ cells are deficient in mitochondrial copper (Figure 6C), yet total cellular copper is normal (data not shown). In addition, supplementation of shy1Δ cultures with exogenous copper partially restores growth on glycerol/lactate medium (Figure 6D). The copper defect observed in coa1Δ and shy1Δ cells suggested that CuB site formation could be impaired. Since Cox11 is implicated in CuB site formation, we tested whether overexpression of COX11 had a growth stimulatory effect. High-copy COX11 neither stimulated respiratory growth of coa1Δ or shy1Δ cells, nor was synergistic with MSS51 in suppressing shy1Δ cells (data not shown). Given the similarities between shy1Δ and coa1Δ strains, Shy1 and Coa1 probably act at a closely related step in CcO assembly.
Coa1 is part of a high-molecular-weight complex and interacts with Mss51
The genetic interaction between COA1 and MSS51 in shy1Δ suggested that the two proteins might interact physically. We constructed a strain in which we integrated 13 copies of the Myc epitope at the 3′end of COA1 and three copies of the HA epitope at the 3′end of MSS51. These tags did not affect the ability of the strain to grow on respiratory medium. Mitochondria isolated from the COA1-13Myc MSS51-3HA cells were solubilized with 0.1% DOC and fractionated on a size-exclusion column (Figure 7A). Analysis of the distribution of Coa1 by immunoblotting of the elution fractions revealed the presence of Coa1-Myc in fractions corresponding to high-molecular-mass components near 400 kDa. Similarly, BN-PAGE of COA1-13Myc mitochondria revealed the presence of a Coa1 complex of approximately 400 kDa (Figure 7B).
Figure 7.
Coa1 and Mss51 form similar sized complexes and interact. (A) Extracts of solubilized mitochondria (0.5 mg protein) from COA1-13Myc MSS51-3HA cells were fractionated over Superdex 200 in solubilization buffer (PBS, 0.1% DOC). The fractions were analyzed by immunoblot. (B) Mitochondria isolated from either wild-type (WT), shy1Δ or cox14Δ strains containing genomically tagged COA1 (COA1-13Myc, lanes 1–3) or MSS51 (MSS51-13Myc, lanes 4–6) were solubilized in buffer containing 1.5% digitonin. Lysates were loaded onto a continuous 4–13% gradient gel and protein complexes were separated by BN-PAGE. The distribution of complexes was analyzed by immunoblotting with mouse monoclonal anti-Myc antibody. Monomeric (V1) and dimeric (V2) forms of respiratory chain complex V served as a control and were visualized using antisera against F1-subunit. (C) Mitochondria (0.3 mg protein) from COA1-13Myc or COA1-13Myc MSS51-3HA cells were solubilized in Tris 20 mM pH 7.4, 100 mM NaCl, 1 mM PMSF, 0.1% lauryl maltoside and clarified extracts were immunoprecipitated with rabbit polyclonal HA antiserum and protein A agarose beads. The load representing 5% of the extracts and the entire fraction of the last wash and bead eluate were analyzed by immunoblotting.
A fraction of Mss51 was found in a similar mass range as Coa1 (Figure 7A; complex A in Figure 7B), in addition to a smaller mass component (complex B in Figure 7B). Immunoprecipitation of Mss51-HA from solubilized mitochondria resulted in the co-adsorption of Coa1-Myc, demonstrating that the two proteins physically interact (Figure 7C). Together, these data show that Coa1 and Mss51 are part of a ∼400-kDa complex.
Coa1 interacts with Cox14 and Cox1
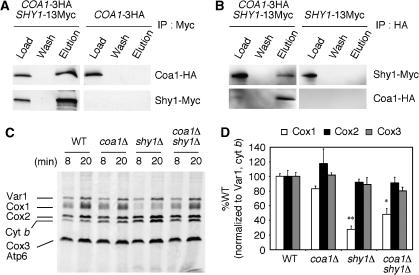
Mss51 was reported previously to interact with Cox14 (Barrientos et al, 2004). To assess whether Coa1 interacts with Cox14 in addition to Mss51, we constructed a strain with 13Myc integrated at the 3′ end of COX14 in addition to 3HA integrated on COA1. Immunoprecipitation of Cox14-Myc resulted in the co-precipitation of Coa1-HA (Figure 8A). Likewise, a strain with a 3HA tag at the 3′ end of COX14 in addition to the integrated 13 Myc tag on COA1 enabled the co-precipitation of Cox14 and Coa1 (Figure 8B). Thus, Coa1 interacts with both Mss51 and Cox14.
