Abstract
Cytochrome c oxidase (complex IV) of the respiratory chain is assembled from nuclear and mitochondrially-encoded subunits. Defects in the assembly process lead to severe human disorders such as Leigh syndrome. Shy1 is an assembly factor for complex IV in Saccharomyces cerevisiae and mutations of its human homolog, SURF1, are the most frequent cause for Leigh syndrome. We report that Shy1 promotes complex IV biogenesis through association with different protein modules; Shy1 interacts with Mss51 and Cox14, translational regulators of Cox1. Additionally, Shy1 associates with the subcomplexes of complex IV that are potential assembly intermediates. Formation of these subcomplexes depends on Coa1 (YIL157c), a novel assembly factor that cooperates with Shy1. Moreover, partially assembled forms of complex IV bound to Shy1 and Cox14 can associate with the bc1 complex to form transitional supercomplexes. We suggest that Shy1 links Cox1 translational regulation to complex IV assembly and supercomplex formation.
Keywords: Leigh syndrome, mitochondria, protein complex assembly, respiratory chain complex, SURF1
Introduction
The respiratory chain of mitochondria plays a central role in energy metabolism of eukaryotic cells. It is comprised of four electron-shuttling multiprotein complexes and the mitochondrial F1FoATPase. Complexes I–IV transfer electrons derived from reducing equivalents to molecular oxygen, thereby generating water and a proton gradient across the inner mitochondrial membrane. The proton gradient is utilized by the F1FoATPase (complex V) to generate ATP. Nuclear genes encode the majority of mitochondrial proteins, among them most subunits of the respiratory chain. These proteins are synthesized on cytosolic ribosomes and imported post-translationally into mitochondria via dedicated protein translocases (Jensen and Dunn, 2002; Koehler, 2004; Rehling et al, 2004; Neupert and Herrmann, 2007). However, a small subset of mostly hydrophobic mitochondrial proteins, eight in the yeast Saccharomyces cerevisiae and 13 in humans, are encoded by mitochondrial DNA (Tzagoloff and Myers, 1986; Grivell et al, 1999; DiMauro and Schon, 2003; Fontanesi et al, 2006). With the exception of the yeast Var1, these proteins are subunits of the respiratory chain complexes and are integrated into the inner mitochondrial membrane by the mitochondrial export machinery (Stuart, 2002; Herrmann and Funes, 2005). Apart from complex II, which does not contain mitochondrially encoded subunits, all respiratory chain complexes are assembled from both nuclear and mitochondrially encoded proteins.
Cytochrome c oxidase (complex IV) is the terminal electron-transferring complex of the respiratory chain. The cytochrome c oxidase of yeast and humans consist of 11 and 13 subunits, respectively. Cox1, Cox2, and Cox3 represent the mitochondrially encoded subunits, forming the catalytic core of the complex and exhibiting high conservation among species (Tsukihara et al, 1996; Herrmann and Funes, 2005; Fontanesi et al, 2006). In order to generate a mature complex IV from individual subunits and to integrate the necessary cofactors during this process, approximately twenty assembly factors are required (McEwen et al, 1986; Tzagoloff and Dieckmann, 1990; Carr and Winge, 2003; Herrmann and Funes, 2005; Fontanesi et al, 2006). Accordingly, the formation of the complex appears to be a highly complicated and regulated process that is thought to occur in a sequential manner via transient forms of subcomplexes. However, the molecular mechanisms of this assembly process are largely enigmatic (Wielburski and Nelson, 1983; Taanman and Williams, 2001; Carr and Winge, 2003; Herrmann and Funes, 2005; Fontanesi et al, 2006).
Defects in complex IV biogenesis or mutations that affect the function of its structural subunits lead to severe neuromuscular diseases in humans. Mutations in the mitochondrial genes for Cox1, Cox2, and Cox3 cause complex IV deficiency in patients. Moreover, deficiency of the protein complex is also apparent when nuclear encoded assembly factors of complex IV are affected (Shoubridge, 2001; Chinnery and Schon, 2003; DiMauro and Schon, 2003; Zeviani and Spinazzola, 2003). Among the latter, Leigh syndrome represents a severe neurodegenerative disorder characterized by necrotic lesions in the subcortical brain (Leigh, 1951; Tiranti et al, 1998; Zhu et al, 1998). The most frequently mutated gene in Leigh syndrome patients that exhibit cytochrome c oxidase deficiency is the nuclear SURF1 (Tiranti et al, 1998; Zhu et al, 1998). The SURF1 protein is highly conserved and its homolog in S. cerevisiae is Shy1 (Mashkevich et al, 1997; Tiranti et al, 1998; Zhu et al, 1998; Poyau et al, 1999; Barrientos et al, 2002). Shy1 is an inner mitochondrial membrane protein mainly exposed to the intermembrane space (Mashkevich et al, 1997). Similar to patients affected in SURF1 function, shy1 mutant mitochondria are characterized by reduced levels of complex IV and show a reduced amount of Cox1 (Nijtmans et al, 2001; Barrientos et al, 2002; Williams et al, 2004). However, although Shy1 does not directly regulate Cox1 translation, it has been implicated in post-translational steps of Cox1 biogenesis, and concomitantly in early steps of complex IV assembly (Mashkevich et al, 1997; Barrientos et al, 2002; Perez-Martinez et al, 2003; Smith et al, 2005; Zambrano et al, 2007). Mss51 and Cox14 of yeast mitochondria act at early stages of Cox1 biogenesis and their inactivation similarly leads to complex IV deficiency. Mss51 represents a Cox1-specific translational activator and assembly factor (Glerum et al, 1995; Barrientos et al, 2002, 2004; Perez-Martinez et al, 2003; Zambrano et al, 2007). The current model suggests that Mss51 binds to Cox1 in a chaperone-like way. In the presence of unassembled Cox1, the assembly factor Cox14 is thought to bind to the Cox1–Mss51 complex, thus holding Mss51 in an inactive state, so that it is unavailable for initiation of another cycle of Cox1 translation (Barrientos et al, 2004; Zambrano et al, 2007). Although a physical interaction between Shy1 and Mss51 or Cox14 has not been found, it has been suggested that Shy1 acts at a rate-limiting assembly step downstream of Mss51 and Cox14, potentially leading to the release of Mss51 from Cox14 (Perez-Martinez et al, 2003; Barrientos et al, 2004; Herrmann and Funes, 2005; Fontanesi et al, 2006).
Here we have analyzed protein-interacting partners of Shy1 through isolation of Shy1 protein complexes, and have found that Shy1 is part of several membrane protein complexes. Shy1 associates with the Cox1 translational regulators Mss51 and Cox14. Moreover, Shy1 can be identified in subcomplexes of complex IV that have incorporated Cox1 and the nuclear encoded Cox5a subunit. The formation of these potential intermediate forms of the complex depends on Coa1, an inner mitochondrial membrane protein that associates with Shy1 and is required for complex IV assembly. Lastly, Shy1 and Cox14 can associate with respiratory chain supercomplexes through partially assembled forms of complex IV. Thus, we conclude that Shy1 functionally couples regulation of Cox1 translation to early steps of complex IV assembly and finally supercomplex formation.