Figure 8.
Coa1 interacts with Cox14, which is required for the Coa1:Mss51 interaction. (A) Mitochondria (0.3 mg protein) from COA1-3HA or COA1-3HA COX14-13Myc cells were solubilized in Tris 20 mM pH 7.4, 100 mM NaCl, 1 mM PMSF, 0.1% lauryl maltoside. Clarified extracts were immunoprecipitated with rabbit polyclonal anti-Myc antiserum and protein A agarose beads. The load representing 7% of the extracts and the entire fraction of the last wash and bead eluate were analyzed by immunoblotting with mouse monoclonal antibodies. (B) Mitochondria (0.3mg protein) from COX14-3HA or COA1-13Myc COX14-3HA cells were treated and analyzed as in panel A. (C) Mitochondria from COA1-3HA MSS51-13Myc or COA1-3HA MSS51-13Myc cox14Δ cells were treated and analyzed as in panel A. (D) Extracts of mitochondria (0.3 mg protein) from MSS51-3HA COX14-13Myc or MSS51-3HA COX14-13Myc coa1Δ cells were treated and analyzed as in panel A.
Deletion of COX14 disrupted the interaction between Coa1 and Mss51 (Figure 8C), whereas the interaction between Mss51 and Cox14 was independent of the presence of Coa1 (Figure 8D). These results suggest that Cox14 may mediate the interaction between Mss51 and Coa1. BN-PAGE confirmed that the 400-kDa Coa1:Mss51 complex is abrogated in cox14Δ cells (Figure 7B). Attempts were made to visualize Cox14-Myc after BN-PAGE, but were unsuccessful. One possibility is that Cox14 occupies a central position in the complex that may mask the Myc tags, therefore precluding detection by immunoblotting under native conditions.
Since both Cox14 and Mss51 were shown to interact with Cox1 (Barrientos et al, 2004), we tested whether Coa1 also interacts with Cox1. Mitochondrial translation products were labeled in vitro in purified mitochondria from COA1-13Myc or WT cells. The mitochondria were then solubilized in digitonin-containing buffer and the soluble fraction was immunoprecipitated with anti c-Myc-agarose beads. Cox1 was specifically detected in the eluate of the beads incubated with the extracts from COA1-13Myc mitochondria (Figure 9). This result shows that like Cox14 and Mss51, Coa1 interacts with newly synthesized Cox1.
Figure 9.
Coa1 interacts with newly translated Cox1. Purified mitochondria from WT cells or cells containing a chromosomally tagged COA1-13Myc were labeled with 35S-Met for 20 min. Mitochondria were solubilized in Tris 20 mM pH 7.4, 100 mM NaCl, 1 mM PMSF, 1% digitonin and clarified extracts were immunoprecipitated with agarose anti c-Myc beads for 3 h at 4°C. The beads were washed three times with 20 volumes of buffer and resuspended in 1 volume of SDS loading dye. The mitochondria (Mito) and the soluble extracts (Soluble) represent 10% of the total sample, the last wash and beads elution correspond to the entire sample. The fractions were run on 12% SDS–PAGE, the gel was dried and exposed to autoradiographic film.
Coa1 interacts with Shy1 and is necessary for the inhibition of Cox1 translation in shy1Δ cells
The similar phenotypes of coa1Δ and shy1Δ cells suggested that Coa1 might also interact with Shy1. The deletion of COA1 did not affect the protein level of Shy1 and vice versa (data not shown). We constructed a strain with 13Myc integrated at the 3′ end of SHY1 in addition to the 3HA tag on COA1. Fractionation of extracts of solubilized mitochondria from this strain showed that Shy1 eluted in fractions similar to those containing Coa1 (Supplementary Figure 2). This result is consistent with the presence of Shy1 in a large-molecular-weight complex (Nijtmans et al, 2001). Immunoprecipitation of Shy1-Myc resulted in the co-precipitation of Coa1 (Figure 10A), and the reverse experiment, immunoprecipitation of Coa1-HA, led to the recovery of Shy1 with the beads (Figure 10B), showing that Coa1 and Shy1 are part of a complex. The ∼400-kDa Coa1:Mss51 complex observed on BN-PAGE remained intact in shy1Δ cells (Figure 7B), suggesting that Shy1 is not part of this complex. Shy1 was postulated to disassemble the Mss51:Cox14 complex during normal Cox1 translation and IM insertion (Barrientos et al, 2004). As predicted from this model, the abundance of the Coa1:Mss51 complex appears to be enhanced in shy1Δ cells (Figure 7B).