Results
Isolation of Shy1-containing complexes
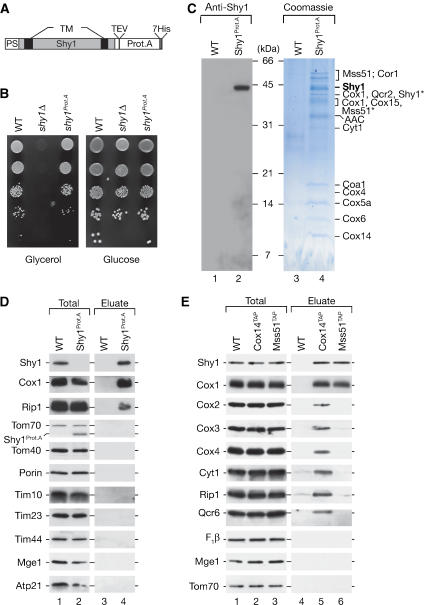
To identify mitochondrial proteins that interact with Shy1, we generated a fusion of Shy1 to protein A from Staphylococcus aureus. A cassette encoding protein A with a cleavage site for the TEV-protease (tobacco etch virus) was integrated in frame into the chromosome of wild-type S. cerevisiae cells downstream of the SHY1 open reading frame, thereby replacing the stop codon (Figure 1A). Functionality of the Shy1Prot.A fusion was assessed by growth of the strain on fermentable and non-fermentable medium. While wild-type yeast cells are capable of growth on both media, shy1Δ cells are respiratory deficient due to the defect in complex IV assembly and thus unable to grow on non-fermentable carbon sources such as glycerol (Mashkevich et al, 1997; Barrientos et al, 2002; Perez-Martinez et al, 2003). Compared to wild-type cells, yeast cells expressing Shy1Prot.A did not display any growth defect on glycerol medium (Figure 1B), indicating that the Shy1Prot.A fusion protein was functional.
Figure 1.
Purification of Shy1 protein complexes. (A) Shy1Prot.A construct used for isolation of Shy1 complexes. Gray box, mature Shy1 protein; black boxes, predicted transmembrane domains (TM); PS, predicted cleavable presequence; Prot.A, Protein A tag; TEV, TEV cleavage site; 7His, heptahistidine tag. (B) Growth test on fermentable and non-fermentable medium. Cells were spotted in serial 10-fold dilutions and incubated at 30°C. WT, wild-type. (C) Mitochondria from wild-type (WT) and Shy1Prot.A were solubilized in 1% digitonin buffer and subjected to IgG chromatography. After TEV-protease cleavage, eluates were separated on tricine-SDS–PAGE, stained with Colloidal Coomassie, or analyzed by Western blotting. Bands were excised and subjected to mass spectrometric analysis. Asterisks denote degradation products identified by mass spectrometry. (D) Eluates from panel C were analyzed by SDS–PAGE and immunodecoration. Total, 1%; eluate, 100%. (E) Cox14TAP and Mss51TAP complexes were isolated from digitonin-solubilized mitochondria via IgG chromatography, proteins separated by SDS–PAGE, and analyzed by Western blotting. Total, 2%; eluate 100%. F1β, β subunit of F1FoATP synthase.
We purified Shy1Prot.A and associated proteins from mitochondria via IgG chromatography. Proteins that were released from the column upon treatment with TEV-protease were separated by SDS–PAGE. Shy1 was recovered in the eluate and no significant degradation products were detected by Western blotting with anti-Shy1 antiserum (Figure 1C, lane 2). After staining of the gel, protein bands were excised and subjected to mass spectrometric analysis (Figure 1C, lane 4). Mss51 and Cox14, the translational regulators of Cox1 (Perez-Martinez et al, 2003; Barrientos et al, 2004), were co-isolated with Shy1. Moreover, we identified subunits of the cytochrome c oxidase (complex IV), the cytochrome bc1 complex (complex III), an uncharacterized predicted protein of ∼22 kDa that we refer to as Coa1 (cytochrome oxidase assembly 1) (YIL157c/Fmp35), as well as the ADP/ATP carrier (AAC), an abundant inner membrane protein. The identification of subunits of complex III of the respiratory chain was surprising, since Shy1 has been implicated in complex IV assembly, but has not been physically linked to complex III thus far.
To analyze if the presence of complex III subunits in the eluate of the Shy1Prot.A purification reflected an impurity of the procedure, we analyzed the eluate by Western blotting. Subunits of complex IV such as Cox1 and the Rieske iron-sulfur protein (Rip1) of complex III were indeed found in the Shy1Prot.A eluate but not in the mock purification. Other mitochondrial proteins did not copurify with Shy1Prot.A in significant amounts (Figure 1D). Thus, purification of Shy1 from mitochondria leads to specific co-isolation of complexes III and IV of the respiratory chain.
Mss51 and Cox14 interact with each other and Cox1 during its early assembly steps (Barrientos et al, 2004). However, an interaction of Mss51 or Cox14 with Shy1 had not been reported so far (Barrientos et al, 2002, 2004; Perez-Martinez et al, 2003). To confirm that Shy1 interacts with Mss51 and Cox14, C-terminal TAP (tandem affinity purification) fusions of these proteins were isolated from mitochondria and eluates were analyzed by Western blotting. Shy1 copurified with both Cox14TAP and Mss51TAP. Surprisingly, Cox14TAP eluates contained subunits of complexes IV and III, while these were not recovered in significant amounts with Mss51TAP or in the mock purification. Among the tested complex IV subunits, only Cox1 was purified together with Mss51TAP (Figure 1E, lane 6), indicating that a pool of Cox1 exists in complex with Mss51, as was suggested in previously analyses (Barrientos et al, 2004). We conclude that Shy1 forms a complex with Cox14 and Mss51. While Mss51 does not appear to significantly interact with mature complexes III or IV of the respiratory chain, Shy1 and Cox14 are both associated with these complexes.
Shy1 associates with respiratory chain supercomplexes
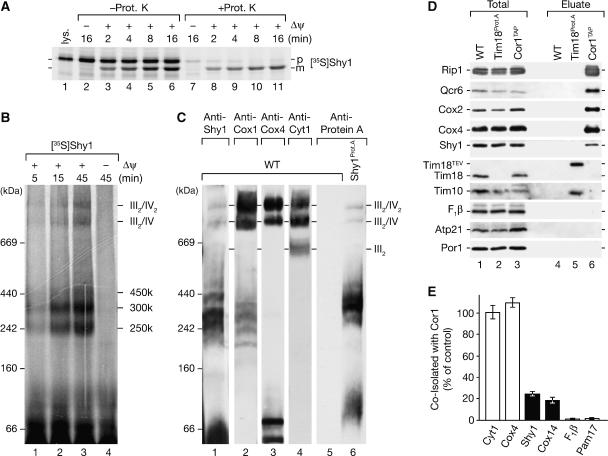
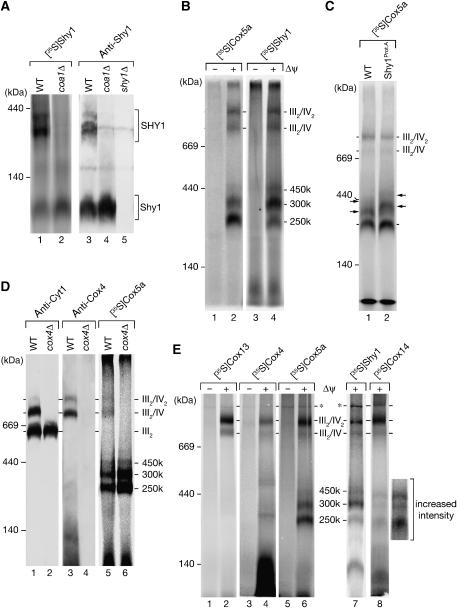
We imported radiolabeled Shy1 precursor protein into energized yeast mitochondria. In the presence of a membrane potential (Δψ), the Shy1 precursor protein was processed into a faster migrating, mature form (Figure 2A). The mature protein was resistant to externally added protease, while the non-imported precursor was largely accessible, inferring translocation of Shy1 into mitochondria. To address if Shy1 associated with larger protein assemblies in mitochondria, we solubilized mitochondria after import of Shy1 and separated protein complexes on a blue-native gel. At least five Shy1-containing complexes could be distinguished, all of which were selectively formed in the presence of a Δψ and protected against protease treatment (Figure 2B). These consisted of three complexes of intermediate size termed 250, 300, and 450 k, and two complexes at roughly 750 kDa and 1 MDa. In addition, a faster migrating form of Shy1 in the range of 100 kDa was present even in the absence of a Δψ. To address if these complexes were similar to the steady-state situation, we analyzed Shy1 complexes by immunodecoration. At steady state, Shy1 was also found in three complexes of intermediate sized and two large complexes (Figure 2C, lane 1). The intermediate-size complexes migrated in the same range as Cox1-containing complexes, but were not detected with antibodies against Cox4 (Figure 2C, lanes 1–3). A similar complex profile, but upshifted in size due to the tag on Shy1, was detected with anti-Protein A antibodies in mitochondria expressing the Shy1Prot.A fusion (Figure 2C, lane 6). Thus, the complexes formed by imported Shy1 reflect stable complexes that are present in organello at steady state. These complexes are unaffected in Shy1Prot.A mitochondria that were used for complex isolation.
Figure 2.
Shy1 associates with respiratory chain supercomplexes. (A) Radiolabeled Shy1 was imported into isolated mitochondria in the presence or absence of a membrane potential (Δψ) for the indicated times. Samples were split and left untreated or treated with proteinase K (Prot. K). Samples were analyzed by SDS–PAGE and digital autoradiography. As control 10% reticulocyte lysate containing Shy1 precursor was applied (lys.); p, precursor; m, mature. (B) 35S-labeled Shy1 precursor was imported into isolated mitochondria as in panel A, samples treated with proteinase K, solubilized in digitonin buffer, and protein complexes separated by blue-native PAGE. Protein complexes were detected by digital autoradiography. (C) Shy1 complexes from wild-type (WT) and Shy1Prot.A mitochondria were separated by blue-native PAGE and analyzed by Western blotting with the indicated antisera. (D) TIM22 complex and respiratory chain supercomplexes were isolated from mitochondria via IgG chromatography using protein A- and TAP-tagged Tim18 and Cor1, respectively. Analysis of the samples was performed by SDS–PAGE and immunodecoration. Total, 10%; eluate, 100%. (E) Proteins co-isolated with Cor1TAP were analyzed by immunoblotting. Quantification of Western blots was performed with Image Quant Software (GE Healthcare). The amount of cytochrome c1 that was coisolated with Cor1TAP was set to 100%. Data represent the means±s.e.m. (n=3). F1β, β subunit of F1FoATP synthase.
In S. cerevisiae the respiratory chain complexes III and IV form supercomplexes that can be separated by blue-native PAGE (Cruciat et al, 2000; Schägger and Pfeiffer, 2000): (i) a dimer of complex III (III2), (ii) a complex III dimer associated with a single complex IV (III2/IV), or (iii) associated with two complex IV units (III2/IV2). The two large Shy1 complexes of 750 kDa and 1 MDa were reminiscent of these respiratory chain supercomplexes. Thus, we compared the migration of the two large Shy1 protein complexes with that of the respiratory chain complexes on blue-native PAGE and observed a migration pattern similar to that of the III2/IV and III2/IV2 supercomplexes (Figure 2C, lanes 1–4). However, no comigration with III2 was found, suggesting that complex IV mediated a putative association of Shy1 with respiratory chain supercomplexes.
To provide evidence for the interaction of Shy1 with supercomplexes, we isolated complex III of the respiratory chain via a TAP tag on the Cor1 protein (Cor1TAP) (van der Laan et al, 2006). Together with Cor1, other subunits of complex III such as Rip1 and Qcr6 were purified. In addition, subunits of complex IV such as Cox2 and Cox4 were recovered in the purified complex fraction, indicating that supercomplexes had been purified (Figure 2D, lane 6). In contrast, other inner membrane proteins, among them subunits of the F1FoATPase and of the carrier translocase (TIM22 complex), intermembrane space proteins such as Tim10, and outer membrane proteins such as Porin (Por1) were not co-isolated. Shy1 was specifically co-isolated with supercomplexes, but not recovered with the carrier translocase (Rehling et al, 2003) or in a mock purification (Figure 2D, lane 6). Quantitative analysis of the amount of Shy1 that co-isolated with Cor1TAP showed that under our experimental conditions, approximately 25% of Shy1 was associated with supercomplexes (Figure 2E). Thus, our analyses show that a significant fraction of Shy1 associates with complex III/IV supercomplexes.
Coa1 is a mitochondrial membrane protein
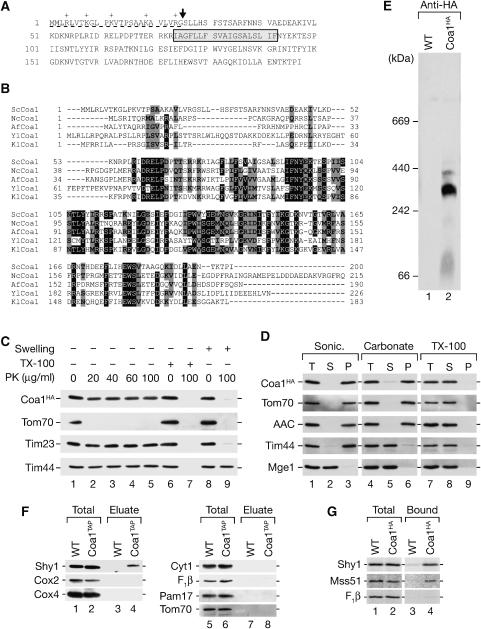
Among the proteins found associated with Shy1, we identified Coa1 (YIL157c/Fmp35), an uncharacterized protein previously localized to mitochondria (Sickmann et al, 2003). The deduced amino-acid sequence predicts a protein of 197 amino acids with a molecular mass of about 22 kDa (Figure 3A). The predicted primary sequence contains a conserved hydrophobic segment (amino acids 74–92) with the potential to form a membrane-spanning helix. Coa1 is highly conserved among lower eukaryotes and clusters of highly conserved residues are located on both sides of the predicted membrane span (Figure 3B). The N-terminus of Coa1 is rich in positively charged amino acids and could form an amphipathic alpha helix, characteristic of a mitochondrial presequence. A cleavage site for the mitochondrial processing peptidase is predicted adjacent to the presequence (Gakh et al, 2002) (Figure 3A). Upon in vitro import into mitochondria, Coa1 is processed to a ∼18-kDa mature form in a Δψ-dependent manner (data not shown), indicating that Coa1 possesses a cleavable presequence.
Figure 3.
Coa1 is a mitochondrial inner membrane protein. (A) Predicted primary structure of S. cerevisiae Coa1. Predicted presequence (underlined); putative mitochondrial processing peptidase cleavage site (arrow); predicted transmembrane segment (gray box). (B) Alignment of Coa1 homologs using Clustal W. Black boxes, identical residues in at least four species, gray boxes, similar amino acids. Sc; S. cerevisiae; Nc, N. crassa; Af, A. fumigatus; Yl, Y. lipolytica; Kl, K. lactis. (C, D) Localization and membrane association of Coa1 was analyzed as described in Materials and methods. (E) Digitonin-solubilized mitochondria from wild-type (WT) and Coa1HA cells were separated by blue-native PAGE and analyzed by immunodecoration using anti-HA antibody. (F) Coa1 was isolated via IgG chromatography using TAP-tagged Coa1 and analyzed by SDS–PAGE and Western blotting. Total, 2%; eluate, 100%. (G) Radiolabeled Shy1, Mss51, and F1β were imported into wild-type and Coa1HA mitochondria. After solubilization of mitochondria, anti-HA antibodies were used for immunoprecipitation of Coa1 from the extracts. Total, 5%; bound, 100%.