Figure 10.
Coa1 interacts with Shy1 and deletion of COA1 relieves the repression of Cox1 translation in shy1Δ cells. (A) Mitochondria (0.3 mg protein) from COA1-3HA or COA1-3HA SHY1-13Myc cells were solubilized in Tris 20 mM pH 7.4, 100 mM NaCl, 1 mM PMSF, 1% digitonin and clarified extracts were immunoprecipitated with rabbit polyclonal anti c-Myc antiserum and protein A agarose beads. The load representing 4% of the extracts and the entire fraction of the last wash and bead eluate were analyzed by immunoblotting. (B) Mitochondria (0.3 mg protein) from SHY1-13Myc or COA1-3HA, SHY1-13Myc cells were solubilized as in panel A. Clarified extracts were immunoprecipitated with rabbit polyclonal anti HA antiserum and protein A agarose beads and analyzed as in panel A. (C) In vivo labeling of mitochondrial translation products. Cells were incubated with 35S-Met for 8 or 20 min before stopping the reaction. The samples were run on 12% SDS–PAGE, the gel was dried and exposed to autoradiographic film. (D) The gel from panel C and two other gels from similar experiments were quantified using the software Quantity One (Biorad). The intensity of the bands corresponding to Cox1, Cox2 and Cox3 were normalized to the intensities of the bands of Var1 and Cyt b. ** Indicates a high statistical significance (P-value<0.0001) of the decrease of Cox1 translation in shy1Δ compared with WT. * Indicates statistical significance (P-value=0.0177) of the increase of Cox1 translation in coa1Δ shy1Δ compared with shy1Δ.
The attenuated Cox1 translation observed in shy1Δ cells was postulated to arise from stabilization of a Mss51:Cox14 complex that effectively titrates out the Mss51 translational activator (Barrientos et al, 2004). Because of the normal translation of Cox1 observed in coa1Δ cells, COA1 was deleted in shy1Δ cells and Cox1 translation was quantified (Figure 10C). The absence of Coa1 in shy1Δ cells partially reversed the Cox1 translation block. Quantification of the gels from three independent in vivo translation experiments showed that only the translation of Cox1 was decreased in shy1Δ cells and that deletion of COA1 in this background significantly restored it, although not to WT levels (Figure 10D). This result suggests that Coa1 has a role in maintaining Mss51 in a state unavailable for translational activation of Cox1 in a shy1Δ background. Despite the significant recovery of Cox1 translation, the deletion of COA1 in shy1Δ did not reverse the respiratory growth defect or the low mitochondrial copper level (Supplementary Figure 3).
Discussion
One of the many challenges that cells face in building active CcO is to coordinate the assembly of the hydrophobic mitochondrial encoded subunits with the subunits derived from the nuclear genome. This task is even greater when one considers that Cox1 and Cox2 have to acquire their metallic cofactors essential for activity. We have identified the gene COA1 as functioning in linking Cox1 translation and the insertion of its cofactors.
CcO biogenesis in the mitochondrial IM commences with the translation of the mitochondrial-encoded subunits mediated by a series of IM-associated translational activators. These activators appear to recruit mRNA to the IM for cotranslational insertion of the nascent polypeptides. Cotranslational insertion of Cox1 is dependent on the Pet309 and Mss51 translational activators (Manthey and McEwen, 1995; Siep et al, 2000; Perez-Martinez et al, 2003). Of the three subunits of CcO encoded by the mitochondrial genome, Cox1 is the first to join the assembly chain. Oxa1 is likely to play a role in the IM insertion of the 12 transmembrane domains of Cox1, as cells lacking Oxa1 show marked Cox1 instability, as seen in a mitochondrial translation assay (Bonnefoy et al, 1994; Barrientos et al, 2004).
Two heme a molecules and a copper atom are buried inside the barrel formed by Cox1 (Tsukihara et al, 1995). Cox1 has an open channel from the IMS side of the IM where heme a insertion may occur. However, this channel is obstructed by the association of Cox2. It is therefore probable that Cox1 acquires its cofactors at an early stage of the assembly process. Heme a is coordinated by axial ligands on helices 2 and 10 of Cox1; thus, heme a insertion could have a significant effect on the IM folding of Cox1, which could trigger the assembly process to proceed to the next step. An intermediate consisting of Cox1, Cox4 and Cox5A accumulates in human cells from patients carrying mutations in a number of genes encoding assembly factors like Sco1 and SURF1 (human Shy1) (Williams et al, 2004; Stiburek et al, 2005). The intermediate fails to accumulate in Cox10- or Cox15-deficient patient cells, consistent with the idea that heme a insertion occurs before the addition of Cox4 and Cox5A (Antonicka et al, 2003a, 2003b; Williams et al, 2004).