When mitochondria containing a C-terminally HA-tagged version of Coa1 (Coa1HA) were treated with proteinase K, Coa1HA was protected against protease. However, upon osmotic disruption of the outer membrane, Coa1HA became accessible to protease, while matrix proteins remained inaccessible (Figure 3C, lane 8 versus 9). Thus, the C-terminus of Coa1 localizes to the intermembrane space. We treated mitochondria by sonication to release soluble proteins into the supernatant. Unlike the soluble Mge1, Coa1HA remained associated with mitochondrial membranes (Figure 3D, lane 3). Similarly, the peripheral inner membrane protein Tim44 and integral membrane proteins (Tom70, AAC) remained in the membrane pellet. After alkaline treatment of mitochondria, Tim44 was released into the supernatant, while Coa1 remained predominantly in the membrane pellet (Figure 3D, lane 6). As a control, treatment of mitochondria with Triton X-100 released all proteins into the supernatant (Figure 3D, lane 8). We conclude that Coa1 is an integral inner mitochondrial membrane protein.
To obtain additional evidence for an interaction between Coa1 and Shy1, we performed blue-native PAGE analysis of Coa1HA mitochondria and identified Coa1 protein complexes around 300 kDa (Figure 3E). In addition, we isolated Coa1TAP and associated proteins from mitochondria. Shy1 was specifically co-isolated with Coa1TAP (Figure 3F, lane 3 versus 4), while other mitochondrial proteins were not recovered with Coa1TAP in significant amounts. Considering that Shy1 was found in association with Mss51, Cox14, and Coa1, we also analyzed if Coa1 interacted with Mss51. Therefore, we imported radiolabeled Shy1, Mss51, and as a control the β-subunit of the F1Fo ATPase (F1β) into Coa1HA mitochondria. After removal of unimported precursors by protease treatment and solubilization of mitochondria, Coa1HA was immunoprecipitated. Shy1 and Mss51 were specifically co-immunoprecipitated with Coa1HA, while F1β was not enriched in the bound fraction (Figure 3G, lane 3 versus 4). Thus, Coa1 interacts with Shy1 and the translational activator Mss51.
Coa1Δ mitochondria are affected in cytochrome c oxidase biogenesis
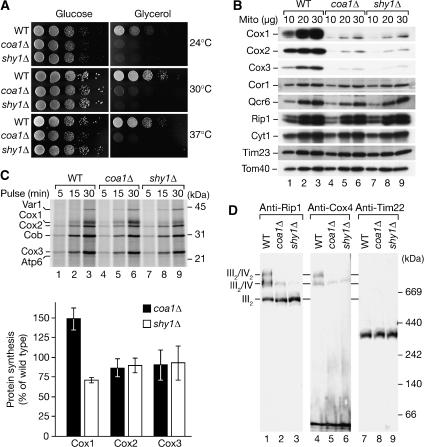
To analyze the role of Coa1 in respiratory chain function, a coa1Δ mutant was generated and analyzed for growth on fermentable and non-fermentable medium. Similar to shy1Δ cells, coa1Δ mutant cells were affected for growth on medium containing glycerol as carbon source (Figure 4A). This phenotype suggested that coa1Δ cells are defective in respiration.
Figure 4.
Coa1 mutant cells are affected in complex IV biogenesis. (A) Yeast cells were spotted in serial 10-fold dilutions and incubated at the indicated temperatures. (B) Indicated amounts of mitochondrial proteins were separated by SDS–PAGE and analyzed by Western blotting. (C) Mitochondrial translation products were labeled for the indicated times in organello, samples were TCA precipitated, separated by SDS–PAGE, and subjected to digital autoradiography. Quantification of radiolabeled proteins was performed with Image Quant (GE Healthcare). The signals of Cox-proteins were standardized to the levels of cytochrome b (Cob), wild-type values were set to 100%. Data represent the means±s.e.m. (n=3). (D) Mitochondrial protein complexes were solubilized in digitonin buffer, separated by blue-native PAGE, and protein complexes detected by immunodecoration. WT, wild-type.
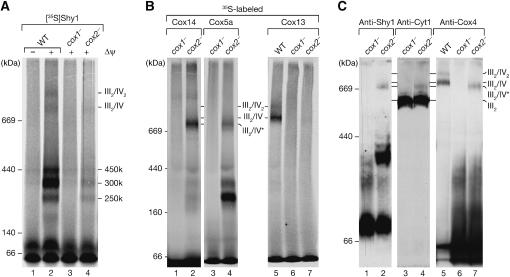
Comparison of the steady-state protein levels of wild-type, coa1Δ, and shy1Δ mitochondria revealed that the amount of the complex IV subunits (Cox1, Cox2, and Cox3) was dramatically reduced in both mutants compared with wild type (Figure 4B, lanes 4–6). This effect was specific for complex IV, since subunits of complex III, and the TIM and TOM translocases were not affected. To determine if the reduced amounts of Cox proteins were due to translational defects, we analyzed the synthesis of mitochondrially encoded proteins. Proteins that are translated in mitochondria can be radiolabeled in organello (Westermann et al, 2001) or in vivo in the presence of cyclohexamide, which induces a specific halt in cytosolic translation. Compared with wild type, shy1Δ mitochondria display a specific defect in the synthesis of Cox1 (Figure 4C, lanes 7–9) (Barrientos et al, 2002, 2004; Fontanesi et al, 2006), while coa1Δ mitochondria showed increased Cox1 translation both in organello (Figure 4C, lanes 4–6 and quantification) and in vivo (data not shown), similar to cox14Δ mitochondria (Barrientos et al, 2004). Hence, reduction in steady-state levels of complex IV subunits in coa1Δ mitochondria cannot be explained by reduced translation of mitochondrially encoded subunits, but rather by instability of unassembled subunits.
Mitochondrial protein complexes from wild type, coa1Δ, and shy1Δ mitochondria were separated by blue-native PAGE and analyzed by Western blotting. In agreement with previous analyses, shy1Δ mitochondria displayed a severe reduction in complex IV levels (Mashkevich et al, 1997; Nijtmans et al, 2001). Similarly, only minute amounts of complex III2/IV supercomplexes were detected in mitochondria from coa1Δ, while the levels of complex III or the TIM22 complex were not affected (Figure 4D). Thus, both mutants display a selective defect in complex IV biogenesis. In agreement with this, compared with wild-type mitochondria, coa1Δ and shy1Δ mitochondria displayed a significantly reduced Δψ (see Supplementary Figure 1).
Shy1–cytochrome c oxidase subcomplex biogenesis
Since coa1Δ and shy1Δ mitochondria displayed a severe reduction of complex IV and consequently of the supercomplexes, we analyzed coa1Δ mitochondria for the presence of intermediate-sized Shy1 complexes by blue-native PAGE. Radiolabeled Shy1 was imported into coa1Δ mitochondria and the low-molecular-weight form of Shy1 was detected in slightly increased amounts in coa1Δ mitochondria compared with the wild-type control. However, Shy1 did not assemble into the intermediate-sized complexes, which were also absent at steady state, as confirmed by immunodecoration (Figure 5A).
Figure 5.