A clear connection between cotranslational insertion of Cox1 and cofactor assembly has not been established to date. We show the gene COA1 is required for efficient CcO assembly in yeast. The respiratory defect of coa1Δ cells is suppressed by high-copy MSS51, MDJ1 and COX10, and the suppression by elevated levels of Mss51 and Cox10 is enhanced when the two proteins are coexpressed. This observed synergistic suppression is consistent with the different functions exerted on Cox1 by Mss51 and Cox10 in translation and heme a biosynthesis, respectively. These results suggest that Coa1 links cotranslational insertion and heme addition to Cox1 (Figure 11). Coa1, like Mss51, only has robust homologs within fungi (Supplementary Figure 4). However, the presence of a functional homolog that would link translation of Cox1 to the assembly of its cofactors in higher eukaryotes is likely, and genetic screens are ongoing in our laboratory to check this possibility.
Figure 11.
Model for Coa1 linking Cox1 translation to cofactors assembly. After the translation of Cox1 mRNA, which depends on Pet309 and Mss51, Mss51 interacts with Cox1 and associates with Cox14 (Barrientos et al, 2004). This ternary complex is recruited by Coa1 and likely forms the ∼400-kDa complex that we detected by BN-PAGE. Membrane insertion of Cox1 probably occurs via Oxa1. Shy1 joins the complex and promotes the dissociation of Cox14 and Mss51 that is recycled to its translational function. The cofactor insertion in Cox1 can then proceed via Cox10, Cox15 (heme a biosynthesis) and Cox11 (CuB insertion) leading to holo-Cox1.
Coa1Δ cells resemble shy1Δ cells in many aspects. First, both strains display residual CcO activity that can be suppressed by elevated levels of Mss51. Second, their respiration is enhanced when Mss51 and Cox10 are combined together. Lastly, mitochondria of each strain have low levels of copper. These data and the physical interaction between Coa1 and Shy1 strongly suggest that the two proteins function at a related step of CcO assembly. The enhancement of the respiratory growth of shy1Δ cells overexpressing Mss51 by COX10 is the first indication that Shy1 is linked to a heme a step in eukaryotes. Accordingly, Shy1 appears to be important for insertion/stabilization of the CuB-heme a3 in R. sphaeroides (Smith et al, 2005).
Mss51 functions in Cox1 translation and elongation of the nascent chain and is known to interact with newly synthesized Cox1 (Siep et al, 2000; Perez-Martinez et al, 2003; Barrientos et al, 2004). Moreover, Mss51 also interacts with Cox14 (Barrientos et al, 2004). Cox1 translation is attenuated in shy1Δ cells and this defect can be rescued either by overexpression of Mss51 or deletion of COX14 (Barrientos et al, 2002b, 2004). Shy1 was postulated to disassemble the Cox1:Cox14:Mss51 complex allowing for Mss51 recycling to its translation/elongation function (Figure 11) (Barrientos et al, 2004). The Cox1:Cox14:Mss51 complex is thought to accumulate in shy1Δ cells, therefore titrating out Mss51 from Cox1 translation/elongation and creating a negative feedback loop that may coordinate translation and membrane insertion (Barrientos et al, 2004). The negative feedback loop model may however be more complex, as the deletion of SHY1 in cells containing a COX1-ARG8 chimera gave normal levels of the Arg8 reporter (Perez-Martinez et al, 2003).
The interaction of Coa1 with Mss51, Cox14 and newly synthesized Cox1 suggests that Coa1 is part of the feedback regulatory loop. Coa1 appears to be a key component of the stalled complex, as Cox1 translation defect in shy1Δ cells is partially reversed by deleting COA1 and Cox1 translation is normal in coa1Δ cells. The stalled complex that inhibits Cox1 translation could correspond to the 400-kDa Mss51:Coa1 complex that we detected by BN-PAGE, since we showed that this complex is stabilized in shy1Δ cells and is disrupted upon deletion of COX14 (Figure 7B). It is likely that the Mss51:Coa1 complex also contains Cox14 and Cox1. We detected a lower molecular weight complex containing Mss51 (Figure 7A and B). This complex is present at higher level in strains where translation of Cox1 occurs optimally (WT and cox14Δ) than in shy1Δ cells where translation of Cox1 is compromised. Therefore, we suggest that the low-molecular-weight Mss51 complex may be involved in Cox1 translation.