Intermediate-sized Shy1 complexes depend on Coa1. (A) Radiolabeled Shy1 was imported into wild-type (WT) and Δcoa1 mitochondria as in Figure 2B. After solubilization with digitonin, complexes were separated by blue-native PAGE and analyzed by digital autoradiography. As a comparison, Shy1 complexes were analyzed by Western blotting using antiserum against Shy1. SHY1, intermediate-sized Shy1 complexes. (B) 35S-labeled precursors of Cox5a and Shy1 were imported into wild-type mitochondria in the presence or absence of Δψ and analyzed as in panel A. (C) In vitro import of 35S-labeled Cox5a precursor into isolated wild-type (WT) and Shy1Prot.A mitochondria as in panel A. Arrows indicate shifted complex species. (D) Radiolabeled Cox5a was imported into isolated wild-type and cox4Δ mitochondria and analyzed by blue-native PAGE and digital autoradiography. For comparison, Western blot analyses of respiratory chain complexes with indicated antibodies were performed. (E) In vitro import of the indicated precursors into mitochondria as in panel B. The asterisks indicate an unspecifically labeled complex species also present in the absence of a Δψ.
Among nuclear encoded subunits, Cox5a (Cox4 in human) is considered to be one of the first that assembles during the process of complex IV maturation (Wielburski and Nelson, 1983; Herrmann and Funes, 2005; Fontanesi et al, 2006). To address if the intermediate-sized 250, 300, and 450 k complexes represent complex IV subcomplexes associated with Shy1, we imported radiolabeled Cox5a and Shy1 into isolated mitochondria and separated protein complexes by blue-native PAGE. Both proteins displayed a similar pattern of supercomplexes and intermediate-sized complexes; however, the proteins were not equally distributed between the complexes. While Shy1 was mainly found in the 300 k complex, Cox5a was predominantly incorporated into the 250 k species (Figure 5B, lanes 2 and 4). When Cox5a was imported into mitochondria that contain Shy1Prot.A, the mobility of the low-molecular-weight Cox5a complex was not altered. In contrast, the 300 and 450 k complexes displayed a slower migration due to the tag on Shy1 (Figure 5C, lane 2). Thus, only the 300 and 450 k Shy1 complexes contain Cox5a, while the 250 k complexes, despite similar mobility, do not contain Shy1 and Cox5a together.
To exclude that the intermediate-sized complexes formed by Shy1, Cox1, and Cox5a resulted from a dissociation of the supercomplexes upon solubilization and blue-native PAGE, we imported Cox5a into wild-type and cox4Δ mutant mitochondria, which lack mature complex IV (Figure 5D, lanes 2 and 4). Cox5a assembled into the intermediate-sized complexes in both strains (Figure 5D, lane 5 versus 6). Moreover, the amount of the intermediate-sized complexes was even slightly increased in cox4Δ mutant mitochondria. This result suggested that the intermediate-sized complexes did not derive from dissociation of mature complex IV. In an alternative approach, we isolated supercomplexes via Cor1TAP. Isolated complexes were then subjected to blue-native PAGE analysis. While both supercomplexes and intermediate-sized complexes could be detected in mitochondrial extracts, the intermediate-sized complexes were not copurified with the supercomplexes. In contrast, a partial dissociation of the supercomplexes, which occurred during the procedure, led to the formation of complex III dimers and monomeric complex IV (see Supplementary Figure 2). Therefore, we conclude that the intermediate-sized protein complexes represent subcomplexes of the biogenesis pathway of cytochrome c oxidase rather than dissociation products of mature complex IV.
Among the tested nuclear encoded, structural subunits of complex IV, Cox13, Cox4, and Cox5a were imported in a Δψ-dependent manner into mitochondria and assembled into respiratory chain supercomplexes (Figure 5E, lanes 1–6). In agreement with previous observations, Cox13 directly assembled into supercomplexes (Brandner et al, 2005), whereas Cox5a assembled also into intermediate-sized complexes. In contrast, Cox4 accumulated mainly in an unassembled form, but also formed intermediate-sized protein complexes clearly distinct from those formed by Cox5a (Figure 5E, lane 4 versus 6). This agrees with the observation that the formation of Cox5a intermediate-sized complexes was not affected in cox4Δ mitochondria (Figure 5D). We conclude that the intermediate-sized complexes formed by Cox5a do not represent mature monomeric complex IV, since they do not contain Cox4 and Cox13. We suggest that the intermediate-sized complexes represent subcomplexes of the complex IV assembly pathway.
When we analyzed Cox14 in a similar way, it was not only found in association with supercomplexes, but also formed intermediate-sized complexes similar to Shy1 and Cox5a (Figure 5E, lane 8). This finding agrees with the observed association of Cox14 with complex III and IV subunits and with Shy1 (Figure 1E). In a quantitative analysis we estimated that close to 20% of Cox14 is associated with supercomplexes under our experimental conditions (Figure 2E).
Shy1/Cox14 association with supercomplexes through partially assembled complex IV
To assess if the formation of Shy1 complexes depends on mitochondrially encoded complex IV subunits, Shy1 was imported into mutant mitochondria that lack either Cox1 or Cox2, termed cox1− and cox2−, respectively. Compared to wild-type mitochondria, assembly of the Shy1 precursor was reduced in cox1− and cox2− mutant mitochondria due to their lower Δψ and reduced import efficiency. However, cox1− and cox2− mutant mitochondria displayed a significant difference with regard to Shy1 complexes. In cox2− mitochondria, the intermediate-sized complexes were still detectable, while they were absent in cox1− (Figure 6A, lane 3 versus 4). Similar to Shy1, imported Cox5a and Cox14 assembled into the intermediate-sized complexes in a Cox1-dependent manner. Both proteins assembled also into a higher molecular weight complex (termed III2/IV*) (Figure 6B, lanes 1–4). In contrast, Cox13 did not assemble into the III2/IV* or other large complexes in cox1− and cox2− mutant mitochondria, but assembled mainly into III2/IV in wild-type mitochondria, which migrated slower in the gel than III2/IV* (Figure 6B, lane 5 versus 7). It should be noted that wild-type, cox1−, and cox2− mutant mitochondria were isolated from cells grown on fermentable carbon sources. In our hands, mitochondria isolated under these conditions exhibit overall reduced amounts of respiratory chain complexes, but also significantly less III2/IV2 supercomplexes (for comparison see also Figure 5D and Supplementary Figure 3).
Figure 6.
Formation of Shy1 complexes depends on Cox1. (A) 35S-labeled Shy1 was imported into isolated mitochondria in the presence or absence of a Δψ. Mitochondria were lysed in digitonin buffer and analyzed by blue-native PAGE and digital autoradiography. (B) Radiolabeled precursors of Cox14, Cox5a, and Cox13 were imported into isolated mitochondria and analyzed as in panel A. (C) Protein complexes of digitonin-lysed mitochondria were subjected to blue-native PAGE and Western blot analysis.
We asked if the incomplete form of complex IV, which lacked Cox2 and Cox13 but contained Cox5a and Cox14, could also be detected under steady-state conditions. Therefore, Western blot analysis of mitochondrial protein complexes was performed. In cox1− mitochondria the intermediate-sized Shy1 complexes were lacking and Shy1 was mainly found in the low-molecular-weight range. In cox2− mitochondria, however, Shy1 was found in the 250, 300, and 450 k complexes as well as in the III2/IV* complex (Figure 6C, lane 2). Anti-Cox4 antibodies detected Cox4 mainly in the low-molecular-weight range in cox1− and cox2− mitochondria. Moreover, a fraction of Cox4 was also detected in the III2/IV* complex in cox2− mitochondria, while it was absent in cox1−. In contrast, in wild-type mitochondria Cox4 was predominantly found in the supercomplexes, migrating slower than the III2/IV* complex, while the intermediate-sized complexes were not detected (Figure 6C, lanes 5–7). In addition, the small fraction of III2/IV* complex in cox2− mitochondria was detected with antiserum against Cyt1 of the bc1 complex, indicating that it contained complex III (Figure 6C). Thus, we conclude that the formation of the intermediate-sized complexes depends on the presence of Cox1, but is independent of Cox2. At steady state these complexes are present in mitochondria and contain at least Shy1, Cox1, Cox14, and Cox5a but not Cox4. Moreover, we find that despite the lack of subunits such as Cox2 or Cox13, a partially assembled form of complex IV in association with Shy1 and Cox14 is already able to oligomerize with complex III. This form of the complex therefore contains all necessary subunits to promote interaction with complex III despite being incomplete.