We propose that Coa1 stabilizes the Mss51:Cox14:Cox1 complex until Shy1 interacts with Coa1, causing Mss51 and Cox14 to disassemble and Cox1 to proceed to the next assembly step involving the insertion of the cofactors (Figure 11). Even though we showed that the interaction between Mss51 and Cox14 was not dependent on Coa1, the absence of Coa1 will likely preclude the formation of the 400-kDa complex, resulting in normal Cox1 translation. Cells lacking COA1 translate Cox1 normally, but are compromised for assembly presumably because Cox1 cannot be presented efficiently to Shy1. The partial suppression of the respiratory defect of coa1Δ cells by overexpression of MSS51 is likely due to the post-translational function of Mss51, which could increase the stability of newly synthesized Cox1 upon binding, allowing eventually more assembly over time. As mentioned, overexpression of MSS51 allows respiratory growth of shy1Δ cells, but the effect is nullified by coexpression of COA1. This dominant-negative effect of COA1 on the MSS51-mediated suppression of shy1Δ cells may arise from titrating out a factor such as Cox14 if formation of the Mss51:Cox14:Coa1 complex occurs in a step-wise manner. High levels of COA1 do not titrate out Mss51, as translation of Cox1 is not attenuated in shy1Δ cells overexpressing COA1 and MSS51 (Figure 6B).
Mdj1 was also recovered as a suppressor of coa1Δ cells. This DnaJ protein associated with mitochondrial Hsp70, may enhance the chaperoning activity of Hsp70. Cells lacking the Mba1 ribosomal receptor or harboring a mutant Oxa1 variant showed an association of Cox1 and Hsp70 (Ott et al, 2006). Thus, the suppression by Mdj1 in coa1Δ cells may occur through an enhanced chaperoning activity of Hsp70 that may stabilize/refold newly synthesized Cox1 polypeptides stalled for insertion. This effect would be similar to the suppression of coa1Δ by MSS51 discussed above. However, since Mdj1 has been shown to interact with mitochondrial nascent translation products (Westermann et al, 1996), a more direct role of Mdj1 in CcO assembly cannot be excluded.
Cells lacking Coa1 or Shy1 have mitochondrial copper and respiratory growth defects that can be partially restored with supplemental copper. Other CcO assembly mutants do not show a similar mitochondrial copper deficiency. The source of copper used for metallation of CcO arises from the matrix copper–ligand complex conserved in eukaryotic cells (Cobine et al, 2004, 2006a). The observed diminution in matrix copper levels in coa1Δ and shy1Δ cells may suggest that the assembly step at which these two proteins are involved could influence the function of the transporter responsible for trafficking the CuL complex. Studies are underway to identify the copper transporter of the IM.
Materials and methods
Yeast strains and vectors
Yeast strains used in this study are described in Supplementary Table I. All FMP deletions screened were in the BY4743 background and were grown in liquid YPD at 30°C. The coa1Δ strains were in the BY4743 background or in W303, created by disruption using KanMX. Cells were cultured in either rich medium or synthetic complete (SC) medium lacking the appropriate nutrients for plasmid selection. The carbon source used was either 2% glucose or a 2% glycerol–2% lactate. A 3′-Myc-tagged version of COA1 was cloned into a yeast low-copy vector under the control of the MET25 promoter and the CYC1 terminator (Mumberg et al, 1994). COA1-Myc was created using PCR, where the 3′ oligonucleotide removed the stop codon and added the coding sequence for a 3′ Myc tag (LEQKLISEEDL). Sequencing was used to confirm cloning products in all created vectors. Yeast strains were transformed using lithium acetate.