Discussion
Cytochrome c oxidase (complex IV) plays a central role in respiratory chain function and cellular bioenergetics. Defects in the function of complex IV affect oxidative phosphorylation and can lead to severe human disorders. In fact, mutations in structural complex IV subunits and in assembly factors were reported to lead to complex IV deficiency and to cause disease (Shoubridge, 2001; Chinnery and Schon, 2003; DiMauro and Schon, 2003; Zeviani and Spinazzola, 2003). Complex IV biogenesis requires a large number of assembly factors, one of which is Shy1, the yeast homolog of human SURF1 (Mashkevich et al, 1997; Tiranti et al, 1998; Zhu et al, 1998; Poyau et al, 1999; Barrientos et al, 2002). Aside from the medical importance, the understanding of how these complicated membrane protein complexes are built from individual subunits and the underlying principles how higher organization is achieved from single molecules is of immense biological interest.
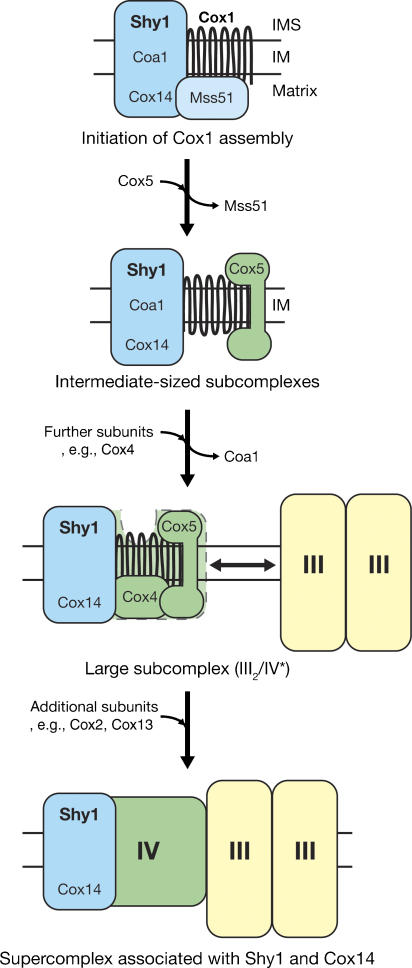
We report that Shy1 associates with different protein modules to promote complex IV assembly: (i) Shy1 interacts with Mss51 and Cox14, which regulate Cox1 expression; (ii) it forms subcomplexes with nuclear and mitochondrially encoded subunits of complex IV that also contain Cox14, thus representing potential assembly intermediates of complex IV; (iii) it interacts through partially and fully assembled forms of complex IV with complex III. Based on these findings we propose that Shy1 and Cox14 accompany Cox1 during early steps of complex IV assembly, and remain associated when nuclear encoded subunits associate with Cox1 (Figure 7). At some stage an intermediate form of complex IV, containing at least Cox1, Cox5a, and Cox4 in association with Cox14 and Shy1, can bind to complex III. This complex is not competent to incorporate late complex IV subunits such as Cox13 when Cox2 is not present. We propose in this model that Cox14 and Shy1 accompany transient forms of complex IV and maintain their competence for the incorporation of additional subunits.
Figure 7.
Model for Shy1 mediated steps of complex IV assembly. Shy1 associates with complex IV during assembly from the initial steps of Cox1 biogenesis to the stage of supercomplex formation: upon membrane integration, Cox1 is bound to the translational regulator complex of Mss51 and Cox14 in a chaperone-like manner. Shy1 and Coa1 are part of this complex. Through incorporation of subunits (e.g., Cox5), complex IV subcomplexes are formed. As more subunits, such as Cox4, get incorporated, Coa1 is released from the potential intermediate states. Partially assembled complex IV still containing Shy1 and Cox14 is already competent to associate to complex III, forming the III2/IV* supercomplex. While complex III is not directly involved in complex IV maturation, late steps of complex IV assembly may take place at the supercomplex level in the presence of complex III, whereby subunits such as Cox2 and Cox13 are incorporated.
Shy1 interacts with Mss51 and Cox14, which regulate Cox1 translation and also associate with Cox1 at the initial stages of complex IV assembly (Perez-Martinez et al, 2003; Barrientos et al, 2004; Fontanesi et al, 2006). In contrast to Mss51, Cox14 and Shy1 associate with respiratory chain supercomplexes (Figure 7). Therefore, a dissociation of Mss51 from Cox14 and Shy1 has to occur. This agrees with previous models on Mss51 regulation (Perez-Martinez et al, 2003; Barrientos et al, 2004). The dissociation of Mss51 from its regulator Cox14, however, cannot solely be induced through Shy1 association with Cox14, since we found Mss51 in complex with Shy1. Thus, it remains to be shown if Shy1 forms a complex with Mss51 and Cox14 at early stages of Cox1 assembly or if two separate complex species of Shy1 exist.
Previous analyses reported that Shy1 is mainly present in a protein complex of 250 kDa on blue-native gels (Nijtmans et al, 2001). When we solubilized mitochondria in the mild detergent digitonin that leaves supercomplexes intact, Shy1 was maintained in three intermediate-sized complexes as well as in association with respiratory chain supercomplexes. We find that formation of these Shy1 complexes requires the mitochondrially encoded structural subunit Cox1 as well as Coa1, a newly identified inner mitochondrial membrane protein (Figure 7). In agreement with the idea that Cox5a assembles early with Cox1 (Wielburski and Nelson, 1983; Taanman and Williams, 2001; Herrmann and Funes, 2005; Fontanesi et al, 2006), imported Cox5a associates with Shy1 in 300 and 450 k complexes, the exact compositions of which are not yet clear. Our experiments suggest that there are two different forms of 250 k complexes; one that contains Shy1 but lacks Cox5a, and vice versa. However, it remains to be addressed what role the latter form has for complex IV biogenesis. Further work will also be required to dissect the dynamics and compositions of the other intermediate-sized protein complexes. However, at this point it is already evident that Cox4 and Cox13 do not associate with any of these three complexes when they are imported into mitochondria, but that they rather assemble at a later stage. This finding supports the idea that the intermediate-sized Shy1 complexes represent partially assembled states of complex IV. While Coa1 function is clearly required for the formation of the intermediate complexes, it is currently open if it directly cooperates with Shy1 or whether it is connected to translational regulation, similar to Cox14. However, since we did not find Coa1HA associated with supercomplexes, we suggest an early role for the protein during complex IV biogenesis.
A surprising finding of our studies was the observed association of Shy1 and Cox14 with respiratory chain supercomplexes. Analyses of cox2− mutant mitochondria show that both Shy1 and Cox14 are bound to a partially assembled complex IV, already associated with complex III. Since complex IV assembly can occur independent of complex III (Supplemtary Figure 3), this interaction reflects that other subunits, which promote association to complex III, have already been incorporated. Thus, Shy1 and Cox14 apparently act also at late steps of complex IV biogenesis. Moreover, this finding suggests that incorporation of new complex IV subunits may still occur in the supercomplex state. This is further supported by the fact that Cox13 seems to assemble directly into supercomplexes (Brandner et al, 2005). However, the association of Shy1 and Cox14 with the III2/IV* supercomplex in cox2− mitochondria does not explain why we also see association of Shy1 and Cox14 with mature supercomplexes in wild type. The III2/IV* supercomplex migrates faster on the gel and is clearly distinguishable from III2/IV, provided it is present in sufficient amounts to be detected. An attractive hypothesis is that at steady state a pool of partially assembled complex IV in association with complex III exists, which has already passed the stage of Cox4 incorporation but still requires Shy1/Cox14 for efficient completion of the assembly process. An alternative hypothetical explanation would be that an exchange of complex IV subunits can occur at the supercomplex level and that Shy1 and Cox14 are required to guard the complex during subunit exchange. Additional experimental work will be required to address these models. Nevertheless, in both cases our finding that Shy1 and Cox14 associate with respiratory chain supercomplexes extends the existing view on the function of these proteins in complex IV biogenesis and sheds new light on the process of respiratory chain assembly.
Materials and methods
Yeast strains and mitochondrial isolation
S. cerevisiae strains used in this study are described in Table I. Mitochondria were isolated from S. cerevisiae cells grown at 30°C in rich liquid medium containing 1% yeast extract, 2% peptone and 3% glycerol, 2% galactose or 2% sucrose in the case of strains affected in respiratory chain function as previously described (Meisinger et al, 2006). For growth tests, yeast cells from liquid cultures were adjusted to an OD600 of 0.3 and serial dilutions spotted onto agar plates containing glucose or glycerol as carbon source and incubated at the indicated temperatures.
Table 1.
List of strains used in this study
| Strains | Genotype | Source |
|---|---|---|
| YPH499 | Mat a, ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801 | Sikorski and Hieter (1989) |
| BY4741 | Mat a, his3-Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Open Biosystems |
| 777-A3 | Mat α, ade1 op1 | Netter et al (1982) |
| DaMY06 (shy1Prot.A) | Mat a, ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; shy1∷shy1-Prot.A | This study |
| Cox14TAP | Mat a, his3-Δ1, leu2Δ0, met15Δ0, ura3Δ0; cox14∷cox14-TAP | Open Biosystems |
| Mss51TAP | Mat a, his3-Δ1, leu2Δ0, met15Δ0, ura3Δ0; mss51∷mss51-TAP | Open Biosystems |
| Tim18Prot.A | Mat a, ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; tim18∷tim18-TEV-ProteinA-7His-HisMX6 | Rehling et al (2003) |
| Cor1TAP | Mat a, his3-Δ1, leu2Δ0, met15Δ0, ura3Δ0; cor1∷cor1-TAP | van der Laan et al (2006) |
| Coa1TAP | Mat a, his3-Δ1, leu2Δ0, met15Δ0, ura3Δ0; coa1∷coa1-TAP | Open Biosystems |
| DaMY23 (coa1HA) | Mat a, ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; coa1∷coa1-HA | This study |
| DaMY21 (coa1Δ) | Mat a, ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; YIL157c∷HisMX6 | This study |
| DaMY22 (shy1Δ) | Mat a, ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; YGR112w∷HisMX6 | This study |
| AFY14 (cor1Δ) | Mat a, ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; YBL045c∷kanMX4 | This study |
| AFY11 (cox4Δ) | Mat a, ade2-101 his3-Δ200 leu2-Δ1 ura3-52 trp1-Δ63 lys2-801; YGL187c∷HisMX6 | Frazier et al (2006) |
| G421 (cox1−) | Mat α, ade1 op1; cox1-G421 | Netter et al (1982) |
| V25 (cox2−) | Mat α, ade1 op1 met3; cox2-V25 | Netter et al (1982) |
In organello labeling of mitochondrial translation products
For in organello labeling of mitochondrial translation products, mitochondria were isolated from yeast cells grown on medium containing 2% galactose. Isolated mitochondria were subjected to the labeling reaction in the presence of [35S]methionine (Westermann et al, 2001). Mitochondria were re-isolated, proteins precipitated with TCA, and processed for separation by SDS–PAGE. Dried gels were analyzed by digital autoradiography and radioactive signals quantified with the Image Quant 5.2 software (GE Healthcare).
Import of radiolabeled precursor proteins
For in vitro translation, open reading frames of interest under the control of the SP6 promotor were cloned into pGEM-4Z (Promega) or amplified by PCR (Wiedemann et al, 2006). Labeling of the preproteins was performed in vitro in rabbit reticulocyte lysate (Promega or GE Healthcare). Import and assembly of the radiolabeled preproteins into isolated mitochondria were performed essentially as described (Wiedemann et al, 2006). For import of the Shy1 precursor the buffer was supplemented with 150 mM KCl.
Protein localization analysis
Isolated mitochondria containing Coa1HA were treated to induce swelling in EM buffer (1 mM EDTA, 10 mM MOPS pH 7.2) or lysed in 1% Triton X-100 and treated with proteinase K. For analysis of membrane association, mitochondria were sonicated using a Branson sonifier (model 250; 3 × 30 s with 40% duty cycle) in 10 mM Tris, pH 7.4, 500 mM NaCl, or subjected to extraction in 0.1 M carbonate buffer at pH 10.8, or 1% Triton X-100 containing 150 mM NaCl. Samples were subjected to ultracentrifugation at 45 000 r.p.m., 4°C for 45 min, in a TLA-45 rotor (Beckmann), TCA precipitated and analyzed by SDS–PAGE and Western blotting.
Miscellaneous
Standard techniques were used for Western blotting to PVDF membranes. Antibody–protein complexes were visualized by enhanced chemiluminescence (GE Healthcare). Isolation of protein complexes via IgG-chromatography and mass spectrometric analyses were performed essentially as described (Rehling et al, 2003; Frazier et al, 2006), with the exception of the solubilization buffer (20 mM Tris, pH 7.4, 60 mM NaCl, 0.5 mM EDTA, 1% digitonin, 10% glycerol). Blue-native electrophoresis analysis was performed according to published procedures (Rehling et al, 2003).
Supplementary Material
Supplemental methods and materials
Acknowledgments
We thank N Pfanner, A Chacinska, N Wiedemann, and C Meisinger for discussions; M Boguta for strains; D Winge and F Pierrel for sharing unpublished observation; S Wiese and C Bunse for technical assistance; and D Stojanovski for critical reading of the paper. This work was supported by the Deutsche Forschungsgemeinschaft, Bundesministerium für Bildung und Forschung, and an EMBO long-term fellowship (MvdL).