Mitochondria purification and assays
Intact mitochondria were isolated from yeast as described previously (Diekert et al, 2001). Total mitochondrial protein concentration was determined by standard Bradford assay (Bradford, 1976). Cytochrome c oxidase activity was assessed by monitoring the oxidation of reduced cytochrome c, and reaction rates were normalized to total mitochondrial protein (Capaldi et al, 1995). The oxygen consumption of the cells grown to stationary phase was determined on a 5300A Biological Oxygen Monitor (Yellow Springs Instrument Co.). Only the linear response was considered in calculating the rate of oxygen consumption (Horng et al, 2005). Metals were quantified in isolated mitochondria using a Perkin-Elmer Optima 3100XL inductively coupled plasma optical emission spectrometer (ICP-OES) after nitric acid digestion at 95°C. A standard curve was determined from commercially available standards (Perkin-Elmer), and buffer samples were run in parallel as a control. Heme analysis was conducted on mitochondria (1–2 mg protein) extracted with 0.5 ml of acetone containing 2.5% HCl, as described previously (Barros et al, 2001). The pH of the extract was adjusted to 4 by addition of 1 μl formic acid and titration of a KOH solution. The sample was clarified by centrifugation at 13 000 r.p.m. for 5 min, and 1 ml was injected onto a 3.9 × 300 mm C18 Bondclone column (Phenomex, USA).
In vitro and in vivo mitochondrial protein synthesis
Purified mitochondria were labeled in vitro as described (Westermann et al, 2001). For in vivo labeling, the cells were grown overnight in selective medium containing 2% raffinose and then re-inoculated in YP-2% raffinose to grow to an absorbance of 1 at 600 nm. The labeling and preparation of the samples for 12% SDS–PAGE was as described in Barrientos et al (2002b). The gel was dried and radiolabeled proteins were visualized by exposing autoradiographic films at −80°C.
Blue native gel electrophoresis
BN-PAGE was performed essentially as described (Wittig et al, 2006), except that 1.5% digitonin was used. After incubation for 20 min on ice and centrifugation (20 000 g for 10 min at 2°C), supernatants were mixed with sample buffer (5% Coomassie brilliant blue G250, 0.5 M 6-aminocaproic acid, pH 7.0) and loaded on 4–13% gradient polyacrylamide gel. Separated complexes were detected by immunoblotting on PVDF membrane.
Immunoblotting
Proteins samples were separated on 15% acrylamide gels and transferred to nitrocellulose. ECL reagents were used to visualize proteins with horseradish peroxidase-conjugated secondary antibodies. Anti-Myc and HA antisera were purchased from Santa Cruz, anti-porin was from Molecular Probes and antisera to Cox1–Cox3 were from Mitosciences. Antiserum to Sod2 was provided by Dr Val Culotta, and antisera to Cyb2, Cyt1 and Cyc1 provided by Dr Carla Koehler.
Localization experiments
Proteinase K treatment was performed as previously described (Diekert et al, 2001). Carbonate extraction experiments used mitoplasts made by diluting intact mitochondria (125 μg protein) 20-fold in 10 mM Hepes, pH 7.4. Mitoplasts were then incubated in 100 mM sodium carbonate, pH 11.3, for 30 min on ice and centrifuged at 100 000 g. The supernatant and pellet were analyzed by immunoblotting. High-salt extraction experiments also used mitoplasts that were sonicated three times for 30 s in 25, 100 or 500 mM NaCl.
Construction of the high-copy library
A 36-μg weight of genomic DNA purified from coa1Δ cells was partially digested with Sau3A. DNA fragments between 3 and 10 kb were purified from a 1% agarose gel. The plasmid YEP352 was digested to completion with SalI. The plasmid DNA was then treated with Klenow polymerase in the presence of dCTP and dTTP, while the genomic DNA was treated with dATP and dGTP. The resulting plasmid DNA was treated with 1 U of calf intestinal alkaline phosphatase for 30 min at 37°C. Ligation of YEP352 with the genomic DNA was conducted overnight at 16°C. The total number of colonies obtained was estimated to be about 20 000, representing a theoretical five-fold coverage of the yeast genome.
Gel filtration
Mitochondria (0.3–0.5 mg protein) were resuspended in solubilization buffer (PBS, 0.1% DOC). After incubation on ice for 10 min, lysates were centrifuged at 16 000 g for 25 min at 4°C, passed through a 0.45 μm filter and loaded onto a Superdex 200 (GE Healthcare) column equilibrated in solubilization buffer.
Immunoprecipitation
Purified mitochondria (0.3–0.5 mg protein) were solubilized by incubation in 0.5 ml Tris 20 mM pH 7.4, 100 mM NaCl, 1 mM PMSF and either 0.1% lauryl maltoside or 1% digitonin (IP buffer) for 30 min at 4°C. Lysates were centrifuged at 16 000 g for 15 min at 4°C. Immunoprecipitation was performed by incubating overnight at 4°C the clarified lysates with 40 μl anti-Myc or HA rabbit polyclonal antisera. The antibodies were precipitated by the addition of 20 μl of recombinant protein A agarose beads (Pierce) for 4 h at 4°C. The beads were pelleted by centrifugation at 2000 g for 2 min and washed with 0.8 ml of IP buffer for 5 min. This step was repeated 3–4 times. Beads were eluted by boiling for 5 min in 50 μl SDS–PAGE loading buffer. The clarified extracts and wash fractions were TCA precipitated before electrophoresis and immunoblotting.
Statistical procedures
Descriptive statistics are represented as average±standard error. The unpaired Student's t-test (two tailed) was used to compare differences between experimental and control groups.
Supplementary Material
Supplementary Figures
Acknowledgments
This work was supported by a grant ES 03817 from the National Institutes of Environmental Health Sciences, NIH to DRW. We acknowledge the support of the CEMH core facility for FPLC chromatography (DK P30 072437) and United Mitochondrial Disease Foundation for support for PAC. We acknowledge Dr Peter Rehling for open discussions on Coa1.
References
- Antonicka H, Leary SC, Guercin GH, Agar JN, Horvath R, Kennaway NG, Harding CO, Jaksch M, Shoubridge EA (2003a) Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum Mol Genet 12: 2693–26702 [DOI] [PubMed] [Google Scholar]
- Antonicka H, Mattman A, Carlson CG, Glerum DM, Hoffbuhr KC, Leary SC, Kennaway NG, Shoubridge EA (2003b) Mutations in COX15 produce a defect in the mitochondrial heme biosynthetic pathway causing early-onset fatal hypertrophic cardiomyopathy. Am J Hum Genet 72: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Barros MH, Valnot I, Rotig A, Rustin P, Tzagoloff A (2002a) Cytochrome oxidase in health and disease. Gene 286: 53–63 [DOI] [PubMed] [Google Scholar]
- Barrientos A, Korr D, Tzagoloff A (2002b) Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J 21: 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A (2004) Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J 23: 3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barros MH, Tzagoloff A (2002) Regulation of the heme a biosynthetic pathway in Saccharomyces cerevisiae. FEBS Lett 516: 119–123 [DOI] [PubMed] [Google Scholar]
- Barros MH, Carlson CG, Glerum DM, Tzagoloff A (2001) Involvement of mitochondrial ferredoxin and Cox15p in hydroxylation of heme O. FEBS Let 492: 133–138 [DOI] [PubMed] [Google Scholar]
- Bonnefoy N, Chalvet F, Hamel P, Slonimski PP, Dujardin G (1994) OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J Mol Biol 239: 201–212 [DOI] [PubMed] [Google Scholar]
- Bradford NM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bratton MR, Hiser L, Anthroline WE, Hoganson C, Hosler JP (2000) Identification of the structural subunits required for formation of the metal centers in subunit I of cytochrome c oxidase of Rhodobacter sphaeroides. Biochemistry 39: 12989–12995 [DOI] [PubMed] [Google Scholar]
- Capaldi RA, Marusich MF, Taanman JW (1995) Mammalian cytochrome c oxidase: characterization of enzyme and immunological detection of subunits in tissue extracts and whole cells. Methods Enzymol 260: 117–132 [DOI] [PubMed] [Google Scholar]
- Carr HS, Winge DR (2003) Assembly of cytochrome c oxidase within the mitochondrion. Acc Chem Res 36: 309–316 [DOI] [PubMed] [Google Scholar]
- Cobine PA, Ojeda LD, Rigby KM, Winge DR (2004) Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J Biol Chem 279: 14447–14455 [DOI] [PubMed] [Google Scholar]
- Cobine PA, Pierrel F, Winge DR (2006b) Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim Biophys Acta (Mol Cell Res) 1763: 759–772 [DOI] [PubMed] [Google Scholar]
- Cobine PA, Pierrel F, Bestwick ML, Winge DR (2006a) Mitochondrial matrix copper complex used in metallation of cytochrome oxidase and superoxide dismutase. J Biol Chem 281: 36552–36559 [DOI] [PubMed] [Google Scholar]
- Diekert K, De Kroon AIPM, Kisplal G, Lill R (2001) Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol 65: 37–51 [DOI] [PubMed] [Google Scholar]
- Green-Willms NS, Butler CA, Dunstan HM, Fox TD (2001) Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene COX2. J Biol Chem 276: 6392–6397 [DOI] [PubMed] [Google Scholar]