References
- Barrientos A, Korr D, Tzagoloff A (2002) Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh's syndrome. EMBO J 21: 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A (2004) Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J 23: 3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner K, Mick DU, Frazier AE, Taylor RD, Meisinger C, Rehling P (2005) Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for barth syndrome. Mol Biol Cell 16: 5202–5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr HS, Winge DR (2003) Assembly of cytochrome c oxidase within the mitochondrion. Acc Chem Res 36: 309–316 [DOI] [PubMed] [Google Scholar]
- Chinnery PF, Schon EA (2003) Mitochondria. J Neurol Neurosurg Psychiatry 74: 1188–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruciat CM, Brunner S, Baumann F, Neupert W, Stuart RA (2000) The cytochrome bc1 and cytochrome c oxidase complexes associate to form a single supracomplex in yeast mitochondria. J Biol Chem 275: 18093–18098 [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA (2003) Mitochondrial respiratory-chain diseases. N Engl J Med 348: 2656–2668 [DOI] [PubMed] [Google Scholar]
- Fontanesi F, Soto IC, Horn D, Barrientos A (2006) Assembly of mitochondrial cytochrome c oxidase, a complicated and highly regulated cellular process. Am J Physiol Cell Physiol 291: C1129–C1147 [DOI] [PubMed] [Google Scholar]
- Frazier AE, Taylor RD, Mick DU, Warscheid B, Stoepel N, Meyer HE, Ryan MT, Guiard B, Rehling P (2006) Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J Cell Biol 172: 553–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakh O, Cavadini P, Isaya G (2002) Mitochondrial processing peptidases. Biochim Biophys Acta 1592: 63–77 [DOI] [PubMed] [Google Scholar]
- Glerum DM, Koerner TJ, Tzagoloff A (1995) Cloning and characterization of COX14, whose product is required for assembly of yeast cytochrome oxidase. J Biol Chem 270: 15585–15590 [DOI] [PubMed] [Google Scholar]
- Grivell LA, Artal-Sanz M, Hakkaart G, de Jong L, Nijtmans LG, van Oosterum K, Siep M, van der Spek H (1999) Mitochondrial assembly in yeast. FEBS Lett 452: 57–60 [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Funes S (2005) Biogenesis of cytochrome oxidase—sophisticated assembly lines in the mitochondrial inner membrane. Gene 354: 43–52 [DOI] [PubMed] [Google Scholar]
- Jensen RE, Dunn CD (2002) Protein import into and across the mitochondrial inner membrane: role of the TIM23 and TIM22 translocons. Biochim Biophys Acta 1592: 25–34 [DOI] [PubMed] [Google Scholar]
- Koehler CM (2004) New developments in mitochondrial assembly. Annu Rev Cell Dev Biol 20: 309–335 [DOI] [PubMed] [Google Scholar]
- Leigh D (1951) Subacute necrotizing encephalomyelopathy in an infant. J Neurol Neurosurg Psychiatry 14: 216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashkevich G, Repetto B, Glerum DM, Jin C, Tzagoloff A (1997) SHY1, the yeast homolog of the mammalian SURF-1 gene, encodes a mitochondrial protein required for respiration. J Biol Chem 272: 14356–14364 [DOI] [PubMed] [Google Scholar]
- McEwen JE, Ko C, Kloeckner-Gruissem B, Poyton RO (1986) Nuclear functions required for cytochrome c oxidase biogenesis in Saccharomyces cerevisiae. Characterization of mutants in 34 complementation groups. J Biol Chem 261: 11872–11879 [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN (2006) Isolation of yeast mitochondria. Methods Mol Biol 313: 33–39 [DOI] [PubMed] [Google Scholar]
- Netter P, Carignani G, Jacq C, Groudinsky O, Clavilier L, Slonimski PP (1982) The cytochrome oxidase subunit I split gene in Saccharomyces cerevisiae: genetic and physical studies of the mtDNA segment encompassing the ‘cytochrome-b-homologous' intron. Mol Gen Genet 188: 51–59 [DOI] [PubMed] [Google Scholar]
- Neupert W, Herrmann JM (2007) Translocation of proteins into mitochondria. Annu Rev Cell Dev Biol 76: 723–749 [DOI] [PubMed] [Google Scholar]
- Nijtmans LG, Artal Sanz M, Bucko M, Farhoud MH, Feenstra M, Hakkaart GA, Zeviani M, Grivell LA (2001) Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett 498: 46–51 [DOI] [PubMed] [Google Scholar]
- Perez-Martinez X, Broadley SA, Fox TD (2003) Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J 22: 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyau A, Buchet K, Godinot C (1999) Sequence conservation from human to prokaryotes of Surf1, a protein involved in cytochrome c oxidase assembly, deficient in Leigh syndrome. FEBS Lett 462: 416–420 [DOI] [PubMed] [Google Scholar]
- Rehling P, Brandner K, Pfanner N (2004) Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol 5: 519–530 [DOI] [PubMed] [Google Scholar]
- Rehling P, Model K, Brandner K, Kovermann P, Sickmann A, Meyer HE, Kühlbrandt W, Wagner R, Truscott KN, Pfanner N (2003) Protein insertion into the mitochondrial inner membrane by a twin-pore translocase. Science 299: 1747–1751 [DOI] [PubMed] [Google Scholar]
- Schägger H, Pfeiffer K (2000) Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19: 1777–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoubridge EA (2001) Cytochrome c oxidase deficiency. Am J Med Genet 106: 46–52 [DOI] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, Joppich C, Zahedi R, Meyer HE, Schönfisch B, Perschil I, Chacinska A, Guiard B, Rehling P, Pfanner N, Meisinger C (2003) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci USA 100: 13207–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith D, Gray J, Mitchell L, Antholine WE, Hosler JP (2005) Assembly of cytochrome c oxidase in the absence of assembly protein Surf1p leads to loss of the active site heme. J Biol Chem 280: 17652–17656 [DOI] [PubMed] [Google Scholar]
- Stuart R (2002) Insertion of proteins into the inner membrane of mitochondria: the role of the Oxa1 complex. Biochim Biophys Acta 1592: 79–87 [DOI] [PubMed] [Google Scholar]
- Taanman JW, Williams SL (2001) Assembly of cytochrome c oxidase: what can we learn from patients with cytochrome c oxidase deficiency? Biochem Soc Trans 29: 446–451 [DOI] [PubMed] [Google Scholar]
- Tiranti V, Hoertnagel K, Carrozzo R, Galimberti C, Munaro M, Granatiero M, Zelante L, Gasparini P, Marzella R, Rocchi M, Bayona-Bafaluy MP, Enriquez JA, Uziel G, Bertini E, Dionisi-Vici C, Franco B, Meitinger T, Zeviani M (1998) Mutations of SURF-1 in Leigh disease associated with cytochrome c oxidase deficiency. Am J Hum Genet 63: 1609–1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S (1996) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 272: 1136–1144 [DOI] [PubMed] [Google Scholar]
- Tzagoloff A, Dieckmann CL (1990) PET genes of Saccharomyces cerevisiae. Microbiol Rev 54: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzagoloff A, Myers AM (1986) Genetics of mitochondrial biogenesis. Annu Rev Biochem 55: 249–285 [DOI] [PubMed] [Google Scholar]
- van der Laan M, Wiedemann N, Mick DU, Guiard B, Rehling P, Pfanner N (2006) A role for Tim21 in membrane-potential-dependent preprotein sorting in mitochondria. Curr Biol 16: 2271–2276 [DOI] [PubMed] [Google Scholar]
- Westermann B, Herrmann JM, Neupert W (2001) Analysis of mitochondrial translation products in vivo and in organello in yeast. Meth Cell Biol 65: 429–438 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Pfanner N, Rehling P (2006) Import of precursor proteins into isolated yeast mitochondria. Methods Mol Biol 313: 373–384 [DOI] [PubMed] [Google Scholar]
- Wielburski A, Nelson BD (1983) Evidence for the sequential assembly of cytochrome oxidase subunits in rat liver mitochondria. Biochem J 212: 829–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SL, Valnot I, Rustin P, Taanman JW (2004) Cytochrome c oxidase subassemblies in fibroblast cultures from patients carrying mutations in COX10, SCO1, or SURF1. J Biol Chem 279: 7462–7469 [DOI] [PubMed] [Google Scholar]
- Zambrano A, Fontanesi F, Solans A, de Oliveira RL, Fox TD, Tzagoloff A, Barrientos A (2007) Aberrant translation of cytochrome c oxidase subunit 1 mRNA species in the absence of Mss51p in the yeast Saccharomyces cerevisiae. Mol Biol Cell 18: 523–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeviani M, Spinazzola A (2003) Mitochondrial disorders. Curr Neurol Neurosci Rep 3: 423–432 [DOI] [PubMed] [Google Scholar]
- Zhu Z, Yao J, Johns T, Fu K, De Bie I, Macmillan C, Cuthbert AP, Newbold RF, Wang J, Chevrette M, Brown GK, Brown RM, Shoubridge EA (1998) SURF1, encoding a factor involved in the biogenesis of cytochrome c oxidase, is mutated in Leigh syndrome. Nat Genet 20: 337–343 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods and materials