- He S, Fox TD (1997) Membrane translocation of mitochondrially coded Cox2p. Distinct requirements for export of N and C termini and dependence on the conserved protein Oxa1p. Mol Biol Cell 8: 1449–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K, Neupert W, Stuart RA (2001) Oxa1p acts as a general membrane insertion machinery for proteins encoded by mitochondrial DNA. EMBO J 20: 1281–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng Y-C, Leary SC, Cobine PA, Young FBJ, George GN, Shoubridge EA, Winge DR (2005) Human Sco1 and Sco2 function as copper-binding proteins. J Biol Chem 280: 34113–34122 [DOI] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci USA 98: 4569–4574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalimonchuk O, Rodel G (2005) Biogenesis of cytochrome c oxidase. Mitochondrion 5: 363–388 [DOI] [PubMed] [Google Scholar]
- Manthey GM, McEwen JE (1995) The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J 14: 4031–4043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMullin TW, Fox TD (1993) COX3 mRNA-specific translational activator proteins are associated with the inner mitochondrial membrane in Saccharomyces cerevisiae. J Biol Chem 268: 11737–11741 [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use of heterologous expression. Nucleic Acids Res 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naithani S, Saracco SA, Butler CA, Fox TD (2003) Interactions among COX1, COX2, and COX3 mRNA-specific translational activator proteins on the inner surface of the mitochondrial inner membrane of Saccharomyces cerevisiae. Mol Biol Cell 14: 324–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijtmans LG, Taanman JW, Muijsers AO, Speijer D, Van den Bogert C (1998) Assembly of cytochrome c oxidase in cultured human cells. Eur J Biochem 254: 389–394 [DOI] [PubMed] [Google Scholar]
- Nijtmans LGJ, Sanz MA, Bucko M, Farhoud MH, Feenstra M, Hakkaart GAJ, Zeviani M, Grivell LA (2001) Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett 498: 46–51 [DOI] [PubMed] [Google Scholar]
- Ott M, Prestele M, Bauerschmitt H, Funes S, Bonnefoy N, Herrmann JM (2006) Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J 25: 1603–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez X, Broadley SA, Fox TD (2003) Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J 22: 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchirico ME, Fox TD, Mason TL (1998) Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J 17: 5796–5804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schonfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siep M, van Oosterum K, Neufeglise H, van der Spek H, Grivell LA (2000) Mss51p, a putative translational activator of cytochrome c oxidase subunit-1 (COX1) mRNA, is required for synthesis of Cox1p in Saccharomyces cerevisiae. Curr Genet 37: 213–220 [DOI] [PubMed] [Google Scholar]
- Smith D, Gray J, Mitchell L, Antholine WE, Hosler JP (2005) Assembly of cytochrome c oxidase in the absence of the assembly protein Surf1p leads to loss of the active site heme. J Biol Chem 280: 17652–17656 [DOI] [PubMed] [Google Scholar]
- Stiburek L, Vesela K, Hansikova H, Pecina P, Tesarova M, Cerna L, Houstek J, Zeman J (2005) Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem J 392: 625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Hakashima R, Yaono R, Yoshikawa S (1995) Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science 269: 1069–1074 [DOI] [PubMed] [Google Scholar]
- Tzagoloff A, Dieckmann CL (1990) PET genes of Saccharomyces cerevisiae. Microbiol Rev 54: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F, Bornhovd C, Neupert W, Reichert AS (2006) Dynamic subcompartmentalization of the mitochondrial inner membrane. J Cell Biol 175: 237–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B, Gaume B, Herrmann JM, Neupert W, Schwarz E (1996) Role of the mitochondrial DnaJ homolog Mdj1p as a chaperone for mitochondrially synthesized and imported proteins. Mol Cell Biol 16: 7063–7071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B, Herrmann JM, Neupert W (2001) Analysis of mitochondrial translation products in vivo and in organello in yeast. Methods Cell Biol 65: 429–438 [DOI] [PubMed] [Google Scholar]
- Williams SL, Valnot I, Rustin P, Taanman J-W (2004) Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1 or SURF1. J Biol Chem 279: 7462–7469 [DOI] [PubMed] [Google Scholar]
- Wittig I, Braun HP, Schagger H (2006) Blue native PAGE. Nat Protoc 418–428 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert AP, Newbold RF, Wang J, Chevrette M, Brown GK, Brown RM, Shoubridge EA (1998) SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet 20: 337–343 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures